Summary
Siglecs (sialic acid binding immunoglobulin (Ig)-like lectins) constitute a group of 15 human and 9 murine cell-surface transmembrane receptors belonging to the I-type lectin family, mostly expressed on innate immune cells and characterized by broadly similar structural features. Here, the prominent inhibitory CD22 (Siglec-2), well known in maintaining tolerance and preventing autoimmune responses on B cells, is studied in its human and murine forms in complex with sialoglycans. In detail, the role of the N-glycolyl neuraminic acid (Neu5Gc) moiety in the interaction with both orthologues was explored. The analysis of the binding mode was carried out by the combination of NMR spectroscopy, computational approaches, and CORCEMA-ST calculations. Our findings provide a first model of Neu5Gc recognition by h-CD22 and show a comparable molecular recognition profile by h- and m-CD22. These data open the way to innovative diagnostic and/or therapeutic methodologies to be used in the modulation of the immune responses.
Subject areas: Biochemistry, Immunology, Structural Biology
Graphical abstract
Highlights
-
•
The structural basis of sialoglycans recognition by h/m CD22 has been investigated
-
•
The binding modes of Neu5Gc-/Neu5Ac-containing ligands to m/h-CD22 were compared
-
•
The bioactive conformation of sialoglycans has been derived
-
•
Our findings may help in the regulation of immune response and cancer prevention
Biochemistry; Immunology; Structural Biology
Introduction
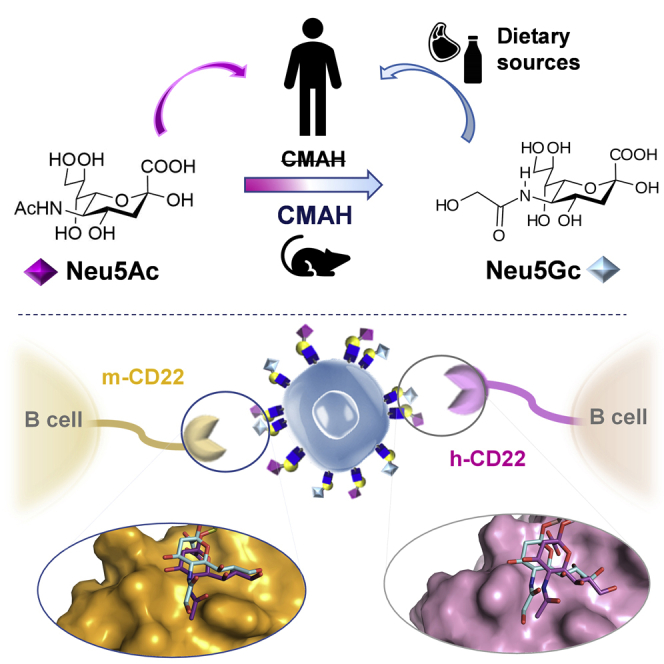
Sialic acids (Sias) comprise a family of nearly 50 members of acidic monosaccharides, expressed by species belonging to the vertebrates and characterized by a particular nine-carbon sugar backbone. The two predominant forms of sialic acid in mammals are the N-acetyl neuraminic acid (Neu5Ac) and the N-glycolyl neuraminic acid (Neu5Gc). The latter is biosynthesized by the enzymatic addition of a hydroxyl group to N-acetyl moiety at 5′-position of Neu5Ac, catalyzed by a hydroxylase/monooxygenase enzyme, the cytidine monophospho-N-acetyl neuraminic acid hydroxylase (CMAH). However, in contrast to mouse or great apes such as chimpanzee, in humans the specific loss of Neu5Gc expression is ascribed to a fixed genomic mutation in CMAH that leads to the gene loss in the hominin lineage (Okerblom and Varki, 2017). Despite the inability to produce Neu5Gc, it can be exogenously introduced from specific dietary sources, such as red meat and cow's milk, and metabolized via the Neu5Ac biochemical pathway (Okerblom and Varki, 2017; Varki, 2017). Different studies reported the presence of Neu5Gc on fetal tissues and on tumor cells, such as melanoma, retinoblastoma, colon cancer, and breast cancer (Samraj et al., 2014). Low levels of Neu5Gc were also detected on the surfaces of human secretory epithelia and small- and large-blood vessels endothelia (Varki, 2017). Therefore, Neu5Gc can be considered a pioneering example of “xeno-autoantigen.” The anti-Neu5Gc antibodies detected in humans contribute to establish “xenosialitis,” a chronic inflammation state in Neu5Gc-enriched tissues that can significantly impact cancer progression, increasing tumor-associated inflammation (Altman and Gagneux, 2019; Zhou et al., 2020).
Sias are considered self-associated molecular patterns (SAMPs), as they function as determinant of “self” through their intrinsic recognition by specific inhibitory receptors belonging to the Siglec family (Häubli and Varki, 2020; Duan and Paulson, 2020). These receptors assist immune cells in the discrimination between “self” and “non-self” and constitute important regulators of the immune system (Macauley et al., 2014). Their functions depend on their unique and precise sialoglycans' binding specificity as well as on their expression pattern within different cellular compartments (Di Carluccio et al., 2021). Siglecs represent a family of immune proteins that can be classified in two different and evolutionary conserved sub-groups; one comprises Sialoadhesin (Siglec-1), CD22 (Siglec-2), MAG (Siglec-4) and Siglec-15, whereas the other includes the so-called CD33 (Siglec-3)-related family, rapidly evolving and displaying high homology with the precursor, namely Siglec-3. Despite this differentiation, Siglecs share the ability to recognize and bind to sialic acid epitopes through the N-terminal V (variable)-set domain. Siglecs also possess a variable number (from 1 to 16) of “C2-set” Ig-like domains followed by a single pass trans-membrane region; most of Siglecs feature cytoplasmic tyrosine motifs, such as ITIM (immunoreceptor tyrosine-based inhibitory motif) and ITIM-like regions, which confer inhibitory signaling properties. A few activatory-type Siglecs, such as Siglecs-14–16, contain instead a positively charged amino acid in their transmembrane region and associate with immunoreceptor tyrosine-based activatory motif (ITAM)-containing adaptor proteins (Ajit and Angata, 2006).
The sialic acids are diversely recognized among the Siglec family, for example MAG only binds to Neu5Ac, human and murine sialoadhesin show a strong preference for Neu5Ac over Neu5Gc, whereas murine and human CD22 bind both, with m-CD22 preferring Neu5Gc over Neu5Ac (Angata, 2018). Interestingly, major human pathogens have evolved the ability to mimic Neu5Ac-containing structures, either synthesized de novo or scavenged from the host, to escape immune surveillance. In contrast, only a few pathogens express Neu5Gc or Neu5Gc-like structures (Varki, 2017).
