Summary
Hypoxia is one of the critical stressors encountered by various cells of the human body under diverse pathophysiologic conditions including cancer and has profound impacts on several metabolic and physiologic processes. Hypoxia prompts internal ribosome entry site (IRES)-mediated translation of key genes, such as VEGF, that are vital for tumor progression. Here, we describe that hypoxia remarkably upregulates RNA Polymerase I activity. We discovered that in hypoxia, rRNA shows a different methylation pattern compared to normoxia. Heterogeneity in ribosomes due to the diversity of ribosomal RNA and protein composition has been postulated to generate “specialized ribosomes” that differentially regulate translation. We find that in hypoxia, a sub-set of differentially methylated ribosomes recognizes the VEGF-C IRES, suggesting that ribosomal heterogeneity allows for altered ribosomal functions in hypoxia.
Subject Areas: Molecular Biology, Molecular Mechanism of Gene Regulation, Proteomics
Graphical Abstract

Highlights
-
•
Chronic hypoxia stimulates RNA Pol I activity
-
•
In hypoxia, a pool of specialized rRNA translates VEGFC IRES
-
•
Hypoxia changes 2′-O-Me modification - epitranscriptomic marks on rRNA
Molecular Biology; Molecular Mechanism of Gene Regulation; Proteomics
Introduction
Hypoxia is a common physiologic stressor. It is one of the noticeable features of solid tumors and is a critical determinant of tumor progression (Gilkes et al., 2014; Nakazawa et al., 2016). Hypoxic tumors are often poorly differentiated, highly invasive, and are resistant to treatment (Samanta et al., 2014). Several evidences indicate that hypoxia supports cancer stem cells (Keith and Simon, 2007; Samanta et al., 2014). Cellular adaptation to hypoxia is predominantly driven by stabilization of the HIF-1α transcription factor. HIF-1α activates transcription of several target genes that profoundly modulate cellular functions (Semenza, 2012). Hypoxia also impacts protein translation (Braunstein et al., 2007). In normoxic conditions, translation of majority of mRNAs is initiated by ribosomes following recognition of the 7-methylguanylate cap (m7GpppN). Cells challenged with hypoxia tend to show a mechanistic switch in the way ribosomes recognize some mRNA transcripts. Hypoxia represses cap-mediated translation by promoting sequestration of eIF4E, as a result of increased binding of eIF4E with 4E-BP1. This causes disruption of the eIF4F-containing translation initiation complex (Gebauer, 2012; van den Beucken et al., 2006). In hypoxic conditions, some genes, such as vascular endothelial growth factor (VEGF-C), utilize an alternative mode of protein translation that engages the internal ribosome entry site (IRES) (Braunstein et al., 2007; Morfoisse et al., 2014). This allows the cells to bypass the inhibition of cap-dependent translation (Holcik and Sonenberg, 2005). This translational switch from cap-dependent to cap-independent (IRES-dependent) translation has been regarded as one of the vital attributes of the adaptive response of cancer cells challenged with a hypoxic microenvironment (Braunstein et al., 2007). Accounting for a broader explanation and divergent evidence, we surmise that hypoxia blocks most of eIF4F-directed cap-dependent translation while activating other eIF4F-independent but cap-dependent pathways in addition to some specific contributors, such as IRES for VEGF-C (Jackson, 2013; Young et al., 2008).
Ribosomes are macromolecular machines involving an intricate assembly of proteins and ribosomal RNA (rRNA), with the sophisticated function of translating messenger RNA (Hussmann et al., 2017). RNA polymerase I (RNA Pol I) is the only RNA polymerase that transcribes ribosomal DNA (rDNA) to make the core rRNA components, 28S, 5.8S, and 18S, for assembly of ribosomes. rDNA exists in tandem repeats (over 200 copies total) located on acrocentric chromosomes (this corresponds to human chromosomes 13,14, 15, and 21, 22) (Weeks et al., 2019). Nucleoli are non-membranous, liquid-liquid droplet-like, intra-nuclear bodies that form around nucleolar organizer regions (NORs), the chromosomal regions that actively code for rRNA (Feric et al., 2016; Leslie, 2014). For cells under homeostasis, only few rDNA loci need to be active; this is reflected as fewer nucleoli. However, physiologically challenged cells or proliferating cells tend to show reliance on increased transcription of rDNA and consequently increased ribosome biogenesis, evident morphologically as increased number of nucleoli (Farley-Barnes et al., 2018; Ferreira et al., 2020). Due to its utmost importance, rDNA transcription is very tightly regulated. Multiple factors influence the binding of RNA Pol I to the rDNA loci (Bywater et al., 2012; Peltonen et al., 2014). Despite its vital role in normal and disease pathophysiologies, the functional and mechanistic impacts of hypoxia on ribosome biogenesis and RNA Pol I activity remain largely unexplored. Here we show our findings that in chronic hypoxic conditions, cells dramatically increase RNA Pol I activity by activating upstream binding factor/upstream binding transcription factor (UBF/UBTF).
Tumor hypoxia triggers an angiogenic switch. Hypoxia prompts vascularization through elevated expression of proangiogenic factors such as VEGF. Angiogenesis is regulated by multiple checks, including at the level of mRNA translation (Bergers and Benjamin, 2003). During hypoxia, VEGF mRNA is translated using an IRES. In the recent past, there have been indications of specialized changes or heterogeneity in ribosomal structure (Genuth and Barna, 2018; Xue and Barna, 2012). However, links to the “functional specialization” of ribosomes are still emerging (Dinman, 2016; Genuth and Barna, 2018). One of the major challenges to defining specialized ribosomes has been a lack of methodology to selectively isolate a specialized subpopulation of ribosomes responsible for the functional diversity (Genuth and Barna, 2018; Haag and Dinman, 2019). Using IRES-based translation of VEGF-C messenger RNA under hypoxia as a specific model, we are able to reveal that ribosomes exist in specialized sub-groups that contain rRNA marked with differential methylation patterns and are entrusted with the specialized task of IRES recognition.
Results
Hypoxia upregulates RNA Pol I activity
Intra-tumoral hypoxia is one of the notable features of poorly differentiated, highly invasive, treatment-resistant solid tumors and is a critical determinant of tumor progression (Semenza, 2012). RNA Pol I activity has been implicated to be very important in tumor growth (Hein et al., 2013; Peltonen et al., 2014). However, the impact of hypoxia on RNA Pol I activity remains largely unknown. To test this, we used two distinct models. The first model comprises T47D breast tumor cells and immortalized human mammary epithelial cells (HMECs) grown under hypoxic (or normoxic) conditions, while the second model is a pair of syngeneic breast epithelial cell lines, MCF10A and MCF10CA1acl.1. In normoxic conditions, there is no detectable HIF-1α protein in MCF10A cells, whereas MCF10CA1acl.1 cells have inherently high levels of HIF-1α protein (Figure S1A). We analyzed RNA Pol I activity by quantifying the short-lived 5′ external transcribed spacer (5′ETS) (Peltonen et al., 2014) (Figure 1A). The results clearly revealed that under hypoxia or in the presence of high levels of HIF-1α, there is a significant increase in RNA Pol I activity, that is evident in elevated levels of the 5′ETS transcripts (Figure 1B). Concurrently, to assess nascent RNA synthesis, most of which is rRNA species, cells were pulsed with 5-Fluorouridine (FUrd). We observed ~2 fold increase in fluorescence intensity in cells cultured in hypoxia, confirming that RNA Pol I activity is elevated in hypoxia (Figures 1C, S1C, and S1D). Consistent with our findings from hypoxic cells, introduction of a stable form of HIF-1α; [with mutations (Pro-402/Pro-564 replaced by Ala) in the recognition sites of E3 ubiquitin ligase VHL], into MCF10A cells, also caused an increase in RNA Pol I activity (Figures S1A and S1B). Furthermore, silencing HIF-1α in MCF10CA1acl.1 cells, induced a significant decrease in RNA Pol I activity as evidenced by a decrease in 5′ETS transcripts and FUrd incorporation (Figures S1A, S1B, and S1E)
Figure 1.
