Summary
Pericytes play a critical role in promoting, regulating, and maintaining numerous vascular functions. Their dysfunction is a major contributor to the progression of vascular and neurodegenerative diseases, making them an ideal candidate for large-scale production for disease modeling and regenerative cell therapy. This protocol describes the rapid and robust differentiation of pericytes from human induced pluripotent stem cells (hiPSCs) while simultaneously generating a population of hiPSC-derived endothelial progenitor cells.
For complete details on the use and execution of this protocol, please refer to Zhang et al. (2017).
Subject areas: Stem cells, Cell differentiation
Graphical abstract
Highlights
-
•
Generate pericytes from hiPSCs using magnetic cell sorting
-
•
Simultaneously generate a population of hiPSC-derived endothelial progenitor cells
-
•
Characterize pericytes via flow cytometry, immunofluorescence, and functional assays
Pericytes play a critical role in promoting, regulating, and maintaining numerous vascular functions. Their dysfunction is a major contributor to the progression of vascular and neurodegenerative diseases, making them an ideal candidate for large-scale production for disease modeling and regenerative cell therapy. This protocol describes the rapid and robust differentiation of pericytes from human induced pluripotent stem cells (hiPSCs) while simultaneously generating a population of hiPSC-derived endothelial progenitor cells.
Before you begin
hiPSCs are obtained and used according to legal and ethical guidelines. All procedures are performed in a Class II biological hood using sterile technique. hiPSCs and derivatives thereof are cultured in a humidified 37°C incubator and 5% CO2.
hiPSCs must be thawed at least 7 days prior to differentiation and passaged regularly before the beginning of the differentiation procedure. Different hiPSC lines may proliferate at different rates, therefore, the protocol may need to be adapted for each cell line.
Prior to beginning the differentiation protocol, prepare media, solutions, and matrix-coated culture plates. Cell-culture treated dishes or plates must be coated with extracellular matrix proteins or protein mixes for enhanced attachment of cells.
Note: E8 and Versene are used for hiPSC culture and passaging in this protocol. Other commercial reagents such as mTesR1 and ReLeSR may be suitable alternatives, however, have not been tested. We cannot guarantee similar results using these alternative reagents, and additional modifications of the protocol may be necessary.
Fibronectin-collagen coating of culture plates
-
1.
Thaw fibronectin (1 mg/mL) and collagen IV (3.35 mg/mL) on ice.
-
2.
Prepare coating solution by combining fibronectin and collagen IV to DME/F12 for a final concentration of 15 μg/mL and 15 μg/mL, respectively.
-
3.
Add coating solution (15 μg/mL fibronectin, 15 μg/mL collagen IV in DME/F12) to a 10 cm dish for a final concentration of 0.25 μg/cm2 fibronectin and 0.25 μg/cm2 collagen IV. Ensure the entire surface is covered.
-
4.
Incubate for 60 min at 37°C.
Note: Fibronectin-collagen coated dishes can be used after 60 min or can be stored for up to 1 week at 37°C with media to prevent drying. Coated dishes should not be used if the dish has dried out.
Matrigel coating of culture plates
-
5.
Thaw Matrigel (7–10 mg/mL) on ice.
-
6.
Resuspend Matrigel in cold DME/F12 (1:120 dilution).
-
7.
Add 1 mL of diluted Matrigel per well of a 6-well plate (6–9 μg/cm2) and 1 mL DME/F12, ensuring the entire surface is covered.
-
8.
Incubate for 60 min at 37°C.
Note: Matrigel-coated dishes can be used after 60 min or can be stored for up to 1 week at 37°C with media to prevent drying. Coated dishes should not be used if the dish has dried out.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE mouse anti-human CD13 | BD Pharmingen | 555394 |
| PE mouse anti-human CD140b | BD Pharmingen | 558821 |
| Anti-α-smooth muscle actin (1A4) mouse mAb | Cell Signaling Technology | 48938 |
| Anti-CD31 hamster mAb | Abcam | ab119341 |
| Anti-TAGLN (SM22) rabbit polyclonal antibody | Thermo Fisher | PA5-27463 |
| Alexa Fluor 488 donkey anti-mouse IgG | Life Technologies | A21202 |
| Alexa Fluor 568 donkey anti-rabbit IgG | Life Technologies | A10042 |
| Alex Fluor 657 goat anti-hamster IgG | Life Technologies | A21451 |
| Chemicals, peptides, and recombinant proteins | ||
| Accutase | Corning | 25-058-CI |
| Essential 8 Medium | Thermo Fisher | A1517001 |
| Versene | Life Technologies | 15040-066 |
| Matrigel | Corning | 354277 |
| Geltrex Matrigel | Gibco | A14133-02 |
| Tocris Bioscience DAPI | Fisher Scientific | 5748 |
| PBS | Fisher Scientific | 1419144 |
| EDTA | Invitrogen | 130-036-702 |
| Fibronectin | Corning | 356008 |
| Collagen IV | Corning | 354245 |
| EGM-2 Medium Kit | Promocell | C-22111 |
| TrypLE | Life Technologies | 12563-029 |
| PM Media Kit | ScienCell | 1201,1252,0010 |
| FBS | Gibco | 16000044 |
| Normal Donkey Serum | Fisher Scientific | 50-413-115 |
| DMSO | Fisher Scientific | BP231 |
| BMP4 | R&D Systems | 314-BP-010/CF |
| Activin A | R&D Systems | 338-AC-50/CF |
| CHIR 99021 | R&D Systems | 4423 |
| DF3S medium | Gibco | A14535DJ |
| SB431542 | R&D Systems | 1614 |
| VEGFA165 | R&D Systems | 293-VE-500/CF |
| FGF2 | R&D Systems | 233-FB-500/CF |
| Transferrin | R&D Systems | 2914-HT |
| Insulin | Sigma | I9278 |
| Paraformaldehyde | Fisher Scientific | 18612139 |
| Rock Inhibitor Y27632 | R&D Systems | 1254/10 |
| Critical commercial assays | ||
| Miltenyi Biotech MACS 34 microbead kit | Miltenyi Biotech | 130046702 |
| MACS LS Column | Miltenyi Biotech | 130042401 |
| MACS MS Column | Miltenyi Biotech | 130042201 |
| Experimental models: cell lines | ||
| iPSC line WTC-11 (GM25256) | Coriell Institute | GM25256 |
| iPSC line WT-83 | University of California San Diego | N/A |
| iPSC line Q83X | University of California San Diego | N/A |
| iPSC line M2 | University of California San Diego | N/A |
| Human Brain Vascular Pericytes | ScienCell | 1200 |
| Other | ||
| 6-well tissue culture plate | Corning | 3516 |
| 100 mm tissue culture dish | Corning | 353003 |
| Angiogenesis Plate | Ibidi | 89646 |
Materials and equipment
All media and solutions are filter sterilized through a membrane of 0.2 μm or smaller pore size.
DAPI 5 mg/mL
Dissolve 1 mg of DAPI in 200 μL of DMSO to obtain a stock solution of 5 mg/mL. Aliquot and store at −20°C for up to 12 months.
Fibronectin 1 mg/mL
Add 1 mL of sterile distilled water to 1 mg of fibronectin and incubate for 30 min at RT without agitation to obtain a stock solution of 1 mg/mL. Aliquot and store at −20°C for up to 2 weeks.
Collagen IV 3.35 mg/mL
Aliquot and store at −80°C for up to 12 months.
BMP4 100 μg/mL
Dissolve 100 μg of BMP4 in 4 mM HCl and 0.1% BSA to obtain a stock solution of 100 μg/mL. Aliquot and store at −80°C for up to 3 months.
Activin A 100 μg/mL
Dissolve 50 μg of Activin A in 500 μL of 4 mM HCl to obtain a stock solution of 100 μg/mL. Aliquot and store at −80°C for up to 3 months.
CHIR 99021 10 mM
Dissolve 10 mg of CHIR 99021 in 2,149 μL of DMSO to obtain a stock solution of 10 mM. Aliquot and store at −20°C for up to 6 months.
SB431542 10 mM
Dissolve 1 mg of SB431542 in 260.152 μL of DMSO to obtain a stock solution of 10 mM. Aliquot and store at −80°C for up to 1 month.
VEGFA165 100 μg/mL
Dissolve 100 μg of VEGFA165 in 1 mL of PBS and 0.1% BSA to obtain a stock solution of 100 μg/mL. Aliquot and store at −20°C for up to 3 months.
FGF2 100 μg/mL
Dissolve 100 μg in 1 mL of PBS and 0.1% BSA to obtain a stock solution of 100 μg/mL. Aliquot and store at −20°C for up to 3 months.
Transferrin 10.7 mg/mL
Dissolve 100 mg of Transferrin in 9,350 μL of sterile distilled water to obtain a stock solution of 10.7 mg/mL. Aliquot and store at −80°C for up to 3 months.
Y27632 5 mM
Dissolve 1 mg of Y27632 in 624.492 μL of sterile distilled water to obtain a stock solution of 5 mM. Aliquot and store at −20°C for up to 1 month.
hiPSC culture medium (essential 8 (E8))
| Reagent | Final concentration | Volume |
|---|---|---|
| Essential 8 Media | n/a | 50 mL |
| E8 Supplement | 2% | 1 mL |
When plating cells from thaw or passaging, add Rock inhibitor Y27632 to media for a final concentration of 10 μM for the initial 24 h.
Store at 4°C for up to 2 weeks.
E8BAC medium
| Reagent | Final concentration | Volume |
|---|---|---|
| Essential 8 Media | n/a | 50 mL |
| E8 Supplement | 2% | 1 mL |
| BMP4 | 5 ng/mL | 2.5 μL |
| Activin A | 25 ng/mL | 12.5 μL |
| CHIR 99021 | 1 μM | 5 μL |
When plating cells from thaw or passaging, add Rock inhibitor Y27632 to media for a final concentration of 10 μM for the initial 24 h.
Store at 4°C for up to 2 weeks.
E7Vi medium
| Reagent | Final concentration | Volume/Mass |
|---|---|---|
| DF3S | n/a | 100 mL |
| SB431542 | 5 μM | 50 μL |
| VEGFA165 | 50 ng/mL | 50 μL |
| FGF2 | 100 ng/mL | 100 μL |
| Transferrin | 10.7 μg/mL | 50 μL |
| Insulin | 20 μg/mL | 100 μL |
Store at 4°C for up to 2 weeks.
EGM-2 medium
| Reagent | Final concentration | Volume/Mass |
|---|---|---|
| Endothelial Cell Growth Basal Medium 2 | n/a | 500 mL |
| Epidermal Growth Factor | 5 ng/mL | Kit aliquot |
| bFGF | 10 ng/mL | Kit aliquot |
| Insulin-like Growth Factor (R3 IGF-1) | 20 ng/mL | Kit aliquot |
| VEGF 165 | 0.5 ng/mL | Kit aliquot |
| Ascorbic Acid | 1 μg/mL | Kit aliquot |
| Heparin | 22.5 ug/mL | Kit aliquot |
| Hydrocortisone | 0.2 ug/mL | Kit aliquot |
Store at 4°C for up to 6 weeks.
PM medium (without antibiotic solution)
| Reagent | Final concentration | Volume/Mass |
|---|---|---|
| Pericyte Media (PM) | n/a | 500 mL |
| FBS | 2% | 10 mL |
| Pericyte growth supplement (PGS) | 1% | 5 mL |
Store at 4°C for up to 4 weeks.
E7V medium
| Reagent | Final concentration | Volume/Mass |
|---|---|---|
| DF3S | n/a | 100 mL |
| Insulin | 20 μg/mL | 100 μL |
| VEGFA165 | 50 ng/mL | 50 μL |
| FGF2 | 100 ng/mL | 100 μL |
| Transferrin | 10.7 μg/mL | 50 μL |
Store at 4°C for up to 2 weeks.
Cryopreservation medium
| Reagent | Final concentration | Volume/Mass |
|---|---|---|
| FBS | 90% | 9 mL |
| DMSO | 10% | 1 mL |
| Total | n/a | 10 mL |
Store at 4°C for up to 2 weeks.
MACS buffer
| Reagent | Final concentration |
|---|---|
| PBS | n/a |
| BSA | 5 mg/mL |
| EDTA | 2 mM |
Store at 4°C for up to 4 weeks.
FACS Buffer
| Reagent | Final concentration |
|---|---|
| PBS | n/a |
| FBS | 2% |
Store at 4°C for up to 4 weeks.
IF blocking buffer
| Reagent | Final concentration |
|---|---|
| PBS | n/a |
| Normal Donkey Serum | 10% |
| BSA | 1% |
| Triton X | 0.25% |
Store at −20°C for up to 8 weeks.
IF antibody buffer
| Reagent | Final concentration |
|---|---|
| PBS | n/a |
| Normal Donkey Serum | 1% |
| BSA | 1% |
| Triton X | 0.25% |
Store at −20°C for up to 8 weeks.
Step-by-step method details
hiPSCs passaging and maintenance
Timing: 2 days
This step is required to ensure cells are in an active growth phase before beginning the differentiation protocol on day 0.
-
1.Day −2. Pass hiPSCs using Versene.
-
a.Wash each well with PBS.
-
b.1 mL of warm Versene was added per well and incubated for 7 min at 37°C.
-
c.Remove Versene via aspiration and add 3 mL of media to each well. Pipet up and down vigorously.
-
d.Split cells at a 1:3 split and plate onto a Matrigel-coated 6 well tissue culture plate in E8 media with 10 μM Rock Inhibitor Y27632.
-
a.
-
2.
Day −1. Feed cells with E8 media.
Note: We suggest a confluency of 50%–60% by day 0 to ensure that colonies of hiPSCs are not too large and that cells maintain an active growth phase before beginning differentiation. We have observed this step to be important for differentiation and obtaining a high yield of CD34− cells.
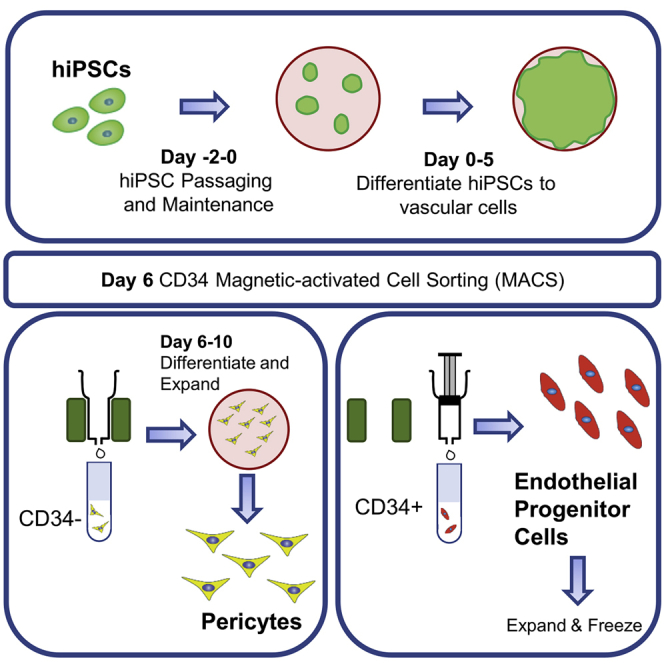
Differentiating hiPSCs to vascular cells
Timing: 6 days
This step differentiates hiPSCs to a mixed population of CD34+ and CD34− vascular cells through a mesoderm intermediate step (Figure 1).
-
3.Day 0. hiPSCs are passaged using Versene.
-
a.Prior to the addition of Versene, wash each well with PBS.
-
b.Add 1 mL of warm Versene per well and incubate for 7 min at 37°C.
-
c.Remove Versene via aspiration and add 3 mL of media to each well. Cells should remain in small colonies and should not be singular.
-
d.Plate cells at a seeding density of 1.0–1.5 × 105 cells per well onto Matrigel-coated 6 well tissue culture plates in 2 mL/well E8BAC media with the addition of 10 μM Rock Inhibitor Y27632.
-
a.
-
4.
Day 1. Feed cells with 2 mL per well E8BAC media.
-
5.
Day 2. Once cells have reached 100% confluency (40–48 h post seeding), aspirate the E8BAC media and replace with E7Vi media.
-
6.
Day 3. Feed cells with 3 mL per well of E7Vi media.
-
7.
Day 4. Feed cells with 4 mL per well of E7Vi media.
-
8.
Day 5 Feed cells with 5 mL per well of E7Vi media.
-
9.
Day 6. Follow CD34 MACS sorting protocol.
Note: Volumes reported are for one well of a 6-well tissue culture plate and will need to be adjusted for other sizes.
CRITICAL: We suggest the media change on day 2 (step 5) occur between 40 and 48 h post seeding, when the cells have reach 100% confluency. It is important to not allow the cells to become overly confluent, as it may result in a lower yield of CD34− cells.
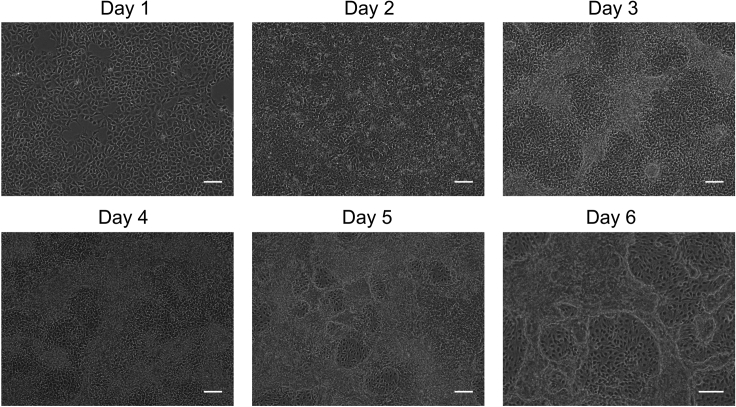
Figure 1.
hiPSCs (WTC11) undergoing differentiation to vascular cells through a mesoderm intermediate step
hiPSCs are differentiated into mesoderm in chemically defined E8BAC medium (days 0–2). At day 3, chemically defined E7Vi medium induces the differentiation of CD34+ and CD34− vascular cells (days 3–6). Scale bar represents 200 μm.
CD34 magnetic-activated cell sorting (MACS)
Timing: 2–3 h
This step sorts the CD34+ endothelial progenitor cells from the CD34− vascular cells. CD34 is widely recognized as a marker of vascular endothelial progenitor cells (Fina et al., 1990; Hristov and Weber, 2008; Sidney et al., 2014). Pericytes, including cerebral pericytes, umbilical pericytes, skeletal muscle pericytes, and microvascular cardiac pericytes, do not express CD34 (Cathery et al., 2018; Faal et al., 2019; Wang et al., 2019).
-
10.
Day 6. Dissociate cells using Accutase for 10 min at 37°C to produce a single cell suspension.
-
11.
Collect cells using an equal volume of 2% FBS in PBS, centrifuge at 300 × g for 10 min and resuspend cells at 1 × 108 total cells (live and dead) in 300 μL of cold MACS buffer.
-
12.
Add 100 μL of FcR blocking reagent and 100 μL of CD34 magnetic microbeads to the cell suspension.
-
13.
Mix well and incubate at 4°C for 30 min.
-
14.
Wash cells with 10 mL MACS buffer and centrifuge at 300 × g for 10 min.
-
15.
Remove supernatant completely and resuspend in 500 μL MACS buffer.
CRITICAL: Keep cells on ice before and after MACS sorting and in between steps when necessary to ensure greater cell viability.
-
16.
Place the LS column on the MACS magnet.
-
17.
Wash the LS column carefully with 3 mL MACS buffer to avoid bubbles.
-
18.Collect CD34− cells
-
a.Add the cell suspension to the column.
-
b.Collect unbound, CD34− cells in the flow through.
-
c.Wash the cell suspension tube with 500 μL MACS buffer to remove any residual cells and add to the column.
-
d.Wash the column with an additional 9 mL MACS buffer.
-
e.Collect the flow through and place on ice.
-
a.
-
19.Collect CD34+ cells
-
a.Remove the column from the magnet.
-
b.Add 5 mL MACS buffer and slowly push the column plunger to flush out the attached CD34+ cells.
-
c.Add an additional 5 mL of buffer to the column and plunge to remove any residual CD34+ cells.
-
a.
Pause point: The CD34+ cells (endothelial progenitor cells) can continue culture or be cryopreserved and stored in liquid nitrogen.
CRITICAL: It is important to have a single-cell suspension to prevent blocking of the column. If blocking does occur, the flow through will be slow and could lead to cell death. Similarly, any bubbles present in the column could also block the column and slow the flow through. When plunging the column, it is important to maintain a steady flow through. Too slow or too fast could lead to cell death and decrease the yield of CD34+ cells.
Note: The Miltenyi Biotech MACS CD34 microbead kit protocol recommends using 2 × 109 total cells per LS column. We have found that this cell density can clump and block the column and have reduced the number to 1 × 108 total cells per column. If cell numbers are low MS columns (Miltenyi Biotech #130042201) can be used in place of the LS columns.
Differentiating CD34− cells to pericytes
Timing: 4 days
-
20.
Day 6. After MACS sorting, count and plate the CD34− cells at 2 × 104 cells/cm2 on to fibronectin-collagen coated 10 cm tissue culture dishes in EGM-2 media.
-
21.
Day 7. Feed cells with EGM-2 media.
-
22.Day 8. Passage pericyte precursors
-
a.Dissociate cells with warm TrypLE for 5 min at 37°C. Dilute TrypLE with media (1:1), collect, centrifuge at 300 × g for 5 min, remove supernatant, and resuspend in PM media.
-
b.Plate cells on to fibronectin-collagen coated 10 cm tissue culture dishes at 1.5 × 104 cells/cm2
-
a.
-
23.
Day 9. Feed cells with PM media.
-
24.
Day 10. Characterize, cryopreserve, or continue cells for further experiments.
Expanding and maintaining hiPSC-derived pericytes
Timing: up to passage 10
hiPSC-derived pericytes can be expanded and maintained for at least ten passages.
-
25.Passage pericytes
-
a.At approximately 80% confluency, dissociate cells with warm TrypLE for 5 min at 37°C. Dilute TrypLE with media (1:1), collect cells, centrifuge at 300 × g for 5 min, remove supernatant, and resuspend in PM Media.
-
b.Plate pericytes on fibronectin-collagen coated tissue culture plates at a 1:3 split.
-
a.
-
26.
Maintain pericytes by feeding every other day with PM medium.
-
27.
When 80% confluent, passage cells up to passage 10.
Cryopreserving hiPSC-derived pericytes
Pause point: hiPSC-derived pericytes can be cryopreserved for future use. Cells can successfully be preserved as pericyte precursors (day 6 immediately following column sorting) or as pericytes (day 10 and later).
-
28.For hiPSC-derived pericytes, freeze when approximately 80% confluent (generally day 10).
-
a.Dissociate cells from the plate using warm TrypLE for 5 min at 37°C. Dilute TrypLE in media (1:1), collect cells, and centrifuge at 300 × g for 5 min.
-
b.Remove supernatant and in a dropwise manner add Cryopreservation media.
-
c.Transfer cells into cryogenic vials (1 × 106 cells per vial in 0.5–1 mL Cryopreservation Media).
-
d.Freeze at −80°C in a Mr. Frosty Freezing Container for 2–24 h, then store in a liquid nitrogen freezer.
-
a.
-
29.For hiPSC-derived pericyte precursors, freeze cells immediately after sorting following the same procedure.
-
a.Remove supernatant and in a dropwise manner add Cryopreservation media.
-
b.Transfer cells into cryogenic vials (2 × 106 cells per vial in 0.5–1 mL Cryopreservation Media).
-
c.Freeze at −80°C in a Mr. Frosty Freezing Container for 2–24 h, then store in a liquid nitrogen freezer.
-
a.
Recovering hiPSC-derived pericytes
Cryopreserved hiPSC-derived pericyte precursors (cryopreserved at day 6 immediately after column sorting) and hiPSC-derived pericytes (day 10) can be recovered as described.
-
30.hiPSC-derived pericyte precursors are recovered from cryopreservation by thawing 1 vial (approximately 2 × 106 cells) onto a fibronectin-collagen coated 10 cm tissue culture dish in EGM-2 media.
-
a.After 24 h, feed with EGM-2 medium.
-
b.At day 2, passage cells (as done on day 8 of differentiation protocol) and plate into PM medium.
-
c.Continue expanding in PM media up to passage 10.
-
a.
-
31.hiPSC-derived pericytes are recovered from cryopreservation by thawing 1 vial (approximately 1 × 106 cells) onto a fibronectin-collagen coated 10 cm tissue culture dish in PM media.
-
a.Feed cells every other day with PM medium.
-
b.Passage at 80% confluency
-
c.Continue expanding in PM media up to passage 10.
-
a.
Note: The recovery rate of pericyte precursors (cryopreserved at day 6) is estimated to be between 50% and 70%. The recovery rate of pericytes (day 10 or later) is estimated to be between 60%–90%. Recovery rates may vary between cell lines.
Characterizing cells: flow cytometry
Timing: 1–2 h
Flow cytometry is used to characterize the cells and confirm their differentiation to pericytes. CD140b, also known as PDGFRβ, is a ubiquitous marker of pericytes and its expression is essential for pericyte function (Armulik et al., 2011; Crisan et al., 2009; Geevarghese and Herman, 2014). CD13 is also a marker of pericytes and it plays a vital role in cell migration, survival, and angiogenesis (Armulik et al., 2011; Cathery et al., 2018; Ramsauer et al., 1998).
-
32.
Detach pericytes from the fibronectin-collagen coated tissue culture plates using warm TrypLE for 5 min at 37°C. Deactivate TrypLE with 2× volume of FACS buffer. Centrifuge cells for 5 min at 300 × g and resuspend in FACs buffer at 5 × 105 cells/100 μL per tube.
-
33.
Incubate cells with primary conjugated antibodies CD13-PE and CD140b-PE at 20°C–25°C for 30 min.
-
34.
Wash cells, centrifuge, and resuspend in FACS buffer.
-
35.
Run flow cytometry.
Note: A LSRII Flow Cytometer was used in these experiments.
Characterizing cells: immunofluorescence
Timing: 2 days
Immunofluorescence can also be used to characterize the differentiated pericytes. Here, cells were stained for the contractile proteins, αSMA (Bergers and Song, 2005; Crisan et al., 2009; Verbeek et al., 1994) and SM22 (Orlova et al., 2014a; Pierantozzi et al., 2016), which are involved in the vascular remodeling and vasoconstriction functions of pericytes. CD31, a widely regarded marker for endothelial cells, was also used as a negative control to ensure effective sorting of pericytes from CD34+ endothelial progenitor cells (Lertkiatmongkol et al., 2016; Liu and Shi, 2012; van Mourik et al., 1985).
-
36.
Remove all media from cell culture via aspiration.
-
37.
Fix cells in 4% paraformaldehyde (PFA) for 1 h at 20°C–25°C.
-
38.
Wash with PBS.
-
39.
Block cells with IF blocking buffer for 1 h at 20°C–25°C.
-
40.
Apply primary antibodies diluted in IF antibody buffer (Table 1) and incubate for 16–24 h at 4°C.
-
41.
Wash cells three times 15 min with PBS
-
42.
Apply secondary antibodies diluted in IF antibody buffer and incubate for 1.5 h at 20°C–25°C the dark. Counterstain with DAPI at 5 μg/mL for the last 15 min of incubation.
