Abstract
Background
During early HIV-1 infection, immunodominant T cell responses to highly variable epitopes lead to the establishment of immune escape virus variants. Here we assessed a type 1-polarized monocyte-derived dendritic cell (MDC1)-based approach to selectively elicit cytotoxic T lymphocyte (CTL) responses against highly conserved and topologically important HIV-1 epitopes in HIV-1-infected individuals from the Thailand RV254/SEARCH 010 cohort who initiated antiretroviral therapy (ART) during early infection (Fiebig stages I-IV).
Methods
Autologous MDC1 were used as antigen presenting cells to induce in vitro CTL responses against HIV-1 Gag, Pol, Env, and Nef as determined by flow cytometry and ELISpot assay. Ultra-conserved or topologically important antigens were respectively identified using the Epigraph tool and a structure-based network analysis approach and compared to overlapping peptides spanning the Gag proteome.
Findings
MDC1 presenting either the overlapping Gag, Epigraph, or Network 14–21mer peptide pools consistently activated and expanded HIV-1-specific T cells to epitopes identified at the 9–13mer peptide level. Interestingly, some CTL responses occurred outside known or expected HLA associations, providing evidence of new HLA-associated CTL epitopes. Comparative analyses demonstrated more sequence conservation among Epigraph antigens but a higher magnitude of CTL responses to Network and Gag peptide groups. Importantly, CTL responses against topologically constrained Gag epitopes contained in both the Network and Gag peptide pools were selectively enhanced in the Network pool-initiated cultures.
Interpretation
Our study supports the use of MDC1 as a therapeutic strategy to induce and focus CTL responses toward putative fitness-constrained regions of HIV-1 to prevent immune escape and control HIV-1 infection.
Funding
A full list of the funding sources is detailed in the Acknowledgment section of the manuscript.
Key Words: HIV-1 cure, Immunotherapy, Dendritic cell, Cytotoxic T cell, Epitopes
Research in context.
Evidence before this study
A major hurdle in the development of a successful HIV-1 immunotherapy is the capacity of the virus to evade the immune response by efficiently establishing epitope variants in response to selective pressure. While effective at suppressing viremia, current regimens of antiretroviral therapy (ART) are not curative. Therefore, achieving immune control of HIV-1 upon cessation of ART, like that observed in ‘elite controllers’ (EC), has been a major therapeutic goal toward a functional cure. Such immune control is realized through the actions of antigen-specific cytotoxic T cell lymphocytes (CTL) capable of specifically targeting sequence-conserved HIV-1 epitopes.
Added value of this study
In this study, a specialized antigen presenting dendritic cell (DC)-based vaccine strategy was used to elicit HIV-1-specific CTL responses in vitro against carefully selected, ultraconserved and topologically important epitopes. The 14–21mer peptide pools selected for afferent induction of T cell immune responses are universally applicable as they cover such a broad range of possible HLA-associated haplotypes. Importantly, this DC-based approach yielded broad effector responses against 9–13mer peptide epitopes of both known and unknown HLA associations, demonstrating cross-presentation and the uncovering of potentially novel epitopes. Importantly, we demonstrate that CTL responses can be re-directed or focused toward potentially more fitness-constrained regions of the virus in people initiating ART during early HIV-1 infection.
Implications of all the available evidence
This study highlights the potential for DC-based therapies to drive immune responses against select antigenic targets critically important to HIV fitness as a means to control the infection while circumventing the potential for viral adaptation that could otherwise lead to immune escape.
Alt-text: Unlabelled box
1. Introduction
Adaptive immune pressure and viral fitness restrictions in untreated HIV-1 infection result in distinct regions of low and high diversity in the viral genome, with the low diversity regions being a preferred antigenic target of immunotherapy [1]. Beginning during acute HIV-1 infection (AHI), immunodominant T cell responses skew towards highly variable viral epitopes leading to the rapid establishment of immune escape variants [2,3]. However, in individuals initiating ART during the early stages of infection, compared with progressive chronic infection, the HIV-1 population is characterized by less antigenic diversity and fewer cytotoxic T lymphocyte (CTL) escape variants [4]. Therefore, implementing a ‘shock and kill’ or ‘kick and kill’ immunotherapeutic approach [5] in individuals who begin ART during earlier stages of infection could effectively target latently infected CD4+ T cells harboring replication-competent HIV-1.
A major challenge to the ‘kick and kill’ hypothesis as a therapeutic modality is identifying a safe and efficient approach for eliciting functional CTL responses to fitness-constrained viral epitopes. Our strategy for immunotherapy of HIV-1 infection centers on the use of myeloid dendritic cells (DC), which are professional antigen presenting cells (APC) that we have shown to be capable of inducing highly potent CTL responses to HIV-1 against a broad array of MHC class I epitopes in vitro [6,7]. Moreover, DC have been proven safe and effective for inducing antigen-specific T cell responses in immunotherapy trials for end-stage cancers [8], [9], [10], and they have been used to treat HIV-1 [11,12], with the current form of DC immunotherapy resulting in a significant, if temporary, delay in HIV-1 rebound after ART interruption [11,13].
We hypothesize that the DC used in HIV-1 immunotherapy trials to date have not been adequately equipped with the characteristics needed to specifically direct and support effective type 1-biased cellular immune responses that are required to successfully combat cancers and intracellular infections such as HIV-1 [6,[14], [15], [16], [17]]. In fact, the methods commonly used to generate mature DC for immunotherapies, including the use of maturation factors such as prostaglandin E2 (PGE2) and CD40L, typically give rise to mature DC that quickly become deficient in their capacity to produce IL-12p70 [18], a critical Th1 and CTL driving factor [19]. Indeed, we have found that naïve CD8+ T cells from individuals with chronic HIV-1 infection [20] and uninfected individuals [21,22] can be primed ex vivo with autologous, high IL-12p70-producing, type 1-polarized monocyte-derived DC (MDC1) to become efficient CTL effectors. Although pre-existing memory CD8+ T cells present during chronic HIV-1 infection are capable of recognizing CD4+ T cell targets expressing established variant HIV-1 epitopes to produce inflammatory cytokines, they are predominantly dysfunctional in their killing capacity [20,21,23,24] and display signatures of immune exhaustion [25]. However, these newly MDC1-primed CTL are effective at killing autologous HIV-1-infected CD4+ T cells [20,22].
Achieving a functional cure for HIV-1 infection will require overcoming the emergence of early CTL escape variants and immune exhaustion. Our overarching hypothesis is that MDC1 can be utilized in individuals initiating ART during AHI as an immunotherapeutic tool to effectively correct or focus CTL activity toward highly conserved or topologically important regions of the HIV-1 proteome that are structurally and functionally critical to viral fitness [26,27].
Here we applied two diverse but complementary methods to select CTL antigenic targets. The first method used was the Epigraph90 [27], [28], [29] graph-theory-based, computational approach, which enabled us to identify conserved HIV-1 peptide libraries to optimize vaccine coverage of potential CD8+ T cell epitope (PTE) variants found in the diverse HIV-1 population. The algorithm allows for exploration of epitope features relevant to an immunotherapeutic DC vaccine design that were previously intractable, such as balancing the costs in PTE coverage with rare epitope exclusion and optimizing coverage of in vivo diversity. The Epigraph approach was thus used to define short regions (14–21 amino acids in length) of the proteome with extremely high conservation levels at the global population level; the included regions contained multiple known and/or predicted CD8+ T cell epitopes and conserved regions for with-in subject targeting [27]. The focus on extremely conserved but short peptide fragments is particularly well suited to MDC1-priming for CTL induction [21]. Similar vaccine antigen design strategies have shown that the immune response can be refocused towards highly conserved elements using DNA delivery [30,31]. Also, longer regions of the proteome that contain relatively conserved regions (balancing the inclusion of more potential epitopes with less stringent conservation requirements) can also help focus the immune response on more conserved regions that are beneficial in terms of clinical outcomes [32], [33], [34], using vector delivery strategies or self-amplifying mRNA [32,35].
Our second method for selecting peptide antigens employed a structure-based network analysis to identify structurally and functionally constrained epitopes [26]. Structural data were used to build networks of noncovalent interactions between amino acid side chains and subsequently analyzed by graph theory metrics to quantify the sum contribution of each residue to the protein's global architecture. The scientific premise and rationale of this network theory is to identify amino acid residues of topological importance, which are critical to a protein's structure and function [26]. Thus, effective immune targeting of these highly networked regions of the viral proteome would greatly and negatively impact viral fitness.
The main objective of this study was to test our MDC1-based cellular vaccine approach as a pre-clinical assessment of selected peptides representing ultra-conserved and topologically constrained HIV-1-associated epitopes as immunogens for therapeutic application as part of the I4C (Immunotherapy for Cure) Martin Delany Collaboratory. The study participants for this pre-clinical assessment were selected randomly from the well-defined RV254/SEARCH 010 cohort in Thailand who initiated ART during very early stages of HIV-1 infection (defined as Fiebig stages I-IV) [36] and represent a specific target population of interest for future clinical trials.
2. Materials and methods
2.1. Study cohort participants
The RV254/SEARCH 010 (NCT00796146 and NCT00796263) cohort enrolls adults diagnosed with AHI at the time of presentation at an HIV-1 screening site at the Thai Red Cross Anonymous Clinic and who were offered immediate ART [37], [38], [39]. The Chulalongkorn University Institutional Review Board and the Walter Reed Army Institute of Research, USA, approved this study. AHI is defined as either non-reactive 4th generation immunoassay with positive nucleic acid test or reactive 4th generation immunoassay together with non-reactive 2nd generation immunoassay [40]. The procedures of staging AHI have been described previously [36,37,40]. For this study, peripheral blood mononuclear cells (PBMC) were obtained from 10 HIV-1-infected individuals in the Thailand/MHRP RV254 cohort who initiated ART during acute/early infection [Fiebig I (n = 2), II (n = 2), III (n = 4), IV (n = 2); Table 1].
Table 1.
Demographic Characteristics of RV254 Cohort.
|
Week 0a |
Week of sample tested |
HLA haplotype |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Age | Fiebig | HIV subtype | CD4+ count | CD8+ count | Plasma VL (copies/ml) | Week # | CD4+ count | CD8+ count | Plasma VL (copies/mL) | HLA-A | HLA-B | HLA-C |
| 4156 | 42 | 1 | CRF01_AE | 641 | 442 | 4452 | 96 | 660 | 425 | <50 | 01:01, 02:07 | 44:03, 46:01 | 01:02, 06:02 |
| 9129 | 46 | 1 | CRF01_AE | 447 | 399 | 31,970 | 96 | 768 | 457 | <20 | 33:03, 33:03 | 38:02, 58:01 | 03:02, 03:04 |
| 7905 | 29 | 2 | CRF01_AE | 213 | 127 | 7,263,860 | 240 | 621 | 461 | <20 | 02:07, 11:01 | 15:25, 40:01 | 04:03, 07:02 |
| 9887 | 28 | 2 | CRF01_AE | 464 | 496 | 2,332,038 | 96 | 590 | 531 | <20 | 02:03, 33:03 | 38:02, 58:01 | 03:02, 07:02 |
| 5113 | 33 | 3 | CRF01_AE | 386 | 570 | 358,198 | 96 | 466 | 402 | <50 | 02:06, 11:01 | 40:06, 58:01 | 03:02, 08:01 |
| 7466 | 27 | 3 | CRF01_AE | 182 | 509 | 22,516,400 | 144 | 377 | 438 | <20 | 33:03, 33:03 | 58:01, 58:01 | 03:02, 03:02 |
| 5497 | 25 | 3 | CRF01_AE | 602 | 1232 | 681,176 | 96 | 1175 | 668 | <20 | 02:07, 33:03 | 46:01, 58:01 | 01:02, 03:02 |
| 4446 | 24 | 3 | CRF01_AE | 278 | 511 | 7,388,080 | 96 | 486 | 573 | <20 | 02:03, 02:03 | 40:02, 48:01 | 08:01, 15:02 |
| 9720 | 29 | 4 | B | 298 | 426 | 7,934,700 | 96 | 463 | 589 | <50 | 29:01, 33:03 | 44:03, 58:01 | 03:02, 07:06 |
| 6038 | 28 | 4 | CRF01_AE | 389 | 973 | 2,507,072 | 48 | 527 | 615 | <20 | 02:06, 11:01 | 40:01, 40:06 | 07:02, 08:01 |
Pre-ART treatment.
2.2. Human leukocyte antigen (HLA) genotyping
HLA genotyping was performed using a multi-locus individual tagging-next generation sequencing (MIT-NGS) method as described previously [41]. Briefly, DNA was extracted from PBMC and full-length HLA genes were sequenced by NGS on the MiSeq platform (Illumina, San Diego, CA). FASTQ files generated by MiSeq Reporter were analyzed by NGSengine v2.16.2 (GenDX, Utrecht, The Netherlands).
2.3. 14–21mer peptides used to initiate T cell cultures
Peptide antigen used to initiate the T cell cultures ranged in length from 14 to 21 amino acids and were collectively termed as the “afferent” stimulator peptides. Highly conserved peptide antigens, referred to as “Epigraph” peptides, were identified by selecting regions spanning only the most conserved PTE in the HIV-1 proteome, based on M group database alignment using the Los Alamos HIV-1 database and the Epigraph tool [27]. The selected regions were cross checked for extremely high conservation with the CRF01 clade endemic to Thailand, as well as to all major clades of HIV-1 group M. While some highly conserved regions were identified in Env and Nef, we did not include them in this study as our original intent was to focus on the 5′ side of the genome to facilitate sequencing of clinical samples in future studies. We lifted this constraint for structure-based network analysis using graph theory metrics to identify structurally and functionally conserved epitopes [26] (Fig. 1). Peptides associated with this identification method were referred to as “Network” peptides. Overlapping peptides spanning the entire Gag proteome, referred to as “Gag” peptides, were used as control antigens. The library of peptides was synthesized by Sigma-Aldrich (St. Louis, MO), and each peptide was resuspended at a concentration of 5 mg/ml using either DMSO (for peptides with negative polarity) or DI water (for peptides with positive polarity). Resuspended peptides were aliquoted and stored at −80 °C until use.
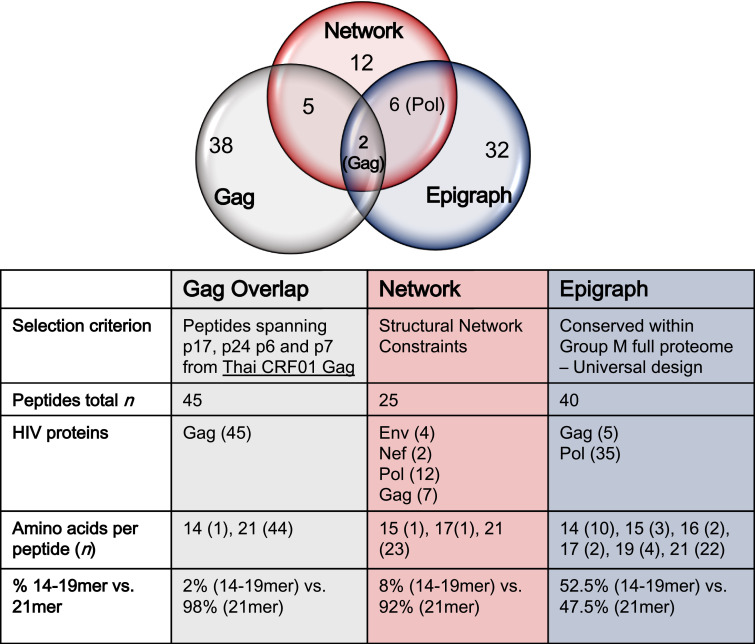
Fig 1.
Description of the three peptide pools used in the study. The full-length HIV-1 Gag protein peptide pool (Gag Overlap) was comprised of both conserved and non-conserved regions of Gag. Two distinct methods used to identify HIV-1 peptides for topologically important and highly conserved CTL antigenic targets (Network and Epigraph) are described in materials and methods. Selection of conserved regions was initially focused in the 5′ half of the genome (Gag and Pol) using the Epigraph method, as to facilitate downstream sequencing of clinical samples. However, this constraint was lifted when using the Network method, and some conserved regions within Env and Nef (3′ half of the genome) were added to this peptide pool.
2.4. Selection of 9–13mer epitopes within larger 14–21mer peptides for use as readout stimulating antigen
For selection of 9–13mer epitopes deriving from the larger afferent 14–21mer sequences (Gag, Network, and Epigraph), we used the Immune Epitope Database (IEDB) and the Los Alamos National Laboratory (LANL) database to identify known and predicted CD8+ T cell epitopes and their HLA associations, based on MHC class I binding predictions (IC50<500). We then selected epitopes contained within each 14–21mer that were predicted to provide maximum coverage of the different HLA types represented in our study participants as our readout antigens. These smaller peptide antigens were collectively referred to as “efferent” stimulator peptides throughout.