Hypoxic stress upregulates RNA Pol I activity
(A) Schematic of rDNA 45S locus: Red arrows indicate the location of ChIP primers ‘c’ used in this study. ETS = External transcribed spacer; ITS = Internal Transcribed Spacer; IGS = Intergenic Sequence.
(B) RT-qPCR performed using primers specific to two distinct ETS regions show increased steady state levels of rRNA corresponding to the 5′ETS, indicating increased Pol I activity in hypoxia (T47D or HMEC) or in MCF10CA1acl.1(cells with high endogenous levels of HIF-1α).
(C) Increased FUrd incorporation in hypoxia (T47D or HMEC). Graphs indicate fold change in mean fluorescence intensity, 120 cells quantified in each group and experiments repeated once.
(D and E) (D) Increased occupancy of rDNA loci by Pol I transcription co-factor UBF (UBTF) measured by ChIP RT-qPCR, evaluated under hypoxia (T47D or HMEC) or in MCF10CA1acl.1 (E) Under hypoxia (T47D cells) as well as in MCF10CA1acl.1, rRNA locus shows enrichment of H3Kme3, H3K9ac and H3K27ac; histone marks corresponding to open chromatin.
(F) Various cell lines of breast epithelial origin show significantly increased number of nucleoli/cell in hypoxia.
(G) Intra-tumoral hypoxic regions corresponding to positive pimonidazole staining show significantly increased nucleoli/nucleus compared to the normoxic regions from the same tumor.
Data are represented as mean ± SEM.
∗ indicates p < 0.05 and ∗∗∗∗ indicates p < 0.001.
RNA Pol I is a multi-subunit enzyme. RPA194 is the catalytic and largest subunit of the RNA Pol I complex. RNA Pol I activity is tightly controlled by a series of molecular events to ensure precise transcription of rDNA. UBTF/UBF plays a prominent role in recruiting RNA Pol I to rDNA and elongation of rRNA transcripts (Potapova and Gerton, 2019; Russell and Zomerdijk, 2005). Thus, elevated RNA Pol I activity will need an increased presence of UBF as well as RPA194 at rDNA loci. Using ChIP primers (positions designated in Figure 1A), we observed that under hypoxia, UBF as well as RPA194 show a remarkably increased occupancy along the rDNA (Figures 1D and S1F). In agreement with this observation, MCF10A cells transfected with stable HIF-1α, showed a significantly increased occupancy of UBF (Figure S1G). Increased transcription rates are supported by open chromatin structure. Histone markers H3K4Me3, H3K9ac, and H3K27ac are indicative of open chromatin in rDNA (Yu et al., 2015). Using ChIP-qPCR, we observed an overall increase in these marks at rDNA in hypoxia, confirming elevated RNA Pol I activity (Figure 1E).
Increased RNA Pol I activity is a requisite for upregulated ribosome biogenesis. Increased ribosome biogenesis is evident as increased number of nucleoli (Farley-Barnes et al., 2018). Supporting this notion, when we cultured multiple breast cell lines in hypoxia, we observed a significant increase in the number of nucleoli per nucleus (Figure 1F). We were interested in determining if this increased nucleolar number will also be seen in the hypoxic region of tumors. For this, we injected spontaneous tumor bearing MMTV-neu mice with pimonidazole and mapped intra-tumoral hypoxic regions by immunohistochemical detection for pimonidazole adducts (Ragnum et al., 2015). Simultaneously, consecutive sections were stained for nucleoli using AgNOR staining. Upon digital overlay of the sections, we observed a noticeable increase in the number of nucleoli per nucleus in the hypoxic region of tumors compared to the normoxic region (Figure 1G). Thus overall, there is robust activation of rRNA synthesis when cells are challenged with hypoxic stress.
Ribosomes show an altered rRNA methylation pattern under hypoxia
Hypoxia reconfigures translational controls (van den Beucken et al., 2006; van den Beucken et al., 2011). Under normoxic conditions, most eukaryotic mRNA translations are initiated by ribosomes upon recognition of the 5′ cap structure. Additionally, translation initiation involves synergistic action of the 5′ cap and the 3′ poly(A) tail of the mRNAs. However, ribosome binding to some mRNAs in certain physiological situations is end-independent and occurs at an IRES (Braunstein et al., 2007). Hypoxic stress compromises cap-dependent translation and prompts a cap-dependent to cap-independent translation switch that utilizes IRES (Braunstein et al., 2007). For most IRESes, the precise molecular determinants of internal ribosome entry are unknown.
VEGF-C mRNA has a well characterized IRES that is active under hypoxia (Arcondeguy et al., 2013; Bornes et al., 2007). We used a bicistronic reporter to confirm the VEGF-C IRES activation. In this system, from a single RNA message, renilla luciferase is translated in a cap-dependent mechanism and a VEGF-C IRES drives translation of firefly luciferase. Increased VEGF-C IRES-driven translation is registered as increased ratio of firefly to renilla luciferase activity (Morfoisse et al., 2014). Hypoxia as well as stabilized HIF-1α, remarkably induced VEGF-C IRES-initiated translation (Figure 2A); whereas HIF-1α silencing dramatically suppressed VEGF-C IRES-dependent translation (Figure S2A). VEGF-A, has a well described independent IRES element that is influenced by hypoxia (Hantelys et al., 2019; Morfoisse et al., 2014, 2015). We utilized a VEGF-A bicistronic reporter which confirmed our findings of hypoxic activation of the VEGF-C IRES (Figure S2B). Consistent with respective IRES activation, protein levels of both VEGF-C and A increased in hypoxic conditions (Figures S2C and S2D).
Figure 2.
Ribosomes show an altered rRNA methylation pattern under hypoxia
(A) Bicistronic luciferase reporter of cap vs VEGF-C IRES-driven translation was used to measure the increase in VEGF-C IRES translation under hypoxia.
(B) Experimental schema: Biotinylated VEGF-C IRES sequence is used to isolate VEGF-C IRES binding ribosomes. Ribosomal RNA isolated VEGF-C was queried for methylation at methylation sites of specific BOX C/D snoRNAs for studies depicted in (C–E).
(C) (I) Methylation pattern of total cellular ribosomes from T47D cells in normoxic conditions was compared to methylation pattern of total cellular ribosomes from T47D cells in hypoxic condition. Similarly, methylation pattern of total cellular ribosomes from MCF10A cells was compared with methylation pattern of total cellular ribosomes from MCF10CA1acl.1 cells (isogenic cells with naturally high levels of HIF-1α). (II) Table listing consistent rRNA methylation changes in total cellular rRNA in hypoxia compared to normoxia in both models (repeated twice).