Within this context, we here considered an interesting Siglec member, the inhibitory CD22 (h-Siglec-2 and its murine ortholog m- Siglec-2), expressed on the surface of B cells and to a lesser extent of T cells, able to modulate B cell tolerance by counteracting B cell receptor (BCR) signaling. On resting B-cells, CD22 is “masked” by cis interactions with sialoglycans exposed on the same plasma membrane, resulting in the formation of CD22 homo-oligomers (Han et al., 2005). Conversely, upon BCR activation, CD22 clusters are recruited to the BCR and, together with Siglec-10, negatively regulate the B cell signaling. CD22 malfunctioning and the resulting lack of appropriate BCR inhibition has been linked to several B-cell-related pathologies, such as hairy cell leukemia, marginal zone lymphoma, chronic lymphocytic leukemia, and non-Hodgkin lymphoma (Müller and Nitschke, 2014; Mahajan and Pillai, 2016; Dörner et al., 2012).
Because Siglecs, and indeed CD22, are considered effective glyco-immuno checkpoints within cancer immunotherapy (Duan and Paulson, 2020), and because changes in the Neu5Gc/Neu5Ac ratio can potentially modulate Siglecs' binding and signaling properties, understanding the basis of Neu5Gc - Siglec interaction may have therapeutic implications. Thus, also considering that the molecular details of Neu5Gc recognition by Siglecs are still far from being explored in-depth, we here aimed to understand the differences in the ability of human and murine CD22 to recognize and bind to various sialoglycans. To achieve this aim, we elucidated the molecular mechanisms of Neu5Gc recognition by, and binding to, human CD22/Siglec-2 (h-CD22) and compared it with the murine ortholog m-Siglec-2 (m-CD22). We combined NMR spectroscopy to computational approaches such as Molecular Dynamics, docking, and CORCEMA-ST methods, thus collecting pivotal information concerning the binding epitope and the conformational behavior of Neu5Gc containing glycans.
Results
The interaction between different sialoglycans bearing acetylated or glycolylated sialic acid, namely Neu5Ac and Neu5Gc with human and murine CD22, was investigated as follows.
Binding specificity of m- and h- CD22 toward Neu5Ac and Neu5Gc ligands
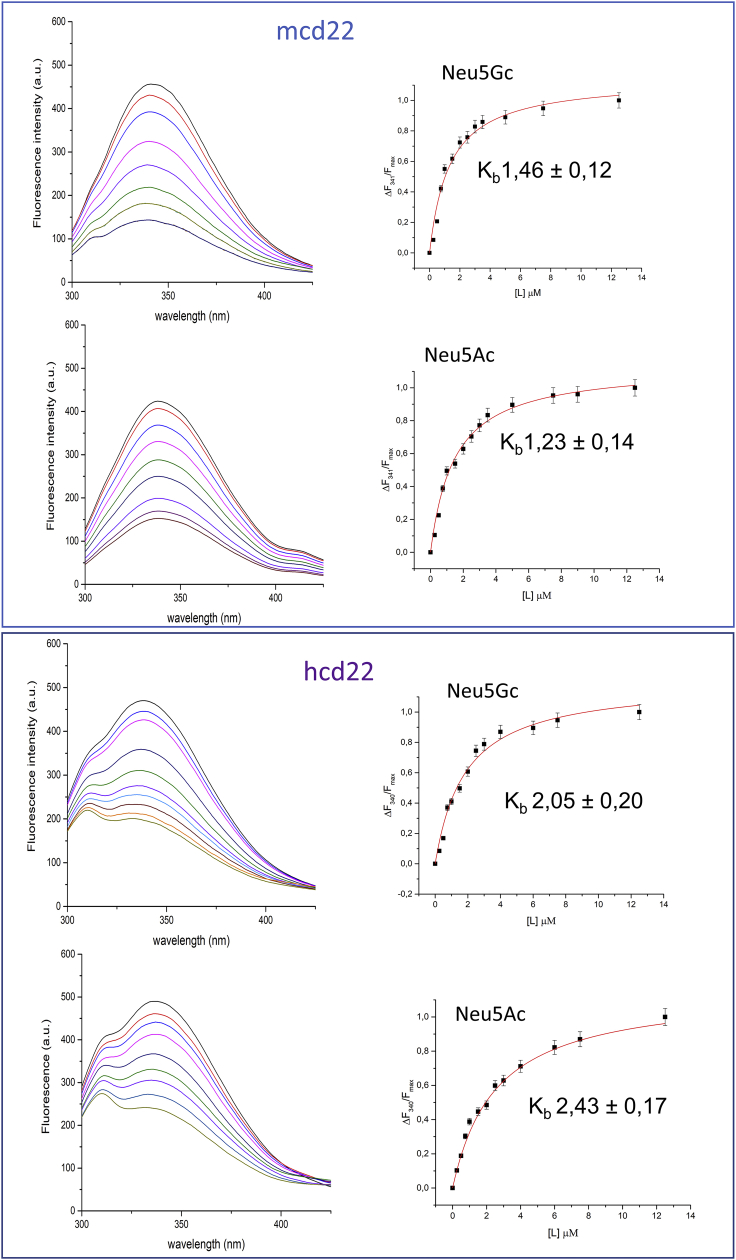
The binding affinities of both m-CD22 and h-CD22 toward Neu5Gc- and Neu5Ac-containing ligands were evaluated by fluorescence analyses; in detail, fluorescence titrations of increasing amounts of sialoglycans into a fixed concentration of the proteins were performed. The results demonstrated the ability of m- and h-CD22 to similarly recognize acetylated and glycolylated sialoglycans (Figure 1), as supported by the derived values of binding constants (Kb), all in the micromolar range. Thus, human and murine CD22 recognized the examined sialoconjugates comparably.
Figure 1.
Fluorescence titrations
Fluorescence spectra of m-CD22 (upper panel, black lines) or hCD22 (lower panel, black lines) in the presence of increasing amounts of Neu5Gc-containing ligand (colored lines) or Neu5Ac-containing ligand (colored lines), respectively. The binding isotherm and the values of the binding constants (Kb) are also reported. For each data point, 10% Y error bars are shown.