2.5. Isolation of peripheral blood monocytes and lymphocytes
PBMC from study participants were collected, aliquoted and frozen at a concentration of 40 × 106 PBMC per vial. Cells were shipped to our facility and stored in liquid nitrogen until use. PBMC were thawed, and monocytes and peripheral blood lymphocytes (PBL) were separated using human anti-CD14 Ab-coated microbeads to positively select monocytes (Miltenyi Biotec Cat# 130–05-201; RRID: AB_2665482) according to the manufacturer's instructions. Negatively isolated PBL were cryopreserved for future use.
2.6. Generation of human monocyte-derived DC (MDC1)
Isolated monocytes were cultured for 5 days in Iscove's Modified Dulbecco's Media (IMDM; Gibco Cat# 12440-053) containing 10% fetal bovine serum and 0.5% gentamicin in the presence of 1000 IU/ml of recombinant human (rh) GM-CSF (Sanofi-aventis Cat# NAC2004–5843–01) and 1000 IU/ml of rhIL-4 (R&D Systems, Cat# 204–1 L) to differentiate them into immature dendritic cells (iDC) in a 24-well plate. On day 5, iDC were divided into 4 groups of treatment, i.e., untreated (Empty; no peptide) or loaded with either the afferent stimulator Gag-overlapping (n = 45), Network (n = 25), or Epigraph (n = 40) peptide pools at a final concentration of 1μg/ml for each peptide. After a 2 h incubation at 37 °C, a previously described alpha-type-1 polarizing cytokine cocktail [42,43], consisting of IFN-α (1000 U/mL; Schering Corporation Cat# NDC:0085–1110–01), IFN-γ (1000 U/mL; R&D Systems, Cat# 285–1F), IL-1β (10 ng/mL; R&D Systems, Cat# 201-LB), TNF-α (25 ng/mL; R&D Systems, Cat# 210-TA), and polyinosinic:polycytidylic acid (Poly I:C; 20 ng/mL; Sigma-Aldrich Cat# P9582–5MG), was added to the iDC cultures for 48 h to yield mature MDC1. MDC1 were harvested and exposed again to the 14–21mer peptide pools for 2 h prior to being used for T cell stimulation.
2.7. In vitro stimulation of t cells
Antigen-loaded MDC1 from the 4 different groups described above (Empty, Gag, Network, and Epigraph) were counted and plated separately at a concentration of 7.5 × 104 MDC1 per well in a 24-well plate. Bulk T cells were negatively selected using the EasySep™ Human T Cell Enrichment Kit (STEMCELL Technologies, Cambridge, MA), and 7.5 × 105 T cells were added per well to the MDC1-containing wells (MDC1 to T cell ratio = 1:10). After an incubation of 45 min at 37 °C, soluble rhCD40L was added at a concentration of 0.25 μg/ml (MEGACD40L® protein, ENZO Life Sciences, Farmingdale, NY). After a 4 to 5 d culture period, rhIL-2 (250 IU/ml) and rhIL-7 (10 ng/ml) were added to the cultures and every 3 d thereafter. At day 21 in culture, each MDC1-stimulated T cell culture was tested for responses against their respective afferent (14–21mer) or efferent (9–13mer) Gag, Network, or Epigraph stimulator peptides as determined by IFNγ ELISpot and by CD107a flow cytometry staining where specifically mentioned. The non-peptide treated (Empty) T cell cultures were also tested for responses to each of the stimulator peptide groups and served as additional non-specific activated T cell controls.
2.8. Surface and intracellular staining and flow cytometry
Expanded T cells were harvested after 21 d in culture, counted, and plated in a V-bottom 96-well plate at a concentration of 1 × 105 cells per well and rested overnight before stimulation with the relevant 9–13mer peptide pools. Antigen-specific T cell responses were assessed by surface CD107a and IFNγ intracellular cytokine staining (ICS) flow cytometry analysis. Cells were resuspended in media containing CD107a-FITC labeled antibody (clone H4A3; BD Biosciences Cat# 555800; RRID: AB_396134) and BD GolgiStop™ (protein transport inhibitor containing monensin, BD Bioscience Cat# 554724) according to the manufacturer's instructions. Peptide pools containing 9–13mer peptide sequences were added to respective wells and incubated for 6 h at 37 °C. Wells without peptide were used as controls. After incubation, cells were washed with 1x PBS and stained for viability with LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit (Invitrogen™ Molecular Probes™) for 20 min at room temperature in the dark. Cells were subsequently stained with CD3 (APC-H7, clone SK7; BD Biosciences Cat# 641397; RRID: AB_1645731), CD4 (Pacific Blue, clone RPA-T4; BD Biosciences Cat# 558116; RRID: AB_397037), and CD8 (PerCP-Cy5.5, clone SK1; BD Biosciences, Cat# 341051; RRID: AB_400298) antibodies and incubated for 30 min at room temperature in FACS buffer. After surface staining, cells were washed, fix, and permeabilized using the BD Cytofix/Cytoperm™ Fixation/Permeabilization Kit (BD Bioscience), and stained with IFNγ monoclonal antibody (AlexaFluor® 700, clone B27; BD Bioscience Cat# 557995; RRID: AB_396977) for 45 min in the dark. Sample data were acquired using an LSR Fortessa II (BD Bioscience) flow cytometer and subsequently analyzed with the FlowJo software (Tree Star).
2.9. ELISpot for detecting IFNγ-secreting cells
In vitro expanded T cells were harvested, counted, and immediately tested for IFNγ secretion by ELISpot. The IFNγ ELISpot assay was performed following the Mabtech Human IFNγ ELISpotBasic protocol (Mabtech, Cincinnati, OH) using anti-human IFN-γ and biotin monoclonal antibodies (clones 1-D1K and 7-B6-1; Mabtech Cat# 3420–6–1000) and 96-well PVDF ELISpot plates from Millipore, as previously described [20]. Briefly, 3 × 104 T cells (100μl) were transferred to anti-IFNγ antibody-coated 96-well ELISpot plates. Individual 9mer peptide dilutions were prepared at 2μg/ml, and added (100μl) to T cell-containing wells to give a final peptide concentration of 1μg/ml. All ELISpot assays included negative-control wells with expanded T cells without peptide stimulation (Media only). T cells expanded using control MDC1 without peptide were also tested for responses to the respective 9–13mer peptide pools but yielded no antigen-specific responses (data not shown). IFNγ responses to each peptide were tested in duplicate wells. The enumeration of spots was done using the Autoimmun Diagnostika GmbH (AID) ELISpot reader and counting software (AID, Strassberg, Germany). ELISpot data, calculated as the mean of spots in duplicate wells minus the mean and 2 standard deviations (SD) of the negative control values, were shown as IFNγ spot forming units (SFU)/106 cells. As an additional level of scrutiny, we defined a response as positive only when the calculated ELISpot value was greater than 50 SFU/106 cells above the initial 2 SD cutoff described above.
2.10. Statistical analysis
The statistical analyses and plotting of data were performed using GraphPad Prism 8 version 8.0.2. The nonparametric Wilcoxon matched-pairs signed rank test was used to determine statistical significance for comparisons within each peptide pool (Fig 2c, 2e), between 21mer and 9mer efferent peptides (Fig 3b), and between Gag and Network Gag peptide pools (Fig 7). Where the mean rank of each condition was compared with the mean rank of a single control condition, statistical significance was assessed by the Kruskal-Wallis test, followed by Dunnett's test for multiple comparisons (Fig 2b). The Friedman test was used to measure the significance of differences between three or more matched groups, with correction for multiple comparisons by Dunnett's test (Fig 4c, 5b, 5c). Unless otherwise indicated, data are presented as the median with interquartile range.
Fig 2.
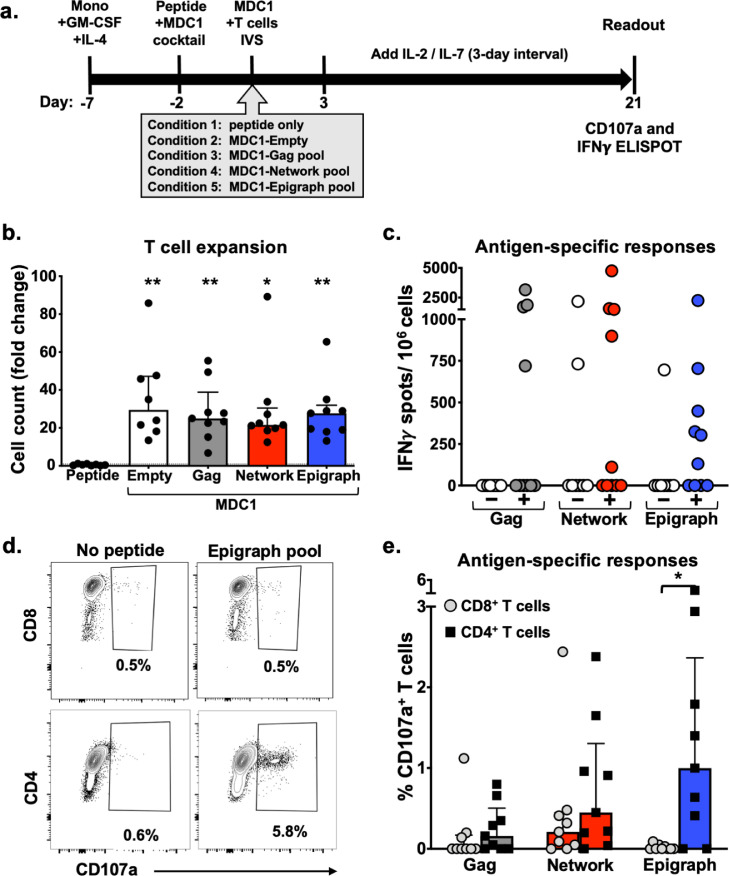
Unrefined evaluation of efferent HIV-1-specific T cell responses initially induced by antigen presenting autologous MDC1. a) Timeline of experimental conditions where monocytes were isolated from PBMC (Day −7) and treated with GM-CSF and IL-4. After 5 days (Day −2), the iDC were treated with the MDC1 Th1-polarizing cocktail and exposed to either DMSO (Empty, Condition 2) or to one of 3 peptide pools (Gag, Network and Epigraph), shown as Conditions 3, 4 and 5, respectively. T cells stimulated with peptide pool only (without DC) served as an additional control (Condition 1). After 48 h (Day 0), the differentially treated MDC1 were cocultured with autologous T cells. After 21 days, the T cells were assessed for net expansion and antigen-specific responses to a secondary exposure to the respective peptide pools. b) T cell expansion was determined by counting the in vitro sensitized T cells at day 21. Results are shown as fold change above the number of T cells used to initiate the cultures at day 0. c) T cell cultures that were expanded in the presence of MDC1 loaded with the Gag, Network or Epigraph peptide pools (represented by + symbol) were tested for induced IFNγ responses to secondary exposure to the respective peptide pools by ELISpot assay. T cells non-specifically expanded by the ‘Empty’ control MDC1 (represented by the – symbol) were also tested for responsiveness to each of the respective peptide pools. Results are shown as IFNγ SFU/106 cells. d) Representative flow cytometry plots of one participant illustrating the gating on Epigraph peptide pool-responding CD107a+CD8+ T cells (top panels) and CD107a+CD4+ T cells (bottom panels). e) Graphical representation of the percent of specific peptide-induced CD107a-expressing CD8+ T cells and CD4+ T cells in 9 study participants tested. **p<0.01, *p<0.05 [determined by Kruskal-Wallis test (b), and Wilcoxon matched-pairs signed rank test (c, e)].
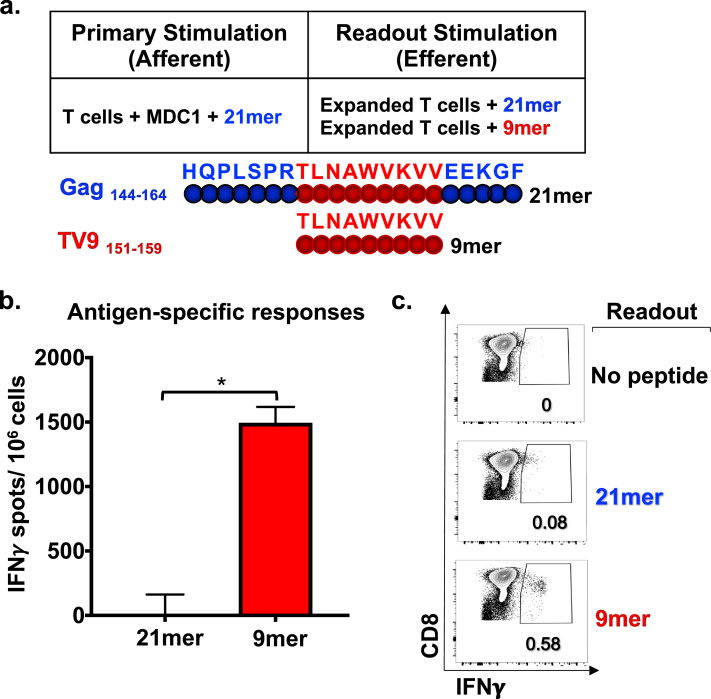
Fig 3.
Efferent CD8+ T cell responses become evident with refined analysis using 9–13mer peptide epitopes. a) Schematic representation of primary (afferent) and secondary assay readout (efferent) in vitro stimulation conditions of T cells from an HLA-A2+ study participant. MDC1 loaded with a 21mer Gag peptide (Gag 144–164) was the afferent stimulator, and either the same 21mer peptide or the known HLA-A2-restricted 9mer epitope TV9 (Gag 151–159) contained within that 21mer sequence was used as the efferent stimulator in readout assays. b) IFNγ ELISpot assay results showing T cell responses induced by the 9mer and 21mer efferent peptide stimulators, recorded as IFNγ SFU/106 cells. c) Peptide antigen-induced IFNγ production by CD8+ T cells determined by ICS flow cytometry analysis. *p<0.05 [determined by Wilcoxon matched-pairs signed rank test].
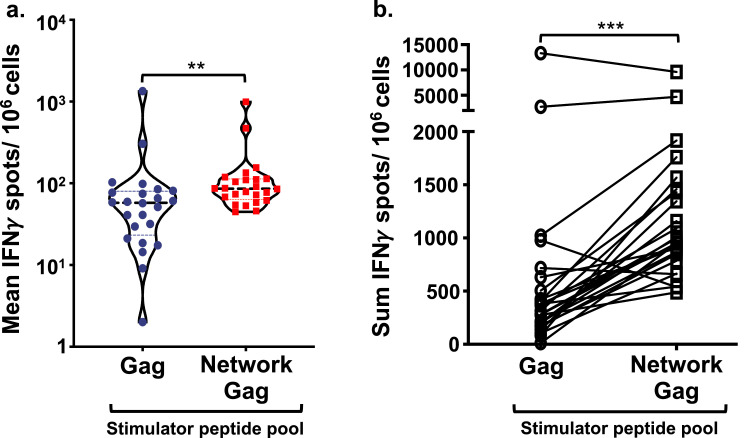
Fig 7.
MDC1 facilitate immune focusing toward conserved and topologically important epitopes. MDC1 were loaded with a pool of 14–21mer peptides containing either a mix of overlapping full-length HIV-1 Gag epitopes (Gag) or with the pool of Network peptides also containing Gag-associated epitopes (Network Gag), and each were used separately to initiate the activation (afferent) and long-term expansion of responsive T cells. Each dot represents the mean (a) or the sum (b) of the efferent readout responses (IFNγ SFU/106 cells) of each study participant induced against the individual 9–13mer Gag CTL epitopes derived from stimulator peptides common to both afferent stimulator peptide pools. ***p<0.001, **p<0.01 [determined by Wilcoxon matched-pairs signed rank test].
Fig 4.
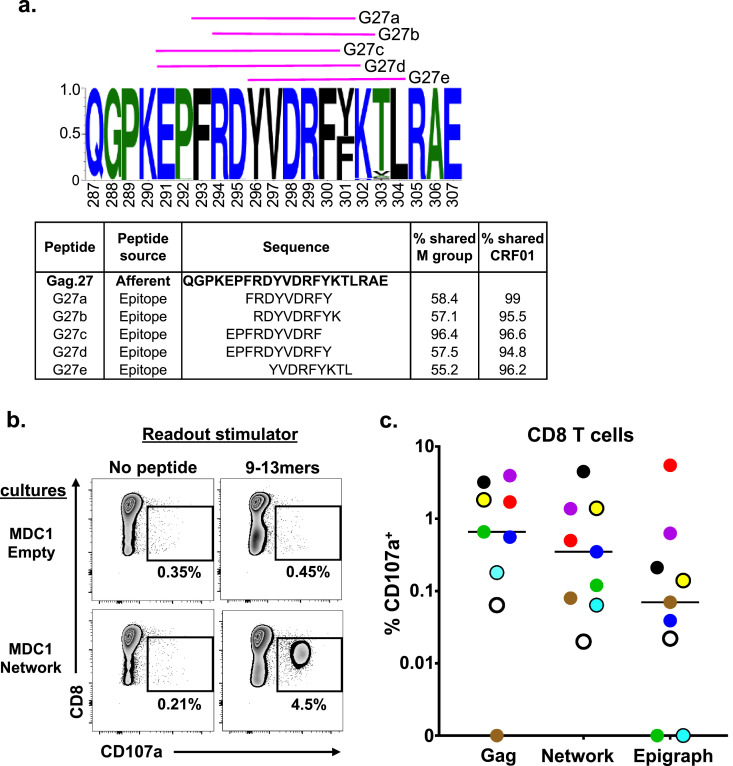
Efferent CD8+ T cell responses to 9–13mer HIV-1 antigen peptide pools across all study participants as evaluated by flow cytometry. a) Example of a sequence logo (Gag.27) summarizing the amino acid frequency within the afferent 21mer peptide. Pink lines represent the shorter 9–13mer peptide epitopes used to test efferent responses (also listed in the table). The table lists the peptides and the percentage of their exact sequence match with the HIV-1 M group and CRF01 clade. b) Representative flow cytometry data plots generated from one study participant illustrating 9–13mer (efferent) peptide antigen-induced expression of CD107a in responding CD8+ T cells generated from cultures initiated using MDC1 loaded with the Network (afferent) antigen pool. c) Graphical representation of each study participant's antigen-specific CD8+ T cell responses generated against the individual Gag, Network and Epigraph peptide pools determined by CD107a expression above background (n = 9). Each color represents an individual participant. The lines represent the median of the responses. Statistical significance was determined by the Friedman test, but the differences were not significant.