(D) (I) Methylation pattern of VEGF-C IRES-bound ribosomes from T47D cells, under normoxia was compared to methylation pattern of VEGF-C IRES-bound ribosomes from T47D cells grown under hypoxia. Similarly, methylation pattern of VEGF-C IRES-bound ribosomes from MCF10A cells was compared with methylation pattern of VEGF-C IRES-bound ribosomes from MCF10CA1acl.1 cells. (II) Table listing consistent rRNA methylation changes in IRES-bound ribosomes in hypoxia compared to normoxia in both models (repeated twice).
(E) (I) Methylation pattern of VEGF-C IRES-bound ribosomes from T47D cells, under hypoxia was compared to methylation pattern of total cellular ribosomes from T47D cells in hypoxic condition. Similarly, methylation pattern of VEGF-C IRES-bound ribosomes from MCF10CA1acl.1 cells was compared with methylation pattern of total cellular ribosomes from MCF10CA1acl.1 cells. (II) Table listing consistent rRNA methylation changes in IRES-bound ribosomes in hypoxia compared to rRNA methylation of total cellular pools of ribosomes in both models (repeated twice).
(F) VEGF-C IRES reporter activity was measured inT47D or HMEC in hypoxia 48 hr after silencing fibrillarin expression.
NOTE: In all tables, orange color indicates changes specific to sites on 28S. Black color indicates changes specific to sites on 18S. Data are represented as mean ± SEM.
∗ indicates p < 0.05 and ∗∗∗∗ indicates p < 0.001
We hypothesized that hypoxic stress would provide a new pool of rRNA to undergo distinct modifications for recognizing the VEGF-C IRES. Ribose 2′-O-methylation (2′-O-Me) is the most abundant rRNA secondary modification and is essential for accurate and efficient protein synthesis in eukaryotic cells (Erales et al., 2017; Krogh et al., 2016). To test our hypothesis, we used VEGF-C IRES as an affinity tool. We employed biotin-labeled RNA from a bona fide IRES sequence in the 5′ UTR of VEGF-C mRNA (Morfoisse et al., 2014) to pulldown ribosomes that specifically bind to this sequence, in the presence or absence of hypoxic stress. We used a site-specific RT-qPCR-based approach to assess 2′-O-Me at 14 different sites in functionally important domains of the rRNA isolated from these enriched ribosomes (Belin et al., 2009; Marcel et al., 2013) (schematic Figure 2B). These rRNA methylation sites are distributed along the 18S and 28S rRNAs and are localized within key functional domains of rRNAs, i.e., the decoding center (DC) in the 18S rRNA, the peptidyl transferase center, and the helix 69 (H69) of 28S rRNA (Marcel et al., 2013). The extent of methylation was determined as methylation ratio, as explained in the Transparent methods section. It is important to note that we also analyzed the total cellular pool of rRNA, under normoxia or hypoxia, for these 2′-O-Me sites. The total cellular rRNA pool showed consistent changes in both, the T47D and MCF10 models, in the level of methylation at seven out of fourteen of these sites indicative of a distinct 2′-O-Me pattern in hypoxia (Figures 2CI and 2CII). We then analyzed rRNA from VEGF-C IRES-bound ribosomes under hypoxia for the 2′-O-Me sites in both, the T47D and MCF10 models. We noticed a distinct rRNA methylation pattern in VEGF-C IRES-bound ribosomes under hypoxia when compared to that of IRES-bound ribosomes in normoxia (Figures 2DI and 2DII). This implies that in hypoxia, a specific pool of ribosomes with a separate 2′-O-Me pattern is enriched at the VEGF-C IRES. Most importantly, the rRNA from IRES-bound ribosomes in hypoxia showed a discrete 2′-O-Me pattern compared to the methylation pattern of the total cellular pool of rRNA under hypoxia (Figures 2EI and 2EII).
Essentially our observations indicate that under hypoxia, the 2′-O-Me methylation patterns of total rRNA change. VEGFC-IRES allowed us to isolate a subpopulation of these ribosomes in hypoxia and that population had a characteristic methylation pattern. To further test the importance of 2′-O-Me on modulating IRES activity in hypoxia, we silenced the rRNA 2′-O-methyltransferase, fibrillarin (Figures S2E and S2F). Fibrillarin was silenced in cells for 48 hr and then cells were incubated in hypoxia prior to assaying IRES activity. We recorded a convincing decrease in VEGF-C IRES reporter activity when fibrillarin was silenced in cells cultured in hypoxia (Figure 2F). Furthermore, VEGF-C and VEGF-A protein levels were considerably decreased despite hypoxia when fibrillarin was silenced (Figures S2E and S2F). Overall, these data reveal that there is a characteristic 2′-O-Me methylated sub-pool of rRNA in hypoxia that constitutes IRES-bound ribosomes that ultimately influences IRES translation initiation.
NMI binds to SNORDs and mediates rRNA methylation
It is exciting to find that rRNA methylation is altered in hypoxia. Moving forward, we sought to unravel clues to underlying mechanisms that control changes in the rRNA methylation patterns under hypoxia. A group of non-coding RNA residing in the nucleolus, called small nucleolar RNAs (snoRNA), play an important role in guiding chemical modifications of rRNA. C/D box snoRNA (SNORDs), guide rRNA 2′-O-Methyltransferase, fibrillarin, to methylate specific bases of the rRNA (van Nues and Watkins, 2017). There are over 106 SNORDs in human cells (D'Souza et al., 2019). However, clues to which SNORDs decide the methylation of rRNA in conditions such as hypoxia are still to be unraveled.
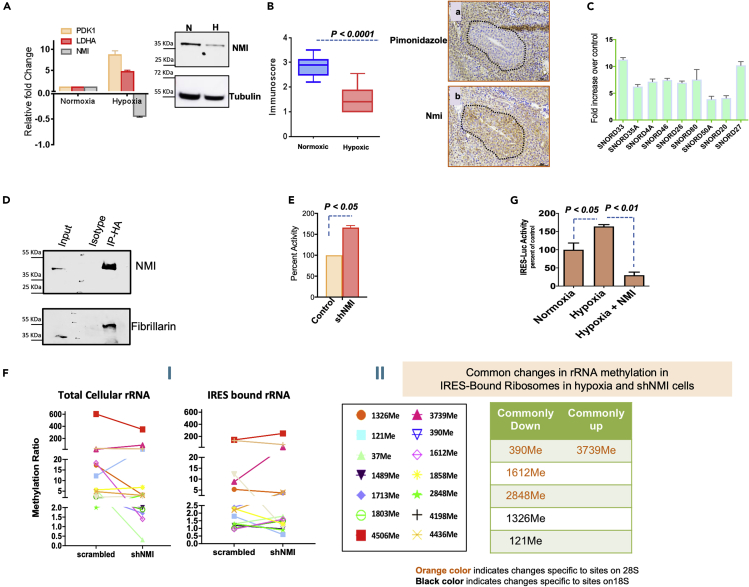
Hypoxia is one of the well-established drivers of epithelial-mesenchymal-transition (EMT). Multiple studies from our group have highlighted the importance of NMI (N-Myc and STAT interactor) protein in negatively impacting EMT (Devine et al., 2014; Pruitt et al., 2018). NMI protein contains a previously uncharacterized RNA recognition motif (RRM) at aa. 157–236. RRMs recognize single stranded RNAs. We explored if hypoxia influenced NMI protein levels. As seen in Figures 3A and S3A we observed that in hypoxic conditions, NMI expression is remarkably compromised at protein, as well as mRNA levels. Moreover, we observed that in MMTV-Neu derived spontaneous tumors, intra-tumoral hypoxic regions showed a noticeable loss of Nmi protein expression compared to the normoxic regions (Figure 3B).
