With the growing interest in using phages to combat antibiotic-resistant infections or manipulate the human microbiome, establishing approaches for the modification of phage host range has become an important research topic. Tfa proteins are a large family of proteins known previously to function as chaperones for the folding of phage fibers, which are crucial determinants of host range for long-tailed phages. Here, we reveal that some Tfa proteins are bi-functional, with the additional activity of binding to LPS, the surface binding receptor for many phages. This discovery opens up new potential avenues for altering phage host range through engineering of the surface binding specificity of Tfa proteins.
KEYWORDS: chaperone, fiber, host range, lipopolysaccharide, phage, phage tail
ABSTRACT
To initiate their life cycle, phages must specifically bind to the surface of their bacterial hosts. Long-tailed phages often interact with the cell surface using fibers, which are elongated intertwined trimeric structures. The folding and assembly of these complex structures generally requires the activity of an intra- or intermolecular chaperone protein. Tail fiber assembly (Tfa) proteins are a very large family of proteins that serve as chaperones for fiber folding in a wide variety of phages that infect diverse species. A recent structural study showed that the Tfa protein from Escherichia coli phage Mu (TfaMu) mediates fiber folding and stays bound to the distal tip of the fiber, becoming a component of the mature phage particle. This finding revealed the potential for TfaMu to also play a role in cell surface binding. To address this issue, we have here shown that TfaMu binds to lipopolysaccharide (LPS), the cell surface receptor of phage Mu, with a similar strength as to the fiber itself. Furthermore, we have found that TfaMu and the Tfa protein from E. coli phage P2 bind LPS with distinct specificities that mirror the host specificity of these two phages. By comparing the sequences of these two proteins, which are 93% identical, we identified a single residue that is responsible for their distinct LPS-binding behaviors. Although we have not yet found conditions under which Tfa proteins influence host range, the potential for such a role is now evident, as we have demonstrated their ability to bind LPS in a strain-specific manner.
IMPORTANCE With the growing interest in using phages to combat antibiotic-resistant infections or manipulate the human microbiome, establishing approaches for the modification of phage host range has become an important research topic. Tfa proteins are a large family of proteins known previously to function as chaperones for the folding of phage fibers, which are crucial determinants of host range for long-tailed phages. Here, we reveal that some Tfa proteins are bi-functional, with the additional activity of binding to LPS, the surface binding receptor for many phages. This discovery opens up new potential avenues for altering phage host range through engineering of the surface binding specificity of Tfa proteins.
INTRODUCTION
Phages, viruses that infect bacteria, are present in all environments where bacteria occur. Phages often outnumber coexisting bacteria and are important drivers of the ecology and evolution of microbial communities (1). The most commonly occurring phages are composed of a double-stranded-DNA-filled icosahedral head attached to a long tail, which can be either contractile or noncontractile. The tail serves as a channel through which phage DNA is delivered into its host, and also mediates specific recognition of the bacterial cell surface (2). Understanding how phages recognize their specific hosts is crucial, as this knowledge can be used to optimize the design of phages and other bactericidal agents for targeted therapeutic applications (3).
Tail fibers are long rod-shaped proteins positioned at the tip of the tail and bind specifically to proteins or carbohydrates exposed on the bacterial surface. They are diverse multidomain protein trimers composed of extended intertwined subunits (4). The complicated nature of fiber structures necessitates the participation of intra- or intermolecular chaperones for their proper folding and assembly (5–7). Tail fiber assembly (Tfa) proteins are implicated in the folding and function of many phage fibers. These proteins hold great biological importance, as there are more than 10,000 members of the Tfa protein family (Pfam Caudo_TAP [PF02413]) found in the UniprotKB protein database (8).
In a previous study, we determined the structure of the Tfa protein from E. coli phage Mu (TfaMu) in a complex with the assembled fiber from the same phage (9). We also elucidated the mechanism by which TfaMu mediates fiber assembly. In the course of that study, we established that TfaMu forms a stable complex with the tail fiber in vitro and that TfaMu is a component of the mature phage particle. These results were surprising, as the Tfa protein from E. coli phage T4 had been previously shown to act as a true chaperone, mediating fiber assembly without forming a stable complex with the fiber (5). Since TfaMu binds to the distal tip of the tail fiber, which is the site of cell surface interaction, we postulated that TfaMu and/or other Tfa family proteins may be involved in cell surface binding and host range determination. The goal of this study was to determine whether TfaMu possesses any activities consistent with a role in determining host range. To this end, we investigated its binding activity toward lipopolysaccharide (LPS), the cell surface receptor for phage Mu. Supporting the potential for Tfa proteins to influence host range, we found that TfaMu and a closely related homologue from E. coli phage P2 (TfaP2) bind to LPS and do so in a strain-specific manner.
RESULTS
Tfa protein from phage Mu binds E. coli LPS.