Fig 5.
MDC1 induce CTL responses of high heterogeneity against HIV-1 antigenic peptides. a) T cell cultures were initiated in 10 study participants using autologous MDC1 cells loaded with either the Gag, Network or Epigraph pools of 14–21mer peptides. Antigen-induced IFNγ T cell responses generated from each participant were determined at a single peptide level using a matrix of individual 9–13mer peptides derived from the Gag (n = 62, black circles), Network (n = 50, red circles) and Epigraph (n = 58, blue circles) pools. Results are shown as IFNγ SFU/106 cells with each circle representing a response to one peptide. One participant was not tested for Epigraph peptide responses due to insufficient cell numbers at the onset of the experiment. b) Breadth of the T cell response was quantified by the number of positive peptide responses (≥50 IFNγ SFU/106 cells) within each participant out of the total number of peptides (Gag, n = 62; Network, n = 50 and Epigraph, n = 58). c) Magnitude of the T cell response was quantified by compiling the sum of individual peptide-induced responses generated within each peptide group for each participant. Each color represents an individual participant, with the black line representing the median. *p<0.05 [determined by Friedman test].
2.11. Disclaimer and ethics statement
The views expressed are those of the authors and should not be construed to represent the positions of the U.S. Army or the Department of Defense. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70–25. The study had IRB approvals from Chulalongkorn University, Thailand, the Walter Reed Army Institute of Research, USA, and the University of Pittsburgh, USA, with informed consent from all study participants being obtained prior to the study.
3. Results
3.1. Characteristics of the RV254/SEARCH 010 study cohort
The specimens utilized in our study were provided from participants of the well-characterized RV254/SEARCH 010 study cohort of adults who were diagnosed with AHI based on HIV-1 screening at the Thai Red Cross Anonymous Clinic as described in the materials and methods [37]. The participants in our study were all men who started virus-suppressive ART during Fiebig I (n = 2), Fiebig II (n = 2), Fiebig III (n = 4), or Fiebig IV (n = 2) stages of early HIV-1 infection based on the Fiebig staging system [44] (Table 1). The HLA alleles of the respective participants are listed in Table 1. Samples used in our experiments were from blood specimens collected between weeks 48 to 240 post-ART initiation, from which PBMC were isolated and stored for future use. Plasma viremia loads at these time points were all bellow 20–50 copies/ml. The median CD4+ T cell count was 559 cells/ml (IQR 465–687), and the median CD8+ T cell count was 496 (IQR 435–596) (Table 1).
3.2. Distinct methods used to identify highly conserved and topologically important CTL antigenic targets
We used 3 distinct approaches for selecting the different sets of peptide immunogens for our study. The first set consisted of a full-length HIV-1 Gag protein peptide pool (referred to as Gag for simplicity) comprised of both conserved and non-conserved regions of Gag, which served as a reference point for our study because Gag-specific responses are associated with viral control in natural infection, including those in the highly variable p17 protein [45](Fig 1). In order to refine immunogen design and to select peptides representing conserved regions of HIV-1 Gag, Env and Pol, two approaches were used as described in the materials and methods. We defined one method as the Network-based design, which is founded on a structure-based network analysis that identifies topologically important epitopes within a protein [26]. The second method was defined as Epigraph, which is based on a highly efficient algorithm that can be used to define conserved HIV-1 peptides as PTE and a complementary set of antigens that can provide optimal population coverage of potential epitopes across diverse viruses [27], [28], [29]. The 3 peptide pools that were tested in our antigenicity studies consisted of peptides ranging from 14 to 21 amino acids in length (14–21mer; Fig 1). The full-length Gag peptide pool consisted of 45 peptides that overlapped by 10 amino acids and spanned Gag p17, p24, p7 and p6 proteins of the HIV-1 subtype CRF01-AE, the predominant strain in Asia. Of these 45 peptides, 44 were 21mers and 1 was a 14mer. Network peptides consisted of 25 15–21mer peptides that were combinations of Env (n = 4), Nef (n = 2), Pol (n = 12) and Gag (n = 7). Epigraph peptides consisted of 40 14–21mers comprised of Gag (n = 5) and Pol (n = 35). It is important to note that the peptide lengths for the three groups differed, with the Gag group having 98% 21mer vs. 2% 14–19mer, the Network group with 92% 21mer vs. 8% 15–17mer, and the Epigraph group with 47.5% 21mer vs. 52.5% 14–19mer. For in vitro T cell stimulation studies, MDC1 were loaded with the pooled peptides and used as APC for inducing autologous CD4+ and CD8+ T cell responses.
3.3. Unrefined evaluation of efferent HIV-1-specific T cell responses initially induced by antigen presenting autologous MDC1
To test for induction and long-term expansion of antigen-specific T cells responsive to the peptide pools described above, we performed a 21-day in vitro stimulation assay using previously described high IL-12p70-producing, antigen presenting MDC1 [46]. The MDC1 were either left untreated (Empty) or loaded with the pool of Gag, Network, or Epigraph peptides, and subsequently used for in vitro stimulation and expansion of isolated autologous T cell responders in long-term cocultures. Using the same peptide antigen pool used to initiate the DC:T cell cocultures, the expanded T cells were tested for their respective recall responsiveness to secondary antigenic stimulation by IFNγ ELISpot assay and flow cytometry analysis (see materials and methods) (Fig 2a).
While T cell expansions were comparable in all of the culture conditions that contained MDC1, even in the absence of exogenous peptide (Empty), T cells failed to expand in culture in the absence of MDC1 (Fig 2b). Antigen-induced IFNγ ELISpot responses were detected in all of the peptide groups tested (Gag, Network, Epigraph), with no significant differences in the cumulative magnitude of responses noted among these pools, although it is important to note that each peptide contained a different number of epitopes (Gag > Epigraph > Network) (Fig 2c). Importantly, T cell cultures that were non-specifically expanded using non-peptide loaded MDC1 (Empty) yielded few antigen responsive cells, with the exception of 2 participants whose cells responded to the Network peptides during the assay readout (Fig 2c), highlighting the importance of both MDC1 and peptide antigen for the selective induction and long-term survival of HIV-1 antigen-specific T cells. Since the expanded cultures included both CD4+ and CD8+ T cells, the ELISpot assay could not distinguish the relative contribution of the responses made by each T cell subset. Therefore, to differentiate between CD4+ and CD8+ T cell responses, we used flow cytometry analysis to test the relative responsiveness of these individual T cell subsets to the HIV-1 peptides based on their induced expression of CD107a after a 6 h stimulation with their respective peptide pool (Fig 2d, 2e). We found a higher percentage of antigen-responsive CD4+ T cells in all groups tested compared to CD8+ T cells, as shown in the representative flow cytometry plots of one study participant's responses to the entire Epigraph peptide pool, where the CD4+ T cell response reached 5.8% compared to a 0.5% response in the CD8+ T cell fraction (Fig 2d). In particular, the frequency of antigen-responsive CD8+CD107a+ T cells was lowest for those cultures generated using MDC1 loaded with either the Gag or Epigraph peptide pools. Cultures generated using MDC1 loaded with the conserved Network peptides also yielded relatively low CD8+ T cell responses (Fig 2e). Interestingly and in contrast to our results with the CD8+ T cells, the highest percentage of antigen-responsive CD4+CD107a+ T cells was generated using MDC1 loaded with the Epigraph peptide pool, which was significantly higher than responses induced among the CD8+T cells (Fig 2e).
3.4. Efferent CD8+ T cell responses become evident with refined analysis using 9–13mer peptide epitopes
We hypothesized that the observed overall higher responses found among CD4+ T cells compared to CD8+ T cells were due to the use of longer peptide antigens as direct stimulators in these short-term efferent readout assays. In accordance with previous findings [47], we reasoned that the longer peptides were more readily presented in the context of MHC class II as compared to MHC class I, especially since no professional APC were present to facilitate presentation in the assay, thus possibly reflecting an inefficient stimulation and detection of the antigen-specific CD8+ T cells in the short-term assays rather than their absence in the expanded T cell cultures. To demonstrate this, we first generated T cells from a representative HLA-A2+ study participant using autologous MDC1 loaded with one of the 21mer Gag (Gag144–164) peptides included in the Gag peptide pool, which contained a known HLA-A2-restricted 9mer epitope TV9 (Gag151–159) (Fig 3a). We used the same MDC1-based afferent stimulation strategy as described before and tested secondary efferent responses to either the 21mer Gag (Gag144–164) peptide or the 9mer Gag TV9 (Gag151–159) peptide epitope, measuring antigen-induced IFNγ production by ELISpot assay and ICS flow cytometry analysis (Fig 3b, 3c). Use of the 9mer TV9 peptide as the efferent readout stimulator revealed a significantly higher frequency of antigen-responsive IFNγ-producing CD8+ T cells by ELISpot (Fig 3b). We also observed by flow cytometry analysis that 0.08% and 0.58% of the CD8+T cells specifically produced IFNγ when stimulated with the 21mer and 9mer peptides, respectively (Fig 3c).