Figure 3.
NMI binds to SNORDs and changes methylation in hypoxia
(A) Analysis of mRNA and protein isolated from T47D cells incubated in hypoxia show that expression of NMI mRNA (bar graph) is decreased in hypoxia. The adjacent Western blot shows that protein level of NMI is also reduced in hypoxia.
(B) Analysis of hypoxic regions from tumors from MMTV-Neu mice shows that NMI protein expression is abundant in areas with less hypoxia as measured by pimonidazole staining. This data is from multiple regions of three independent tumors from three different mice. The insets a and b are representative images showing example of NMI staining pattern compared with pimonidazole staining.
(C) Real-time PCR validation of SNORDs bound to NMI identified by RNA IP-seq.
(D) MDA-MB-231 HA-tagged NMI cells co-immunoprecipitationusing HA-tagged beads and subsequent detection of fibrillarin using Western blot analysis.
(E) Bicistronic luciferase reporter vector for VEGF-C IRES-driven translation showed significant increase in IRES-driven translation in T47D cells silenced for NMI expression.
(F) (I) Methylation pattern of total cellular ribosomes from T47D cells with non-targeting shRNA shows a distinctive profile when compared with total methylation pattern of total cellular ribosomes from NMI silenced T47D cells. Similarly, methylation pattern of VEGF-C IRES-bound ribosomes from T47D cells with non-targeting shRNA shows clear changes in methylation pattern of VEGF-C IRES-bound ribosomes from NMI silenced T47D cells. (II) Common rRNA methylation changes in IRES-bound ribosomes in hypoxia compared to T47D cells silenced for NMI expression.
(G) VEGF-C-IRES reporter activity shows significant increase in hypoxia. This increased activity is reduced by exogenous expression of NMI. The experiments were conducted in T47D cells. Each group was measured in triplicate. Data represented are from two biological repeats.
We conducted RNA IP-seq (RIP-seq) using HA-tagged NMI to determine RNA species that can physically bind to NMI protein. To our excitement, we found that many of the top-ranking RNA entities bound to NMI were SNORDs (Figure S3B). This prompted us to undertake a targeted query for all 14 SNORDs that guide 2′-O-Me at 14 sites that we previously queried for differential methylation patterns. Figure S3C shows the SNORDS with corresponding 2′-O-Me sites. We queried NMI-bound RNA by real-time PCR using specific primers to these SNORDs. We found that nine out of fourteen SNORDs bound NMI protein (Figure 3C). Moreover, seven of the nine SNORDs recognized by NMI corresponded to the rRNA 2′-O-Me sites that were notably reduced for methylation in VEGF-C IRES-recognizing ribosomes in hypoxia. These SNORDs (33, 35A, 4A, 26, 80, 50A and 20) and corresponding methylation sites are shown Figure S3C. We wanted to explore if NMI mediates the binding between SNORDs and the rRNA 2′-O-methyltransferase, fibrillarin. We immunoprecipitated NMI and observed that fibrillarin was co-immunoprecipitated with it (Figure 3D). Silencing NMI expression resulted in an increase in VEGF-C IRES-mediated translation, similar to hypoxia induced IRES translation (Figure 3E). We then sought to determine the effect of silencing NMI on rRNA methylation pattern of VEGF-C IRES-recognizing ribosomes. Site-specific RT-qPCR analysis of rRNA bound to VEGF-C IRES showed that loss of NMI yielded changes in methylation pattern common to hypoxia (Figures 3FI and 3FII). Moreover, the bicistronic reporter of cap vs VEGF-C IRES translation showed that exogenous restoration of NMI in hypoxia treated cells significantly decreased cap-independent translation (Figure 3G). Overall, these results highlight loss of NMI as an important event that shapes rRNA methylation in hypoxic conditions.
A distinct set of proteins is recruited to VEGF-C IRES under hypoxia
We further explored if hypoxic conditions instigate recruitment of specific proteins to the VEGF-C IRES. We used the biotin-labeled VEGF-C IRES sequence to capture IRES-bound ribosomes and associated proteins using two different cell systems (i) T47D cultured in normoxic or hypoxic conditions and (ii) MCF10A, MCF10CA1acl.1. We conducted proteomics analysis to identify the captured IRES-bound proteins. Combined datasets from two independent runs were analyzed using Scaffold 4 and unique proteins were identified. Unsurprisingly, mass spectrometry (MS)-based identification revealed that more than 80% of the proteins were commonly present in IRES-bound ribosomes under hypoxic and normoxic conditions (Figure S4A). Many of these proteins are ribosome-associated proteins or structural constituents of ribosomes as revealed by KEGG Analysis. A gene ontology search revealed that the most predominant proteins that were pulled down were ribosomal proteins involved in ribosomal subunit assembly, rRNA processing, splicing, and translational elongation (Figures S4B–S4D). Five proteins were distinctly enriched at the VEGF-C IRES under hypoxia. These were Eukaryotic initiation factor 4A-I (EIF4A1), Rho/rac guanine nucleotide exchange factor 2 (ARHGEF2), Nuclear fragile X mental retardation-interacting protein 2 (NUFIP2), Polyadenylate-binding protein 1 (PABPC1), and ribosomal protein L6 (RPL6) (Figure 4A).
Figure 4.
Under hypoxia a distinct set of proteins is recruited to VEGF-C IRES
(A) Mass spectrometry analysis results of proteins isolated from ribosomes bound to VEGF-C IRES in normoxia were compared with proteins isolated from ribosomes bound to VEGF-C IRES in hypoxia. Volcano plot reveals specific enrichment of PABPC1, RPL6, EIF4A1, ARHGEF2, and NUFIP2 as IRES-bound proteins significantly enriched under hypoxia.
(B) Western blot analysis of protein extracts from VEGF-C IRES-bound proteins in hypoxia or MCF10CA1acl.1 confirms specific enrichment of PABPC1 and ITAFs (hnRNPL and hnRNPE1).
(C) Bicistronic reporter vector for VEGF-C IRES-driven translation showed significant reduction in IRES-driven translation in hypoxia under limiting availability of ITAFs (hnRNPE1 and hnRNPL); scrambled siRNA was used as a control.
(D) Schematic representation of IRES translation switch under hypoxia and contribution of specialized ribosomes: Under normoxic condition, CAP-dependent translation proceeds when ribosomal subunit 40S, in collaboration with PABPC1(PABP), and EIF4 complex (Including EIF4A) establishes a loop-like structure that encompasses cap and polyA tail. In hypoxia, ribosomal subunit 40S with specific methylation pattern, in collaboration with PABP, EIF4A, and ITAFs constitutes an alternative loop at the IRES site that will allow IRES-driven translation.
Data are represented as mean SEM ∗ indicates p < 0.05.
Presence of RPL6, a protein constituent of the large ribosomal subunit, points to the recruitment of ribosomes for translation via the IRES. EIF4A1 is a member of the DEAD-box family of RNA helicases that unwinds the secondary structure of mRNAs to permit ribosomal recruitment. It is required at high abundance (three copies per ribosome) for translation initiation, specifically to unwind and prepare mRNA for translation. It is an essential factor for 5′ cap-dependent translation but is also required for IRES-dependent translation initiations (Bordeleau et al., 2006). The 3′ poly(A) tail of eukaryotic mRNAs functions as an important regulator of protein synthesis and mRNA stability. The poly(A) tail is recognized by PABPC1, and its recruitment improves the efficiency of translation initiation. It is suggested that the “closed-loop” structure formed from the interaction of PABPC1 with poly(A) is key to recruitment of ribosomes (Lacerda et al., 2017; Shatsky et al., 2018). PABPC1 has also been proposed to associate with the 60S subunit of ribosome (Goss and Kleiman, 2013; Lacerda et al., 2017). Overall, the enrichment of RPL6, EIF4A1 and importantly, PABPC1 at the VEGF-C IRES indicates active VEGF-C IRES mediates translation under hypoxia.
IRES-transacting factors (ITAFs) belong to the group of heterogeneous nuclear ribonucleoproteins (hnRNPs). These are proteins specifically involved in IRES-mediated translation but not in cap-dependent translation. ITAFs mediate binding between IRESes, ribosomes, and translation initiation factors (Lacerda et al., 2017). Thus, under conditions that impair canonical, cap-dependent translation, the synthesis of select proteins is enabled by ITAF-directed alternative mechanisms. These include a cache of proteins required for cell survival and stress recovery. Thus, it would be logical to expect that the MS based analysis will reveal certain ITAFs enriched at IRES under hypoxia. However, we did not see ITAFs among the distinctly enriched proteins (Figure 4A). This was due to the stringent exclusion criteria used by us for proteomics analysis. However, in-depth examination of proteomics data at reduced stringency revealed that ITAFs, hnRNPL, and hnRNPE1 (PCBP1), were enriched at IRES in hypoxia. To confirm the recruitment of ITAFs, we performed Western blot analysis of VEGF-C IRES-bound proteins under hypoxia. The results clearly revealed enrichment of hnRNPL and hnRNPE1 in IRES-bound proteins under hypoxia (Figure 4B). Both these ITAFS are implicated in IRES recognition (King et al., 2010). In order to address their relevance to VEGF-C IRES, we used bicistronic reporter of cap vs VEGF-C IRES-driven translation (Morfoisse et al., 2014). We found that silencing the expression of any one of these ITAFs (hnRNPE1 or hnRNPL) reduced IRES-driven translation activity under hypoxia, confirming the requirement of these ITAFs in VEGF-C IRES-driven translation (Figure 4C).
Thus, in addition to the enrichment of RPL6, EIF4A1 and PABPC1 at the VEGF-C IRES, its confirmed association with ITAFs, hnRNPL and hnRNPE1 suggests hypoxia-driven active VEGF-C IRES. Overall, these data suggest that in hypoxia the VEGF-C IRES recognizes ribosomes with an altered methylation pattern.
Discussion
Tumorigenesis involves molecular mechanisms that enable cells to endure or adapt to stress. Several growth factors and oncogenes have IRES elements in their 5′UTRs. Use of the VEGF-C IRES allowed us to categorically segregate an IRES-bound ribosomal pool. The sequence and secondary and tertiary structures of rRNAs are well conserved. Recent studies have brought to light heterogeneity in secondary modifications of rRNA (Eisenstein, 2017; Krogh et al., 2016; Roundtree et al., 2017). Also recently, the concept of ribosome-mediated regulation of translation has emerged, which proposes that cells produce sub-sets of ribosomes of different composition, and each sub-set displays selective translational properties (Erales et al., 2017). These changes seem to be in response to various physiologic activities of cells, including mechanisms necessary to endure cell stress. For example, nutrient limitation-induced stress in E. coli changes the relative expression of rDNA operons to alter the rRNA composition within the actively translating ribosome pool (Kurylo et al., 2018). rRNA methylation is critical for proper assembly of ribosomal complex and efficient translation initiation through IRES (Basu et al., 2011). Observations like these have given rise to the concept of “ribosome heterogeneity” and “specialized ribosomes” (Genuth and Barna, 2018; Shi et al., 2017; Xue and Barna, 2012). Ribosomal RNA modification is emerging as a major contributor of ribosomal heterogeneity (Monaco et al., 2018). However, the specific functions of modified nucleotides in rRNA and how they program ribosomal heterogeneity are still being deciphered. The nucleotides queried in this study are located in evolutionarily conserved and functionally important regions of rRNA (Kufel and Grzechnik, 2019; Marcel et al., 2013). The role of snoRNAs in controlling cell behavior and tumor biology is gaining attention (Liu et al., 2020). Considering that each human ribosome contains around 100 ribose methylations and 100 pseudouridines and that each position is subject to individual regulation through specific snoRNA guides, the potential for heterogeneity within ribosomal subpopulations is enormous (D'Souza et al., 2019; Sloan et al., 2017).
Cumulatively, our observations suggest that rRNA transcription is upregulated in hypoxia. Hypoxia also enables generation of rRNA “coded” by differential methylation. Overall, increased RNA Pol I activity combined with methylation pattern differences, driven by SNORDs causes a shift in rRNA subpopulations in response to hypoxic stress. It is likely that this shift in the methylation patterning of rRNA ultimately leads to recruitment of proteins with the ribosome to activate VEGF-C IRES, pivoting the cells ability from cap-dependent to cap-independent translation. Molecular details of how hypoxia affects tumor cell fate are still evolving (Choudhry and Harris, 2018; Qiu et al., 2017; Rankin et al., 2016). Responses of a cell to acute vs chronic hypoxia are different (Chee et al., 2019; Han et al., 2017). Additionally, some of the response is tissue, organ and pathology specific, simply because what is considered as hypoxic condition in one context is not directly applicable to the other (Rundqvist and Johnson, 2013). Activation of HIF-1α has a major transcriptional impact in activating expression of multiple genes. In addition to transcription, regulation of mRNA stability plays a key role in hypoxia. For instance, under hypoxia VEGFA mRNA is stabilized (Goldberg-Cohen et al., 2002). Furthermore, hypoxia indirectly regulates mRNA stability and translation through miRNAs (Camps et al., 2014; Voellenkle et al., 2012). Thus, there are several variables in the resultant cellular response to hypoxia. It is critical to note that detailed studies by Tiana et al. show that rRNA is degraded in hypoxia, which seems counter intuitive. A major difference between Tiana et al., and this manuscript is the length of hypoxic incubation. Tiana et al., use significantly shorter periods of hypoxia (8–16 hr) whereas work presented here uses 72 hr (Tiana et al., 2020). However, an interesting likelihood emerges that integrates these apparent discrepancies. It is possible that hypoxic cells choose to degrade mRNAs that are not being translated, together with the ribosomes that contain unsuitably methylated rRNA, as those may no longer be needed for translation. While the cell is making a go-no go survival decision in response to hypoxic stress, it tries to curtail unnecessary energy expenditure and thus contextually tries to reduce translation and possibly adopt cap-independent means of translation for some key genes. Whether this translation is IRES-dependent is highly debatable (Young et al., 2008). However, there are certain IRESes such as VEGF-A and C that are characterized in depth and widely accepted to be driven downstream of hypoxia (Hantelys et al., 2019; Morfoisse et al., 2014, 2015). Our observations suggest that this alternative translation may be driven by rRNA biogenesis that at least, in part, provides a modified pool of rRNA. Our observations may look counter intuitive in the light of published reports that show that hypoxic protein synthesis is mostly cap-dependent (Uniacke et al., 2012; Yi et al., 2013; Young et al., 2008). We surmise that the need for cap-dependent translation is obvious as a result of robust cellular response to hypoxia and there may possibly be a complex chronology between the choices of translation routes that a cell makes. Taken together these observations in-fact suggest the need for the diverse pools of (specialized) ribosomes (Dalla Venezia et al., 2019; Fujii et al., 2018; Genuth and Barna, 2018).