-
43.
Wash cells three times 15 min with PBS and store in PBS at 4°C until imaged.
Table 1.
Antibodies for immunofluorescence
| Antibody | Dilution | Supplier |
|---|---|---|
| Conjugated antibody | ||
| PE Mouse anti-human CD31 | 1:5 | BD Pharmingen |
| PE Mouse anti-human CD140b | 1:5 | BD Pharmingen |
| Primary antibody | ||
| Anti-α-Smooth Muscle Acting (1A4) monoclonal mouse | 1:200 | Cell Signaling Technology |
| Anti-CD31 monoclonal hamster | 1:250 | Abcam |
| Anti-TAGLN (SM22) polyclonal rabbit | 1:200 | Thermo Fisher |
| Anti-collagen IV monoclonal mouse | 1:62.5 | R&D Systems |
| Anti-PDGFR beta monoclonal rabbit | 1:100 | Abcam |
| Secondary antibody | ||
| Alex Fluor 488 donkey anti-mouse | 1:200 | Life Technologies |
| Alexa Fluor 568 donkey anti-rabbit | 1:200 | Life Technologies |
| Alexa Fluor 647 goat anti-hamster | 1:200 | Life Technologies |
Characterizing cell function: angiogenesis assay
Timing: 5 days for assay, 2 days for immunofluorescence staining
-
44.
Add hiPSC-derived endothelial cells only (1.5 × 104) or hiPSC-derived endothelial cells (1.5 × 104) and hiPSC-derived pericytes (3 × 103) from the same cell line per well of a Geltrex-coated angiogenesis plate and culture in E7V media at standard culture conditions (5% CO2, 37°C).
-
45.
Day 1–4. Feed cells daily with E7V media.
-
46.
At day 5 fix cells with 4% paraformaldehyde for 1 h at 20°C–25°C.
-
47.
Wash cells with PBS and block with blocking buffer for 2 h at 20°C–25°C.
-
48.
Apply primary antibodies diluted in IF antibody buffer (Table 1) for 16–24 h at 4°C.
-
49.
Wash cells three times 15 min in PBS
-
50.
Apply fluorescence-conjugate secondary antibodies diluted in IF antibody buffer for 1.5 h at 20°C–25°C in the dark.
-
51.
Counterstain with DAPI at 5 μg/mL for the last 15 min of incubation.
-
52.
Wash cells with PBS and store in PBS at 4°C until imaged.
Expected outcomes
This protocol for pericyte differentiation of hiPSCs has been adapted and modified from previously published protocols (Orlova et al., 2014b; Xu et al., 2017; Zhang et al., 2017). In our previous work, endothelial cells were differentiated from hPSCs through a mesoderm intermediate step using serum-free, chemically defined media and without the use of feeder cells or embryoid bodies (Zhang et al., 2017). It was reported that endothelial cells (CD34+CD31+) were present at day 5 of differentiation, and higher purity was achieved by day 7. Here, the protocol employs a similar approach, but with the addition of CD34 MACS at day 6 of differentiation to separate the CD34+ endothelial progenitor cells from the remaining CD34− vascular cell population. In contrast to alternative protocols that require 2–3 weeks to generate a substantial pericyte population (Orlova et al., 2014b), this protocol requires 10 days. Approximately 50%–75% of cells recovered from MACS are CD34−. By the 10th day of differentiation, approximately 4–6 million pericytes are generated from a single 10 cm tissue culture dish of plated hiPSCs. At which point, pericytes can either be cryopreserved or expanded for at least ten passages.
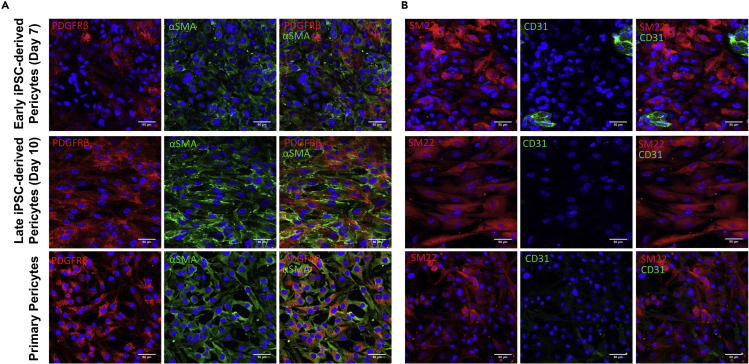
Protein expression of early stage (day 7), late stage (day 10) hiPSC-derived pericytes, and primary human brain vascular pericytes was evaluated via immunofluorescence (Figure 2). Early stage hiPSC-derived pericytes (day 7) stained positively for the pericyte markers SM22 and αSMA. However, only a fraction expressed the pericyte marker PDGFRβ (Figure 2A) and several cells expressed the endothelial cell marker CD31 (Figure 2B). Following additional differentiation and culture in PM media, all late stage hiPSC-derived pericytes (day 10) expressed SM22, αSMA, as well as PDGFRβ and did not express CD31. This was similar to primary human brain vascular pericytes which stained positively for SM22, αSMA and PDGFRβ and negatively for CD31.
Figure 2.
Immunofluorescent images of early (day 7) and late (day 10) hiPSC-derived pericytes and primary human brain vascular pericytes
Cells were stained for the pericyte markers (A) PDGFRβ (red), αSMA (green), and (B) SM22 (red) and the endothelial cell marker (B) CD31 (green). Nuclei are counterstained with DAPI (blue). Scale bar represents 50 μm.
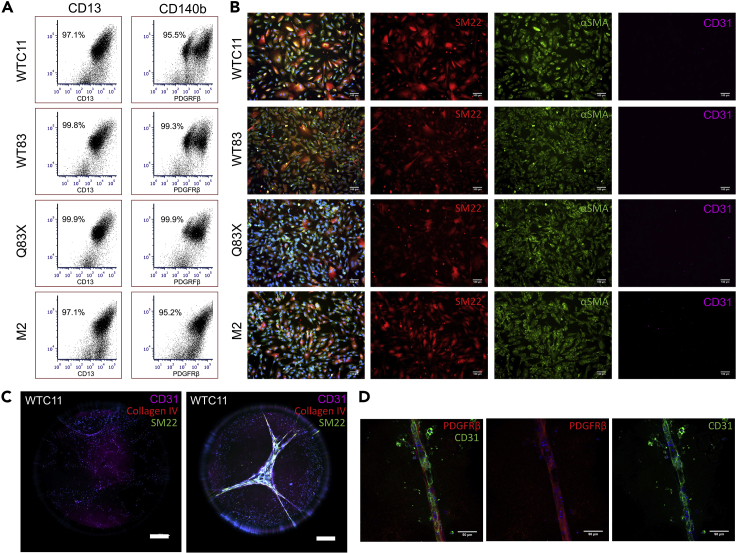
The pericyte differentiation protocol has been applied successfully to four different hiPSC lines including two wildtype cell lines (WTC11 and WT83) and two cell lines derived from patients with neurodevelopmental disorders: Rett syndrome caused by a mutation in the MECP2 gene (Q83x) and MECP2 duplication syndrome caused by a duplication of the MECP2 gene (M2). This protocol robustly yields greater than 95% CD13+PDGFRβ+ pericyte populations (Figure 3A) and cells stain positively for both pericyte markers smooth muscle-specific protein 22 (SM22) and alpha-smooth muscle actin (α-SMA) (Figure 3B). Additionally, cells did not stain positively for the endothelial marker CD31.
Figure 3.
Characterization of hiPSC-derived pericytes from WTC11, WT83, Q83X, and M2 hiPSC cell lines
(A) Flow cytometry of pericytes for pericyte markers CD13 and CD140b.
(B) Representative immunofluorescent images of hiPSC-derived pericytes stained positively for markers SM22 (red) and αSMA (green) and negatively for the endothelial marker CD31 (purple). Nuclei are counterstained with DAPI (blue). Scale bar represents 100 μm.
(C and D) Representative immunofluorescent images of hiPSC (WTC11)-derived endothelial cells (C) and hiPSC (WTC11)-derived endothelial cells and hiPSC (WTC11)-derived pericytes cocultured (D) at day 5 of an angiogenesis assay. hiPSC-derived endothelial cells stained positively for the endothelial marked CD31 (C, purple) (D, green) and hiPSC-derived pericytes stained positively for the pericyte markers SM22 (C, green), collagen IV (C, red) and PDGFRβ (D, red). Nuclei are counterstained with DAPI (blue). Scale bars represent 500 μm and 50 μm.
An angiogenesis assay can be used to assess the function of the hiPSC-derived pericytes. Alone, hiPSC-derived endothelial cells remained singularized and did not form an interconnected vascular network after five days (Figure 3C). Conversely, when cultured with pericytes that were derived following this protocol, endothelial cells and pericytes aligned together to form vascular tubules (Figure 3D). The tubules stained positively for the endothelial cell marker CD31 and the pericyte marker PDGFRβ. These results suggest pericytes differentiated using the protocol described herein express pericyte-specific markers and aid in vascular network formation.
Limitations
Differentiation efficiencies and cell yields can vary between experiments and cell lines. Additionally, the MACS cell sorting technique can result in low cell viability if guidelines are not followed due to blocking of the MACS column.
Troubleshooting
Problem 1
Poor hiPSC viability, proliferation, and/or pluripotency prior to beginning the differentiation protocol.
Potential solution
Confirm that that Rock inhibitor Y27632 is added to the cells at every passage to ensure cell attachment and enhance viability. Additionally, the pluripotent status of the starting hiPSCs can be evaluated through the use of commercially available stem cell characterization kits that assess for gene and protein expression of pluripotent markers such as NANOG, OCT4, and SOX2.
Problem 2
Low yield of CD34− cells following CD34 MACS sorting on day 6 of the differentiation protocol.
Potential solution
It is likely that the CD34 MACS column is blocked. To reduce blocking, ensure cells are in a single cell suspension. Multiple columns can be used to reduce the number of cells per column. Additional washings with MACS buffer of the column may increase yield.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. William Murphy (wlmurphy@ortho.wisc.edu).
Materials availability
This study did not generate any unique materials or reagents.
Data and code availability
This study did not generate any unique datasets or code.
Acknowledgments
This research was funded by the US Environmental Protection Agency (STAR grant no. 83573701), the US National Institutes of Health (award nos. 1U01TR002383, R01HL093282, and 1R01NS109427). E.A.A. acknowledges funding by the US National Institutes of Health (T32HL110853).
Author contributions
Conceptualization, W.D. and W.L.M.; investigation, E.A.A., E.T., H.J., and C.S.; writing, E.A.A., E.T., and W.L.M.; supervision, W.D. and W.L.M.
Declaration of interests
W.L.M. is co-founder and chief scientific officer at Stem Pharm, Inc. and Dianomi Therapeutics, Inc. Patents are pending for intellectual property described in this report.
Contributor Information
Elizabeth A. Aisenbrey, Email: eaisenbrey@wisc.edu.
William L. Murphy, Email: wlmurphy@ortho.wisc.edu.
References
- Armulik A., Genové G., Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell. 2011;21:193–215. doi: 10.1016/j.devcel.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Bergers G., Song S. The role of pericytes in blood-vessel formation and maintenance. Neuro Oncol. 2005;7:452–464. doi: 10.1215/S1152851705000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathery W., Faulkner A., Maselli D., Madeddu P. Concise review: the regenerative journey of pericytes toward clinical translation. Stem Cells. 2018;36:1295–1310. doi: 10.1002/stem.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisan M., Chen C.-W., Corselli M., Andriolo G., Lazzari L., Péault B. Perivascular multipotent progenitor cells in human organs. Ann. N. Y. Acad. Sci. 2009;1176:118–123. doi: 10.1111/j.1749-6632.2009.04967.x. [DOI] [PubMed] [Google Scholar]
- Faal T., Phan D.T.T., Davtyan H., Scarfone V.M., Varady E., Blurton-Jones M., Hughes C.C.W., Inlay M.A. Induction of mesoderm and neural crest-derived pericytes from human pluripotent stem cells to study blood-brain barrier interactions. Stem Cell Reports. 2019;12:451–460. doi: 10.1016/j.stemcr.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fina L., Molgaard H.V., Robertson D., Bradley N.J., Monaghan P., Delia D., Sutherland D.R., Baker M.A., Greaves M.F. Expression of the CD34 gene in vascular endothelial cells. Blood. 1990;75:2417–2426. [PubMed] [Google Scholar]
- Geevarghese A., Herman I.M. Pericyte-endothelial crosstalk: implications and opportunities for advanced cellular therapies. Transl. Res. 2014;163:296–306. doi: 10.1016/j.trsl.2014.01.011. Regenerative Medicine: The Hurdles and Hopes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristov M., Weber C. Endothelial progenitor cells in vascular repair and remodeling. Pharmacol. Res. 2008;58:148–151. doi: 10.1016/j.phrs.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Lertkiatmongkol P., Liao D., Mei H., Hu Y., Newman P.J. Endothelial functions of PECAM-1 (CD31) Curr. Opin. Hematol. 2016;23:253–259. doi: 10.1097/MOH.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Shi G.-P. CD31: beyond a marker for endothelial cells. Cardiovasc. Res. 2012;94:3–5. doi: 10.1093/cvr/cvs108. [DOI] [PubMed] [Google Scholar]
- Orlova V.V., Drabsch Y., Freund C., Petrus-Reurer S., van den Hil F.E., Muenthaisong S., Dijke P.T., Mummery C.L. Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts. Arterioscler. Thromb. Vasc. Biol. 2014;34:177–186. doi: 10.1161/ATVBAHA.113.302598. [DOI] [PubMed] [Google Scholar]
- Orlova V.V., van den Hil F.E., Petrus-Reurer S., Drabsch Y., ten Dijke P., Mummery C.L. Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nat. Protoc. 2014;9:1514–1531. doi: 10.1038/nprot.2014.102. [DOI] [PubMed] [Google Scholar]
- Pierantozzi E., Vezzani B., Badin M., Curina C., Severi F.M., Petraglia F., Randazzo D., Rossi D., Sorrentino V. Tissue-specific cultured human pericytes: perivascular cells from smooth muscle tissue have restricted mesodermal differentiation ability. Stem Cells Dev. 2016;25:674–686. doi: 10.1089/scd.2015.0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsauer M., Kunz J., Krause D., Dermietzel R. Regulation of a blood-brain barrier—specific enzyme expressed by cerebral pericytes (pericytic aminopeptidase N/pAPN) under cell culture conditions. J. Cereb. Blood Flow Metab. 1998;18:1270–1281. doi: 10.1097/00004647-199811000-00014. [DOI] [PubMed] [Google Scholar]
- Sidney L.E., Branch M.J., Dunphy S.E., Dua H.S., Hopkinson A. Concise review: evidence for CD34 as a common marker for diverse progenitors. Stem Cells. 2014;32:1380–1389. doi: 10.1002/stem.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mourik J.A., Leeksma O.C., Reinders J.H., de Groot P.G., Zandbergen-Spaargaren J. Vascular endothelial cells synthesize a plasma membrane protein indistinguishable from the platelet membrane glycoprotein IIa. J. Biol. Chem. 1985;260:11300–11306. [PubMed] [Google Scholar]
- Verbeek M.M., Otte-Höller I., Wesseling P., Ruiter D.J., de Waal R.M. Induction of alpha-smooth muscle actin expression in cultured human brain pericytes by transforming growth factor-beta 1. Am. J. Pathol. 1994;144:372–382. [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu J., Chang L., Meyers C.A., Zhang L., Broderick K., Lee M., Peault B., James A.W. Relative contributions of adipose-resident CD146+ pericytes and CD34+ adventitial progenitor cells in bone tissue engineering. NPJ Regen. Med. 2019;4:1–9. doi: 10.1038/s41536-018-0063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Gong T., Heng B.C., Zhang C.F. A systematic review: differentiation of stem cells into functional pericytes. FASEB J. 2017;31:1775–1786. doi: 10.1096/fj.201600951RRR. [DOI] [PubMed] [Google Scholar]
- Zhang J., Schwartz M.P., Hou Z., Bai Y., Ardalani H., Swanson S., Steill J., Ruotti V., Elwell A., Nguyen B.K., Bolin J., Stewart R., Thomson J.A., Murphy W.L. A genome-wide analysis of human pluripotent stem cell-derived endothelial cells in 2D or 3D culture. Stem Cell Reports. 2017;8:907–918. doi: 10.1016/j.stemcr.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any unique datasets or code.