Figure 3.
Relative abundance of eukaryotic plankton viruses is associated with carbon export efficiency in the global ocean
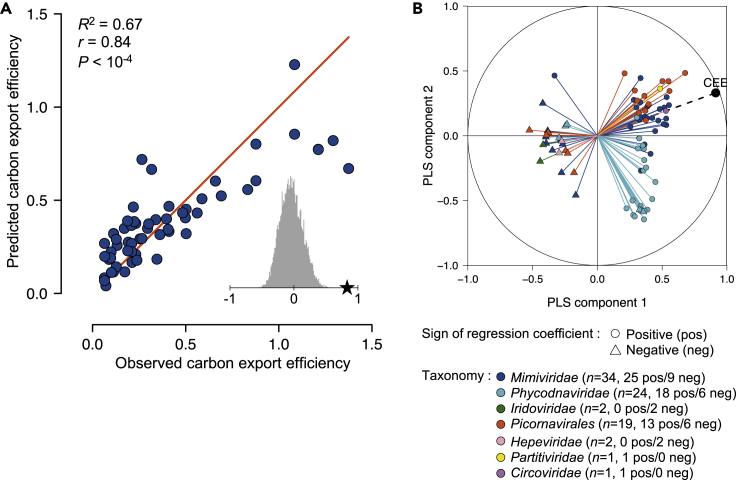
(A) Bivariate plot between predicted and observed values in a leave-one-out cross-validation test for carbon export efficiency. The PLS regression model was constructed using occurrence profiles of 1,523 marker-gene sequences (1,309 PolBs, 180 RdRPs, and 34 Reps) derived from environmental samples. r, Pearson correlation coefficient; R2, the coefficient of determination between measured response values and predicted response values. R2, which was calculated as 1—SSE/SST (sum of squares due to error and total) measures how successful the fit is in explaining the variance of the response values. The significance of the association was assessed using a permutation test (n = 10,000) (gray histogram in (A)). The red diagonal line shows the theoretical curve for perfect prediction.
(B) Pearson correlation coefficients between CEE and occurrence profiles of 83 viruses that have VIP scores >2 (VIPs) with the first two components in the PLS regression model using all samples. PLS components 1 and 2 explained 83% and 11% of the variance of CEE, respectively. Fifty-eight VIPs had positive regression coefficients in the model (shown with circles), and 25 had negative regression coefficients (shown with triangles). See also Figures S6, S7, and S12, Table S1, and Data S1.