These results prompted us to redesign our CTL readout strategy in order to detect optimal CD8+ T cell-specific effector readout responses by selecting shorter 9–13mer assay readout peptide sequences derived from the larger afferent stimulator peptides using the IEDB and LANL database approach to identify MHC class I epitopes with the given HLA associations. We narrowed the peptide library selection based on the presence of their sequences within the longer afferent peptides and their associations with the HLA alleles common to our cohort of Thai study participants (Table 2, Table 3, Table 4). An example of a set of smaller efferent assay readout peptides that were selected and derived from one of the larger Gag-associated Network afferent stimulator peptides is described in Fig 4a. In addition, we determined the degree to which these sequences matched those shared among the entire HIV-1 M group, as well as to the Thailand dominant CRF01 clade specifically, an example of which is shown in Fig 4a and detailed in Fig S2-S5. A complete list of this analysis for all peptides tested is also included in Table 2, Table 3, Table 4. Of note, while the Network-associated epitopes individually showed a variable degree of exact matching among the entire M group (Fig S4), they were more highly matched within the relevant CRF01 clade of this Thai patient population (Fig S5). Given the algorithm used for their selection, the Epigraph selected peptides were more highly and uniformly matched to both the entire HIV-1 M group (Fig S4), as well as the Thailand dominant CRF01 clade (Fig S5). The Gag overlapping peptide group followed a similar pattern to that of the Network group, with a higher degree of variability in exact matching to the entire HIV-1 M group and a relative increase in exact matching to the CRF01 clade.
Table 2.
T cell responses to Gag-overlapping pool epitopes.
| Peptide | Peptide source | Sequence | % shared M group | % shared CRF01 | Protein | Start position | End position | Total Responder | HLA+/Total | #Respond/HLA+ | HLA associations |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gag.1 | Afferent | MGARASVLSGGKLDAWEKIRL | Gag | 1 | 21 | 4/10 | 4/4 | ||||
| G1a | Epitope | LSGGKLDAW | 22.7 | 72.9 | Gag | 8 | 16 | 4/10 | 2/4 | 2/6 | B*5801 |
| G1b | Epitope | RASVLSGGK | 34.1 | 65.3 | Gag | 4 | 12 | 1/10 | 0/1 | 0/3 | A*1101 |
| Gag.3 | Afferent | PGGKKKYRMKHLVWASRELER | Gag | 23 | 43 | 6/10 | 1/6 | ||||
| G3a | Epitope | YRMKHLVWA | 19 | 36.9 | Gag | 29 | 37 | 6/10 | 1/6 | 1/3 | C*0602, C*0701 |
| Gag.4 | Afferent | LVWASRELERFALNPGLLETA | Gag | 34 | 54 | 6/10 | 3/6 | ||||
| G4a | Epitope | WASRELERF | 87.77 | 93.8 | Gag | 36 | 44 | 4/10 | 1/4 | 1/6 | B*5801 |
| G4b | Epitope | ALNPGLLET | 41.6 | 63.4 | Gag | 45 | 53 | 5/10 | 1/5 | 1/2 | A*0201, A*0206 |
| Gag.5 | Afferent | ALNPGLLETAEGCQQIIEQLQ | Gag | 45 | 65 | 5/10 | 1/5 | ||||
| G4b* | Epitope | ALNPGLLET | repeat | Gag | 45 | 53 | 5/10 | 1/5 | 1/2 | A*0201, A*0206 | |
| Gag.6 | Afferent | GCQQIIEQLQSTLKTGSEELK | Gag | 56 | 76 | 3/10 | 1/3 | ||||
| G6a | Epitope | QIIEQLQST | 5.2 | 32.9 | Gag | 59 | 67 | 3/10 | 1/3 | 1/2 | A*0206 |
| Gag.7 | Afferent | TLKTGSEELKSLFNTVATLWC | Gag | 67 | 87 | 3/10 | 0/3 | ||||
| G7a | Epitope | LFNTVATLW | 2.5 | 14.4 | Gag | 78 | 86 | 3/10 | 0/3 | 0/6 | B*5801 |
| Gag.8 | Afferent | LFNTVATLWCVHQRIEVKDTK | Gag | 78 | 98 | 5/10 | 2/5 | ||||
| G8a | Epitope | NTVATLWCV | 46 | 47.6 | Gag | 80 | 88 | 2/10 | 1/2 | 1/2 | A*0201, A*0206 |
| G8b | Epitope | ATLWCVHQR | 6.3 | 27.7 | Gag | 83 | 91 | 2/10 | 0/2 | 0/3 | A*1101 |
| G7a* | Epitope | LFNTVATLW | repeat | Gag | 78 | 86 | 3/10 | 0/3 | 0/6 | B*5801 | |
| Gag.9 | Afferent | HQRIEVKDTKEALDKIEEVQK | Gag | 89 | 109 | 2/10 | 0/2 | ||||
| G9a | Epitope | IEVKDTKEAL | 20.8 | 42.23 | Gag | 92 | 101 | 2/10 | 0/2 | 0/0 | A*0201 |
| Gag.11 | Afferent | SQQKTQQAAAGTGSSSKVSQN | Gag | 111 | 131 | 3/10 | 1/3 | ||||
| G11a | Epitope | AAGTGSSSK | 3.4 | 23.1 | Gag | 119 | 127 | 3/10 | 1/3 | 1/3 | A*1101 |
| Gag.12 | Afferent | TGSSSKVSQNYPIVQNAQGQM | Gag | 122 | 142 | 6/10 | 4/5 | ||||
| G12a | Epitope | KVSQNYPIV | 37.1 | 72.9 | Gag | 127 | 135 | 3/10 | 2/3 | 2/2 | A*0206 |
| G12b | Epitope | SSSKVSQNY | 9.11 | 50.5 | Gag | 124 | 132 | 5/10 | 2/5 | 2/6 | B*5801 |
| Gag.13 | Afferent | PIVQNAQGQMVHQPLSPRTLN | Gag | 133 | 153 | 4/10 | 1/4 | ||||
| G13a | Epitope | MVHQPLSPR | 15 | 38.9 | Gag | 142 | 150 | 4/10 | 1/4 | 1/3 | A*1101 |
| Gag.14 | Afferent | HQPLSPRTLNAWVKVVEEKGF | Gag | 144 | 164 | 5/10 | 4/5 | ||||
| G14a | Epitope | LSPRTLNAW | 97.8 | 97.7 | Gag | 149 | 157 | 4/10 | 2/4 | 2/6 | B*5801 |
| G14b | Epitope | RTLNAWVKV | 96.6 | 99.04 | Gag | 150 | 158 | 5/10 | 4/5 | 4/7 | A*0206 |
| G14c | Epitope | TLNAWVKVV | 49.6 | 93.5 | Gag | 151 | 159 | 2/10 | 2/2 | 2/7 | A*02 |
| Gag.15 | Afferent | WVKVVEEKGFNPEVIPMFSAL | Gag | 155 | 175 | 7/10 | 5/7 | ||||
| G15a | Epitope | EEKGFNPEV | 16.23 | 97.25 | Gag | 160 | 168 | 1/10 | 0/1 | 0/2 | B*44, B*4415, B*4501 |
| G15b | Epitope | KGFNPEVIPMF | 16.3 | 92.4 | Gag | 162 | 172 | 4/10 | 0/4 | 0/1 | A*01, A*0201, B*08, B*0801, B*57, B*5701, B*5703, B*63 |
| G15c | Epitope | EVIPMFSAL | 58.1 | 95.5 | Gag | 167 | 175 | 6/10 | 4/6 | 4/8 | A*0206, A*26, A*2601, A*2602, A*2603, C*01, C*0102, C*02, C*03 |
| Gag.16 | Afferent | PEVIPMFSALSEGATPQDLNM | Gag | 166 | 186 | 6/10 | 6/6 | ||||
| G15c* | Epitope | EVIPMFSAL | repeat | Gag | 167 | 175 | 6/10 | 4/6 | 4/8 | A*0206, A*26, A*2601, A*2602, A*2603, C*01, C*0102, C*02, C*03 | |
| G16a | Epitope | SEGATPQDL | 88 | 95.2 | Gag | 176 | 184 | 4/10 | 3/4 | 3/6 | B*4001, B*4403 |
| G16b | Epitope | LSEGATPQDL | 87.9 | 95.2 | Gag | 175 | 184 | 4/10 | 3/4 | 3/6 | B*40, B*4001, B*42, B*44, B*4403, B*60, B*61 |
| Gag.17 | Afferent | EGATPQDLNMMLNIVGGHQAA | Gag | 177 | 197 | 5/10 | 5/10 | ||||
| G17a | Epitope | NIVGGHQAA | 25.8 | 94.4 | Gag | 189 | 197 | 1/10 | 1/1 | 1/2 | A*0206 |
| G17b | Epitope | ATPQDLNMMLNIV | 22.8 | 94.5 | Gag | 179 | 191 | 3/10 | 0/3 | 0/0 | B*07, B*08, B*08101, B*14, B*4202, B*53, B*5301 |
| G17c | Epitope | TPQDLNMMLNIV | 23.6 | 92.7 | Gag | 180 | 191 | 3/10 | 0/3 | 0/0 | B*07, B*08101, B*14, B*4202, B*53, B*5301 |
| G17d | Epitope | ATPQDLNMML | 23.4 | 95.1 | Gag | 179 | 188 | 3/10 | 0/3 | 0/0 | B*07, B*08101, B*14, B*4202, B*53, B*5301 |
| G17e | Epitope | DLNTMMNIVG | 24.4 | 97.8 | Gag | 183 | 192 | 0/10 | 0/0 | 0/7 | A*02, B*14 |
| Gag.18 | Afferent | LNIVGGHQAAMQMLKETINEE | Gag | 188 | 208 | 5/10 | 2/5 | ||||
| G17a* | Epitope | NIVGGHQAA | repeat | Gag | 189 | 197 | 1/10 | 1/1 | 1/2 | A*0206 | |
| G18a | Epitope | HQAAMQMLK | 96 | 96 | Gag | 194 | 202 | 4/10 | 1/4 | 1/3 | A*1101 |
| Gag.19 | Afferent | QMLKETINEEAAEWDRVHPVH | Gag | 199 | 219 | 5/10 | 4/5 | ||||
| G19a | Epitope | AEWDRVHPV | 16.6 | 57.2 | Gag | 210 | 218 | 5/10 | 2/5 | 2/5 | A*0206, B*4001, B*4403 |
| G19b | Epitope | TINEEAAEW | 88.4 | 89.5 | Gag | 204 | 212 | 2/10 | 1/2 | 1/6 | B*5801 |
| Gag.20 | Afferent | AEWDRVHPVHAGPIPPGQMRE | Gag | 210 | 230 | 5/10 | 2/5 | ||||
| G19a* | Epitope | AEWDRVHPV | repeat | Gag | 210 | 218 | 5/10 | 2/5 | 2/5 | A*0206, B*4001, B*4403 | |
| Gag.21 | Afferent | GPIPPGQMREPRGSDIAGTTS | Gag | 221 | 241 | 7/10 | 1/7 | ||||
| G21a | Epitope | MREPRGSDI | 71.9 | 76.3 | Gag | 228 | 236 | 5/10 | 1/5 | 1/3 | C*0602, C*0701 |
| G21b | Epitope | GPIPPGQM | 18.4 | 71.7 | Gag | 221 | 228 | 1/10 | 0/1 | 0/0 | B*35 |
| G21c | Epitope | GQMREPRGSDI | 71.6 | 76.1 | Gag | 226 | 236 | 5/10 | 0/5 | 0/3 | B*13 |
| Gag.22 | Afferent | RGSDIAGTTSTLQEQIGWMTN | Gag | 232 | 252 | 10/10 | 10/10 | ||||
| G22a | Epitope | TTSTLQEQI | 70.4 | 82.3 | Gag | 239 | 247 | 1/10 | 0/1 | 0/6 | B*5801 |
| G22b | Epitope | STLQEQIGW | 39.2 | 68.9 | Gag | 241 | 249 | 8/10 | 6/8 | 6/6 | B*5801 |
| G22c | Epitope | TSTLQEQIGWM | 39.1 | 59.6 | Gag | 240 | 250 | 8/10 | 8/8 | 8/10 | A*02, B*08, B*0801, B*57, B*5701 |
| Gag.23 | Afferent | LQEQIGWMTNNPPIPVGDIYK | Gag | 243 | 263 | 1/10 | 0/1 | ||||
| G23a | Epitope | MTNNPPIPV | 31.7 | 45.7 | Gag | 250 | 258 | 1/10 | 0/1 | 0/2 | A*0201, A*0206 |
| Gag.24 | Afferent | PPIPVGDIYKRWIILGLNKIV | Gag | 254 | 274 | 3/10 | 0/3 | ||||
| G24a | Epitope | WIILGLNKI | 86.8 | 98.1 | Gag | 265 | 273 | 0/10 | 0/0 | 0/2 | A*0206 |
| G24b | Epitope | IILGLNKIV | 86.9 | 98.4 | Gag | 266 | 274 | 2/10 | 0/2 | 0/2 | A*0206 |
| G24c | Epitope | IYKRWIILGL | 85.2 | 97.3 | Gag | 261 | 270 | 1/10 | 0/1 | 0/0 | A*0201 |
| Gag.25 | Afferent | WIILGLNKIVRMYSPVSILDI | Gag | 265 | 285 | 6/10 | 3/6 | ||||
| G24a* | Epitope | WIILGLNKI | repeat | Gag | 265 | 273 | 0/10 | 0/0 | 0/2 | A*0206 | |
| G25a | Epitope | KIVRMYSPV | 64.2 | 80.5 | Gag | 272 | 280 | 6/10 | 1/6 | 1/2 | A*0201, A*0206 |
| G25b | Epitope | RMYSPVSIL | 61.4 | 67.4 | Gag | 275 | 283 | 2/10 | 2/2 | 2/3 | A*0201, C*0702 |
| G24b* | Epitope | IILGLNKIV | repeat | Gag | 266 | 274 | 2/10 | 0/2 | 0/2 | A*0206 | |
| Gag.27 | Afferent | QGPKEPFRDYVDRFYKTLRAE | Gag | 287 | 307 | 6/10 | 6/6 | ||||
| G27a | Epitope | FRDYVDRFY | 58.4 | 99 | Gag | 293 | 301 | 5/10 | 1/5 | 1/3 | C*0602, C*0701 |
| G27b | Epitope | RDYVDRFYK | 57.1 | 95.5 | Gag | 294 | 302 | 4/10 | 1/4 | 1/3 | A*1101 |
| G27c | Epitope | EPFRDYVDRF | 96.4 | 96.6 | Gag | 291 | 300 | 2/10 | 2/2 | 2/7 | A*02, A*0201 |
| G27d | Epitope | EPFRDYVDRFY | 57.5 | 94.8 | Gag | 291 | 301 | 2/10 | 2/2 | 2/7 | A*0101, A*02, A*0201 |
| G27e | Epitope | YVDRFYKTL | 55.2 | 96.2 | Gag | 296 | 304 | 3/10 | 3/3 | 3/10 | A*02, A*26, A*2601, B*15, B*1503, B*1510, B*70, C*03, C*0303, C*04 |
| Gag.28 | Afferent | DRFYKTLRAEQATQEVKNWMT | Gag | 298 | 318 | 4/10 | 0/4 | ||||
| G28a | Epitope | QATQEVKNW | 27.5 | 83.5 | Gag | 308 | 316 | 4/10 | 0/4 | 0/6 | B*5801 |
| Gag.30 | Afferent | TLLVQNANPDCKSILKALGTG | Gag | 320 | 340 | 4/10 | 2/4 | ||||
| G30a | Epitope | VQNANPDCK | 86.4 | 94.7 | Gag | 323 | 331 | 4/10 | 2/4 | 2/3 | A*1101 |
| Gag.31 | Afferent | KSILKALGTGATLEEMMTACQ | Gag | 331 | 351 | 6/10 | 4/6 | ||||
| G31a | Epitope | KALGTGATL | 13.4 | 82.6 | Gag | 335 | 343 | 1/10 | 1/1 | 1/6 | B*5801 |
| G31b | Epitope | ATLEEMMTA | 81.9 | 90.37 | Gag | 341 | 349 | 5/10 | 1/5 | 1/2 | A*0201, A*0206 |
| Gag.32 | Afferent | TLEEMMTACQGVGGPSHKARV | Gag | 342 | 362 | 5/10 | 4/5 | ||||
| G32a | Epitope | EMMTACQGV | 96.5 | 98.2 | Gag | 345 | 353 | 3/10 | 2/3 | 2/7 | A*02, A*0201, A*0206 |
| G32b | Epitope | ACQGVGGPSHK | 46.8 | 92.8 | Gag | 349 | 359 | 3/10 | 1/3 | 1/3 | A*0201, A*11, A*1101, A*1103 |
| G32c | Epitope | GVGGPSHKAR | 50.1 | 92.3 | Gag | 352 | 361 | 2/10 | 1/2 | 1/3 | A*11 |
| Gag.33 | Afferent | VGGPSHKARVLAEAMSQAQHA | Gag | 353 | 373 | 5/10 | 1/5 | ||||
| G33a | Epitope | VLAEAMSQA | 34 | 35.6 | Gag | 362 | 370 | 5/10 | 1/5 | 1/2 | A*0201, A*0206 |
| Gag.34 | Afferent | AEAMSQAQHANIMMQRGNFKG | Gag | 364 | 385 | 7/10 | 1/7 | ||||
| G34a | Epitope | IMMQRGNFK | 18 | 49.4 | Gag | 376 | 384 | 7/10 | 1/7 | 1/3 | A*1101 |
| Gag.35 | Afferent | IMMQRGNFKGQKRIKCFNCGK | Gag | 376 | 397 | 8/10 | 2/8 | ||||
| G35a | Epitope | RIKCFNCGK | 16.8 | 65.6 | Gag | 388 | 397 | 3/10 | 1/3 | 1/3 | A*1101 |
| G34a* | Epitope | IMMQRGNFK | repeat | Gag | 376 | 384 | 7/10 | 1/7 | 1/3 | A*1101 | |
| Gag.36 | Afferent | KRIKCFNCGKEGHLARNCRAP | Gag | 388 | 408 | 3/10 | 1/3 | ||||
| G35a* | Epitope | RIKCFNCGK | repeat | Gag | 388 | 397 | 3/10 | 1/3 | 1/3 | A*1101 | |
| Gag.39 | Afferent | HQMKDCTERQANFLGKIWPSN | Gag | 421 | 441 | 5/10 | 3/5 | ||||
| G39a | Epitope | QANFLGKIW | 80.8 | 66.3 | Gag | 430 | 438 | 4/10 | 1/4 | 1/6 | B*5801 |
| G39b | Epitope | RQANFLGKI | 79.9 | 66.2 | Gag | 429 | 437 | 3/10 | 0/3 | 0/2 | A*0206 |
| Gag.43 | Afferent | MGEEITSFLKQEQKDKEHPPP | Gag | 465 | 486 | 6/10 | 2/6 | ||||
| G43a | Epitope | TSFLKQEQK | 0.091 | 5.64 | Gag | 470 | 478 | 1/10 | 1/1 | 1/3 | A*1101 |
| G43b | Epitope | EEITSFLKQ | 0.05 | 3.3 | Gag | 467 | 475 | 6/10 | 0/6 | 0/2 | B*4403 |
Table 3.
T cell responses to Network pool epitopes.