Glycolylated 6’SLN displays a comparable molecular behavior in the binding pocket of murine and human CD22
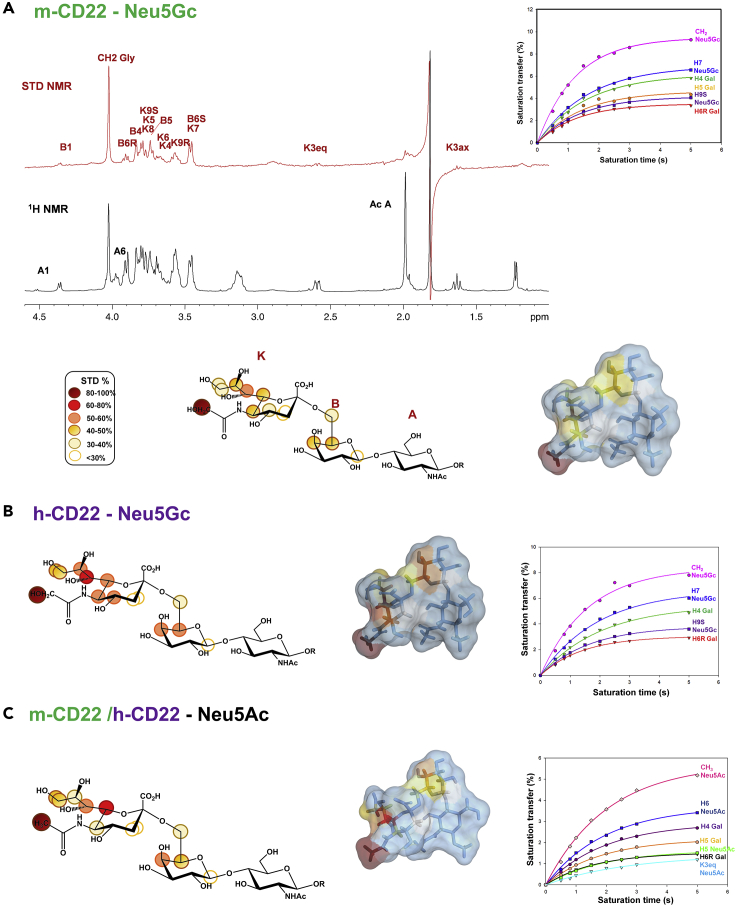
The glycolylated 6’-sialylactosamine [Neu5Gc-α-(2,6)-Gal-β-(1,4)-GlcNAc-β-OR, 6′SLN] was investigated upon binding to human and murine CD22. A detailed STD NMR (Meyer and Peters, 2003; Angulo and Nieto, 2011) analysis allowed us to map the epitopes of the ligand when interacting to both m-CD22 (Figure 2A) and h-CD22 (Figure 2B). The glycolylated ligand was recognized similarly by both receptors, as highlighted by an almost comparable binding epitope (Figures 2A and 2B). The highest STD signal belonged to the glycolyl moiety of Neu5Gc, whose signal was set to 100%. The sialic acid—galactose moiety was the main determinant of the binding to both h-CD22 and m-CD22; in detail, H-7 of Neu5Gc (K7, Figure 2A) was saturated more than 50%, whereas H-5, H-6, and H-8 of Neu5Gc (K5, K6 and K8 protons in Figure 2A), as well as H-5 and H-4 of Gal (B5 and B4, Figure 2A) were in the range of 40%–50%. Furthermore, the STD signals of the diastereotopic H-3eq and H-3ax protons (K3) showed the lowest effects with both murine and human CD22. The GlcNAc residue (A) was completely excluded from the CD22-binding pocket (Figures 2A and 2B), indicating its solvent exposure. The construction of STD build-up curves (Yan et al., 2003) then allowed to accurately define the glycolylated trisaccharide epitope excluding potential artifacts caused by differences in the longitudinal relaxation time T1 of the ligand protons (Marchetti et al., 2016) (Figures 2A and 2B, Tables S1 and S2).
Figure 2.
STD NMR analysis of glycolylated and acetylated Sia-α-(2,6)-Gal-β-(1,4)-GlcNAc-β−OCH2CH2NH2 interacting with murine and human CD22
(A) Reference 1H NMR spectrum (black) and STD 1D NMR spectrum (red) with a molecular ratio m-CD22/glycolylated ligand of 1:100 at saturation time of 2s. On the right, STD build-up curves are reported. STD build-up curves are calculated using the following monoexponential equation: , where STD (tsat) is the STD signal intensity of each proton at tsat saturation time, STDmax is the asymptotic maximum of the curve and ksat represents the observed saturation rate constant measuring the speed of STD build-up. The highest STD signal intensity is referred to the glycolyl/acetyl group of sialic acid, set to 100%, whereas the other protons were normalized to this value. The STD-derived epitope mapping on the molecular envelope of Neu5Gc ligand in the bioactive conformation with color code according to the observed STD effects is also shown.
(B) On the left, epitope map of the glycolylated ligand interacting with h-CD22, calculated from the ratio (I0−Isat)/I0, where (I0−Isat) is the STD signal and I0 is the peak intensity of the unsaturated reference spectrum. STD effects lower than 10% are not indicated. In the middle, the STD-derived epitope mapping on the molecular envelope of the ligand in its bioactive conformation is shown. On the right, STD build up curves are reported.(C) On the left, epitope map of the acetylated ligand interacting with m/h-CD22, calculated from the ratio (I0−Isat)/I0, where (I0−Isat) is the STD signal and I0 is the peak intensity of the unsaturated reference spectrum. STD effects lower than 10% are not indicated. In the middle, the STD-derived epitope mapping on the molecular envelope of the ligand in its bioactive conformation is shown. On the right, STD build up curves are reported.H6R, H6S, H9R, and H9S protons refer to H6-proR, H6-proS, H9-proR, and H9-proS, respectively.
The topology and conformation adopted by 6′SLN when interacting with h- and m-CD22 (the bioactive conformation) was achieved by transferred NOESY (tr-NOESY) analyses (Meyer and Peters, 2003). The stability of φ and ψ dihedral angles of the glycosidic linkages in the free state was monitored during 100 ns Molecular Dynamic simulation in explicit solvent, carried out using the Amber18 package (Case et al., 2018) (See also Methods and Figure S2). Differently from the Neu5Ac trisaccharide (Di Carluccio et al., 2019; Forgione et al., 2020), that in solution explores different populations depending on the values of φ torsion angle (−60°/180°), the Neu5Gc glycan preferentially adopts a conformation with φ around −60° (see Table S3).
As for the bound state, tr-NOESY analyses confirmed the preference of the glycolylated glycan for the energetic minimum characterized by φ/ψ dihedral angles of −60°/180°. The absence of NOE contacts between the H6-proR of galactose and the diastereotopic (axial and equatorial) H-3 protons of sialic acid and the key NOE established between the acetyl group of GlcNAc and H-5 of sialic acid (Table S3) observed in the tr-NOESY spectra (Figures S1B and S1C) revealed a bent conformation of the ligand, which assumed a shape characterized by an umbrella-like topology when bound either to h-CD22 and m-CD22 (Chandrasekaran et al., 2008).