Recent studies by Prakash V et al. have uncovered a novel connection between changes in ribosome biogenesis and the execution of the EMT program, that is pivotal during tumor development and metastasis (Godet et al., 2019; Prakash et al., 2019). They identified Snail as a mediator of ribosome biogenesis that accompanies EMT. Even though there is an absence of studies detailing effects of hypoxia on ribosome biogenesis, multiple studies have defined roles for hypoxia in regulating a program of epithelial-mesenchymal-plasticity (Prakash et al., 2019; Williams et al., 2019). Studies from our lab have previously defined a critical role of NMI in tumor progression and metastasis via alterations in the EMT program (Devine et al., 2014; Pruitt et al., 2018). Based on these studies, NMI presents as a critical regulator of epithelial-mesenchymal-plasticity. Our current study identifies a new functional responsibility for NMI in mediating a translation switch that occurs in hypoxia. RNA-IP-seq analysis and targeted queries revealed selective binding of NMI with small nucleolar RNAs. NMI also binds to rRNA methyl transferase, fibrillarin. We speculate that NMI acts to regulate the available pool of fibrillarin and SNORDs that can interact, which most likely will affect rRNA methylation patterns. Studies have proposed partial methylation of rRNA, and these sites may be important for highly variable and regulatable positions, which could give rise to increased rRNA heterogeneity (Falaleeva et al., 2017; Krogh et al., 2016). Interactions of SNORDs and fibrillarin with proteins like NMI may offer fine-tune control over partial methylation patterning. As NMI protein expression is compromised in hypoxia, it likely results in altered availability of fibrillarin to certain SNORDs. Collectively, our observations of NMI loss in hypoxia coupled with overall repatterning of rRNA methylation, highlight NMI as a possible regulatory bridge in determining the methylation status of rRNA. Ultimately, these changes in the rRNA methylation drive differential recruitment of proteins to sites of IRES-mediated translation. Namely, we found ITAFs, hnRNPE1 and hnRNPL, to be preferentially associated with VEGF-C IRES during hypoxic stress, and depletion of these proteins shifts the balance to cap-independent translation in hypoxia (Figure 4D). These observations are consistent with the emerging importance of ITAFs such as Vasohibin1 in regulation of translation in hypoxia (Hantelys et al., 2019). These results solidify that epitranscriptomic modifications of rRNA prompt a compositional change in the ribosome-associated translational machinery to give rise to ribosomes for specialized tasks, most notably in hypoxia. These results add a new dimension of “ribosomal diversity” to the field and provide a foundation for further investigation of the specialized ribosomes that provide translational adaptability in response to various environmental and physiologic stresses.
Limitations of the study
This study offers a rationalization that the increased rRNA in hypoxia allows the cells to code it (rRNA) differently than in normoxia by epitranscriptomic modifications (such as 2′-O-Me). Our study supports a possibility that epitranscriptomic modifications allow formation of a distinct pool of rRNA and implies that it supports formation of ribosomes that are specialized to perform a task of translation of a specific transcript. The study uses VEGF-C IRES as a tool.
The limitation of this study is that it uses a specific IRES. Thus, the 2′-O-Me modifications of rRNA may not be identical for other IRESes that may work in hypoxia or other pathophysiologic conditions.
It is important to note that these conclusion are only based on an artifactual construct that is highly controversial(Jackson, 2013) with little data to support a role for endogenous translation regulation IRES.
Also, the study is focused on a critical sub-set of methylations on rRNA but does not account for entire profile of 2'-O-Me modifications on the rRNA. Thus, our study does not allow to precisely determine if and which partial 2′-O-Me rRNA modifications are regulated by NMI. Also, NMI may just be one example of hypoxia regulated guide proteins that regulate mature 2′-O-Me complex and thus offers a partial explanation of hypoxia-driven changes.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Rajeev Samant Ph.D (rsamant@uab.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019238 and 10.6019/PXD019238.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We acknowledge insightful discussions with the attendees of the ASBMB Nucleolus 2019 Meeting.
We acknowledge the following grant support: Cancer Center Core grant CA013148 for UAB & CCC Mass Spectrometry & Proteomics Shared Facility, UAB Transgenic & Genetically Engineered Models Core (TGEM) and the Heflin Genomics facility.
The work is supported in part by Merit Review Award number I01 BX003374 (from the U.S. Department of Veterans Affairs BLRD service and CA194048 (NCI/NIH) & funding from the Breast Cancer Research Foundation of Alabama to RSS & CA169202 (NCI/NIH) to LAS.
H.C.P. was an HHMI-Med-To-Grad Fellow (UAB). H.C. Pruitt is currently at Johns Hopkins University; Institute for Nanobiotechnology; Department of Chemical and Biomolecular Engineering.
Author contributions
B.J.M., S.C.K., and R.S.S. designed experiments and analyzed the data. B.J.M., S.C.K., and H.C.P. performed the experiments. L.A.S. provided reagents and expertise. B.J.M, L.A.S., and R.S.S wrote and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 22, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.102010.