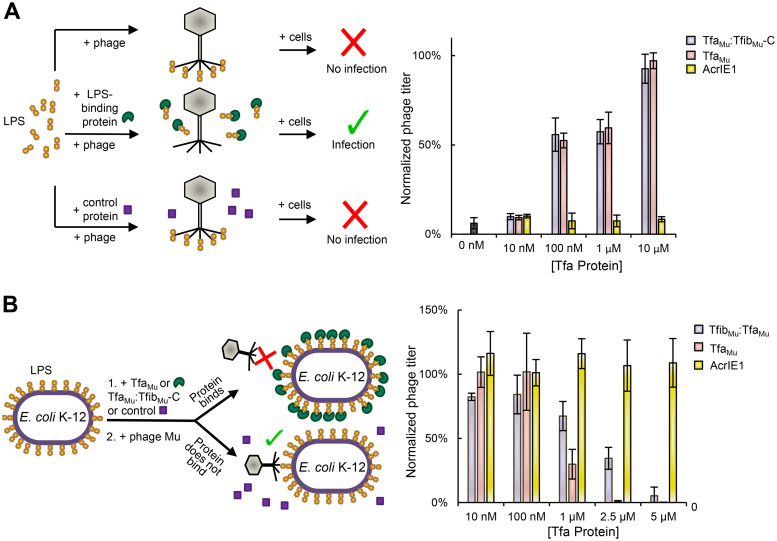
We previously showed that a complex comprising the C-terminal 140 residues of the Mu tail fiber (TfibMu-C) and TfaMu binds to LPS (9), which is the established receptor for this phage (10). To assess the LPS-binding ability of the TfaMu protein, we used our previously developed LPS-binding assay (9). This assay is based on the dose-dependent decrease of phage Mu infectivity caused by the addition of E. coli K-12 LPS to the phage particles. This inhibition is likely caused by LPS binding to the phage fibers, thereby preventing their interaction with LPS on the cell surface. This inhibitory effect can be relieved by premixing of the LPS with the TfaMu:TfibMu-C complex, which binds to the LPS and prevents it from binding to the phage particles. Strikingly, addition of 6×-His tagged TfaMu alone to the LPS also alleviated the inhibitory activity of LPS in a similar dose-dependent manner to the TfaMu:TfibMu-C complex (Fig. 1A). In contrast, premixing of the LPS with a control 6×-His-tagged protein (anti-CRISPR protein AcrIE1, a 100-residue CRISPR-Cas inhibitor [11]) did not relieve the inhibitory effect of the LPS (Fig. 1A). These data imply that TfaMu by itself binds to LPS.
FIG 1.
TfaMu binds E. coli LPS and cell surface. (A) As illustrated by the diagram on the left, a Mu phage lysate was added to 50 μg of E. coli K-12 LPS alone or LPS premixed for 1 h with increasing amounts of TfaMu, TfibMu-C:TfaMu complex, or control protein (AcrIE1). After a further 1-h incubation, the number of infective particles was determined by plaquing assays. The phage titer was normalized to an untreated lysate (left y axis) and the actual number of PFU counted is also shown (right y axis). (B) As illustrated by the diagram on the left, E. coli K-12 cells were mixed for 1 h with increasing amounts of TfaMu, TfibMu-C:TfaMu complex, or control protein (AcrIE1), then the cells were mixed with a lysate of phage Mu. This mixture was plated and the number of resulting plaques was determined. The phage titer was normalized to that obtained on cells that were not premixed with protein (left y axis) and the actual number of PFU counted is also shown (right y axis). The values shown are the mean of three replicates with error bars representing the standard deviation. The actual reduction in PFU under conditions of maximal inhibition by TfaMu protein or LPS was approximately 300-fold.
Since TfaMu displayed LPS-binding activity in vitro, we hypothesized that it could bind to LPS on the surface of E. coli cells and thereby inhibit phage infection. As shown in Fig. 1B, the premixing of TfaMu to E. coli K-12 cells led to a dramatic dose-dependent decrease in the ability of phage Mu to infect these cells (Fig. 1B), implying that TfaMu binds to LPS on the cell surface and blocks phage adsorption. An inhibitory effect on phage replication was not seen when cells were mixed with the AcrIE1 control protein (Fig. 1B). The inhibitory activity of TfaMu was somewhat more potent than that of the TfaMu:TfibMu-C complex (e.g., at 2.5 μM protein), indicating that the binding of TfaMu to the cell surface is as strong or possibly even stronger than the established receptor-binding protein of phage Mu. Overall, these experiments provide the first evidence that a Tfa protein can bind to a phage cell surface receptor.
Tfa proteins from phages Mu and P2 display distinct LPS-binding specificities.
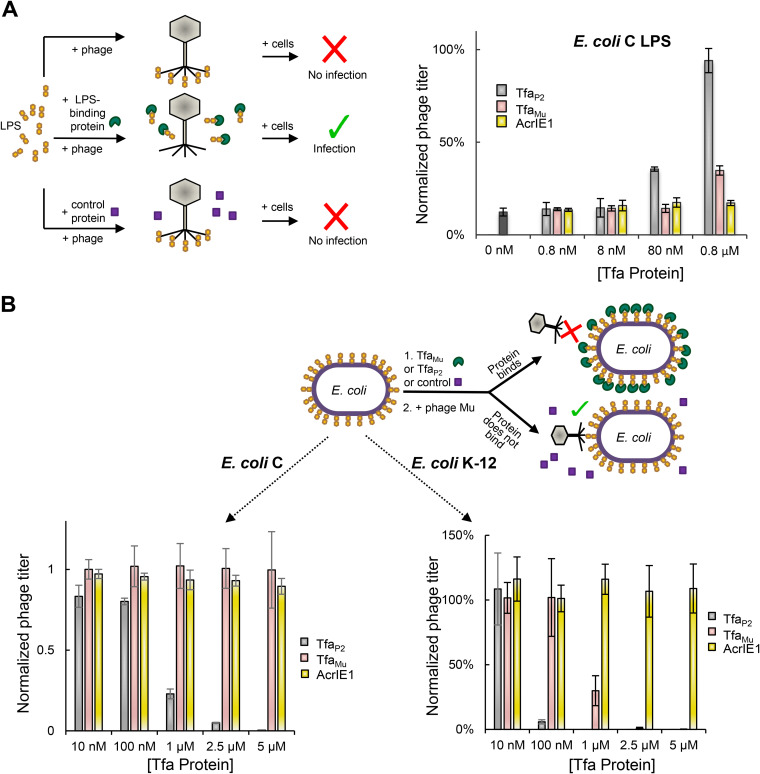
To investigate the specificity of LPS binding by Tfa proteins, we compared the behavior of TfaMu to TfaP2. Phage P2 is able to infect and replicate on both E. coli K-12 and E. coli C strains, while phage Mu can replicate only on E. coli K-12. We previously showed that LPS purified from an E. coli C strain does not inhibit phage Mu infection, implying that phage Mu does not bind to this LPS (9). In contrast, the infectivity of phage P2 particles was reduced by 10-fold after mixing with E. coli C LPS (Fig. 2A, black bar). This inhibitory effect was completely relieved by premixing of the E. coli C LPS with TfaP2 at 0.8 μM and was partially relieved by premixing with 0.08 μM TfaP2 (Fig. 2A). On the other hand, TfaMu did not at all relieve inhibition at 0.08 μM and provided only partial relief at 0.8 μM. Consistent with these results, addition of TfaMu to E. coli C cells did not block infection of these cells by phage P2 at any concentration tested, while TfaP2 partially inhibited infection at 1 μM and completely inhibited it at 5 μM (Fig. 2B). Phage P2 replication on E. coli K-12 cells was blocked by both TfaMu and TfaP2, with TfaP2 fully inhibiting at a concentration of 100 nm and TfaMu inhibiting fully at a 2.5 μM concentration (Fig. 2C). Taken together, these experiments show that TfaMu and TfaP2 both bind to LPS, but their LPS-binding specificity is distinct and mirrors the host specificity of these two phages.
FIG 2.
TfaMu and TfaP2 show distinct LPS-binding specificities. (A) As illustrated by the diagram on the left, a P2 phage lysate was added to 50 μg of E. coli C LPS alone or LPS premixed for 1 h with increasing amounts of TfaMu, Tfa P2, or control protein (AcrIE1). After a further 1-h incubation, the number of infective particles was determined by plaquing assays. The phage titer was normalized to an untreated lysate. (B) As illustrated by the diagram, E. coli C cells (left graph) or E. coli K-12 cells (right graph) were mixed for 1 h with increasing amounts of TfaMu, Tfa P2, or control protein (AcrIE1). The cells were then mixed with a lysate of phage P2 and plated. The number of resulting plaques was normalized to that obtained on cells that were not premixed with protein. The data shown for TfaMu in Fig. 1B are also shown here in the right graph for the purpose of comparison. The values shown are the mean of three replicates with error bars representing the standard deviation. The actual reduction in PFU under conditions of maximal inhibition by Tfa proteins or LPS was approximately 300-fold.
It should be noted that phage Mu actually contains a second set of genes encoding a different fiber and Tfa protein. These genes become expressed through a stochastically occurring DNA inversion event catalyzed by a phage-encoded enzyme, and the fibers produced confer a completely different host specificity (12). However, phages carrying the inverted fibers cannot form plaques on E. coli C or E. coli K-12 under the conditions used here (13, 14).
Substituting glutamic acid 10 with glycine extends TfaMu binding specificity.