Molecular modeling showed similar binding features of murine and human CD22 in the interaction with glycolylated ligands
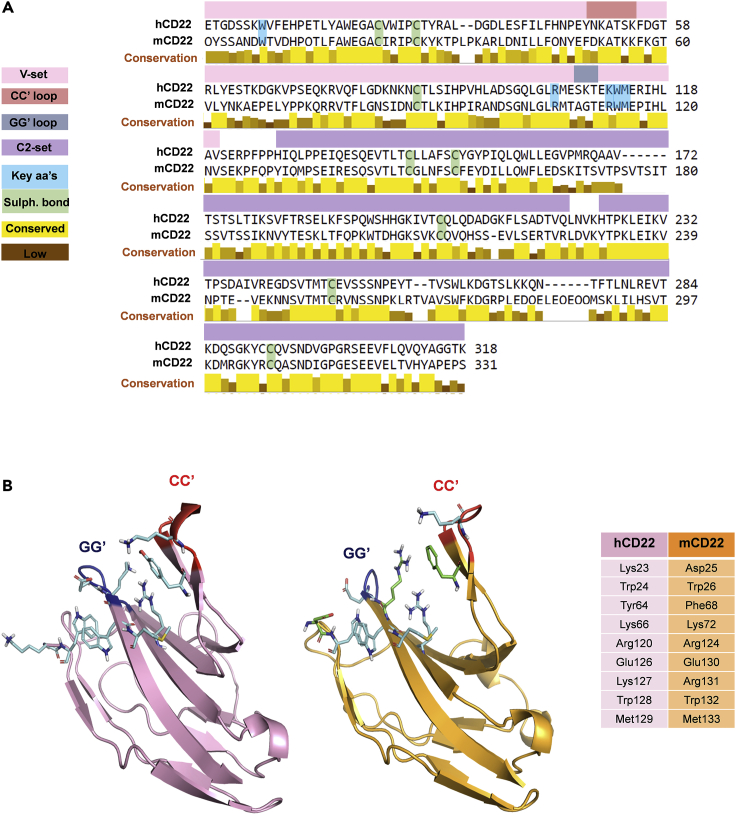
Computational studies including homology modeling, docking, and MD simulations were carried out to describe the binding of CD22 with Neu5Gc ligands. The crystal structure of h-CD22 (PDB: 5VKJ) (Ereño-Orbea et al., 2017) was used as structural template to model m-CD22, whose three-dimensional structure is not available. The sequence encoding for m-CD22 extracellular V-set and C2-set domains was aligned to the template sequence using BLAST (Basic Local Alignment Search Tool) (Altschul et al., 1990). According to the sequence alignment displayed in Figure 3A, m-CD22 expectedly showed significant sequence identity (above 58%) relatively to h-CD22, in agreement with the conserved nature of CD22. The target template alignment was submitted to the SWISS-MODEL (Waterhouse et al., 2018) server to obtain a three-dimensional model of m-CD22; the structure quality was assessed through PROCHECK web server (Laskowski et al., 1993), giving a Ramachandran plot in which 80.3% of model torsional angles were in the mostly favored region and 19.4% in the additional allowed region. The m-CD22 structural model obtained showed the typical Siglec Ig-like folding, with the sialic acid binding site located at the summit of the N-terminal V-set domain (Figure 3B). Similarly to other Siglecs, m-CD22 binding site architecture features a shallow pocket constituted by the A, F, and G strands and bound by the CC′ and GG′ variable loops. Compared with h-CD22, the composition of the binding site residues entitled to sialylated epitopes binding was overall conserved. The most relevant differences lied in the replacement of Lys23h, Tyr64h, and Lys127h with Asp25m, Phe68m, and Arg131m, which slightly affected the polarity of the binding region. Prior to docking calculations, the structural model was subjected to MD simulation, monitoring along the trajectory the backbone RMSD and the RMSF of the whole structure, as well as the RMSD of the CC′ and GG′ loops (Figures S3A–S3D). The analysis of the potential energy along the simulation allowed to establish the m-CD22 structure at lowest energy, which was subsequently employed for docking calculations (Figure S3D).
Figure 3.
Comparison of h-CD22 and m-CD22 structures
(A) BLAST alignment of the extracellular regions of murine CD22 and human CD22. Key amino acids are highlighted in blue and Cys forming disulfide bridges in green. Sequence corresponding to the V-set domain is evidenced in pink, sequence of C2-set domains in purple. Conservation between the two sequences is evaluated using Jalview (Waterhouse et al., 2009).
(B) Comparison of the N-terminal V-set domains of h-CD22 (pink), PDB: 5VKM, and m-CD22 homology model (orange). Common residues constituting the binding sites are highlighted in cyan. Residues of m-CD22 pocket differing from h-CD22 are colored in green. A direct comparison of the binding site residues can be found in the table on the right.
To analyze its binding mode, the Neu5Gc ligand was docked into h-CD22- and m-CD22-binding sites by means of Autodock 4.20 (Morris e al., 2009). The energies and populations of the top clusters were very similar for both receptors, ranging from −3.5 to −2 kcal mol−1 (Table S4). Thus, from analysis of the docking results, the h-CD22/and m-CD22/ligand complexes displaying lower relative energy and higher cluster populations were selected to run MD simulations. Notably, in the aforementioned poses Neu5Gc ligand displayed a similar binding mode inside the receptors pocket, in accordance with NMR data, showing the involvement of the following major determinants of sialylated ligands binding, i.e. the conserved arginine (Arg120h and Arg124m ) and aromatic residues (Trp24h, Trp128h and Trp26m, Trp132m) (Ereño-Orbea et al., 2017). It is worth noting that with both h-CD22 and m-CD22, the ligand assumed an umbrella-like conformation in the chosen clusters.
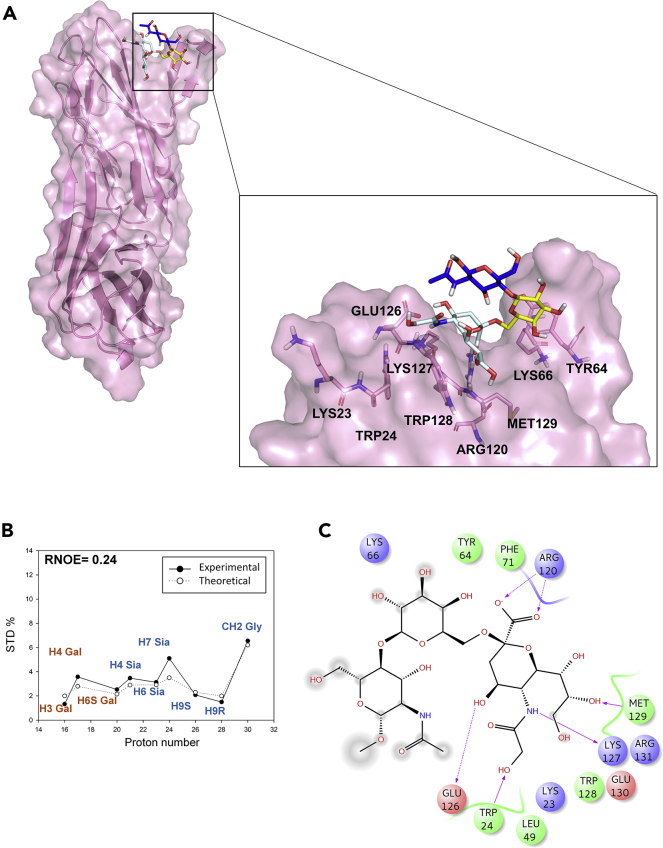
To finely describe the complexes between Neu5Gc-containing glycans and h- and m-CD22, the aforementioned structures were used as starting point to run MD simulations throughout 100 ns. Along the trajectory, the receptor, and ligand RMSD, the ligand dihedral angles fluctuations, as well as hydrogen bonds and contacts between the ligand and the receptor were monitored (Figures S3–S5). In either complexes, the ligand remained anchored to both h-CD22 and m-CD22 receptors until the end of the simulation, as demonstrated by the ligand RMSD values within ∼1.5/2 Å, suggesting the stability of the binding poses (Figures S4A and S4C). Representative complexes selected on the basis of the cluster analysis of the dynamics (see Methods section for details) were then analyzed by means of the CORCEMA-ST program (Jayalakshmi and Krishna, 2002) that allowed the comparison between the theoretical and the