| Peptide | Peptide source | Sequence | % match M group | % match CRF01 | Protein | Start position | End Position | Total Respond | HLA+/Total | #Respond/HLA+ | HLA associations |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Env.1 | Afferent | LWDQSLKPCVKLTPLCVTLKC | gp160 | 111 | 131 | 3/10 | 3/3 | ||||
| E1a | Epitope | KLTPLCVTL | 82.9 | 85.8 | gp160 | 121 | 129 | 3/10 | 3/3 | 3/7 | A*02, A*0201 |
| Env.2 | Afferent | QCTHGIRPVVSTQLLLNGSLA | gp160 | 246 | 266 | 2/10 | 0/2 | ||||
| E2a | Epitope | RPVVSTQLLL | 30.1 | 91.8 | gp160 | 252 | 261 | 2/10 | 0/2 | 0/0 | B*07, B*08, B*35, B*3501 |
| Env.3 | Afferent | GGDPEIVTHSFNCGGEFFYCN | gp160 | 366 | 386 | 4/10 | 2/4 | ||||
| E3a | Epitope | PEIVTHS | 39 | 0 | gp160 | 369 | 375 | 4/10 | 2/4 | 2/7 | A*02 |
| E3b | Epitope | HSFNCGGEFFY | 2.7 | 0.19 | gp160 | 374 | 384 | 4/10 | 0/4 | 0/2 | A*03, A*29, B*08, B*15, B*1516, B*63, C*04, C*0401, C*0407 |
| Env.4 | Afferent | LLRAIEAQQHLLQLTVWGTKQ | gp160 | 555 | 575 | 5/10 | 5/5 | ||||
| E4a | Epitope | LLRAIEAQQHL | 41.4 | 54.3 | gp160 | 555 | 565 | 5/10 | 5/5 | 5/9 | A*11, B*1501, B*51, B*5101, B*15, B*51, B*57, B*58, B*63, C*03, C*0304, C*12, C*15, C*03, C*08 |
| Nef.1 | Afferent | GYFPDWQCYTPGPGVRYPLTF | Nef | 119 | 139 | 4/10 | 0/4 | ||||
| N1a | Epitope | YFPDWQCYTP | 1.2 | 0.2 | Nef | 120 | 129 | 1/10 | 0/1 | 0/7 | A*01, A*29, A*2902, A*3002, B*58, B*3501, B*37, B*3701, B*51, B*5401, B*57, B*5701, B*5801, B*63, C*06 |
| N1b | Epitope | YTPGPGVRY | 28.1 | 10.7 | Nef | 127 | 135 | 4/10 | 0/4 | 2/6 | A*24, B*07, B*08, B*57, B*58, B*63 |
| Nef.2 | Afferent | VRYPLTFGWCFKLVPVEPDLV | Nef | 133 | 153 | 4/10 | 3/4 | ||||
| N2a | Epitope | RYPLTFGWCF | 61.7 | 0.2 | Nef | 134 | 143 | 4/10 | 3/4 | 3/8 | A*11, A*2301, A*2402, A*33, B*27, B*35, B*53 |
| N2b | Epitope | PLTFGWCFKLV | 43.6 | 0.2 | Nef | 136 | 146 | 4/10 | 3/4 | 3/7 | A*02, A*0201, B*1517, B*57, B*63 |
| Pol.17 | Afferent | YWQATWIPEWEFVNTPPLVKL | Pol | 560 | 580 | 4/10 | 3/4 | ||||
| P17a* | Epitope | FVNTPPLVK | 93.1 | 96.9 | Pol | 571 | 579 | 4/10 | 3/4 | 3/7 | A*02, A*11, A*1101 |
| Pol.22 | Afferent | ILVAVHVASGYIEAEVIPAET | Pol | 788 | 808 | 7/10 | 1/7 | ||||
| P22a* | Epitope | GYIEAEVIPAET | 84.9 | 96 | Pol | 797 | 808 | 4/10 | 1/4 | 1/1 | A*2402, B*4002 |
| P22b* | Epitope | HVASGYIEA | 79.8 | 97.2 | Pol | 793 | 801 | 7/10 | 0/7 | 0/0 | B*5401 |
| Pol.4 | Afferent | GPKVKQWPLTEEKIKAL | Pol | 173 | 189 | 5/10 | 0/5 | ||||
| P2b* | Epitope | GPKVKQWPLT | 82.9 | 90.46 | Pol | 173 | 182 | 5/10 | 0/5 | 0/0 | B*07, B*08, B*0801, B*4202 |
| Pol.5 | Afferent | RKLVDFRELNKRTQDFWEVQL | Pol | 227 | 247 | 4/10 | 2/4 | ||||
| P5a* | Epitope | KLVDFRELNK | 97.3 | 96.3 | Pol | 228 | 237 | 4/10 | 2/4 | 2/5 | A*03, A*0301, B*08, A*34, A*29, B*40 |
| Pol.8 | Afferent | NNETPGIRYQYNVLPQGWKGS | Pol | 291 | 311 | 4/10 | 1/4 | ||||
| P8a* | Epitope | NNETPGIRYQY | 83.9 | 92.1 | Pol | 291 | 301 | 4/10 | 0/4 | 0/0 | B*18, B*1801 |
| P8b* | Epitope | TPGIRYQYNVL | 87.1 | 92.6 | Pol | 294 | 304 | 2/10 | 1/2 | 1/7 | A*02, B*1401, B*4202 |
| Pol.9 | Afferent | NVLPQGWKGSPAIFQ | Pol | 302 | 316 | 2/10 | 0/2 | ||||
| P9a* | Epitope | LPQGWKGSPAI | 94.5 | 92.6 | Pol | 304 | 314 | 2/10 | 0/2 | 0/0 | B*3910, B*5101, B*5401 |
| Pol.31 | Afferent | RKYTAFTIPSINNETPGIRYQ | Pol | 280 | 300 | 5/10 | 3/5 | ||||
| P31a | Epitope | KYTAFTIPSI | 55.3 | 67.7 | Pol | 281 | 290 | 5/10 | 3/5 | 3/7 | A*02, A*0201, A*0205, A*0217, B*51, B*5101 |
| Pol.32 | Afferent | FTIPSINNETPGIRYQYNVLP | Pol | 285 | 305 | 7/10 | 0/7 | ||||
| P32a | Epitope | NETPGIRYQYNVL | 83.8 | 92.6 | Pol | 292 | 304 | 5/10 | 0/5 | 0/0 | B*18, B*1801, B*1401, B*4202 |
| P32b | Epitope | NETPGIRYQY | 84.7 | 85.8 | Pol | 292 | 301 | 4/10 | 0/4 | 0/0 | B*18 |
| P32c | Epitope | IRYQYNVL | 88.9 | 93.5 | Pol | 297 | 304 | 6/10 | 0/6 | 0/0 | B*1401 |
| Pol.33 | Afferent | ETWETWWTEYWQATWIPEQEF | Pol | 551 | 571 | 3/10 | 0/3 | ||||
| P33a | Epitope | TWETWWTEYW | 16.7 | 23.4 | Pol | 552 | 561 | 3/10 | 0/3 | 0/2 | B*44, B*49 |
| Pol.34 | Afferent | QATWIPEWEFVNTPPLVKLWY | Pol | 562 | 582 | 4/10 | 2/4 | ||||
| P34a | Epitope | FVNTPPLVK | repeat | Pol | 571 | 579 | 4/10 | 2/4 | 2/7 | A*02, A*11, A*1101 | |
| Pol.35 | Afferent | KEALLDTGADDTVLEEMNLPG | Pol | 76 | 96 | 4/10 | 3/4 | ||||
| P35a | Epitope | LLDTGADDTVL | 94.5 | 97.9 | Pol | 79 | 89 | 4/10 | 3/4 | 3/7 | A*02, A*0201 |
| Pol.36 | Afferent | PTPVNIIGRNLLTQIGCTLNF | Pol | 135 | 155 | 3/10 | 2/3 | ||||
| P36a | Epitope | TQIGCTLNF | 53.1 | 67.4 | Pol | 147 | 155 | 3/10 | 2/3 | 2/6 | B*1501, B*1503, B*62, C*03 |
| Gag.15 | Afferent | WVKVVEEKGFNPEVIPMFSAL | Gag | 155 | 175 | 4/10 | 3/4 | ||||
| G15a | Epitope | EEKGFNPEV | 18.01 | 97.3 | Gag | 160 | 168 | 2/10 | 0/2 | 0/2 | B*44, B*4415, B*4501 |
| G15b | Epitope | KGFNPEVIPMF | 16.3 | 97 | Gag | 162 | 172 | 4/10 | 0/4 | 0/1 | A*01, A*0201, B*08, B*0801, B*57, B*5701, B*5703, B*63 |
| G15c | Epitope | EVIPMFSAL | 58.7 | 95.5 | Gag | 167 | 175 | 4/10 | 3/4 | 3/8 | A*0206, A*26, A*2601, A*2602, A*2603, C*01, C*0102, C*02, C*03 |
| Gag.16 | Afferent | PEVIPMFSALSEGATPQDLNM | Gag | 166 | 186 | 5/10 | 5/5 | ||||
| G15c* | Epitope | EVIPMFSAL | repeat | Gag | 167 | 175 | 4/10 | 3/4 | 3/8 | A*0206, A*26, A*2601, A*2602, A*2603, C*01, C*0102, C*02, C*03 | |
| G16a | Epitope | SEGATPQDL | 86.7 | 95.2 | Gag | 176 | 184 | 4/10 | 0/4 | 0/4 | B*4001, B*4403 |
| G16b | Epitope | LSEGATPQDL | 86.5 | 95.2 | Gag | 175 | 184 | 3/10 | 1/3 | 1/6 | B*40, B*4001, B*42, B*44, B*4403, B*60, B*61 |
| Gag.17 | Afferent | EGATPQDLNMMLNIVGGHQAA | Gag | 177 | 197 | 7/10 | 6/7 | ||||
| G17a | Epitope | NIVGGHQAA | 26.5 | 95.05 | Gag | 189 | 197 | 5/10 | 2/5 | 2/2 | A*0206 |
| G17b | Epitope | ATPQDLNMMLNIV | 23.8 | 94.5 | Gag | 179 | 191 | 5/10 | 0/5 | 0/0 | B*07, B*08, B*08101, B*14, B*4202, B*53, B*5301 |
| G17c | Epitope | TPQDLNMMLNIV | 24.3 | 95.3 | Gag | 180 | 191 | 2/10 | 0/2 | 0/0 | B*07, B*08,101, B*14, B*4202, B*53, B*5301 |
| G17d | Epitope | ATPQDLNMML | 24.7 | 94.1 | Gag | 179 | 188 | 7/10 | 0/7 | 0/0 | B*07, B*08101, B*14, B*4202, B*53, B*5301 |
| G17e | Epitope | DLNTMMNIVG | 25.6 | 96.6 | Gag | 183 | 192 | 4/10 | 3/4 | 3/7 | A*02, B*14 |
| Gag.21 | Afferent | GPIPPGQMREPRGSDIAGTTS | Gag | 221 | 241 | 6/10 | 1/6 | ||||
| G21a | Epitope | MREPRGSDI | 70.7 | 76.3 | Gag | 228 | 236 | 5/10 | 1/5 | 1/3 | C*0602, C*0701 |
| G21b | Epitope | GPIPPGQM | 19.5 | 71.6 | Gag | 221 | 228 | 4/10 | 0/4 | 0/0 | B*35 |
| G21c | Epitope | GQMREPRGSDI | 70.6 | 76.1 | Gag | 226 | 236 | 3/10 | 0/3 | 0/3 | B*13 |
| Gag.22 | Afferent | RGSDIAGTTSTLQEQIGWMTN | Gag | 232 | 252 | 8/10 | 7/8 | ||||
| G22a | Epitope | TTSTLQEQI | 69.8 | 75 | Gag | 239 | 247 | 3/10 | 2/3 | 2/6 | B*5801 |
| G22b | Epitope | STLQEQIGW | 36.8 | 68.9 | Gag | 241 | 249 | 6/10 | 5/6 | 5/6 | B*5801 |
| G22c | Epitope | TSTLQEQIGWM | 36.5 | 68.6 | Gag | 240 | 250 | 7/10 | 7/7 | 7/10 | A*02, B*08, B*0801, B*57, B*5701 |
| Gag.27 | Afferent | QGPKEPFRDYVDRFYKTLRAE | Gag | 287 | 307 | 7/10 | 6/7 | ||||
| G27a | Epitope | FRDYVDRFY | 51.6 | 95.5 | Gag | 293 | 301 | 5/10 | 1/5 | 1/3 | C*0602, C*0701 |
| G27b | Epitope | RDYVDRFYK | 50.5 | 95.5 | Gag | 294 | 302 | 3/10 | 1/3 | 1/3 | A*1101 |
| G27c | Epitope | EPFRDYVDRF | 96.1 | 96.9 | Gag | 291 | 300 | 4/10 | 3/4 | 3/7 | A*02, A*0201 |
| G27d | Epitope | EPFRDYVDRFY | 50.7 | 94.8 | Gag | 291 | 301 | 4/10 | 3/4 | 3/7 | A*0101, A*02, A*0201 |
| G27e | Epitope | YVDRFYKTL | 48.7 | 96.2 | Gag | 296 | 304 | 4/10 | 4/4 | 4/10 | A*02, A*26, A*2601, B*15, B*1503, B*1510, B*70, C*03, C*0303, C*04 |
| Gag.32 | Afferent | TLEEMMTACQGVGGPSHKARV | Gag | 342 | 362 | 5/10 | 4/5 | ||||
| G32a | Epitope | EMMTACQGV | 95.5 | 98.2 | Gag | 345 | 353 | 3/10 | 2/3 | 2/7 | A*02, A*0201, A*0206 |
| G32b | Epitope | ACQGVGGPSHK | 47.3 | 92.8 | Gag | 349 | 359 | 3/10 | 1/3 | 1/3 | A*0201, A*11, A*1101, A*1103 |
| G32c | Epitope | GVGGPSHKAR | 47.3 | 92.3 | Gag | 352 | 361 | 3/10 | 0/3 | 0/3 | A*11 |
Table 4.
T cell responses to Epigraph pool epitopes.
| Peptide | Peptide source | Sequence | % match M group | % match CRF01 | Protein | Start position | End Position | Total Respond | HLA+/Total | #Respond/HLA+ | HLA associations |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gag.46 | Afferent | REPRGSDIAGTTST | Gag | 229 | 242 | 1/9 | 0/1 | ||||
| G46a | Epitope | SDIAGTTST | 95.1 | 97.1 | Gag | 234 | 242 | 1/9 | 0/1 | 0/1 | A*01 |
| Gag.47 | Afferent | REPRGSDIAGTTSN | variant | Gag | 229 | 242 | 3/9 | 1/3 | |||
| G47a | Epitope | SDIAGTTSN | variant | Gag | 234 | 242 | 3/9 | 1/3 | 1/1 | A*01 | |
| Gag.48 | Afferent | IYKRWIILGLNKIVRMYSP | Gag | 261 | 279 | 4/9 | 4/9 | ||||
| G24a* | Epitope | WIILGLNKI | 86.9 | 98.1 | Gag | 265 | 273 | 1/9 | 1/1 | 1/7 | A*0206 |
| G24b* | Epitope | IILGLNKIV | 86.1 | 98.4 | Gag | 266 | 274 | 3/9 | 2/3 | 2/2 | A*0206 |
| G48a | Epitope | KRWIILGLNK | 84.6 | 97.4 | Gag | 263 | 272 | 0/9 | 0/0 | 0/6 | A*02, A*0201,A*03,A*24,A*30, B*81, B*27, B*2705 |
| G48b | Epitope | IILGLNKIVR | 86.8 | 97.8 | Gag | 266 | 275 | 3/9 | 2/3 | 2/7 | A*0201, A*03, A*11, A*33,B*27 |
| G48c | Epitope | GLNKIVRMY | 96.8 | 98.4 | Gag | 269 | 277 | 1/9 | 0/1 | 0/1 | B*1501, B*27, B*3502, B*3503, B*5301, C*01 |
| Gag.49 | Afferent | QGPKEPFRDYVDRF | Gag | 287 | 300 | 3/9 | 2/3 | ||||
| G27c* | Epitope | EPFRDYVDRF | 96.6 | 96.6 | Gag | 291 | 300 | 3/9 | 2/3 | 2/6 | A*02, A*0201 |
| Gag.50 | Afferent | LEEMMTACQGVGGP | Gag | 343 | 356 | 2/9 | 1/2 | ||||
| G32a* | Epitope | EMMTACQGV | 95.6 | 98.2 | Gag | 345 | 353 | 2/9 | 1/2 | 1/6 | A*02, A*0201, A*0206 |
| Pol.1 | Afferent | WKPKMIGGIGGFIKV | Pol | 98 | 112 | 2/9 | 1/2 | ||||
| P1a | Epitope | WKPKMIGGI | 88.6 | 91.4 | Pol | 98 | 106 | 0/9 | 0/0 | 0/5 | C*03 |
| P1b | Epitope | KMIGGIGGFI | 91.8 | 92.3 | Pol | 101 | 110 | 2/9 | 1/2 | 1/6 | A*02, A*0201, B*1501, B*62 |
| Pol.2 | Afferent | TVPVKLKPGMDGPKVKQWPLT | Pol | 162 | 182 | 2/9 | 2/2 | ||||
| P2a | Epitope | PGMDGPKVKQ | 83.8 | 90.8 | Pol | 169 | 178 | 1/9 | 1/1 | 1/3 | A*11 |
| P2b | Epitope | GPKVKQWPL | 84.8 | 90.8 | Pol | 173 | 181 | 2/9 | 0/2 | 0/0 | B*07, B*08, B*0801, B*4202 |
| Pol.3 | Afferent | TVPVTLKPGMDGPKVKQWPLT | Pol | 162 | 182 | 2/9 | 2/2 | ||||
| P2a* | Epitope | PGMDGPKVKQ | repeat | Pol | 169 | 178 | 1/9 | 1/1 | 1/3 | A*11 | |
| P2b* | Epitope | GPKVKQWPL | repeat | Pol | 173 | 181 | 2/9 | 0/2 | 0/0 | B*07, B*08, B*0801, B*4202 | |
| Pol.4 | Afferent | GPKVKQWPLTEEKIKAL | Pol | 173 | 189 | 2/9 | 0/2 | ||||
| P2b* | Epitope | GPKVKQWPLT | 84.3 | 90.5 | Pol | 173 | 182 | 2/9 | 0/2 | 0/0 | B*07, B*08, B*0801, B*4202 |
| Pol.5 | Afferent | RKLVDFRELNKRTQDFWEVQL | Pol | 227 | 247 | 3/9 | 2/3 | ||||
| P5a | Epitope | KLVDFRELNK | 97.3 | 96.3 | Pol | 228 | 237 | 3/9 | 2/3 | 2/5 | A*03, A*0301, B*08, A*34, A*29, B*40 |
| Pol.6 | Afferent | RTQDFWEVQLGIPHPAGLKKK | Pol | 238 | 258 | 2/9 | 1/2 | ||||
| P6a | Epitope | GIPHPAGLKK | 85.8 | 89.9 | Pol | 248 | 257 | 2/9 | 1/2 | 1/3 | A*03, A*0301, A*11, B*07, C*12 |
| Pol.7 | Afferent | SVTVLDVGDAYFSVPLD | Pol | 260 | 276 | 3/9 | 1/3 | ||||
| P7a | Epitope | SVTVLDVGDAY | 94.5 | 91.7 | Pol | 260 | 270 | 3/9 | 1/3 | 1/3 | A*0206, A*1101 |
| P7b | Epitope | TVLDVGDAYFS | 96.3 | 93.2 | Pol | 262 | 272 | 1/9 | 1/1 | 1/3 | A*0206, A*1101 |
| P7c | Epitope | LDVGDAYFSVP | 92.8 | 91.4 | Pol | 264 | 274 | 1/9 | 0/1 | 0/0 | Unknown |
| P7d | Epitope | VGDAYFSVPLD | 80.6 | 88.3 | Pol | 266 | 276 | 1/9 | 0/1 | 0/0 | Unknown |
| P7e | Epitope | TVLDVGDAY | 96.5 | 93.9 | Pol | 262 | 270 | 1/9 | 1/1 | 1/3 | A*0206, A*1101 |
| Pol.8 | Afferent | NNETPGIRYQYNVLPQGWKGS | Pol | 291 | 311 | 3/9 | 2/3 | ||||
| P8a | Epitope | NNETPGIRYQY | 83.9 | 92.3 | Pol | 291 | 301 | 1/9 | 0/1 | 0/6 | B*18, B*1801 |
| P8b | Epitope | TPGIRYQYNVL | 87.1 | 92.6 | Pol | 294 | 304 | 2/9 | 2/2 | 2/6 | A*02, B*1401, B*4202 |
| Pol.9 | Afferent | NVLPQGWKGSPAIFQ | Pol | 302 | 316 | 2/9 | 0/2 | ||||
| P9a | Epitope | LPQGWKGSPAI | 94.5 | 97.9 | Pol | 304 | 314 | 2/9 | 0/2 | 0/0 | B*3910, B*5101, B*5401 |
| Pol.10 | Afferent | IYQYMDDLYVGSDLEIGQHR | Pol | 335 | 354 | 3/9 | 2/3 | ||||
| P10a | Epitope | YQYMDDLYV | 92.8 | 93.2 | Pol | 336 | 344 | 3/9 | 2/3 | 2/6 | A*02, A*0201 |
| Pol.11 | Afferent | TTPDKKHQKEPPFLWMGYELHP | Pol | 370 | 391 | 2/9 | 1/2 | ||||
| P11a | Epitope | TTPDKKHQKE | 92.6 | 96.6 | Pol | 370 | 379 | 2/9 | 1/2 | 1/3 | A*11 |
| Pol.12 | Afferent | EPPFLWMGYELHPD | Pol | 379 | 392 | 2/9 | 0/2 | ||||
| P12a | Epitope | EPPFLWMGY | 96.7 | 96.9 | Pol | 379 | 387 | 2/9 | 0/2 | 0/0 | Unknown |
| Pol.13 | Afferent | SWTVNDIQKLVGKLNWASQIY | Pol | 406 | 426 | 3/9 | 1/3 | ||||
| P13a | Epitope | KLVGKLNWA | 97.6 | 99 | Pol | 414 | 422 | 2/9 | 1/2 | 1/6 | A*02, A*0201 |
| P13b | Epitope | KLNWASQIY | 98.6 | 99.4 | Pol | 418 | 426 | 2/9 | 0/2 | 0/1 | A*24, A*29, A*30, A*3002, B*1501, B*1502, B*3502, B*3503, B*5301, C*02, C*14 |
| Pol.14 | Afferent | EAELELAENREILK | Pol | 453 | 466 | 3/9 | 1/3 | ||||
| P14a | Epitope | EAELELAENRE | 92.7 | 92.9 | Pol | 453 | 463 | 2/9 | 0/2 | 0/0 | Unknown |
| P14b | Epitope | ELELAENREIL | 91.2 | 94.5 | Pol | 455 | 465 | 1/9 | 0/1 | 0/0 | Unknown |
| P14c | Epitope | LELAENREILK | 90.3 | 91.4 | Pol | 456 | 466 | 1/9 | 1/1 | 1/3 | A*1101 |
| P14d | Epitope | LAENREILK | 92.1 | 91.7 | Pol | 458 | 466 | 0/9 | 0/0 | 0/3 | A*1101 |
| Pol.15 | Afferent | EAELELAENREILR | variant | Pol | 453 | 466 | 3/9 | 2/3 | |||
| P15a | Epitope | LELAENREILR | variant | Pol | 456 | 466 | 2/9 | 1/2 | 1/4 | B*4001, B*4403 | |
| P15b | Epitope | LELAENREI | 92.1 | 94.2 | Pol | 456 | 464 | 3/9 | 2/3 | 2/4 | B*4001, B*4403 |
| Pol.16 | Afferent | YQEPFKNLKTGKYA | Pol | 497 | 510 | 0/9 | 0/0 | ||||
| P16a | Epitope | FKNLKTGKY | 82.3 | 91.7 | Pol | 501 | 509 | 0/9 | 0/0 | 0/0 | Unknown |
| Pol.17 | Afferent | YWQATWIPEWEFVNTPPLVKL | Pol | 560 | 580 | 2/9 | 1/2 | ||||
| P17a | Epitope | FVNTPPLVK | 93.1 | 96.9 | Pol | 571 | 579 | 2/9 | 1/2 | 1/6 | A*02, A*11, A*1101 |
| Pol.18 | Afferent | FVNTPPLVKLWYQLEK | Pol | 571 | 586 | 2/9 | 1/2 | ||||
| P17a* | Epitope | FVNTPPLVK | repeat | Pol | 571 | 579 | 2/9 | 1/2 | 1/6 | A*02, A*11, A*1101 | |
| P18a | Epitope | PLVKLWYQL | 95.6 | 98.2 | Pol | 576 | 584 | 2/9 | 1/2 | 1/6 | A*02, A*0201 |
| Pol.19 | Afferent | EVNIVTDSQYALGIIQAQPD | Pol | 647 | 666 | 2/9 | 1/2 | ||||
| P19a | Epitope | VTDSQYALGI | 92.5 | 93.9 | Pol | 651 | 660 | 2/9 | 1/2 | 1/2 | A*24, A*6802, B*81, B*14, B*1402, B*1503, B*1517, B*3502, B*3503, B*52, B*5301, C*08, C*12 |
| Pol.20 | Afferent | WVPAHKGIGGNEQVDKLVS | Pol | 690 | 708 | 2/9 | 0/2 | ||||
| P20a | Epitope | WVPAHKGIGGN | 98.6 | 97.9 | Pol | 690 | 700 | 1/9 | 0/1 | 0/0 | Unknown |
| P20b | Epitope | PAHKGIGGNEQ | 98.9 | 96.6 | Pol | 692 | 702 | 1/9 | 0/1 | 0/0 | Unknown |
| P20c | Epitope | HKGIGGNEQVD | 88.4 | 95.1 | Pol | 694 | 704 | 1/9 | 0/1 | 0/0 | Unknown |
| P20d | Epitope | GIGGNEQVDKL | 87.6 | 94.5 | Pol | 696 | 706 | 2/9 | 0/2 | 0/0 | Unknown |
| P20e | Epitope | GGNEQVDKLVS | 87.7 | 94.2 | Pol | 698 | 708 | 2/9 | 0/2 | 0/0 | Unknown |
| Pol.21 | Afferent | HGQVDCSPGIWQLDCTHLEGK | Pol | 766 | 786 | 2/9 | 0/2 | ||||
| P21a | Epitope | QVDCSPGI | 89 | 97.2 | Pol | 768 | 775 | 0/9 | 0/0 | 0/0 | Unknown |
| P21b | Epitope | QLDCTHLEGK | 94.3 | 97.5 | Pol | 777 | 786 | 2/9 | 0/2 | 0/0 | A*03 |
| Pol.22 | Afferent | ILVAVHVASGYIEAEVIPAET | Pol | 788 | 808 | 3/9 | 0/3 | ||||
| P22a | Epitope | GYIEAEVIPAET | 80.1 | 96.3 | Pol | 797 | 808 | 1/9 | 0/1 | 0/1 | A*2402, B*4002 |
| P22b | Epitope | HVASGYIEA | 84.9 | 97.2 | Pol | 793 | 801 | 3/9 | 0/3 | 0/0 | B*5401 |
| Pol.23 | Afferent | IEAEVIPAETGQETAYQETAY | Pol | 799 | 819 | 2/9 | 0/2 | ||||
| P23a | Epitope | IPAETGQETAY | 90 | 92.6 | Pol | 804 | 814 | 2/9 | 0/2 | 0/0 | A*2601, B*07, B*3501, B*56 |
| Pol.24 | Afferent | QEFGIPYNPQSQGVVESMNKE | Pol | 852 | 872 | 1/9 | 0/1 | ||||
| P24a | Epitope | GIPYNPQSQ | 98.6 | 99.1 | Pol | 855 | 863 | 1/9 | 0/1 | 0/0 | Unknown |
| Pol.25 | Afferent | QGVVESMNKELKKIIGQVR | Pol | 863 | 881 | 3/9 | 2/3 | ||||
| P25a | Epitope | ELKKIIGQVR | 98.6 | 72.3 | Pol | 872 | 881 | 3/9 | 2/3 | 2/4 | A*33, A*3301 |
| Pol.26 | Afferent | QAEHLKTAVQMAVFIHNFKRK | Pol | 883 | 903 | 2/9 | 1/2 | ||||
| P26a | Epitope | HLKTAVQMAVF | 90.4 | 95.4 | Pol | 886 | 896 | 1/9 | 0/1 | 0/6 | A*02, B*0801, B*1524, B*3501, B*3502, B*3503, B*40, B*57, B*5701, B*5703 |
| P26b | Epitope | AVQMAVFIHNFK | 93.2 | 98.2 | Pol | 890 | 901 | 1/9 | 0/1 | 0/3 | A*03, A*0301, A*1101 |
| P26c | Epitope | AVFIHNFKRK | 89.7 | 95.7 | Pol | 894 | 903 | 2/9 | 1/2 | 1/3 | A*03, A*0301, A*11, A*1101, A*24, A*68 |
| Pol.27 | Afferent | AVFIHNFKRKGGIGGYSAGER | Pol | 894 | 914 | 2/9 | 2/2 | ||||
| P26c* | Epitope | AVFIHNFKRK | repeat | Pol | 894 | 903 | 2/9 | 1/2 | 1/3 | A*03, A*0301, A*11, A*1101, A*24, A*68 | |
| P27a | Epitope | FKRKGGIGGY | 91.8 | 94.5 | Pol | 900 | 909 | 2/9 | 1/2 | 1/7 | B*15, B*1503, B*27, B*2705, C*01, C*03 |
| Pol.28 | Afferent | AVFIHNFKRKGGIGEYSAGER | variant | Pol | 894 | 914 | 2/9 | 1/2 | |||
| P26c* | Epitope | AVFIHNFKRK | repeat | Pol | 894 | 903 | 2/9 | 1/2 | 1/3 | A*03, A*0301, A*11, A*1101, A*24, A*68 | |
| Pol.29 | Afferent | KIQNFRVYYRDSRDP | Pol | 934 | 948 | 0/9 | 0/0 | ||||
| P29a | Epitope | KIQNFRVYYR | 82.9 | 96.9 | Pol | 934 | 943 | 0/9 | 0/0 | 0/8 | A*01, A*03, A*11, A*30, A*3002, A*32, A*3303 |
| Pol.30 | Afferent | WKGPAKLLWKGEGAVVIQDNS | Pol | 950 | 970 | 2/9 | 1/2 | ||||
| P30a | Epitope | LLWKGEGAV | 98.5 | 97.9 | Pol | 956 | 964 | 2/9 | 1/2 | 1/3 | A*02, A*0201 |
We repeated the primary in vitro stimulation of T cells as described earlier in Fig 2 and subsequently tested the effector CD8+ T cell responses at day 21 against the relevant 9–13mer efferent stimulator peptides. The results for one representative participant's efferent secondary CD8+ T cell responses to the Network peptide 9–13mer peptide pool based on the induced expression of CD107a is shown in Fig 4b. We observed a substantial increase in the percentage of antigen-responsive CD107a+CD8+ T cells using this approach, with 4.5% of the CD8+ T cells responding to the Network peptide pool compared to the unstimulated background of 0.21% (Fig 4b). Importantly, these responses were not observed in the T cell cultures expanded by MDC1 in the absence of antigen (MDC1-Empty) (Fig 4b). When we analyzed all participants for antigen-induced CD107a+CD8+ T cell responses, we found a range of responses to each peptide pool, with most participants reacting to the Gag group (median: 0.70%) and all participants reacting to the Network group (median: 0.35%). The Epigraph group elicited the lowest levels of antigen-specific responses (median: 0.07%) (Fig 4c). Indeed, MDC1 are capable of processing and cross-presenting the larger HIV-1 associated peptides in the context of MHC class I to induce HIV-1-specific CD8+ T cell responses to highly conserved and topologically important regions of the HIV-1 proteome. These data also demonstrate that smaller, 9–13mer peptides are required for more accurate quantification of antigen-specific CD8+ T cell responses.
3.5. MDC1 induce CTL responses of high heterogeneity against HIV-1 antigenic peptides
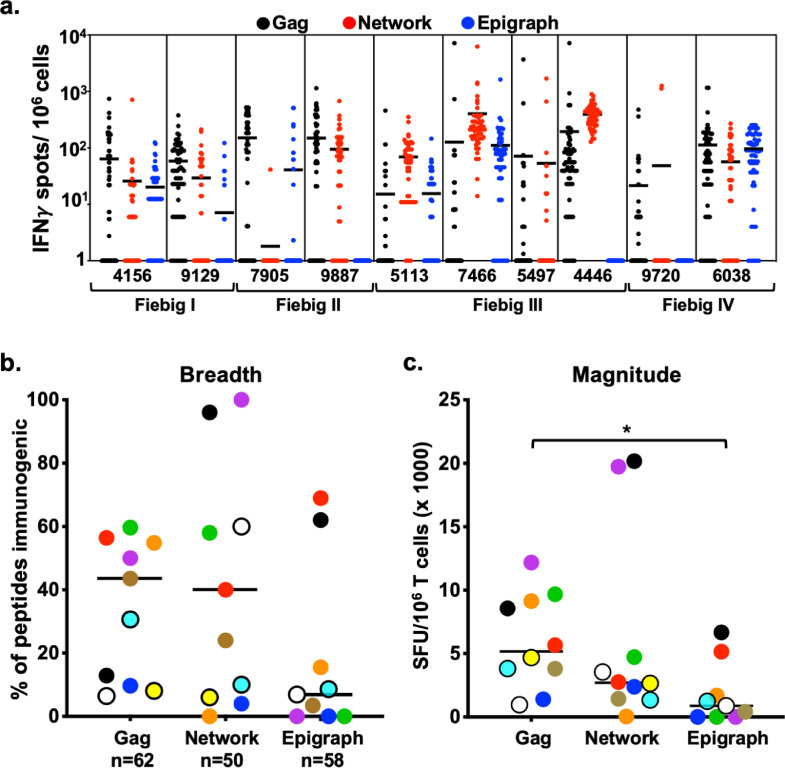
Each study participant had individual T cell cultures generated using autologous MDC1 stimulator cells loaded with either the larger Gag, Network, or Epigraph peptides. To further analyze the breadth and magnitude of the expanded antigen-specific T cells, the efferent CD8+ T cell responses were tested at a single peptide level by evaluating antigen-induced IFNγ secretion by ELISpot using a matrix of 62, 50, and 58 individual 9–13mer peptides for the Gag, Network, and Epigraph groups, respectively. As mentioned above, we selected known and predicted CD8+ T cell epitopes (by searching the LANL database) contained within the larger 14–21mer sequences that spanned a maximum number of HLA associations representative of the HLA types of the participants in our cohort. This was done to minimize the number of peptides needed to yield maximum results for each peptide group tested, as the cell number was ultimately a limiting factor. One participant was not tested by the Epigraph pool due to insufficient PBMC availability at the time of initiation of the cultures.
The participants had a broad range of T cell responses to the peptide antigens, with some participants responding to all 3 HIV-1 peptide groups, and others responding to peptides within at least 2 of the groups (Fig 5a). Interestingly, there were particularly high T cell responses generated against several of the 9–13mer peptides among the Fiebig stage III participants, which were especially apparent with the Network peptides. When analyzing participant ID 7466 and 4446 (Fiebig III) in particular, we found these 2 participants had relatively higher viral loads at week 0, before initiation of ART (Table 1), compared to the other Fiebig III participants (Table 1). Moreover, we observed a positive correlation between viral load at initiation of ART (week 0) and the magnitude of the responses against the Network pool of peptides (Fig S1). The median breadth of the CD8+ T cell responses to the individual Gag, Network, and Epigraph peptides was 44% (27/62), 40% (20/50), and 7% (4/58), respectively (Fig 5b). When analyzing the magnitude of the cumulative responses to each peptide group, the Gag group generated significantly higher values than the Epigraph group (Fig 5c). The Network peptides also elicited higher responses than the Epigraph group although this trend did not reach statistical significance (Fig 5c).
3.6. Unveiling HLA-associated effector T cell responses to 9–13mer HIV-1 peptides
We analyzed the efferent responses to each of the individual 9–13mer peptides and compartmentalized these epitopes based on the respective larger peptides used during the afferent arm of the MDC1-mediated stimulation from which they were derived, as well as to their known or predicted HLA associations. By doing this, we could predict which of the epitopes were more likely to induce a response based on the individual's HLA genotype (Fig 6; Table 2, Table 3, Table 4). We first quantified the number of individuals that generated antigen-specific effector responses relevant to each 14–21mer peptide used during the initial afferent MDC1-mediated induction of the T cell cultures. We determined the individuals with T cells responding to any of relevant 9–13mer epitopes derived from that larger afferent stimulator peptide by IFNγ ELISpot assay. A value of ≥50 IFNγ SFU/106 cells was used as a cutoff for an individual to be considered a responder to that epitope (Fig 6). These results were then matched with the participant's HLA types (Table 2, Table 3, Table 4).
Fig 6.
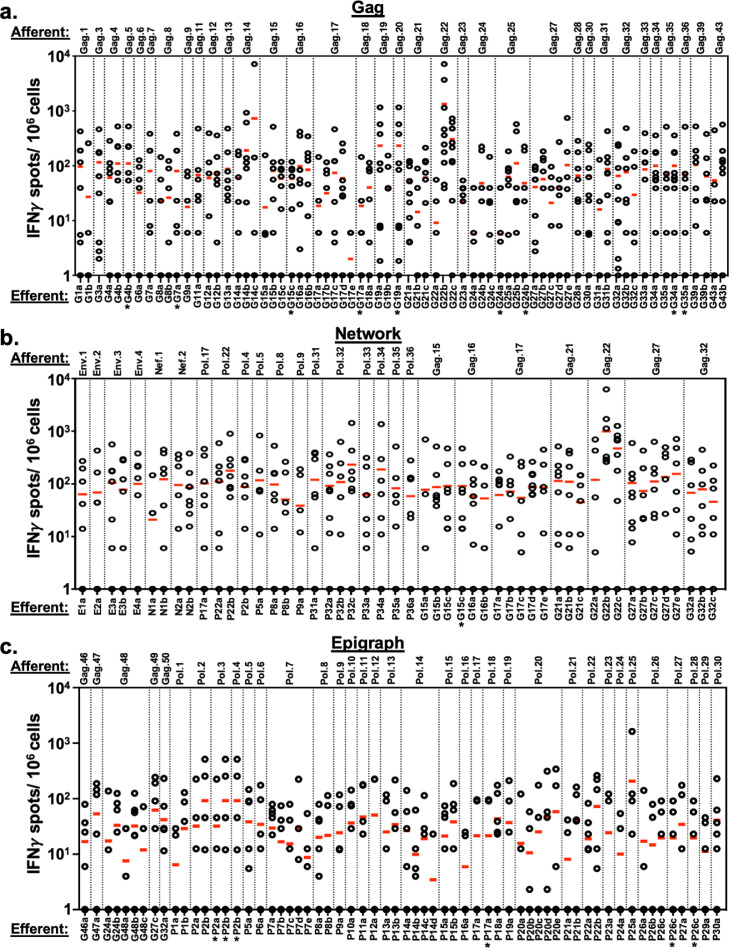
Unveiling 9–13mer peptide HLA-restricted T cell responses to HIV-1 antigen pools. T cell responses to 9–13mer single epitopes in the Gag peptide pool (a), Network peptide pool (b), and Epigraph peptide pool (c) were analyzed by IFNγ ELISpot. Responses to individual 9–13mer peptides from all 10 study participants were plotted and organized based on the respective larger afferent stimulator peptides used during the initiation of the MDC1:T cell cocultures. The 14–21mer afferent peptides (top of graphs) and their corresponding efferent assay readout 9–13mer peptides (bottom of graphs) used in the study are listed in Tables 2, 3 and 4. Each plotted circle represents a value generated from 1 of the 10 study participants tested in response to that particular efferent peptide stimulator. A value of ≥50 IFNγ SFU/106 cells was used as a cutoff for an individual to be considered a responder to that epitope.
Of the 34 14–21mer peptides contained in the Gag peptide pool used in the initial MDC1-based T cell stimulation that were assessed, 10 effectively generated cultures yielding antigen-specific effector responses to relevant 9–13mer efferent peptides in at least 5 or more of the 10 study participants tested (Fig 6a; Table 2). One afferent peptide, Gag.22 (sequence RGSDIAGTTSTLQEQIGWMTN), which was present in both the Gag and Network peptide pools, was particularly immunogenic and generated efferent epitope responses in all 10 study participants. Importantly, these responsive participants had restricting HLA alleles representative of those capable of binding the Gag.22-associated 9–13mer epitopes based on MHC class I binding predictions (IC50<500). Interestingly, we found that 13 of the larger afferent stimulator peptides from the Gag pool used in the initiation of the T cell cultures yielded specific effector responses to epitopes outside of those previously reported or expected to be associated with the HLA types of the individual participants based on their poor predicted MHC class I binding potential (all having IC50>500) (Table 2), suggesting potentially novel epitopes or unreported HLA associations. In the cultures generated using the Network peptide group (Fig 6b; Table 3), we found 11 of the 25 afferent MDC1 stimulator peptides yielded responses to relevant efferent 9–13mer epitopes in at least 50% of the participants. We also observed 4 afferent peptide antigens that drove efferent responses in 50% or more of the participants to peptides outside of expected HLA associations, again indicating the potential discovery of new HLA-associated epitopes. Finally, the Epigraph group also elicited a broad range of responses (Fig 6c; Table 4). While the overall response rate among the participants was not as high as to the Gag and Network peptide groups, with only 1 afferent peptide (Gag.48) from the Epigraph pool approaching a 50% efferent response rate (4 out of 9), 12 of the 35 afferent peptides induced responses to their associated 9–13mer peptides in more than 30% of the study participants.
3.7. MDC1 facilitate immune focusing toward subdominant and topologically important epitopes
We hypothesized that HIV-1-specific T cell responses naturally dominate or become skewed toward immunodominant epitopes in HIV-1, which can be highly variable, allowing the virus the capacity to easily adapt and escape CTL immune pressure. This can also lead to the establishment of adapted epitopes that drive ineffective cross-reactive memory CTL responses, characterized by their release of cytokines and chemokines in the absence of target killing, thus promoting an inflammatory environment favoring viral dissemination [20,21,23]. Alternatively, some mutations associated with immune selection pressure impair viral fitness. We posited that MDC1 can be used to facilitate immune focusing of CTL activity toward subdominant HIV-1 epitopes or to sequences that are most important to maintain protein structures critical to the overall fitness of the virus. Importantly, the Gag peptide pool covered the entire Gag proteome and therefore was comprised of both variable and highly conserved Gag-associated epitopes, which included epitopes shared in the select Network peptide pool. This allowed us to directly compare the output (efferent) responses against the same MHC class I epitopes from T cells derived from cultures that were initiated using MDC1 loaded with either the full-length Gag peptide pool or the select Network peptides. The notion here was to test whether elimination of the variable epitopes from the afferent antigen pool used to load the MDC1 stimulators during coculture initiation would result in enhanced and focused responses toward the conserved and topologically important regions of the virus.
We therefore limited exposure of the MDC1 stimulators during the initiation of the T cell cultures to those select Gag peptides contained within the Network pool, as compared to when they comprised a fraction of the larger pool of overlapping Gag peptides. This resulted in the selective expansion of effector T cells having a significantly enhanced capacity to respond, in both breadth and magnitude, to the same 9–13mer Gag epitopes in the readout assays (Fig 7a). This enhancing effect was noted in total T cell responses generated among all the participants against 21 out of 24 common 9–13mer Gag CTL epitopes tested (Fig 7b). These results demonstrate the utility and potential of using MDC1 to generate and focus effector CTL responses toward conserved and topologically important regions of HIV-1 in those who initiate ART during early HIV-1 infection as a therapeutic strategy to prevent HIV-1 immune escape and control viral replication.
4. Discussion
Major effort has been made toward the design of novel therapeutic strategies to achieve a functional cure for HIV-1 infection. This would allow HIV-1-infected individuals to immunologically restrain or inhibit the virus without ART, similar to that observed with HIV-1 EC [48]. Research that has focused on understanding the mechanism of natural viral control in EC has highlighted the importance of generating effective HIV-1-specific CD8+ T cell responses to manage the virus [49,50]. Such individuals appear to control viremia by targeting sequence-conserved epitopes derived from topologically important regions of the viral proteome critical to viral fitness [26,27,[32], [33], [34],51,52]. Moreover, early ART initiation provides additional benefit for the induction of strong CTL immunity through preservation of CD4+ T cell support [53].
In this study, we investigated the induction of HIV-1-specific T cell responses in early ART treated HIV-1-infected individuals, applying two different analytical approaches to select optimal peptide antigens for immunotherapy, based on their presumed importance to viral fitness of the virus. In one approach, a set of peptides derived from Gag and Pol were selected on the basis of extreme sequence conservation and coverage of a wide range of HLA associations. In the other approach, targeted Gag-, Pol-, Env-, and Nef-associated epitopes were selected based on topological analysis and structural importance. As a more conventional design, we also tested a pool of overlapping peptides spanning the Gag proteome, which consisted of both conserved and highly variable epitopes from the Thailand strain CRF01. The Gag peptide pool served as a control verifying that HIV-1-specific responses could indeed be induced in those initiating ART during the very early stages of HIV-1 infection using the MDC1-based approach. As we expected, peptide antigen alone was unable to induce the activation and expansion of isolated T cells but instead required the presence of the MDC1. Moreover, we did not see enhancement of HIV-1-specific responses when T cells were expanded in the presence of MDC1 in the absence of peptide. In addition to inducing responses against the overlapping Gag epitopes, MDC1 were also notably consistent in their capability to activate and expand HIV-1-specific T cells responsive to both the highly conserved and the structurally important peptide pools. While we did not extend our analysis to test for true killing activity of all of the participants’ CTL due to insufficient cell numbers, we were able to test the killing activity of MDC1 Network peptide-induced CTL generated from study participant 5497 (Fig S6). This indicated that these CTL have the capacity to specifically eliminate HIV-1-infected targets.
Importantly, we showed that MDC1 can efficiently process the longer exogenous peptides for cross-presentation to drive a broad range of MHC class I-restricted CTL responses. This is in line with previous studies demonstrating the benefit of exposure of DC to longer exogenous peptides and/or proteins rather than shorter peptides to achieve prolonged cross-presentation on MHC class I and efficient afferent induction of CTL immunity [54], [55], [56], [57]. However, we also found that the larger afferent peptide T cell stimulators were ineffective for detecting CD8+ T cell responses in the readout assays. This was not that surprising given that the test samples lacked the presence of APC, and it is in accordance with previous reports demonstrating that longer sequences are not ideal for identifying and enumerating MHC class I-restricted CD8+ T cell responses [47].
In order to more accurately evaluate the induced effector CD8+ T cell responses by IFNγ ELISpot, we assessed a group of smaller 9–13mer stimulator peptides that were derived from the larger peptide sequences used to initiate the cultures. To select these smaller peptides for testing, we defined a number of optimal MHC class I epitopes derived from the larger 14–21mer sequences that also had broad HLA associations specific to those haplotypes present in our study cohort. This allowed us to identify a wider range of CD8+ T cell responses than would have otherwise been revealed. However, this dramatically increased the number of peptide epitopes to be tested, the number of cells needed to carry out the assays, and the overall difficulty of monitoring the effectiveness of the MDC1-based vaccine strategy. Moreover, by selecting only those peptides associated with the HLA haplotypes within our cohort for the readout assays, we understand that a number of unknown but relevant epitope responses could have been missed and thus exposes a limitation of our study. This is supported by our finding of a number of unexpected responses to certain epitopes in participants with HLA haplotypes that differed from those known to be associated with the peptide being tested, suggesting the discovery of novel MHC class I-associated epitopes using the MDC1 stimulation strategy. Nevertheless, our study highlights the difficulties of accurately assessing or monitoring the responsiveness to a vaccine that has the potential of eliciting responses to such a broad range of antigenic targets spanning a large number of HLA associations, through the use of standard monitoring techniques, as well as the need to be cognizant of these points when designing a vaccine study.
Detailed analysis of the data generated in this study led to additional noteworthy findings. One outcome was the observed higher magnitude of effector T cell responses focused against topologically important Gag epitopes when they were introduced as part of the afferent Network peptide pool as compared to when they were part of the overlapping Gag set of peptides. It is important to note that the Network peptide set contained a fewer number of peptides than the Gag peptide pool (25 vs. 45, respectively). Therefore, it is unclear if the noted enhancement in immune focusing using the Network strategy was due to the specific removal of more immunogenic variable epitopes from the stimulator pool or if this was instead due to the fewer number of peptides targets in the pool, allowing for a decrease in competition for MHC class I peptide loading. Another notable observation was that a higher magnitude of responses was generated against several of the 9–13mer Network peptides, particularly in those individuals initiating ART in the Fiebig III stage of infection. In addition, a positive correlation was found between a study participant's viral load upon initiation of ART and the magnitude of responses against the Network peptide pool. Although the sample size of individuals tested was too small to make definitive conclusions, we speculate that both the time to treatment and the antigen burden (viral load) at ART initiation had considerable impact on the potential to induce primary and/or memory CD8+ T cell responses against topologically important and conserved epitopes. This subsequently affects the quality of the anamnestic responses generated in vitro, which are likely what were measured here. These results are in accordance with a previous report showing that individuals in stage 3 (equivalent to Fiebig III) undergo full T cell differentiation during AHI and are therefore able to respond more effectively than those who begin ART before or after this window of peak viremia [58].
While the two methods employed to select the antigenic targets were strategically different, we hypothesized that both would share a high degree of conservation within the HIV-1 M group and, in particular, within the CRF01 clade most relevant to the cohort. However, when the sequences were analyzed for their degree of exact matching to viruses found among the entire M group and those specific to the CRF01 clade, the Network peptides were not as relatively conserved as the Epigraph peptides (Fig S4, S5). However, this apparent variability within the Network epitopes was primarily observed in peptides that were selected at an interim stage of development of the structure-based network analysis approach and were in fact not classified as being topologically important upon finalization of the algorithm [9]. The modest polymorphism observed for topologically important epitopes that precludes them from being classified as exact matches in HIV-1 M group is likely the result of their underlying immunogenicity, which we observed in this study. Moreover, mutations within highly networked epitopes have been shown to impair viral fitness, so these responses may be effective in mediating immune control despite mutational changes [26,[59], [60], [61]]. The finding that the Network epitopes display some sequence variation and were targeted more robustly than the Epigraph peptides could indicate that these are more immunogenic in vivo and that these topologically important regions would indeed be part of the peptide repertoire presented on infected target cells. Moreover, while the extreme level of conservation among the Epigraph peptide sequences could mean that they are important for viral fitness and less likely to be subjected to CTL immune escape, they may not be as readily presented on HIV-1-infected cells. On the other hand, the lower responses directed at the Epigraph peptides may be related to the lower proportion of 21mers present in that set, which if prepared in a longer format these peptides might be more effectively processed by the MDC1 for MHC class I cross-presentation as previously suggested [54], [55], [56], [57]. Nevertheless, we believe that both antigen selection strategies for vaccine development have their distinct advantages and therefore propose a mosaic model, where antigenic peptides representing both highly conserved and topologically important viral sequences are targeted.
Our study demonstrates that MDC1 can induce HIV-1-specific T cells against highly conserved and topologically important epitopes in individuals initiating ART during early stages of infection. The rationale for focusing on this patient population was based on the notion that early ART may help to intercept the development of CTL immune dysfunction that often results due to chronic activation or the establishment of CTL escape variants. We posited that this would also provide a better chance to induce de novo CTL or to re-direct memory CTL activity toward critical regions of the virus, thus making this patient population attractive for testing this vaccine approach. One limitation of our current study is that we do not provide distinct evidence to determine whether the CTL responses induced were a result of de novo priming, as previously demonstrated [20], [21], [22], or if the responses were a result of recall responses of memory T cells that originated through natural infection. Most importantly, rather than targeting the viral proteome as a whole, by carefully targeting only those regions of the virus that contain ultra-conserved and topologically important epitopes, the competition for MHC binding and presentation that may otherwise favor memory T cell responses toward highly variable immunodominant epitopes can be minimized. By prospectively avoiding such ‘immunologic noise’, we demonstrate a level of ‘immune focusing’, i.e., where HIV-1 antigen-loaded MDC1 have a therapeutic competence to selectively drive and focus CTL responses toward highly conserved epitopes that are less likely to lead to viral escape from CTL pressure and more likely to prove critical to viral fitness [26]. Together, this study highlights the potential of implementing this MDC1-based approach for selective immune targeting as an integral part of a successful ‘kick and kill’ strategy to control chronic HIV-1 infection.
Contributors
TM Garcia-Bates co-wrote the manuscript and performed experiments and data analyses. ML Palma and PA Piazza provided experimental data and technical assistance. RR Anderko contributed to statistical analyses and manuscript editing. BT Korber, GD Gaiha, and BD Walker participated in the study design, provided intellectual input, and editing of the manuscript. DC Hsu, J Ananworanich, N Phanuphak, R Thomas, S Tovanabutra, E Kroon, NL Michael, SA Riddler, and JW Mellors, contributed to the study design and logistics related to involvement of the Thailand RV254/SEARCH 010 cohort. CR Rinaldo and RB Mailliard provided primary leadership and supervision for the study, contributed to the study design, data interpretation, and writing of the manuscript. All authors read and approved the final version of the manuscript.
Declaration of Competing Interests
JWM reports grants from the NIH and Gilead Sciences, personal fees (consultant) from Gilead Sciences, Merck, Accelevir Diagnostics, and others from Co-Crystal Pharmaceuticals, Inc., Infectious Diseases Connect, and Abound Bio, Inc., outside the submitted work; JA reports grants from The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., the U.S. Department of Defense, and others from ViiV Healthcare, Gilead, Merck, Roche, and AbbVie, outside the submitted work; SAR reports grants from the NIH; RBM and CRR report a patent PCT/US2020/039843, Pitt Ref: 04973 (pending) for use of antigen presenting cells in HIV therapy: GDG reports a patent PCT/US2020/022403 (pending), Network Immunogen Composition; and BTK reports several provisional patents not directly related to the present work but to HIV vaccines in general. The most recent of these patents is US 2020/0055901, Signature-based human immunodeficiency virus envelope trimer vaccines and methods of using the same.
Acknowledgments
Acknowledgments
We would like to thank the RV254/SEARCH 010 study participants and support staff who committed their time and effort for this study, and we thank Holly A. Bilben and Jan Kristoff for technical support.