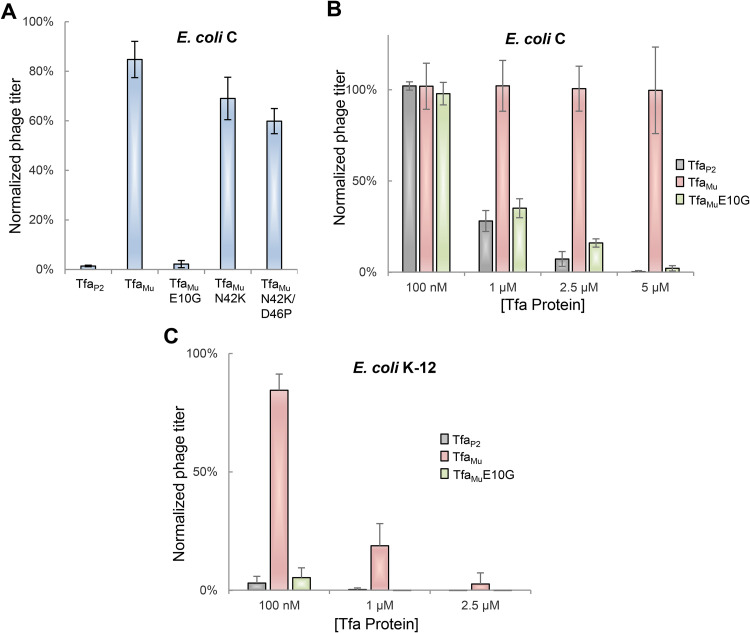
Our discovery of different LPS-binding specificities for TfaMu and TfaP2 was surprising because these proteins are 93% identical, differing at only 12 amino acid positions (Fig. S1 in the supplemental material). To determine which of these sequence differences was responsible for the distinct specificities of these Tfa proteins, we performed site-directed mutagenesis to substitute residues in TfaMu with those found in TfaP2. Tfa is a two-domain protein with the N-terminal domain involved in fiber binding and the C-terminal domain involved in fiber folding (9). Since the structure of the N-terminal TfaMu domain bound to Mu fiber is known (9), we focused on the solvent-exposed residues in this domain that differed between TfaMu and TfaP2 (E10, N42, and D46). Substitution of these positions with the residues found in TfaP2 revealed that TfaMu with just a single amino acid substitution (E10G) was able to inhibit phage P2 infection of an E. coli C strain to the same degree as TfaP2 (Fig. 3A). The other TfaMu mutants tested (N42K and N42K/D46P) displayed only a small increase in their ability to inhibit P2 infection. Testing of various concentrations of the TfaMuE10G mutant showed that it inhibited P2 infection of E. coli C with a very similar concentration dependence as TfaP2 (Fig. 3B). The TfaMuE10G mutant also inhibited P2 infection of E. coli K-12 more strongly than did TfaMu, but with a similar potency to TfaP2 (Fig. 3C). Although we have not delineated the mechanism by which Tfa proteins bind LPS, we can conclude that the difference in LPS-binding activities of TfaP2 and TfaMu is primarily determined by the identity of the residue at position 10.
FIG 3.
Substituting Glu10 with Gly extends TfaMu binding specificity. (A) E. coli C cells were mixed for 1 h with 100 μl of 5 μM Tfa P2, TfaMu, or the indicated TfaMu mutants. The cells were then mixed with a lysate of phage P2 and plated. The number of resulting plaques was normalized to that obtained on cells that were not premixed with protein. (B) This experiment was performed in the same manner as Fig. 3A, except E. coli C cells were mixed for 1 h with increasing levels of TfaMuE10G. Data from Fig. 2B obtained for TfaMu and Tfa P2 are shown for comparison. (C) This experiment was performed in the same manner as Fig. 3B except that E. coli K-12 was used as the host strain. Data from Fig. 2B obtained for TfaMu and Tfa P2 are shown for comparison. The values shown are the mean of three replicates with error bars representing the standard deviation. The actual reduction in PFU under conditions of maximal inhibition by Tfa proteins was approximately 300-fold.
Swapping TfaMu for TfaP2 does not enable Mu to infect E. coli C.
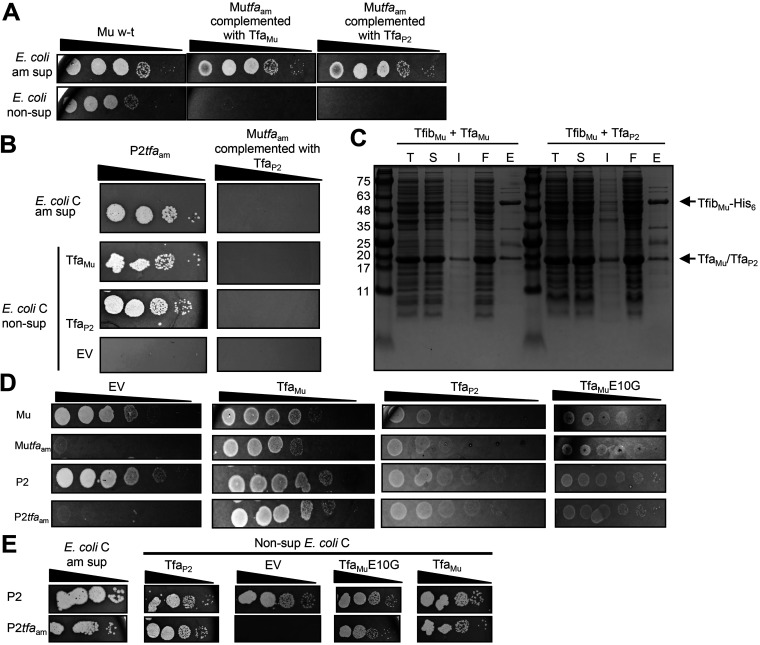
We previously showed that TfaMu is a component of mature phage Mu particles (9). Our finding here that Tfa proteins bind LPS in a strain-specific manner led us to the hypothesis that Tfa proteins might influence phage host range. To address this issue, we determined whether the addition of TfaP2 to phage Mu would allow it to infect E. coli C. To produce hybrid phage Mu particles containing TfaP2, we induced a mutant Mu prophage bearing an amber mutation in the tfa gene (Mutfaam) within cells bearing a plasmid expressing untagged TfaP2. In this way, we aimed to produce phage Mu particles bound to TfaP2. The resulting lysate contained infectious phage particles, as was shown by spotting on QD5003, an amber suppressor E. coli K-12 strain (Fig. 4A). This production of infectious particles, which was similar to the number produced in the presence of plasmid-expressed TfaMu, proved that the expression of TfaP2 efficiently complemented the tfaam mutation. Although the hybrid Mu:TfaP2 particles were able to infect E. coli K-12 cells, they were not able to infect an E. coli C amber suppressor strain or nonsuppressor E. coli C strains in which TfaP2 or TfaMu were expressed from plasmids (Fig. 4B). All three of these strains could be infected by a P2tfaam mutant, demonstrating the functionality of the suppressor strain and the ability of the plasmids to complement a phage lacking Tfa. These results imply that the addition of TfaP2 to Mu phage particles is not sufficient to expand their host range to E. coli C.
FIG 4.
Effects of Tfa expression on phage replication. (A) Serial 10-fold dilutions of lysates of Mu wild type, Mutfaam mutant phage complemented by TfaMu, or Mutfaam mutant phage complemented by TfaP2 were spotted on a lawn of E. coli K-12 amber suppressor and nonsuppressor cells. The images shown depict the zones of clearing produced on the bacterial lawns as a result of phage-mediated cell lysis. The infecting Mutfaam mutant particles contain TfaMu or TfaP2, but the zones of clearing observed on the suppressor strain are due to suppression of the amber mutation in subsequent rounds of phage growth. Although replication in these cells is due to suppression of the tfaam mutation, the initial infection of the cells would potentially be affected by the identity of the Tfa protein added by plasmid-based complementation. Data are representative of at least three biological replicates. (B) Serial 10-fold dilutions of lysates of P2tfaam, or Mutfaam complemented by TfaP2 were spotted on a lawn of E. coli C amber suppressor cells (top), or nonsuppressor cells transformed with an empty vector (EV, bottom), or a plasmid expressing either TfaMu or TfaP2 (middle). The infecting Mutfaam mutant particles contain TfaP2, the zones of clearing observed on the suppressor strain are due to suppression of the amber mutation in subsequent rounds of phage growth, and the zones of clearing observed on nonsuppressor strains result from complementation of the amber mutation with a plasmid-encoded TfaP2 or TfaMu. Although replication in these cells is due to suppression of the tfaam mutation, the initial infection of the cells would potentially be affected by the identity of the Tfa protein added by plasmid-based complementation. Data are representative of at least three biological replicates. (C) Cell lysates and fractions from protein purification experiments using Ni-NTA affinity chromatography were analyzed by SDS polyacrylamide gel electrophoresis stained by Coomassie blue. The indicated proteins were coexpressed and total cell lysate (T) and soluble (S) and insoluble (I) fractions of the lysate were assessed. The 6×-His-containing protein complexes were purified from the soluble fraction of the cell lysate. Samples from the eluted fraction from the purification (E) and the flowthrough fraction (F) are shown. (D) Serial 10-fold dilutions of Mu wild type, Mutfaam, P2 wild type, or P2tfaam phage lysate were spotted on a lawn of E. coli K-12 cells transformed with an empty vector (EV) or a plasmid expressing TfaMu, TfaP2, or TfaMuE10G. Data are representative of at least three biological replicates. (E) Serial 10-fold dilutions of P2 wild type or P2tfaam phage lysate were spotted on a lawn of E. coli C amber suppressor cells or nonsuppressor cells transformed with an empty vector (EV) or a plasmid expressing TfaP2, TfaMuE10G, or TfaMu. Data are representative of at least three biological replicates.
One possible explanation for the above results is that TfaP2 does not bind stably to the Mu tail fiber (TfibMu). To assess this possibility, we used the same experimental approach as we previously used to show that TfaMu binds stably to TfibMu (9). Two plasmids were used to coexpress full-length 6×-His-tagged TfibMu with untagged TfaP2 within the same E. coli cells. This coexpression resulted in the production of soluble TfibMu, which could subsequently be purified using Ni-affinity chromatography (Fig. 4C). When TfibMu is expressed on its own, it is insoluble and cannot be purified (9). We found that the untagged TfaP2 coeluted with TfibMu in the same manner that TfaMu coeluted with TfibMu. Neither untagged TfaMu nor TfaP2 bound to a Ni-affinity column when expressed on its own (Fig. S2). These experiments indicate that TfaP2 does indeed interact stably with TfibMu.
Expression of TfaP2 or TfaMu E10G inhibits phage infectivity.
In searching for other in vivo effects that could distinguish TfaMu and TfaP2, we noticed that plasmid-based expression of TfaP2 or TfaMuE10G in the E. coli K-12 strain caused a reproducible attenuation of phage plaquing efficiency. For example, expression of TfaP2 caused an approximately 10-fold reduction in plaquing efficiency of wild-type P2 and Mu phage lysates compared to the empty vector control, and the zones of clearing were much fainter at all lysate dilutions (Fig. 4D). Although expression of TfaP2 led to complementation of both P2tfaam and Mutfaam mutants, a comparable attenuation of plaquing was observed (Fig. 4D). The expression of TfaMuE10G also attenuated plaquing of all four phages tested. In contrast, expression of TfaMu from the same plasmid vector had no effect on replication of the wild-type phages and the tfaam mutant phages were fully complemented. Plasmid-based expression of TfaP2, TfaMu, or TfaMuE10G in E. coli C had no effect on the plaquing ability of phage P2 (Fig. 4E).
DISCUSSION
To effectively predict or manipulate the host range of phages, a full understanding of the phage proteins that determine this property is required. Here, we show that Tfa proteins from phages Mu and P2 bind to LPS, which is the receptor for these phages. Furthermore, despite these proteins differing in sequence by only 12 amino acids, their ability to bind to LPS mirrors the host specificity of Mu and P2 phages. Phage Mu, which infects E. coli K-12 but not E. coli C, encodes a Tfa protein that binds strongly only to LPS from E. coli K-12, while phage P2 infects both of these strains and encodes a Tfa that binds to the LPS from both strains. These results indicate that Tfa proteins have the potential to influence host specificity since they are found as components of phage particles in some cases (9, 15). The role of Tfa proteins in recognition of the cell surface had not been previously investigated because the first well-characterized Tfa, that from phage T4, does not bind stably to the tail fiber and is not a component of the phage particle (5). Thus, our work supports a new role for this large family of proteins and shows that some members of this family may be bi-functional, being required both for fiber folding and host range specificity.
Incorporation of TfaP2 into phage Mu particles did not expand their host range and allow them to infect E. coli C. However, we demonstrated that TfaMu and TfaP2 elicited different in vivo effects when expressed from plasmids. Namely, TfaP2 caused an attenuation of phage P2 or Mu replication when grown on E. coli K-12. The same phenomenon was observed upon expression of TfaMuE10G, indicating that it resulted from the LPS-binding properties of these proteins. This effect may result from the release of excess Tfa protein upon phage-induced cell lysis. This released Tfa may then bind to the surface of neighboring cells and inhibit the adsorption of phage particles. It is clear that both TfaP2 and TfaMuE10G bind to E. coli K-12 LPS more strongly that TfaMu (Fig. 3C), which may explain why these Tfa proteins inhibit phage infectivity and TfaMu does not. Supporting this idea, TfaP2 and TfaMuE10G did not appear to bind E. coli C as strongly as they do E. coli K-12 (Fig. 3B), and plasmid-based expression of these proteins in E. coli C does not inhibit phage replication (Fig. 4E). Although these experiments involve expression of the Tfa proteins at nonphysiological levels, they nevertheless demonstrate that the different LPS-binding properties of TfaP2 and TfaMu can have a biological effect.
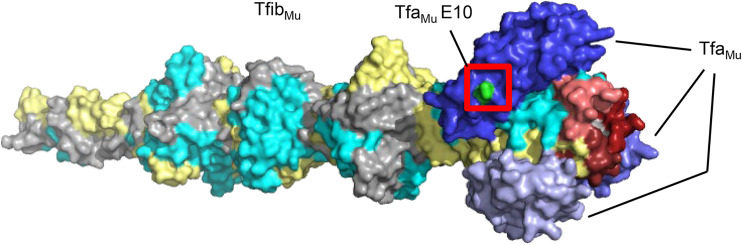
While Tfa did not affect host range within the context of this assay, Tfa proteins may affect host range in other phages or when phage Mu or P2 are infecting different strains. Assuming that the E10 position is part of the LPS-binding interface of Tfa, it can be seen that this region of Tfa is distinct from the cell surface binding loops of the phage Mu fiber (Fig. 5) (9). Thus, Tfa can provide an additional Tfa-interacting surface that, in some cases, may strengthen interaction with the same LPS moieties bound by the fiber, or, in other cases, interact with distinct portions of the LPS. In either case, a phage particle bearing two distinct LPS-interacting surfaces may have a fitness advantage at low cell concentrations, or an added evolutionary flexibility in being able to experiment with amino acid substitutions on one surface without abrogating binding by the other surface. Interestingly, the fibers of phages T2 and T6 are bound by a small “adhesin” protein that is required for recognition of the host cell surface (16). These proteins, which are unrelated to Tfa in sequence and structure (17), are encoded immediately downstream of the fiber-encoding genes but are not required for fiber folding. From the work presented here, Tfa proteins are now seen to act purely as chaperones as in phage T4, and also potentially as chaperone/adhesins in phages P2 and Mu. Tfa proteins appear to lie in an evolutionary middle ground between these two functions.
FIG 5.
Position of the TfaMuE10 residue in the TfibMu:TfaMu structure. A surface representation of the previously solved crystal structure (9) of the TfibMu:TfaMu complex is shown. The three monomers of the intertwined fiber structure are shown in gray, yellow, and cyan. The three bound Tfa proteins are shown in shades of blue. The residues at the distal tip of the fiber that are likely involved in cell surface binding are colored in shades of red. The E10 residue of one TfaMu monomer is colored green and highlighted by a red box.
MATERIALS AND METHODS
Cloning, protein expression, and purification.