experimental STD data and the validation of 3D models of the complexes (Figures 3 and 4). Thus, as for the h-CD22/Neu5Gc model, several contacts between the ligand and the receptor binding pocket residues were observed; the majority of them were retained for most of the simulation time (Figure S4B). In particular, the polar network between h-CD22 and the glycolylated ligand was similar to that already described for the corresponding Neu5Ac ligand. (Di Carluccio et al., 2019) Indeed, the highly conserved Arg120 established a salt bridge between its guanidine group and Neu5Gc carboxylate. Neu5Gc glycerol moiety was involved in hydrogen bonds with Met129 backbone oxygen and amide, as well as CH-π contacts with Trp128. Also, the Neu5Gc OH-4 formed a polar interaction with Glu126. Most importantly, the N-glycolyl group of Neu5Gc engaged a stable hydrogen bond with Lys127 and hydrophobic contacts between its methylene protons and both Trp24 and Trp128 aromatic moieties (present 82% and 100% of the simulation time, respectively, as shown in Figure S4B). The Gal unit contributed to the receptor binding by means of CH–π interaction with Tyr64 aromatic residue (present for 100% of the simulation time). On the contrary, the GlcNAc unit was far from the h-CD22 surface for most part of the simulation, and this sugar moiety displayed higher RMSD with respect to the other sugar units (Figure S5A). Concerning the conformational behavior along the MD simulation, the bound ligand maintained the umbrella topology for 82% of the simulation (Figure S5B). Indeed, the distance between the CH3 group of the N-acetyl glucosamine and the H-5 of sialic acid assumed an average value of 4.9 Å, in accordance with the NOE derived distance (Figure S5C and Table S3).
Figure 4.
Interaction between h-CD22 and Neu5Gc ligand
(A) Three-dimensional model of Neu5Gc ligand bound to h-CD22 V-set domain as derived by STD, tr-NOESY, and MD.
(B) The three-dimensional h-CD22/Neu5Gc complex showing the best fit between theoretical (solid line) and experimental (dashed line) STD data derived by CORCEMA-ST analysis. (R-NOE values of 0.24).
(C) Two-dimensional plots representing the interactions between the glycolylated trisaccharide and h-CD22-binding site residues, derived from a representative frame of the MD simulation. Dotted arrows represent hydrogen bonds with functional groups from side chains and solid arrows those with functional groups of the backbone. The interaction diagram was produced by Maestro 10.4.018 program.
The CORCEMA-ST highlighted a good agreement between the theoretical and experimental values (Figure 4B). Indeed, the strongest STD effects in the h-CD22 complex were predicted for protons belonging to Neu5Gc unit; significant saturation was also estimated for some protons of the Gal unit; conversely no saturation was predicted for protons of GlcNAc units, in full agreement with the experimental STD data (Figures 2B and S1A, Table S2). The high STD value of the glycolyl group of the ligand was consistent with the close contacts with Trp24, Trp128, and Glu126 side chains. Regarding STD data of the Neu5Gc glycerol moiety, the higher STD effect observed for the H-7 is due to the strong CH-π interaction of this proton with Trp128 indole group, beyond the contacts between the entire moiety and Met129. Concerning the hydroxymethylene group, only the H-9S was oriented toward Trp128, thus exhibiting a higher STD effect with respect to H-9R. Also, H-4 and H-6 of Neu5Gc displayed significant STD effects for their vicinity to the receptor surface. For the Gal unit, considerable saturation was predicted especially for the proton at position 4, due to its close contacts with Tyr64 aromatic ring.
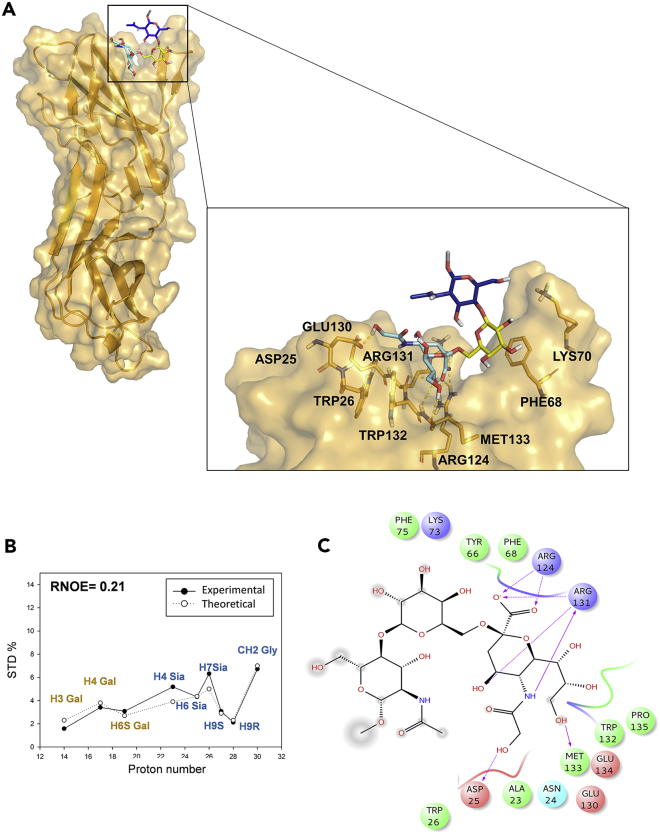
As for m-CD22/Neu5Gc-validated model, the ligand interaction pattern showed many similarities with its human ortholog. Still, Neu5Gc majorly contributed to the binding, interacting with Arg124, Arg131, Trp132, and Met133 receptor residues, whereas no participation of GlcNAc residue was observed (Figures 5 and S4D). Specifically, the carboxylate of Neu5Gc formed the key electrostatic interactions with the Arg124 guanidinium group. Met133 established numerous polar interactions with the hydroxyl groups of the ligand glycerol moiety. In addition, the Arg131 played the same role of Lys127 in h-CD22 receptor, thus forming a stable hydrogen bond between its backbone oxygen and the amide nitrogen of Neu5Gc N-glycolyl moiety, which was also in close contact with Trp26 and Trp132 aromatic residues (present for 93% and 100% of simulation time, respectively).
Figure 5.
Interaction between m-CD22 and Neu5Gc ligand
(A) Three-dimensional model of Neu5Gc ligand bound to m-CD22 V-set domain as derived by STD, tr-NOESY, and MD.
(B) The three-dimensional m- CD22/Neu5Gc complex showing the best fit between theoretical (solid line) and experimental (dashed line) STD data derived by CORCEMA-ST analysis. (R-NOE values of 0.21).