Supplemental Information
References
- Arcondeguy T., Lacazette E., Millevoi S., Prats H., Touriol C. VEGF-A mRNA processing, stability and translation: a paradigm for intricate regulation of gene expression at the post-transcriptional level. Nucleic Acids Res. 2013;41:7997–8010. doi: 10.1093/nar/gkt539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A., Das P., Chaudhuri S., Bevilacqua E., Andrews J., Barik S., Hatzoglou M., Komar A.A., Mazumder B. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular internal ribosome entry sites. Mol. Cell. Biol. 2011;31:4482–4499. doi: 10.1128/MCB.05804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin S., Beghin A., Solano-Gonzalez E., Bezin L., Brunet-Manquat S., Textoris J., Prats A.C., Mertani H.C., Dumontet C., Diaz J.J. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS One. 2009;4:e7147. doi: 10.1371/journal.pone.0007147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergers G., Benjamin L.E. Tumorigenesis and the angiogenic switch. Nat. Rev. Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- Bordeleau M.E., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- Bornes S., Prado-Lourenco L., Bastide A., Zanibellato C., Iacovoni J.S., Lacazette E., Prats A.C., Touriol C., Prats H. Translational induction of VEGF internal ribosome entry site elements during the early response to ischemic stress. Circ. Res. 2007;100:305–308. doi: 10.1161/01.RES.0000258873.08041.c9. [DOI] [PubMed] [Google Scholar]
- Braunstein S., Karpisheva K., Pola C., Goldberg J., Hochman T., Yee H., Cangiarella J., Arju R., Formenti S.C., Schneider R.J. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Bywater M.J., Poortinga G., Sanij E., Hein N., Peck A., Cullinane C., Wall M., Cluse L., Drygin D., Anderes K. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps C., Saini H.K., Mole D.R., Choudhry H., Reczko M., Guerra-Assuncao J.A., Tian Y.M., Buffa F.M., Harris A.L., Hatzigeorgiou A.G. Integrated analysis of microRNA and mRNA expression and association with HIF binding reveals the complexity of microRNA expression regulation under hypoxia. Mol. Cancer. 2014;13:28. doi: 10.1186/1476-4598-13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee N.T., Lohse I., Brothers S.P. mRNA-to-protein translation in hypoxia. Mol. Cancer. 2019;18:49. doi: 10.1186/s12943-019-0968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry H., Harris A.L. Advances in hypoxia-inducible factor biology. Cell Metab. 2018;27:281–298. doi: 10.1016/j.cmet.2017.10.005. [DOI] [PubMed] [Google Scholar]
- D'Souza M.N., Gowda N.K.C., Tiwari V., Babu R.O., Anand P., Dastidar S.G., Singh R., James O.G., Selvaraj B., Pal R. FMRP interacts with C/D box snoRNA in the nucleus and regulates ribosomal RNA methylation. iScience. 2019;12:368. doi: 10.1016/j.isci.2019.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalla Venezia N., Vincent A., Marcel V., Catez F., Diaz J.J. Emerging role of eukaryote ribosomes in translational control. Int. J. Mol. Sci. 2019;20:1226. doi: 10.3390/ijms20051226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine D.J., Rostas J.W., Metge B.J., Das S., Mulekar M.S., Tucker J.A., Grizzle W.E., Buchsbaum D.J., Shevde L.A., Samant R.S. Loss of N-Myc interactor promotes epithelial-mesenchymal transition by activation of TGF-beta/SMAD signaling. Oncogene. 2014;33:2620–2628. doi: 10.1038/onc.2013.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J.D. Pathways to specialized ribosomes: the Brussels Lecture. J. Mol. Biol. 2016;428:2186–2194. doi: 10.1016/j.jmb.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein M. Epitranscriptomics: mixed messages. Nat. Methods. 2017;14:15–17. [Google Scholar]
- Erales J., Marchand V., Panthu B., Gillot S., Belin S., Ghayad S.E., Garcia M., Laforets F., Marcel V., Baudin-Baillieu A. Evidence for rRNA 2'-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. U S A. 2017;114:12934–12939. doi: 10.1073/pnas.1707674114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaleeva M., Welden J.R., Duncan M.J., Stamm S. C/D-box snoRNAs form methylating and non-methylating ribonucleoprotein complexes: Old dogs show new tricks. Bioessays. 2017;39 doi: 10.1002/bies.201600264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley-Barnes K.I., McCann K.L., Ogawa L.M., Merkel J., Surovtseva Y.V., Baserga S.J. Diverse regulators of human ribosome biogenesis discovered by changes in nucleolar number. Cell Rep. 2018;22:1923–1934. doi: 10.1016/j.celrep.2018.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R., Schneekloth J.S., Jr., Panov K.I., Hannan K.M., Hannan R.D. Targeting the RNA polymerase I transcription for cancer therapy comes of age. Cells. 2020;9:266. doi: 10.3390/cells9020266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K., Susanto T.T., Saurabh S., Barna M. Decoding the function of expansion segments in ribosomes. Mol. Cell. 2018;72:1013–1020.e6. doi: 10.1016/j.molcel.2018.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F. Versatility of the translational machinery during stress: changing partners to keep dancing. Cell Res. 2012;22:1634–1636. doi: 10.1038/cr.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuth N.R., Barna M. The discovery of ribosome heterogeneity and its implications for gene regulation and organismal life. Mol. Cell. 2018;71:364–374. doi: 10.1016/j.molcel.2018.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes D.M., Semenza G.L., Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat. Rev. Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godet I., Shin Y.J., Ju J.A., Ye I.C., Wang G., Gilkes D.M. Fate-mapping post-hypoxic tumor cells reveals a ROS-resistant phenotype that promotes metastasis. Nat. Commun. 2019;10:4862. doi: 10.1038/s41467-019-12412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg-Cohen I., Furneauxb H., Levy A.P. A 40-bp RNA element that mediates stabilization of vascular endothelial growth factor mRNA by HuR. J. Biol. Chem. 2002;277:13635–13640. doi: 10.1074/jbc.M108703200. [DOI] [PubMed] [Google Scholar]
- Goss D.J., Kleiman F.E. Poly(A) binding proteins: are they all created equal? Wiley Interdiscip. Rev. RNA. 2013;4:167–179. doi: 10.1002/wrna.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag E.S., Dinman J.D. Still searching for specialized ribosomes. Dev. Cell. 2019;48:744–746. doi: 10.1016/j.devcel.2019.03.005. [DOI] [PubMed] [Google Scholar]
- Han J., Li J., Ho J.C., Chia G.S., Kato H., Jha S., Yang H., Poellinger L., Lee K.L. Hypoxia is a key driver of alternative splicing in human breast cancer cells. Sci. Rep. 2017;7:4108. doi: 10.1038/s41598-017-04333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantelys F., Godet A.C., David F., Tatin F., Renaud-Gabardos E., Pujol F., Diallo L.H., Ader I., Ligat L., Henras A.K. Vasohibin1, a new mouse cardiomyocyte IRES trans-acting factor that regulates translation in early hypoxia. Elife. 2019;8:e50094. doi: 10.7554/eLife.50094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein N., Hannan K.M., George A.J., Sanij E., Hannan R.D. The nucleolus: an emerging target for cancer therapy. Trends Mol. Med. 2013;19:643–654. doi: 10.1016/j.molmed.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Holcik M., Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Hussmann J.A., Osadnik H., Gross C.A. Ribosomal architecture: constraints imposed by the need for self-production. Curr. Biol. 2017;27:R798–R800. doi: 10.1016/j.cub.2017.06.080. [DOI] [PubMed] [Google Scholar]
- Jackson R.J. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb. Perspect. Biol. 2013;5:a011569. doi: 10.1101/cshperspect.a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith B., Simon M.C. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King H.A., Cobbold L.C., Willis A.E. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans. 2010;38:1581–1586. doi: 10.1042/BST0381581. [DOI] [PubMed] [Google Scholar]
- Krogh N., Jansson M.D., Hafner S.J., Tehler D., Birkedal U., Christensen-Dalsgaard M., Lund A.H., Nielsen H. Profiling of 2'-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res. 2016;44:7884–7895. doi: 10.1093/nar/gkw482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J., Grzechnik P. Small nucleolar RNAs tell a different tale. Trends Genet. 2019;35:104–117. doi: 10.1016/j.tig.2018.11.005. [DOI] [PubMed] [Google Scholar]
- Kurylo C.M., Parks M.M., Juette M.F., Zinshteyn B., Altman R.B., Thibado J.K., Vincent C.T., Blanchard S.C. Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep. 2018;25:236–248.e6. doi: 10.1016/j.celrep.2018.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda R., Menezes J., Romao L. More than just scanning: the importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell Mol. Life Sci. 2017;74:1659–1680. doi: 10.1007/s00018-016-2428-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie M. Central command. Science. 2014;345:506–507. doi: 10.1126/science.345.6196.506. [DOI] [PubMed] [Google Scholar]
- Liu Y., Ruan H., Li S., Ye Y., Hong W., Gong J., Zhang Z., Jing Y., Zhang X., Diao L. The genetic and pharmacogenomic landscape of snoRNAs in human cancer. Mol. Cancer. 2020;19:108. doi: 10.1186/s12943-020-01228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V., Ghayad S.E., Belin S., Therizols G., Morel A.P., Solano-Gonzalez E., Vendrell J.A., Hacot S., Mertani H.C., Albaret M.A. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 2013;24:318–330. doi: 10.1016/j.ccr.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco P.L., Marcel V., Diaz J.J., Catez F. 2'-O-methylation of ribosomal RNA: towards an epitranscriptomic control of translation? Biomolecules. 2018;8:106. doi: 10.3390/biom8040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfoisse F., Kuchnio A., Frainay C., Gomez-Brouchet A., Delisle M.B., Marzi S., Helfer A.C., Hantelys F., Pujol F., Guillermet-Guibert J. Hypoxia induces VEGF-C expression in metastatic tumor cells via a HIF-1alpha-independent translation-mediated mechanism. Cell Rep. 2014;6:155–167. doi: 10.1016/j.celrep.2013.12.011. [DOI] [PubMed] [Google Scholar]
- Morfoisse F., Renaud E., Hantelys F., Prats A.C., Garmy-Susini B. Role of hypoxia and vascular endothelial growth factors in lymphangiogenesis. Mol. Cell Oncol. 2015;2:e1024821. doi: 10.1080/23723556.2015.1024821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa M.S., Keith B., Simon M.C. Oxygen availability and metabolic adaptations. Nat. Rev. Cancer. 2016;16:663–673. doi: 10.1038/nrc.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen K., Colis L., Liu H., Trivedi R., Moubarek M.S., Moore H.M., Bai B., Rudek M.A., Bieberich C.J., Laiho M. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell. 2014;25:77–90. doi: 10.1016/j.ccr.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapova T.A., Gerton J.L. Ribosomal DNA and the nucleolus in the context of genome organization. Chromosome Res. 2019;27:109–127. doi: 10.1007/s10577-018-9600-5. [DOI] [PubMed] [Google Scholar]
- Prakash V., Carson B.B., Feenstra J.M., Dass R.A., Sekyrova P., Hoshino A., Petersen J., Guo Y., Parks M.M., Kurylo C.M. Ribosome biogenesis during cell cycle arrest fuels EMT in development and disease. Nat. Commun. 2019;10:2110. doi: 10.1038/s41467-019-10100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt H.C., Metge B.J., Weeks S.E., Chen D., Wei S., Kesterson R.A., Shevde L.A., Samant R.S. Conditional knockout of N-Myc and STAT interactor disrupts normal mammary development and enhances metastatic ability of mammary tumors. Oncogene. 2018;37:1610–1623. doi: 10.1038/s41388-017-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G.Z., Jin M.Z., Dai J.X., Sun W., Feng J.H., Jin W.L. Reprogramming of the tumor in the hypoxic niche: the emerging concept and associated therapeutic strategies. Trends Pharmacol. Sci. 2017;38:669–686. doi: 10.1016/j.tips.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Ragnum H.B., Vlatkovic L., Lie A.K., Axcrona K., Julin C.H., Frikstad K.M., Hole K.H., Seierstad T., Lyng H. The tumour hypoxia marker pimonidazole reflects a transcriptional programme associated with aggressive prostate cancer. Br. J. Cancer. 2015;112:382–390. doi: 10.1038/bjc.2014.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin E.B., Nam J.M., Giaccia A.J. Hypoxia: signaling the metastatic cascade. Trends Cancer. 2016;2:295–304. doi: 10.1016/j.trecan.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundqvist H., Johnson R.S. Tumour oxygenation: implications for breast cancer prognosis. J. Intern. Med. 2013;274:105–112. doi: 10.1111/joim.12091. [DOI] [PubMed] [Google Scholar]
- Russell J., Zomerdijk J.C. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D., Gilkes D.M., Chaturvedi P., Xiang L., Semenza G.L. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc. Natl. Acad. Sci. U S A. 2014;111:E5429–E5438. doi: 10.1073/pnas.1421438111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatsky I.N., Terenin I.M., Smirnova V.V., Andreev D.E. Cap-independent translation: what's in a name? Trends Biochem. Sci. 2018;43:882–895. doi: 10.1016/j.tibs.2018.04.011. [DOI] [PubMed] [Google Scholar]
- Shi Z., Fujii K., Kovary K.M., Genuth N.R., Rost H.L., Teruel M.N., Barna M. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell. 2017;67:71–83.e77. doi: 10.1016/j.molcel.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan K.E., Warda A.S., Sharma S., Entian K.D., Lafontaine D.L.J., Bohnsack M.T. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017;14:1138–1152. doi: 10.1080/15476286.2016.1259781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiana M., Acosta-Iborra B., Hernandez R., Galiana C., Fernandez-Moreno M.A., Jimenez B., Del Peso L. Metabolic labeling of RNA uncovers the contribution of transcription and decay rates on hypoxia-induced changes in RNA levels. RNA. 2020;26:1006–1022. doi: 10.1261/rna.072611.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke J., Holterman C.E., Lachance G., Franovic A., Jacob M.D., Fabian M.R., Payette J., Holcik M., Pause A., Lee S. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486:126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Beucken T., Koritzinsky M., Wouters B.G. Translational control of gene expression during hypoxia. Cancer Biol. Ther. 2006;5:749–755. doi: 10.4161/cbt.5.7.2972. [DOI] [PubMed] [Google Scholar]
- van den Beucken T., Magagnin M.G., Jutten B., Seigneuric R., Lambin P., Koritzinsky M., Wouters B.G. Translational control is a major contributor to hypoxia induced gene expression. Radiother. Oncol. 2011;99:379–384. doi: 10.1016/j.radonc.2011.05.058. [DOI] [PubMed] [Google Scholar]
- van Nues R.W., Watkins N.J. Unusual C/D motifs enable box C/D snoRNPs to modify multiple sites in the same rRNA target region. Nucleic Acids Res. 2017;45:2016–2028. doi: 10.1093/nar/gkw842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellenkle C., Rooij J., Guffanti A., Brini E., Fasanaro P., Isaia E., Croft L., David M., Capogrossi M.C., Moles A. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. RNA. 2012;18:472–484. doi: 10.1261/rna.027615.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks S.E., Metge B.J., Samant R.S. The nucleolus: a central response hub for the stressors that drive cancer progression. Cell Mol. Life Sci. 2019;76:4511–4524. doi: 10.1007/s00018-019-03231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E.D., Gao D., Redfern A., Thompson E.W. Controversies around epithelial-mesenchymal plasticity in cancer metastasis. Nat. Rev. Cancer. 2019;19:716–732. doi: 10.1038/s41568-019-0213-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S., Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T., Papadopoulos E., Hagner P.R., Wagner G. Hypoxia-inducible factor-1alpha (HIF-1alpha) promotes cap-dependent translation of selective mRNAs through up-regulating initiation factor eIF4E1 in breast cancer cells under hypoxia conditions. J. Biol. Chem. 2013;288:18732–18742. doi: 10.1074/jbc.M113.471466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.M., Wang S.J., Gordan J.D., Ji X., Liebhaber S.A., Simon M.C. Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. J. Biol. Chem. 2008;283:16309–16319. doi: 10.1074/jbc.M710079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Shen X., Fan L., Yu Z. Analysis of histone modifications at human ribosomal DNA in liver cancer cell. Sci. Rep. 2015;5:18100. doi: 10.1038/srep18100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019238 and 10.6019/PXD019238.