This study was in part supported by the U.S. NIH, National Institute of Allergy and Infectious Diseases Grants R21-AI131763, R21-AI138716, UM1-AI126603, and U01-AI35041. The RV254/SEARCH 010 is supported by cooperative agreements (WW81XWH-18-2-0040) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense and by an intramural grant from the Thai Red Cross AIDS Research Centre. Antiretroviral therapy for RV254/SEARCH 010 participants was supported by the Thai Government Pharmaceutical Organization, Gilead, Merck, and ViiV Healthcare. The funding sources did not play a role in the study design, data generation and analysis, interpretation of the findings, manuscript preparation, or decision for publishing.
Data sharing
Data collected for the study, including deidentified participant data, experimental data, statistical analysis, and reagent information will be made available when possible upon request made to the corresponding author RBM.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103175.
Appendix. Supplementary materials
References
- 1.Gaschen B., Taylor J., Yusim K., Foley B., Gao F., Lang D. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296(5577):2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 2.Robb M.L., Ananworanich J. Lessons from acute HIV infection. Curr Opin HIV AIDS. 2016;11(6):555–560. doi: 10.1097/COH.0000000000000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu M.K., Hawkins N., Ritchie A.J., Ganusov V.V., Whale V., Brackenridge S. Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. J Clin Invest. 2013;123(1):380–393. doi: 10.1172/JCI65330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ananworanich J., Dubé K., Chomont N. How does the timing of antiretroviral therapy initiation in acute infection affect HIV reservoirs? Curr Opin HIV AIDS. 2015;10(1):18–28. doi: 10.1097/COH.0000000000000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deeks S.G. HIV: shock and kill. Nature. 2012;487(7408):439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 6.Smith K.N., Mailliard R.B., Larsen B.B., Wong K., Gupta P., Mullins J.I. Dendritic cells restore CD8+ T cell reactivity to autologous HIV-1. J Virol. 2014;88(17):9976–9990. doi: 10.1128/JVI.01275-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang X.-.L., Fan Z., Borowski L., Mailliard R.B., Rolland M., Mullins J.I. Dendritic cells reveal a broad range of MHC class I epitopes for HIV-1 in persons with suppressed viral load on antiretroviral therapy. PLoS ONE. 2010;5(9) doi: 10.1371/journal.pone.0012936. e12936-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mellman I. Dendritic cells: master regulators of the immune response. Cancer Immunol Res. 2013;1(3):145–149. doi: 10.1158/2326-6066.CIR-13-0102. [DOI] [PubMed] [Google Scholar]
- 9.Dillman R.O., Nistor G.I., Cornforth A.N. Dendritic cell vaccines for melanoma: past, present and future. Melanoma Manag. 2016;3(4):273–289. doi: 10.2217/mmt-2016-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudek A.M., Martin S., Garg A.D., Agostinis P. Immature, semi-mature, and fully mature dendritic cells: toward a DC-cancer cells interface that augments anticancer immunity. Front Immunol. 2013;4:438. doi: 10.3389/fimmu.2013.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García F., Climent N., Guardo A.C., Gil C., León A., Autran B. A dendritic cell-based vaccine elicits T cell responses associated with control of HIV-1 replication. Sci Transl Med. 2013;5(166) doi: 10.1126/scitranslmed.3004682. 166ra2-ra2. [DOI] [PubMed] [Google Scholar]
- 12.Lévy Y., Thiébaut R., Montes M., Lacabaratz C., Sloan L., King B. Dendritic cell-based therapeutic vaccine elicits polyfunctional HIV-specific T-cell immunity associated with control of viral load. Eur J Immunol. 2014;44(9):2802–2810. doi: 10.1002/eji.201344433. [DOI] [PubMed] [Google Scholar]
- 13.Andrés C., Plana M., Guardo A.C., Alvarez-Fernández C., Climent N., Gallart T. HIV-1 reservoir dynamics after vaccination and antiretroviral therapy interruption are associated with dendritic cell vaccine-induced T cell responses. J Virol. 2015;89(18):9189–9199. doi: 10.1128/JVI.01062-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mailliard R.B., Egawa S., Cai Q., Kalinska A., Bykovskaya S.N., Lotze M.T. Complementary dendritic cell-activating function of CD8+ and CD4+ T cells: helper role of CD8+ T cells in the development of T helper type 1 responses. J Exp Med. 2002;195(4):473–483. doi: 10.1084/jem.20011662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mailliard R.B., Wankowicz-Kalinska A., Cai Q., Wesa A., Hilkens C.M., Kapsenberg M.L. alpha-type-1 polarized dendritic cells: a novel immunization tool with optimized CTL-inducing activity. Cancer Res. 2004;64(17):5934–5937. doi: 10.1158/0008-5472.CAN-04-1261. [DOI] [PubMed] [Google Scholar]
- 16.Kalinski P., Nakamura Y., Watchmaker P., Giermasz A., Muthuswamy R., Mailliard R.B. Helper roles of NK and CD8+ T cells in the induction of tumor immunity. Polarized dendritic cells as cancer vaccines. Immunol Res. 2006;36(1–3):137–146. doi: 10.1385/IR:36:1:137. [DOI] [PubMed] [Google Scholar]
- 17.Smith K.N., Mailliard R.B., Rinaldo C.R. Programming T cell killers for an HIV cure: teach the new dogs new tricks and let the sleeping dogs lie. For Immunopathol Dis Therap. 2015;6(1–2):67–77. doi: 10.1615/ForumImmunDisTher.2016014160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langenkamp A., Messi M., Lanzavecchia A., Sallusto F. Kinetics of dendritic cell activation: impact on priming of TH1, TH2 and nonpolarized T cells. Nat Immunol. 2000;1(4):311–316. doi: 10.1038/79758. [DOI] [PubMed] [Google Scholar]
- 19.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3(2):133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 20.Smith K.N., Mailliard R.B., Piazza P.A., Fischer W., Korber B.T., Fecek R.J. Effective cytotoxic T lymphocyte targeting of persistent HIV-1 during antiretroviral therapy requires priming of naive CD8+ T cells. MBio. 2016;7(3) doi: 10.1128/mBio.00473-16. e00473-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mailliard R.B., Smith K.N., Fecek R.J., Rappocciolo G., Nascimento E.J., Marques E.T. Selective induction of CTL helper rather than killer activity by natural epitope variants promotes dendritic cell-mediated HIV-1 dissemination. J Immunol. 2013;191(5):2570–2580. doi: 10.4049/jimmunol.1300373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristoff J., Palma M.L., Garcia-Bates T.M., Shen C., Sluis-Cremer N., Gupta P. Type 1-programmed dendritic cells drive antigen-specific latency reversal and immune elimination of persistent HIV-1. EBioMedicine. 2019;43:295–306. doi: 10.1016/j.ebiom.2019.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin K., Boppana S., Du V.Y., Carlson J.M., Yue L., Dilernia D.A. CD8 T cells targeting adapted epitopes in chronic HIV infection promote dendritic cell maturation and CD4 T cell trans-infection. PLoS Pathog. 2019;15(8) doi: 10.1371/journal.ppat.1007970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel S., Hanajiri R., Grant M., Saunders D., Van Pelt S., Keller M. HIV-specific T cells can be generated against non-escaped T cell epitopes with a GMP-compliant manufacturing platform. Mol Ther Methods Clin Dev. 2020;16:11–20. doi: 10.1016/j.omtm.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day C.L., Kaufmann D.E., Kiepiela P., Brown J.A., Moodley E.S., Reddy S. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443(7109):350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 26.Gaiha G.D., Rossin E.J., Urbach J., Landeros C., Collins D.R., Nwonu C. Structural topology defines protective CD8(+) T cell epitopes in the HIV proteome. Science. 2019;364(6439):480–484. doi: 10.1126/science.aav5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korber B., Fischer W. T cell-based strategies for HIV-1 vaccines. Hum Vaccin Immunother. 2020;16(3):713–722. doi: 10.1080/21645515.2019.1666957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theiler J., Yoon H., Yusim K., Picker L.J., Fruh K., Korber B. Epigraph: a vaccine design tool applied to an HIV therapeutic vaccine and a pan-filovirus vaccine. Sci Rep. 2016;6:33987. doi: 10.1038/srep33987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theiler J., Korber B. Graph-based optimization of epitope coverage for vaccine antigen design. Stat Med. 2018;37(2):181–194. doi: 10.1002/sim.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu X., Valentin A., Rosati M., Manocheewa S., Alicea C., Chowdhury B. HIV Env conserved element DNA vaccine alters immunodominance in macaques. Hum Vaccin Immunother. 2017;13(12):2859–2871. doi: 10.1080/21645515.2017.1339852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu X., Lu Z., Valentin A., Rosati M., Broderick K.E., Sardesai N.Y. Gag and env conserved element CE DNA vaccines elicit broad cytotoxic T cell responses targeting subdominant epitopes of HIV and SIV Able to recognize virus-infected cells in macaques. Hum Vaccin Immunother. 2018;14(9):2163–2177. doi: 10.1080/21645515.2018.1489949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ondondo B., Murakoshi H., Clutton G., Abdul-Jawad S., Wee E.G., Gatanaga H. Novel conserved-region T-cell mosaic vaccine with high global HIV-1 coverage is recognized by protective responses in untreated infection. Mol Ther. 2016;24(4):832–842. doi: 10.1038/mt.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakoshi H., Zou C., Kuse N., Akahoshi T., Chikata T., Gatanaga H. CD8(+) T cells specific for conserved, cross-reactive Gag epitopes with strong ability to suppress HIV-1 replication. Retrovirology. 2018;15(1):46. doi: 10.1186/s12977-018-0429-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou C., Murakoshi H., Kuse N., Akahoshi T., Chikata T., Gatanaga H. Effective suppression of HIV-1 replication by cytotoxic T lymphocytes specific for pol epitopes in conserved mosaic vaccine immunogens. J Virol. 2019;93(7) doi: 10.1128/JVI.02142-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyo N., Vogel A.B., Buus S., Erbar S., Wee E.G., Sahin U. Efficient induction of T cells against conserved HIV-1 regions by mosaic vaccines delivered as self-amplifying mRNA. Mol Ther Methods Clin Dev. 2019;12:32–46. doi: 10.1016/j.omtm.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Souza M.S., Pinyakorn S., Akapirat S., Pattanachaiwit S., Fletcher J.L., Chomchey N. Initiation of antiretroviral therapy during acute HIV-1 infection leads to a high rate of nonreactive HIV serology. Clin Infect Dis. 2016;63(4):555–561. doi: 10.1093/cid/ciw365. [DOI] [PubMed] [Google Scholar]
- 37.De Souza M.S., Phanuphak N., Pinyakorn S., Trichavaroj R., Pattanachaiwit S., Chomchey N. Impact of nucleic acid testing relative to antigen/antibody combination immunoassay on the detection of acute HIV infection. AIDS. 2015;29(7):793–800. doi: 10.1097/QAD.0000000000000616. [DOI] [PubMed] [Google Scholar]
- 38.Ananworanich J., Schuetz A., Vandergeeten C., Sereti I., de Souza M., Rerknimitr R. Impact of multi-targeted antiretroviral treatment on gut T cell depletion and HIV reservoir seeding during acute HIV infection. PLoS ONE. 2012;7(3):e33948. doi: 10.1371/journal.pone.0033948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuetz A., Deleage C., Sereti I., Rerknimitr R., Phanuphak N., Phuang-Ngern Y. Initiation of ART during early acute HIV infection preserves mucosal Th17 function and reverses HIV-related immune activation. PLoS Pathog. 2014;10(12) doi: 10.1371/journal.ppat.1004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ananworanich J., Fletcher J.L.K., Pinyakorn S., van Griensven F., Vandergeeten C., Schuetz A. A novel acute HIV infection staging system based on 4th generation immunoassay. Retrovirology. 2013;10:56. doi: 10.1186/1742-4690-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ehrenberg P.K., Geretz A., Thomas R. High-throughput contiguous full-length next-generation sequencing of HLA class I and II genes from 96 donors in a single MiSeq run. Methods Mol Biol. 2018;1802:89–100. doi: 10.1007/978-1-4939-8546-3_6. [DOI] [PubMed] [Google Scholar]
- 42.Kalinski P., Wieckowski E., Muthuswamy R., de Jong E. Generation of stable Th1/CTL-, Th2-, and Th17-inducing human dendritic cells. Methods Mol Biol. 2010;595:117–133. doi: 10.1007/978-1-60761-421-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaccard C.R., Watkins S.C., Kalinski P., Fecek R.J., Yates A.L., Salter R.D. CD40L induces functional tunneling nanotube networks exclusively in dendritic cells programmed by mediators of type 1 immunity. J Immunol. 2015;194(3):1047–1056. doi: 10.4049/jimmunol.1401832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fiebig E.W., Wright D.J., Rawal B.D., Garrett P.E., Schumacher R.T., Peddada L. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17(13):1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 45.Mothe B., Llano A., Ibarrondo J., Daniels M., Miranda C., Zamarreno J. Definition of the viral targets of protective HIV-1-specific T cell responses. J Transl Med. 2011;9:208. doi: 10.1186/1479-5876-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Bates T.M., Palma M.L., Shen C., Gambotto A., Macatangay B.J.C., Ferris R.L. Contrasting roles of the PD-1 signaling pathway in dendritic cell-mediated induction and regulation of HIV-1-specific effector T cell functions. J Virol. 2019;93(5) doi: 10.1128/JVI.02035-18. e02035-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiore-Gartland A., Manso B.A., Friedrich D.P., Gabriel E.E., Finak G., Moodie Z. Pooled-peptide epitope mapping strategies are efficient and highly sensitive: an evaluation of methods for identifying human T cell epitope specificities in large-scale HIV vaccine efficacy trials. PLoS ONE. 2016;11(2) doi: 10.1371/journal.pone.0147812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang O.O., Cumberland W.G., Escobar R., Liao D., Chew K.W. Demographics and natural history of HIV-1-infected spontaneous controllers of viremia. AIDS. 2017;31(8):1091–1098. doi: 10.1097/QAD.0000000000001443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker B.D., Yu X.G. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol. 2013;13(7):487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- 50.Betts M.R., Nason M.C., West S.M., De Rosa S.C., Migueles S.A., Abraham J. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107(12):4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barton J.P., Goonetilleke N., Butler T.C., Walker B.D., McMichael A.J., Chakraborty A.K. Relative rate and location of intra-host HIV evolution to evade cellular immunity are predictable. Nat Commun. 2016;7:11660. doi: 10.1038/ncomms11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferguson A.L., Mann J.K., Omarjee S., Ndung’u T., Walker B.D., Chakraborty A.K. Translating HIV sequences into quantitative fitness landscapes predicts viral vulnerabilities for rational immunogen design. Immunity. 2013;38(3):606–617. doi: 10.1016/j.immuni.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ndhlovu Z.M., Kazer S.W., Nkosi T., Ogunshola F., Muema D.M., Anmole G. Augmentation of HIV-specific T cell function by immediate treatment of hyperacute HIV-1 infection. Sci Transl Med. 2019;11(493) doi: 10.1126/scitranslmed.aau0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Montfoort N., Camps M.G., Khan S., Filippov D.V., Weterings J.J., Griffith J.M. Antigen storage compartments in mature dendritic cells facilitate prolonged cytotoxic T lymphocyte cross-priming capacity. Proc Natl Acad Sci U S A. 2009;106(16):6730–6735. doi: 10.1073/pnas.0900969106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melief C.J., van der Burg S.H. Immunotherapy of established (pre)malignant disease by synthetic long peptide vaccines. Nat Rev Cancer. 2008;8(5):351–360. doi: 10.1038/nrc2373. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H., Hong H., Li D., Ma S., Di Y., Stoten A. Comparing pooled peptides with intact protein for accessing cross-presentation pathways for protective CD8+ and CD4+ T cells. J Biol Chem. 2009;284(14):9184–9191. doi: 10.1074/jbc.M809456200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosario M., Bridgeman A., Quakkelaar E.D., Quigley M.F., Hill B.J., Knudsen M.L. Long peptides induce polyfunctional T cells against conserved regions of HIV-1 with superior breadth to single-gene vaccines in macaques. Eur J Immunol. 2010;40(7):1973–1984. doi: 10.1002/eji.201040344. [DOI] [PubMed] [Google Scholar]
- 58.Takata H., Buranapraditkun S., Kessing C., Fletcher J.L., Muir R., Tardif V. Delayed differentiation of potent effector CD8(+) T cells reducing viremia and reservoir seeding in acute HIV infection. Sci Transl Med. 2017;9(377) doi: 10.1126/scitranslmed.aag1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorin A.M., Du Y., Liu F.Y., Zhang T.H., Ng H.L., Hofmann C. HIV-1 epitopes presented by MHC class I types associated with superior immune containment of viremia have highly constrained fitness landscapes. PLoS Pathog. 2017;13(8) doi: 10.1371/journal.ppat.1006541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai M.C., Singh S., Adland E., Goulder P. Impact of HLA-B*52:01-driven escape mutations on viral replicative capacity. J Virol. 2020;94(13) doi: 10.1128/JVI.02025-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneidewind A., Brockman M.A., Yang R., Adam R.I., Li B., Le Gall S. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81(22):12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.