(C) Two-dimensional plots representing the interactions between the glycolylated trisaccharide and m-CD22 binding site residues, derived from a representative frame of the MD simulation. Dotted arrows represent hydrogen bonds with functional groups from side chains and solid arrows those with functional groups of the backbone. The interaction diagram was produced by Maestro 10.4.018 program.
Differently from h-CD22 complex, the hydroxyl group of the N-glycolyl moiety forms polar interactions with Asp25 and Asn24 residues, which lie in proximity of the binding region, although these contacts were observed for 60% and 40% of the MD simulation, respectively. Furthermore, similarly to h-CD22 binding mode, the Gal unit is mostly engaged in CH–π interaction with Phe-68 residue (contact present for 99% of the simulation time); also, the GlcNAc residue did not interact with m-CD22, exhibiting a higher degree of fluctuation with respect to the other residues (Figure S5D). In accordance with the experimental data, the ligand retained an umbrella-like conformation for 89% of the simulation time, as supported by the average distance between the CH3 group of the N-acetyl glucosamine and the H-5 of Neu5Gc along the simulation (Figures S5E and S5F).
Thus, a comparison of CORCEMA-ST results of m-CD22 and h-CD22 in complex with Neu5Gc-containing trisaccharide highlights how the orientation of the glycan inside the receptors binding pockets is comparable. The strong STD value observed for N-glycolyl moiety of Neu5Gc in the interaction with m-CD22 could be explained by the contacts between the hydroxymethyl group and Asp25 and Asn24 residues, beyond those observed with the Trp26 and Trp132 aromatic residues and the hydrogen bond with Arg131, analogous to that described in the human receptor. The potential involvement of the Asp25 in the recognition of N-glycolyl trisaccharide by m-CD22 was supported by the results of CORCEMA-ST analysis performed on several other structures lacking the hydrogen bond with the Asp25, resulting in higher R-NOE values due to the significantly lower STD value attributed to the N-glycolyl moiety protons (data not shown). Considering the Gal moiety, similar STD effects were predicted for the protons directed toward the residue aromatic side chain, namely H-4 and H-3, in line with conservative mutation of Tyr64h into Phe68m.
STD NMR analysis revealed a comparable recognition profile of the acetylated 6’SLN by murine and human CD22
The STD NMR analysis of the interaction of Neu5Ac-containing trisaccharide [Neu5Ac-α-(2,6)-Gal-β-(1,4)-GlcNAc] with murine CD22 (Figures 2C and S1A, left panel) revealed an epitope map and a binding mode fully comparable to that of human CD22, previously characterized by our group (Di Carluccio et al., 2019). The several changes in the multiplicity and relative intensity of signals observed in the STD NMR spectrum with respect to the corresponding reference (Figure S1A) were diagnostic of the binding specificity. In detail, the sialic acid residue (K) mostly participated to the interaction with m-CD22, with the acetyl group giving the highest STD signal. On the contrary, the acetyl group belonging to the glucosamine residue (A) disappeared from the STD spectrum, highlighting its distance from the binding site. In addition, H-6 proton of sialic acid gave a good STD signal, close to 70%, followed by the protons belonging to the glycerol chain and the H5 (range of 30%–50%). The contribution of the diastereotopic H-3 protons was less remarkable (<30%). Interaction of the galactose unit (B) was also detected, mainly relative to protons H-4, H-5, and H-6. Notably, as further confirmation of the binding specificity, the multiplet around 3.9 ppm in the off-resonance, deriving from the overlapping of H-6 A and H-6 B, was converted into a triplet corresponding to the only H-6 Gal B in the STD spectrum, further evidence that the N-acetylglucosamine moiety was excluded from the recognition process. Additional data gathered from the construction of STD build-up curves (Figure 2C) corroborated the results obtained from the qualitative STD NMR analysis (Table S5). The above experimental data highlighted a totally comparable binding mode of Neu5Ac-containing trisaccharide with human and murine CD22.
Comparison of acetylated and glycolylated glycans interaction with murine and human CD22
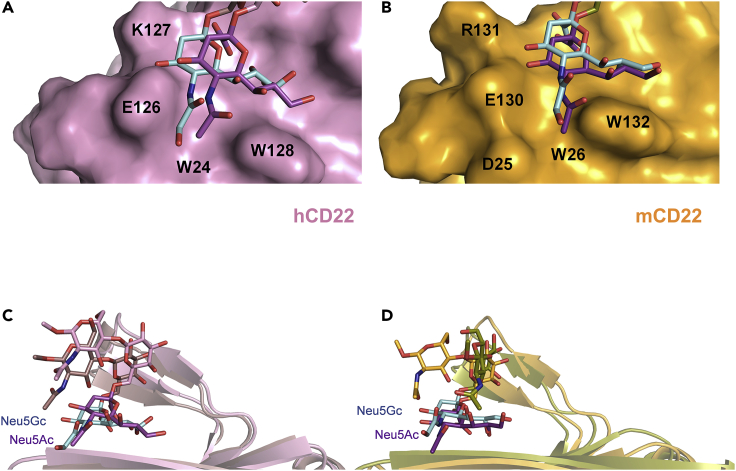
To directly compare the mode of interaction of Neu5Ac/Neu5Gc with m-CD22, a computational approach was performed to establish a three-dimensional complex of m-CD22 and Neu5Ac glycans, thus defining a reliable model of interaction (Figure 6). According to our results, the Neu5Ac ligand displayed a similar orientation with respect to Neu5Gc ligand, establishing the crucial salt bridge with Arg124 through its carboxylate. The hydroxyl groups of the glycerol lateral chain interacted through hydrogen bonds with the Met133 backbone. The N-acetyl group was involved in the binding with Arg131 as well as hydrophobic interactions with Trp26 and Trp132 aromatic residues. The Gal residue, similarly to Neu5Gc ligand, was involved in CH-π interactions with Phe68, and the GlcNAc was far from the binding region. Thus, it can be assessed that m-CD22 interacts with Neu5Ac and Neu5Gc ligands in analogous manner. A comparison of the 3D structures of the models showed slightly different shape and polarity of the receptor cavities, which accommodate the Neu5Ac and Neu5Gc moieties (Figures 6A and 6B). Indeed, although the h-CD22 region responsible for N-acetyl and N-glycolyl binding is essentially constituted by aromatic residues, m-CD22 also comprises the Asp25 residue in optimal position to interact with the longer N-glycolyl chain.
Figure 6.
Comparison of the interaction of Neu5Ac/Neu5Gc ligands with m-CD22 and h-CD22
(A) Close up view of N-Acetyl (purple)- and N-glycolyl (cyan)-binding region of h-CD22, showing the protein surface (pink).
(B) Close up view of N-Acetyl (purple)- and N-glycolyl (cyan)-binding region of m-CD22, showing the protein surface (orange).
(C) Superimposition of h-CD22/Neu5Gc (pink) and h-CD22/Neu5Ac complexes (dirty violet).