For protein expression and purification, genes tfaMu and tfaP2 were amplified by PCR from genomic phage Mu and P2 DNA and cloned into a pCDF-1b (EMD Millipore) plasmid vector. The proteins were untagged for coexpression experiments, or fused at their N terminus with the amino acid sequence “MAHHHHHHVGT” for Tfa purification experiments. Amplified tfibMu, and a tfibMu fragment, tfibMu-C, genes were cloned into a pET15b (Novagen) derivative plasmid vector. These vectors are suited for cotransformation into the same cells and transcription of the cloned genes in each vector is mediated by promoters recognized by phage T7 RNA polymerase. TfaMu, TfaP2, TfaMu:TfibMu-C, and TfaP2:TfibMu protein purification was performed as previously described (9). For the in vivo phage complementation experiments, the tfaMu and tfaP2 genes were also cloned into vectors derived from pAD100 (18). Expression in this context was driven by an isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible Ptrc promoter that did not require T7 RNA polymerase. The Tfa proteins expressed from these plasmids were untagged.
Phage growth assays.
Phage isolates used were Mucts62 (referred to as wild-type Mu, but actually encodes a temperature-sensitive repressor protein [19]), Mucts62Uam1958gin− (referred to as Mutfaam [14]), P2vir1 (referred to as wild-type P2, but is a virulent mutant [20]), and P2vir1Gam1 (referred to as P2tfaam [14)]). Spotting assays were used to assess the phage titer. E. coli cultures were incubated with shaking at 37°C for 16 h. An aliquot of 150 μl of this overnight culture was mixed with 3 ml of molten top agar (containing 0.6% agar) and poured onto prewarmed LB agar plates. The plates were left on the benchtop until the solidification of the top agar lawns (10 min), after which 2 μl of serially diluted phage lysates were spotted on the top agar lawn. The plates were transferred to a stationary incubator set at 30°C and incubated overnight. E. coli K-12 strains used were 594 (wild type) and QD5003 (amber suppressor strain; supF [21]). E. coli C strains used were E. coli C1a (22) and amber suppressor strain C1792 (20).
Production of phage Mu particles containing TfaP2.
E. coli 594 cells containing a Mutfaam mutant prophage were transformed with pAD100-derived plasmids expressing the tfaMu or tfaP2 genes. These cells were grown in 500 ml of LB with shaking at 30°C to an optical density at 600 nm (OD600) of 0.4. Prophage induction was achieved by transferring the cells to 45°C for 15 min (the repressor of this prophage is temperature-sensitive), and then growth was resumed at 37°C until the culture cleared due to phage-mediated cell lysis (1 h). The cell lysate was centrifuged at 4,000 rpm for 15 min, and then the phage-containing supernatant was used for phage spotting assays. It should be noted that the Mutfaam mutant phage used is gin−, so it does not produce the fiber and Tfa proteins resulting from DNA inversion.
Cell surface and LPS-binding assays.
LPS extraction was performed as previously described (23). LPS was extracted from the bacterial cell suspension by heating at 70°C for 1 h. After centrifugation, the supernatant was washed with diethyl ether and LPS was collected by lyophilization. Lyophilized LPS was weighed and resuspended in 20 mM Tris-HCl (pH 7.5), 200 mM NaCl at a concentration of 5 mg ml−1. To assess phage binding to LPS, we used a procedure described previously (9). A Mu or P2 phage lysate was incubated with 50 μg of E. coli K-12 (or E. coli C) LPS for 1 h, after which the phage titer was determined by plaquing assays. To assess protein binding to LPS, purified LPS was first premixed for 1 h with increasing amounts of TfaMu, TfibMu-C:TfaMu complex, TfaP2, or TfaMu mutants. Then, the phage lysate was added to the mixture and it was incubated for one more hour. Finally, the number of infectious particles was determined by plaquing assays and the phage titer was normalized to an untreated lysate.
To assess the binding of Tfa proteins to the cell surface, a culture of an E. coli K-12 or E. coli C strain was incubated with shaking at 37°C for 16 h. An aliquot of 150 μl of this overnight culture was added to 100 μl of various concentrations of TfaMu, TfaP2, TfaMuN42K, TfaMuN42K/D46P, or TfaMuE10G. The cell and protein mixture was incubated for 1 h at room temperature, after which it was mixed with 100 μl of a 4 × 103 PFU/ml lysate of phage Mu or P2. Finally, the mixture was added to 3 ml of molten top agar (0.6%) and poured over a prewarmed LB agar plate. After the top agar solidified, the plates were incubated overnight at 30°C.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding from the Canadian Institutes of Health Research to A.R.D. (Foundation grant FDN-15427).
We thank Martha Howe for providing the Mu phage isolates, and Rich Calendar and Gail Christie for providing the phage P2 isolates.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hampton HG, Watson BNJ, Fineran PC. 2020. The arms race between bacteria and their phage foes. Nature 577:327–336. doi: 10.1038/s41586-019-1894-8. [DOI] [PubMed] [Google Scholar]
- 2.Nobrega FL, Vlot M, de Jonge PA, Dreesens LL, Beaumont HJE, Lavigne R, Dutilh BE, Brouns SJJ. 2018. Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol 16:760–773. doi: 10.1038/s41579-018-0070-8. [DOI] [PubMed] [Google Scholar]
- 3.Dams D, Brondsted L, Drulis-Kawa Z, Briers Y. 2019. Engineering of receptor-binding proteins in bacteriophages and phage tail-like bacteriocins. Biochem Soc Trans 47:449–460. doi: 10.1042/BST20180172. [DOI] [PubMed] [Google Scholar]
- 4.Bhardwaj A, Olia AS, Cingolani G. 2014. Architecture of viral genome-delivery molecular machines. Curr Opin Struct Biol 25:1–8. doi: 10.1016/j.sbi.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartual SG, Garcia-Doval C, Alonso J, Schoehn G, van Raaij MJ. 2010. Two-chaperone assisted soluble expression and purification of the bacteriophage T4 long tail fibre protein gp37. Protein Expr Purif 70:116–121. doi: 10.1016/j.pep.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Marti R, Zurfluh K, Hagens S, Pianezzi J, Klumpp J, Loessner MJ. 2013. Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol Microbiol 87:818–834. doi: 10.1111/mmi.12134. [DOI] [PubMed] [Google Scholar]
- 7.Schulz EC, Dickmanns A, Urlaub H, Schmitt A, Muhlenhoff M, Stummeyer K, Schwarzer D, Gerardy-Schahn R, Ficner R. 2010. Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding. Nat Struct Mol Biol 17:210–215. doi: 10.1038/nsmb.1746. [DOI] [PubMed] [Google Scholar]
- 8.The UniProt Consortium. 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.North OI, Sakai K, Yamashita E, Nakagawa A, Iwazaki T, Buttner CR, Takeda S, Davidson AR. 2019. Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres. Nat Microbiol 4:1645–1653. doi: 10.1038/s41564-019-0477-7. [DOI] [PubMed] [Google Scholar]
- 10.Sandulache R, Prehm P, Kamp D. 1984. Cell wall receptor for bacteriophage Mu G(+). J Bacteriol 160:299–303. doi: 10.1128/JB.160.1.299-303.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pawluk A, Shah M, Mejdani M, Calmettes C, Moraes TF, Davidson AR, Maxwell KL. 2017. Disabling a type I-E CRISPR-Cas nuclease with a bacteriophage-encoded anti-CRISPR protein. mBio 8:e01751-17. doi: 10.1128/mBio.01751-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Putte P, Cramer S, Giphart-Gassler M. 1980. Invertible DNA determines host specificity of bacteriophage mu. Nature 286:218–222. doi: 10.1038/286218a0. [DOI] [PubMed] [Google Scholar]
- 13.Sandulache R, Prehm P, Expert D, Toussaint A, Kamp D. 1985. The cell wall receptor for bacteriophage Mu G(-) in Erwinia and Escherichia coli C. FEMS Microbiol Lett 28:307–310. doi: 10.1111/j.1574-6968.1985.tb00811.x. [DOI] [Google Scholar]
- 14.Grundy FJ, Howe MM. 1984. Involvement of the invertible G segment in bacteriophage mu tail fiber biosynthesis. Virology 134:296–317. doi: 10.1016/0042-6822(84)90299-x. [DOI] [PubMed] [Google Scholar]
- 15.Hendrix RW, Duda RL. 1992. Bacteriophage lambda PaPa: not the mother of all lambda phages. Science 258:1145–1148. doi: 10.1126/science.1439823. [DOI] [PubMed] [Google Scholar]
- 16.Riede I, Drexler K, Schwarz H, Henning U. 1987. T-even-type bacteriophages use an adhesin for recognition of cellular receptors. J Mol Biol 194:23–30. doi: 10.1016/0022-2836(87)90712-1. [DOI] [PubMed] [Google Scholar]
- 17.Dunne M, Denyes JM, Arndt H, Loessner MJ, Leiman PG, Klumpp J. 2018. Salmonella phage S16 tail fiber adhesin features a rare polyglycine rich domain for host recognition. Structure 26:1573–1582.e4. doi: 10.1016/j.str.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 18.Davidson AR, Sauer RT. 1994. Folded proteins occur frequently in libraries of random amino acid sequences. Proc Natl Acad Sci U S A 91:2146–2150. doi: 10.1073/pnas.91.6.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howe MM. 1973. Prophage deletion mapping of bacteriophage Mu-1. Virology 54:93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- 20.Sunshine MG, Thorn M, Gibbs W, Calendar R, Kelly B. 1971. P2 phage amber mutants: characterization by use of a polarity suppressor. Virology 46:691–702. doi: 10.1016/0042-6822(71)90071-7. [DOI] [PubMed] [Google Scholar]
- 21.Yanofsky C, Ito J. 1966. Nonsense codons and polarity in the tryptophan operon. J Mol Biol 21:313–334. doi: 10.1016/0022-2836(66)90102-1. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki I, Bertani G. 1965. Growth abnormalities in Hfr derivatives of Escherichia coli strain C. J Gen Microbiol 40:365–376. doi: 10.1099/00221287-40-3-365. [DOI] [PubMed] [Google Scholar]
- 23.Thaipisuttikul I, Hittle LE, Chandra R, Zangari D, Dixon CL, Garrett TA, Rasko DA, Dasgupta N, Moskowitz SM, Malmstrom L, Goodlett DR, Miller SI, Bishop RE, Ernst RK. 2014. A divergent Pseudomonas aeruginosa palmitoyltransferase essential for cystic fibrosis-specific lipid A. Mol Microbiol 91:158–174. doi: 10.1111/mmi.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.