(D) Superimposition of m-CD22/Neu5Gc (bright orange) and m-CD22/Neu5Ac complexes (olive).
Finally, MM/GBSA and MM/PBSA analysis (Srinivasan et al., 1998) was performed with Amber to have an indication of the relative binding energy of the complexes (Table 1). As result, all four complexes exhibited comparable ΔGb values, in agreement with the similar binding properties of the receptors toward the different forms of sialic acid discussed here (Figures 6C and 6D). Therefore, despite some differences in the binding regions described earlier, it is possible to assess that human and murine CD22 similarly recognize Neu5Ac and Neu5Gc ligands, in accordance with the experimental results.
Table 1.
Relative binding energies of h-CD22 and m-CD22 with acetylated and glycolylated ligands
| Complex | ΔGb (MM/GBSA) | ΔGb (MM/PBSA) |
|---|---|---|
| h-CD22/Neu5Gc | −41.24 ± 0.15 | −12.66 ± 0.16 |
| h-CD22/Neu5Ac | −38.43 ± 0.18 | −10.75 ± 0.18 |
| m-CD22/Neu5Gc | −38.29 ± 0.16 | −11.73 ± 0.19 |
| m-CD22/Neu5Ac | −34.69 ± 0.26 | −10.72 ± 0.18 |
All units are expressed in kcal/mol.
Discussion
The family of Sias includes nearly 50 structurally diverse members deriving from naturally occurring modifications in different positions of the original nonulosonic acid skeleton (Pearce and Läubli, 2016). Usually, Sias cap the terminal moiety of cell surfaces glycoconjugates and glycolipids as well as secreted glycoproteins are attached to the underlying glycan via α-(2→3), α-(2→6), or α-(2→8) linkages. By virtue of their location, diversity, and ubiquity in vertebrates, Sias serve as ligands of endogenous and exogenous glycan-binding proteins, thus representing important regulators of several biologically relevant recognition processes (Varki, 2008). Sialylated glycans from human cells mainly terminate with the 5′-acetylated isoform of neuraminic acid, Neu5Ac, because of the evolutionary loss of the Neu5Gc, which differs from Neu5Ac by one additional oxygen atom. In contrast with the majority of other mammals, humans do not possess the ability to synthesize Neu5Gc; it can, however, be metabolically incorporated from dietary sources, i.e. red meat, and becomes present on some epithelial and endothelial cell surfaces (Alisson-Silva et al., 2018). Also, the presence of Neu5Gc on nontypeable Haemophilus influenzae (NTHi) LOS (lipooligosaccharide) that forages exogenous sialic acids from the host was recently demonstrated (Ng Preston, 2018). Being Neu5Gc a xeno-autoantigen mostly present on malignant cells, the interaction between inhibitory CD22 and Neu5Gc glycans may indeed play a key role in the regulation of the tumor immune response (Samraj et al., 2014), impact on cancer progression, and increase of tumor-associated inflammation. Here, we evaluated the effect of sialic acid glycolylation on the binding with CD22, comparing the behavior of different ligands in complex with murine and human CD22, improving the knowledge of the structural basis of the recognition of sialylated N-glycans from CD22 receptor.
NMR analysis revealed a comparable binding epitope of both murine and human CD22 toward glycolylated glycans (Figures 2A–2C and S1). In both complexes a bent umbrella-like conformation of the ligand was adopted, as supported by the results achieved from the NMR data and MD simulations (Figures S1B, S1C, S5C, and S5F). From molecular modeling, it was confirmed that Neu5Gc/Neu5Ac ligands displayed a similar orientation inside the binding site of murine and human CD22, independently from the Sia nature, as supported by a comparison of the molecular surfaces of h-CD22 and m-CD22 in the interaction with N-acetyl/N-glycolyl Sia chains, in Figure 6. Furthermore, it was evidenced the possibility for m-CD22 of forming additional interactions with Neu5Gc ligand, mainly involving the hydroxymethyl group of N-glycolyl moiety. This was further supported by a comparison of the molecular surfaces of h-CD22 and m-CD22 deputed to interact with N-acetyl/N-glycolyl Sia chains. In addition, the substitution of Asp25m in place of Lys23h in the binding subsite of m-CD22 influences the possibility to establish hydrogen bonds with the hydroxyl group of the glycolyl moiety.
Overall, our studies indicate that, despite the different nature of sialic acid residue, the recognition region of h-CD22 is almost invariant comparing Neu5Ac and Neu5Gc containing glycans. These results agree with the similar affinity of h-CD22 toward Neu5Gc/Neu5Ac structures, as recently reported by Angata (Angata, 2018). In conclusion, the obtained outcomes provide a global vision of how the most diffuse neuraminic acid forms of sialylated N-glycans in mammals are arranged in the binding pocket of CD22. Hence, potential high-affinity analogues of ligands naturally recognized from the CD22 could be specifically designed and synthetized for targeting the receptor protein in order to impede the biological interaction, thus modulating immune responses.
Limitations of the study
This study reports the interactions of the biologically relevant CD22, in human and murine variants, with the two predominant forms of sialic acid in vertebrates. A comparison of the binding mode was carried out mainly by NMR spectroscopy, molecular modeling, and CORCEMA-ST calculations. In the absence of high-resolution coordinates of the murine CD22, we employed homology modeling and different computational approaches to validate the generated models (docking, MD simulations, CORCEMA-ST). Despite the high homology with respect to the template, the structural conclusions we derived from homology modeling will require the definition of the three-dimensional structure of the murine CD22 to avoid misinterpretation of the data.
Resource availability
Lead contact
Roberta Marchetti (roberta.marchetti@unina.it).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate datasets.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This study was supported by PRIN 2017 “Glytunes” (2017XZ2ZBK, 2019-2022) to AS and SS; progetto POR SATIN and Progetto POR Campania Oncoterapia to AM; the European Commission (H2020-MSCA- 814102--SWEET CROSSTALK project) to AM, RM, and AS; and the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program under grant agreement No 851356 to RM. PON Ricerca e Innovazione 2014-2020, Azione I.1 “Dottorati Innovativi con caratterizzazione Industriale” is acknowledged for funding the Ph.D. grant to R.E.F.
This paper is dedicated to Prof. Jesús Jiménez-Barbero for his 60th birthday.
Author contributions
A.S. and R.M. conceived and designed the project. R.M., R.E.F., C.D.C., A.S., and A.M. carried out NMR, docking, and CORCEMA experiments and analyzed the results. R.E.F. performed homology modeling. R.E.F., R.M., A.S., S.S., M.M., and M.C. carried out and analyzed MD simulations. P.R.C. and G.S. produced the proteins. K.F. and Y.M. provided the glycans. All the authors wrote, revised, and reviewed the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2020.101998.
Contributor Information
Roberta Marchetti, Email: roberta.marchetti@unina.it.
Alba Silipo, Email: silipo@unina.it.
Supplemental information
References
- Ajit V., Angata T. Siglecs—the major subfamily of I-type lectins. Glycobiology. 2006;16:1–27. doi: 10.1093/glycob/cwj008. [DOI] [PubMed] [Google Scholar]
- Alisson-Silva F., Liu J.Z., Diaz S.L., Deng L., Gareau M.G., Marchelletta R., Chen X., Nizet V., Varki N., Barrett K.E., Varki A. Human evolutionary loss of epithelial Neu5Gc expression and species-specific susceptibility to cholera. PLoS Pathog. 2018;14:e1007133. doi: 10.1371/journal.ppat.1007133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman M.O., Gagneux P. Absence of Neu5Gc and presence of anti-Neu5Gc antibodies in humans-an evolutionary perspective. Front. Immunol. 2019;10:789. doi: 10.3389/fimmu.2019.00789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Angata T. Possible influences of endogenous and exogenous ligands on the evolution of human Siglecs. Front. Immunol. 2018;10:789. doi: 10.3389/fimmu.2018.02885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo J., Nieto P.M. Std NMR: application to transient interactions between biomolecules—a quantitative approach. Eur. Biophys. J. 2011;40:1357–1369. doi: 10.1007/s00249-011-0749-5. [DOI] [PubMed] [Google Scholar]
- Di Carluccio C., Crisman E., Manabe Y., Forgione R.E., Lacetera A., Amato J., Pagano B., Randazzo A., Zampella A., Lanzetta R. Characterization of the dynamic interactions between complex N-glycans and human CD22. Chembiochem. 2019;21:129. doi: 10.1002/cbic.201900295. [DOI] [PubMed] [Google Scholar]
- Di Carluccio C., Forgione R.E., Molinaro A., Crocker P., Marchetti R., Silipo A. Exploring the fascinating world of sialoglycans in the interplay with Siglecs. Carbohydr. Chem. 2021;44:31–55. [Google Scholar]
- Case D.A., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham III T.E., Cruzeiro V.W.D., Darden T.A., Duke R.E., Ghoreishi D., Gilson M.K. University of California; 2018. AMBER 2018. [Google Scholar]
- Chandrasekaran A., Srinivasan R., Raman K., Viswanathan S., Raguram T.M., Tumpey V., Sasisekharan R. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat. Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- Dörner T., Shock A., Smith K.G. CD22 and autoimmune disease. Int. Rev. Immunol. 2012;31:363–378. doi: 10.3109/08830185.2012.709890. [DOI] [PubMed] [Google Scholar]
- Duan S., Paulson J.C. Siglecs as immune cell checkpoints in disease. Annu. Rev. Immunol. 2020;38:365–395. doi: 10.1146/annurev-immunol-102419-035900. [DOI] [PubMed] [Google Scholar]
- Ereño-Orbea J., Sicard T., Cui H., Mazhab-Jafari M.T., Benlekbir S., Guarné A., L Rubinstein J., Julien J.P. Molecular basis of human CD22 function and therapeutic targeting. Nat. Commun. 2017;8:764. doi: 10.1038/s41467-017-00836-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgione R.E., Di Carluccio C., Guzmán-Caldentey J., Gaglione R., Battista F., Chiodo F., Manabe Y., Arciello A., Del Vecchio P., Fukase K. Unveiling molecular recognition of sialoglycans by human siglec-10. iScience. 2020;23:101–401. doi: 10.1016/j.isci.2020.101401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Collins B.E., Bengtson P., Paulson J.C. Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking Nat. Chem. Biol. 2005;1:93–97. doi: 10.1038/nchembio713. [DOI] [PubMed] [Google Scholar]
- Häubli H., Varki A. Sialic acid–binding immunoglobulin-like lectins (Siglecs) detect self-associated molecular patterns to regulate immune responses. Cell. Mol. Life Sci. 2020;77:593–605. doi: 10.1007/s00018-019-03288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayalakshmi V., Krishna N.R. Complete relaxation and conformational exchange matrix (CORCEMA) analysis of intermolecular saturation transfer effects in reversibly forming ligand-receptor complexes. J. Magn. Reson. 2002;155:106–118. doi: 10.1006/jmre.2001.2499. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. Procheck - a program to check the stereochemical quality of protein structures. J. App. Cryst. 1993;26:283–291. [Google Scholar]
- Macauley M.S., Crocker P.R., Paulson J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014;14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan V.S., Pillai S. Sialic acids and autoimmune diseases. Immunol. Rev. 2016;269:145–161. doi: 10.1111/imr.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti R., Perez S., Arda A., Imberty A., Jimenez-Barbero J., Silipo A., Molinaro A. “Rules of engagement” of protein–glycoconjugate interactions: a molecular view achievable by using NMR spectroscopy and molecular modeling. ChemistryOpen. 2016;5:274–296. doi: 10.1002/open.201600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B., Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chem. Int. Ed. Engl. 2003;21:864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. AutoDock treats the ligand as a flexible unit and the protein as a rigid unit. J. Comput. Chem. 2009;30:2785–2791. [Google Scholar]
- Müller J., Nitschke L. The role of CD22 and Siglec-G in B-cell tolerance and autoimmune disease. Nat. Rev. Rheumatol. 2014;10:422–428. doi: 10.1038/nrrheum.2014.54. [DOI] [PubMed] [Google Scholar]
- Ng Preston S.K. Nontypeable Haemophilus influenzae has evolved preferential use of N-acetylneuraminic acid as a host adaptation. mBio. 2018;10:422–519. doi: 10.1128/mBio.00422-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okerblom J., Varki A. Biochemical, cellular, physiological, and pathological consequences of human loss of N-glycolylneuraminic acid. ChemBioChem. 2017;18:1–18. doi: 10.1002/cbic.201700077. [DOI] [PubMed] [Google Scholar]
- Pearce O.M.T., Läubli H. Sialic acids in cancer biology and immunity. Glycobiology. 2016;26:111–128. doi: 10.1093/glycob/cwv097. [DOI] [PubMed] [Google Scholar]
- Samraj A.N., Läubli H., Varki N., Varki A. Involvement of a non-human sialic Acid in human cancer. Front. Oncol. 2014;4:33. doi: 10.3389/fonc.2014.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J., Cheatham T.E., Cieplak P., Kollman P.A., Case D.A. Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate - DNA helices. J. Am. Chem. Soc. 1998;120:9401–9409. [Google Scholar]
- Varki A. Sialic acids in human health and disease. Trends Mol. Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. Are humans prone to autoimmunity? Implications from evolutionary changes in hominin sialic acid biology. J. Autoimmun. 2017;83:134–142. doi: 10.1016/j.jaut.2017.07.011. [DOI] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:296–303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan A.D.K., Mo H., Shapiro M.J., Zartler E.R. The effect of relaxation on the epitope mapping by saturation transfer difference NMR. J. Magn. Reson. 2003;163:270–276. doi: 10.1016/s1090-7807(03)00106-x. [DOI] [PubMed] [Google Scholar]
- Zhou X., Yang G., Guan F. Biological functions and analytical strategies of sialic acids in tumor. Cells. 2020;9:273. doi: 10.3390/cells9020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets.