Abstract
Viruses from the genus Enterovirus (EV) of the Picornaviridae family are known to cause diseases such as hand foot and mouth disease (HFMD), respiratory diseases, encephalitis and myocarditis. The capsid of EV is an attractive target for the development of direct-acting small molecules that can interfere with viral entry. Some of the capsid binders have been evaluated in clinical trials but the majority have failed due to insufficient efficacy or unacceptable off-target effects. Furthermore, most of the capsid binders exhibited a low barrier to resistance. Alternatively, host-targeting inhibitors such as peptides derived from the capsid of EV that can recognize cellular receptors have been identified. However, the majority of these peptides displayed low anti-EV potency (µM range) as compared to the potency of small molecule compounds (nM range). Nonetheless, the development of anti-EV peptides is warranted as they may complement the small-molecules in a drug combination strategy to treat EVs. Lastly, structure-based approach to design antiviral peptides should be utilized to unearth potent anti-EV peptides.
Keywords: Antiviral compound, Antiviral peptide, Enterovirus, Picornaviridae
Introduction
The genus Enterovirus (EV) belonging to the Picornaviridae family comprises 13 species, of which seven are human viruses [1]. Four of the species are: (1) EV-A such as coxsackievirus (CV)-A6, CV-A10, CV-A16 and EV-A71, (2) EV-B such as the CV-B viruses, echoviruses (ECHO) and CV-A9, (3) EV-C such as polioviruses (PV) and CV-A21, (4) EV-D such as EV-D68 and EV-D70 [1]. The other three species are rhinoviruses RV-A, RV-B and RV-C which comprised over 100 different numbered RVs [1]. EV RNA contains a single open reading frame (ORF) flanked by two untranslated regions (UTRs), 5′ UTR and 3′ UTR [2]. The ORF encodes a single polyprotein that is cleaved into P1, P2 and P3 proteins [3]. The P1 protein is proteolytically cleaved to produce capsid proteins VP1–4 [3]. P2 and P3 are cleaved to produce non-structural (NS) proteins 2A, 2B, 2C and 3A, 3B, 3C, 3D, respectively [3]. The role of the capsid proteins is to enclose the genetic material and to recognize cellular receptors during viral entry [3]. The NS proteins are crucial for replication, translation and subversion of host cell machinery [3]. The capsid proteins are suitable targets for antiviral development due to their role in cellular entry and uncoating of the genetic material [3].
The diverse viruses in the genus EV are known to cause a range of diseases such as hand, foot and mouth disease (HFMD), encephalitis, aseptic meningitis, myocarditis and various respiratory diseases [1]. Although most EV infections are mild, the symptoms can be severe in the very young and immunodeficient individuals [4]. In recent years, viruses such as EV-A71 and CV-A16 have emerged as serious public health threats, as they have caused major outbreaks of HFMD in China and South East Asia [5, 6]. Additionally, EV-D68 has caused a large outbreak of severe lower respiratory infections in North America in 2014 [7]. Therefore, broad-spectrum antiviral drugs that could inhibit multiple EVs across the genus will be instrumental to overcome the public health burden caused by these EVs. In this review, we will summarize the efforts in developing direct-acting antivirals targeting the capsid of EVs and host factor-targeting inhibitors. The low barrier to resistance of the capsid binders will be discussed and the possible strategies to overcome this challenge will be suggested. Lastly, we look at the viability of peptide-based strategy to develop anti-EV therapies.
The architecture of enterovirus capsid
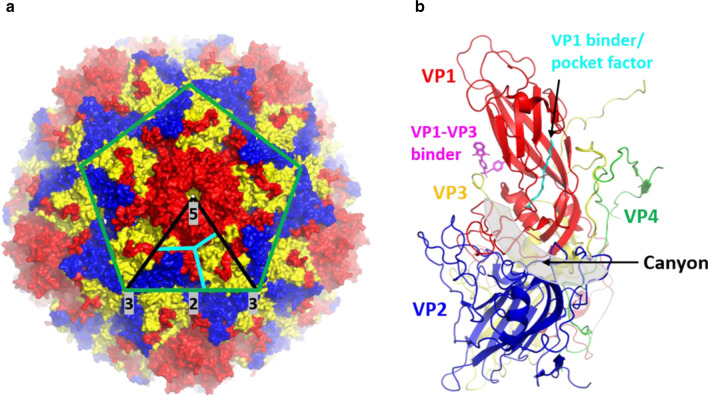
All EVs have a naked icosahedral capsid with five, three, and twofold rotational symmetry formed by 60 identical protomers (Fig. 1a) [8]. Each protomer is composed of VP1, VP2 and VP3 that formed the capsid surface while VP4 is located in the inner surface of the capsid [1]. VP1 to VP3 have a common fold formed by eight-stranded β barrels and two α helices [1]. The main surface features of the external capsid include: (1) star-shaped surface protrusions formed by five copies of VP1, (2) a “canyon” formed by the junction of a “north rim” formed by VP1 and “south rim” formed by VP2 and VP3 encircling the fivefold axes, (3) a protrusion or “puff” formed by VP2 loop, (4) a “knob” formed by a VP3 loop, (5) a large twofold depression and (6) VP1 hydrophobic pockets beneath the canyon bound by lipid molecules known as “pocket factors” (Fig. 1a, b) [1, 8, 9]. The differences in the loops connecting the α helices and β barrels result in the unique surface capsid traits between different EVs [1, 3].
Fig. 1.
Enterovirus capsid organization and features. a Overall view of the enterovirus capsid comprising the VP1 (red), VP2 (blue) and VP3 (yellow). PDB ID: 4RQP [8]. The green lines indicate the boundaries of one pentamer. The black lines indicate the icosahedral symmetric subunit. The five, three, twofold symmetry axes are labeled and highlighted in grey. The cyan lines separate VP1 (red), VP2 (blue) and VP3 (yellow). Black arrows indicate the canyon region and the fivefold axis region formed by five VP1. b Canonical enterovirus protomer formed by VP1 (red), VP2 (blue), VP3 (yellow) and VP4 (green). PDB ID: 6GZV [9]. The canyon is highlighted in grey transparent and is indicated by a black arrow. Antiviral compounds that bind to the two binding pockets which are VP1 hydrophobic pocket and VP1–VP3 interprotomer pocket are shown in cyan and magenta, respectively
Enterovirus cellular attachment and uncoating
EV infections start with viral attachment to cellular receptors [10]. The majority of the cellular receptors belong to the (Ig) superfamily or the integrin receptors [10]. EVs recognize cellular receptors by accommodating the apical Ig domain into the canyon (Fig. 1b) [9]. In contrast, non-Ig fold receptors are recognized by EVs via regions outside of the canyon, such as the vertex of the fivefold axis of viral capsid [11]. The viral canyon-host receptor binding usually triggers the formation of an expanded particle known as the altered (A) particle primed for genome uncoating [12–14]. On the other hand, the interaction of cellular receptors with regions outside of the canyon seldom induce significant conformational changes. Instead, this interaction has been shown to signal for the localization of the attached virus to the main receptors that can bind to the canyon region [15]. For example, CVB3 binds to the co-receptor decay-accelerating factor (DAF) to facilitate the localization of the attached virus to its main receptor, coxsackie and adeno receptor (CAR) [15]. However, some EVs such as EV-A71, CV-A10 and CV-A16 have a shallow canyon indicating that they employ a distinct mechanism for receptor binding and uncoating [16]. Indeed, one of the uncoating receptors for EV-A71 and CV-A16 is the human scavenger receptor class B member 2 (SCARB2) which is a non-Ig fold receptor that binds mainly to the VP1 GH and VP2 EF loops instead of the canyon region [17–19].
Following receptor binding, EVs will empty their pocket factors from the VP1 hydrophobic pockets [10]. This expulsion triggers a cascade of conformational changes to form the A particles characterized by radial expansion and pore formation [1, 20]. The pores allow the release of myristylated VP4 which is inserted into cellular membranes and the exposure of the hydrophobic N-terminus of VP1 which facilitates the shackling of the virion to the cell membranes [1, 21]. These events promote the transport of the viral genome into the cytosol. Currently, our understanding of the mechanism of RNA release is still limited. The 3D structure of an uncoating intermediate of a clinical C4 strain of EV-A71 indicated that a significant capsid rearrangement at the icosahedral two and fivefold axes allowed the formation of large channels for the release of viral RNA [21]. More recently, cryo-electron microscopy study of human ECHO18 and 30 revealed that the release of RNA from the viral particles requires the loss of one, two or three adjacent capsid pentamers [20]. Therefore, stabilization of the pentamers by small molecules or peptides could be a viable strategy to inhibit genome release.
Enteroviral drugs evaluated in clinical trials
One of the highly explored strategies to hinder EV infections is to target their capsids. There are three regions on the EV capsid that have been identified to be viable targets for drug development. The first is the VP1 hydrophobic pocket occupied by the pocket factor [22]. Many direct-acting antivirals targeting this pocket have been identified (Table 1) [23]. These compounds dislodge the pocket factor and bind to the hydrophobic pocket to stabilize the capsid in a rigid and compressed form [23]. This prevents the formation of expanded A particles that is required for genome uncoating [24]. Additionally, there are evidence that demonstrated the binding of compounds to this pocket hindered EV attachment to host cells [25]. Generally, the hydrophobic pocket binders inhibited EV infectivity with half-maximal inhibitory concentration (IC50) or half-maximal effective concentration (EC50) in the nM to pM ranges [26–32]. Five compounds that have been evaluated in phase I and II clinical trials were disoxaril, pleconaril, pirodavir, vapendavir and pocapavir [33–38]. Despite their promising in vitro potencies, the majority of these inhibitors demonstrated insufficient efficacy and unwanted side effects in clinical trials. The unwanted side effects include asymptomatic crystalluria seen in patients receiving disoxaril and the induction of cytochrome P-450 3A (CYP3A4) enzymes by pleconaril that led to menstrual irregularities in pleconaril-treated women taking oral contraceptives [33, 34].
Table 1.
Antivirals targeting the enteroviral proteins evaluated in clinical trials
| Compound | Enterovirus inhibited | In vitro potency | Clinical trial results | Refs |
|---|---|---|---|---|
| Capsid binders | ||||
| Disoxaril | EV-B, C, D70, RV-A, B | nM-μM | The clinical studies were halted due to the appearance of crystalluria in healthy individuals | [33, 44] |
| Pleconaril | EV-B, C, D68, RV-A, B | nM-μM | FDA application for RV colds rejected due to safety concerns | [29] |
| Pirodavir | EV-A, B, C, D, RV-A, B | pM-nM | No clinical benefit in treating RV colds | [35, 45] |
| Vapendavir | EV-A71, C, D68, RV-A, B | nM | Failed to reduced asthma exacerbations in a phase II clinical trial | [46] |
| Pocapavir | EV-B, C | nM | Accelerated the clearance of monovalent oral PV1 vaccine in healthy adults | [31, 32, 38] |
| 3C protease inhibitors | ||||
| Rupintrivir | All | nM | Failed to show significant beneficial effects in clinical trials for RV common colds | [39, 47] |
| AG7404 | EV-A, B, C, D, RV-A, B | nM-μM | Failed to show significant beneficial effects in clinical trials for RV common colds | [40, 48] |
| 3A and/or 3AB inhibitor | ||||
| Enviroxime | EV-A, B, C, D, RV-A, B | nM-μM | Clinical development was discontinued due to insufficient therapeutic effects and gastrointestinal side effects | [41, 43, 44] |
In addition to the capsid binders, the 3C protease inhibitors such as rupintrivir and its analog AG7404 have also been evaluated in clinical trials (Table 1). The 3C proteases are essential for cleaving the polyprotein precursor into structural proteins and non-structural proteins responsible for viral replication. However, these inhibitors failed to show significant beneficial effects in clinical trials involving RV [39, 40]. Enviroxime is another compound that has been evaluated in clinical trials [41]. It inhibited EV infections by targeting the viral proteins 3A and/or 3AB to prevent the formation of the replication complex [42]. Despite showing potent EV replication inhibition in vitro, its clinical development was halted due to gastrointestinal side effects and the lack of therapeutic effect [41, 43, 44].
Anti-enterovirus targeting the VP1 hydrophobic pocket
Despite the failures, significant efforts are being put into identifying novel compounds that target the VP1 hydrophobic pocket (Table 2). For instance, Kim et al. [49] identified a novel series of benzothiophene derivatives and analogues with potent antiviral activities against RV-A and RV-B strains. In particular, compound 6g inhibited RV-A21 (EC50: 0.078 µM), RV-A71 (EC50: 0.015 µM) and RV-B14 (EC50: 0.083 µM) [49]. It also inhibited PV3 (EC50: 0.063 µM), indicating the potential of these compounds to inhibit other EVs as well [49]. Molecular docking study demonstrated the subtle difference between the binding modes of 6g and pleconaril whereby 6g formed a distinct hydrophobic interaction between its 3-methyl group and Leu25 in VP3 [49]. In addition, PR66 which is an imidazolidinone derivative was found to inhibit the uncoating process of EV-A71 by interacting with the VP1 hydrophobic pocket [50]. PR66 was demonstrated to provide complete protection in mice against neurological symptoms induced by EV-A71 [50].
Table 2.
Small molecule compounds targeting viral capsid
| Compound | Enterovirus inhibited | Potency (EC50 or IC50) | Refs |
|---|---|---|---|
| VP1 hydrophobic pocket | |||
| Compound 6g | RV-A21, RV-A71, RV-B14, PV3 | nM | [49] |
| PR66 | EV-A71 | nM | |
| ALD | EV-A71 | nM | [51] |
| NLD | EV-A71 | pM | [51] |
| ICA135 | CV-A10, EV-A71, CV-A16, CV-B3, PV1 and EV-D68 | nM-µM | [16] |
| Compound 10g | CV-B3, RV-A, RV-B | nM-µM | [74] |
| Fivefold vertex | |||
| Suramin | EV-A and EV-B (CV-A9, ECHO20, ECO25) | µM | [60] |
| NF449 | EV-A71 | µM | [61] |
| E151 | EV-A71, CV-A6, CV-A16 | µM | [63] |
| Dendrimer 12 | EV-A71, ECHO11, EV-D68 | pM-µM | [64, 65] |
| HS mimetics | EV-A71 | µM | [68] |
| HS fragments | EV-A71 | µM | [69] |
| Rosmarinic acid | EV-A71 | µM | [70] |
| VP1–VP3 interprotomer | |||
| Compound 12 | Specific for CV-B1, CV-B3, CV-B4, CV-B5, CV-B6 | µM | [72] |
| Compound 1 | Specific for CV-B1, CV-B2, CV-B3, CV-B4, CV-B5, CV-B6 | µM | [72] |
| Compound 17 | CV-B1, CV-B3, CV-B6, CV-B4, CV-B5 and CV-A9 | nM-µM | [9] |
HS Heparan sulfate
Structure-based rational design of VP1 hydrophobic pocket binders has also been pursued. The structural analysis of four pyridyl imidazolidinones derivatives (GPP2, GPP3, GPP4 and GPP12) in complexes with EV-A71 facilitated the design of two highly potent anti-EV-A71 compounds ALD and NLD with notable IC50 values of 8.5 nM and 25 pM, respectively [51]. Both compounds also inhibited a wide range of other EVs including CV-A9, CV-A16, CV-A21, CV-B3, PV1-3, RV-2 and RV-14 with IC50 values ranging from pM to µM [52].
Antivirals targeting the fivefold axis of the capsid
The second region that can be targeted by antiviral compounds is the fivefold axis of the capsid. Many of the EV-A members such as CV-A6, CV-A16 and EV-A71 and EV-B members like CV-A9 and ECHO5 possess the positively charged fivefold axis that is responsible for viral attachment to host cell receptors including PSGL1 and heparan sulfate [53–57]. However, McLeish et al. [55] demonstrated that ECHO6 did not bind to heparan sulfate despite having a positive charge cluster at the fivefold axis. This suggests precise structure and conformation of the positive cluster is critical for the interaction between the fivefold axis and host receptors [55]. As SCARB2 was demonstrated to be the main attachment and uncoating receptor for EV-A viruses [58], it can be speculated that its inhibition could prevent EV infection. However, there is no report of any antiviral agent capable of inhibiting the binding of EVs to SCARB2.
Various compound series have been identified to target the fivefold axis (Table 2). One of the compounds, suramin, is a multi-functional molecule that has been evaluated for potential applications in viral diseases and cancer, despite its manifold adverse effects which have been reported including nephrotoxicity and dermatitis [59]. Ren et al. [60] reported that suramin inhibited several EV-A viruses including CV-A2, 3, 10, 12, and 16, and some EV-B viruses such as CV-A9, ECHO20 and ECHO25. Suramin and its derivatives such as NF449 were proposed to interact with the fivefold axis of the capsid to prevent EV association with PSGL1 and heparan sulfate [60, 61]. In vivo studies revealed that suramin significantly reduced mortality in mice challenged with a lethal dose of EV-A71 and decreased the peak viral load in adult rhesus monkeys [62].
Screening of sulfonated azo dyes against EVs has shown that the majority of the dyes exhibited in vitro inhibitory effects on the infectivity of EV-A71. In particular, brilliant black BN (E151) inhibited three EVs which are EV-A71, CV-A6 and CV-A16 [63]. It had the highest efficacy in blocking virus entry and it protected AG129 mice against EV-A71 lethal challenge. However, the in vitro potency of E151 is low with IC50 values ranging from 2.39 to 28.12 µM for various EV-A71 strains. E151 was identified to interact with the fivefold axis of the capsid and inhibited PSGL1 and cyclophilin A (CyP-A)-mediated EV-A71 entry into host cells [63].
Furthermore, the attachment of EV-A71 to host cells via PSGL1 and heparan sulfate was reported to be inhibited by a series of tryptophan dendrimers that target the fivefold axis of EV capsid [64, 65]. These dendrimers contain different central scaffolds and multiple tryptophan groups that are linked to the dendrimer branches through an amino group. A consensus compound named dendrimer 12 that was synthesized according to the structure–activity relationship analysis of the series was found to inhibit a large panel of EV-A71 clinical isolates with high potency in the nM to pM range [64].
The anti-EV activities of heparan sulfate mimetics have also been evaluated since a number of in vivo studies in mice and monkeys have demonstrated heparan sulfate could specifically interact with the key residue VP1-145G in EV-A71 to inhibit the virus [66, 67]. The mimetics including heparin, heparan sulfate and pentosan polysulfate were shown to exhibit antiviral actions against EV-A71 with low potency in the µM range [68, 69]. In addition, shorter heparan sulfate-based fragments exhibited inhibitory actions against EV-A71 infection [69]. These compounds bind to the capsid of EV-A71 to act as decoy receptors to block viral attachment [69]. A comparison of the in vitro potency between the small molecules and the larger mimetics revealed that the former exhibited a higher potency than the latter [69]. For instance, compound 22 has an IC50 value of 8.5 µg/mL, in comparison to IC50 values of 102.1 µg/mL and 142.8 µg/mL for heparan sulfate and heparin, respectively [69]. Importantly, the shorter heparan sulfate disaccharide mimetics lacked anti-coagulant activities and will not cause unwanted side effects [69].
Rosmarinic acid (RA) which is a compound from herbal medicine Salvia miltiorrhiza (Danshen) was found to target this region as well [70]. Similar to most of the compounds targeting this region, RA inhibited various EV-A71 genotypes with IC50 values in the μM range (Table 2) [70, 71]. In vivo evaluation revealed that RA reduced the mortality of mice infected with mouse-adapted EV-A71 strain [70, 71].
VP1–VP3 interprotomer binding pocket
More recently, a novel druggable pocket within the conserved VP1–VP3 interprotomer interface of the viral capsid has been reported (Table 2) [9, 72]. The novel drug target was initially identified in screening the antiviral activity of 4-dimethylamino benzoic acid (compound 12) and its analogues [72]. Compound 12 displayed weak potency with an EC50 value of 9 μM while its most potent analogue compound 1 displayed antiviral activity with an EC50 of 2.6 μM against CV-B3. These compounds were identified to be highly specific against CV-B viruses as they did not inhibit other EVs such as ECHO11, EV-A71, RV-2 and RV-14. Mutational and molecular modeling studies revealed that compound 12 and its analogues bind to a small cavity surrounded by amino acids Arg219 and Tyr75 from two different units of VP1 of CV-B3, which is distinct to the VP1 hydrophobic pocket targeted by pleconaril. Furthermore, the evaluation of the combinatorial antiviral activity of compound 12 and pleconaril revealed that the mechanism of action of compound 12 is distinct from that of pleconaril, indicating that both drugs targeted different binding pockets.
Subsequently, a benzenesulfonamide derivative, compound 17, was identified as an inhibitor of CV-B3 with an EC50 value of 0.7 μM [9]. It also inhibited the replication of CV-B1, CV-B6, CV-B4, CV-B5 and CV-A9. However, it lacked activity against viruses in the EV-A group (CV-A16 and EV-A71), EV-C group (CV-A21 and PV1), EV-D group (EV-D68) and RV-B group (RVB14). Structural study of compound 17 interaction with the viral capsid revealed that the compound binds to a pocket formed by two VP1 units (amino acids 73, 75–78, 155–157,159–160, 219, and 234) and one VP3 unit (amino acids 233–236) at the interface of an interprotomer. This novel drug target is located 16 Å away from the VP1 hydrophobic pocket. Sequence analysis revealed that the pocket is reasonably conserved across the EV-B group, with 7 of 16 amino acids being identical across the eight CV-B viruses including CV-B1, CV-B2, CV-B3, CV-B4, CV-B5, CV-B6, CV-A9 and ECHO11. Furthermore, the binding site is also conserved across a panel of EVs, in particular the amino acids Arg219 and Arg234 of CV-B3 [73]. The underlying mechanism of EV inhibition by compounds targeting this pocket is yet to be fully elucidated. It was proposed that the binding of compounds in this pocket stabilized the viral particle, which ultimately impeded structural rearrangements that allowed the transition to the A-particle [73].
Abdelnabi et al. [9] initiated hit optimization to develop a broad-spectrum antiviral that could inhibit the replication of multiple EV groups. The skeleton of compound 17 was used as the core structure to design more active analogues. The medicinal chemistry efforts guided by the data from antiviral assays yielded a series of broad-spectrum analogues with activity against EV-B (CV-Bs), EV-C (PV1 and CV-A21), EV-D (EVD68), RV-A (RVA09, RVA59, and RVA63), and RV-B (RVB14). However, the analogues still lacked activity against EV-A viruses such as CV-A16 and EV-A71. Among the broad-spectrum analogues include compound 48 with activities against EV-B and EV-C viruses and compound 77 with activities against RV-A and RV-B groups. Some of the compounds were also active against echoviruses E1 and E7.
The emergence of resistant variants towards antiviral drugs
Antiviral resistance is the major drawback for all the direct-acting antivirals targeting the EV capsid. In some cases, a single amino acid substitution within the binding pocket of EV was sufficient to reduce or completely abolish the antiviral activity of the capsid binders [75]. Many in vitro studies have demonstrated that the propagation of EV in the presence of capsid binders would lead to the emergence of resistant variants [30, 37, 76–78]. For example, serial passaging of EV-A71 and CV-A16 in the presence of ALD or NLD led to mutations in the VP1 of EV-A71 (Ile113 and Val123) and CV-A16 (Leu113) [78]. The amino acids were identified to be substituted with bulkier amino acids such as Ile113Met and Val123Ile in resistant EV-A71 and Leu113Phe in resistant CV-A16. The bulky amino acids hindered the entry of these inhibitors into the VP1 hydrophobic pockets. Resistant variants have also been identified in individuals receiving treatment during clinical trials. For instance, the clinical trials to evaluate pleconaril efficacy to treat cold symptoms in RV-infected individuals have indicated that RVs with reduced susceptibility to pleconaril were identified from 10.7% of the pleconaril-treated patients [79]. Additionally, fully pleconaril-resistant RVs were also recovered from 2.7% of these patients [79].
Although the capsid binders exhibited a low barrier to resistance, the majority of the resistant variants displayed lower fitness and virulence than the wild-type. For instance pleconaril-resistant RV isolated from patients appeared to be non-pathogenic and attenuated in cell cultures [79]. In addition, escape mutants such as NLD-resistant EV-A71 and CV-A16 variants were found to readily revert to the wild-type genotype when passaged in the absence of NLD [78].
Strategies to overcome drug resistance
Multiple strategies have been pursued to overcome the drug resistance problem. Combining antiviral agents with synergistic antiviral effects is a proven approach to increase antiviral potency, exemplified by the success in the combinatorial treatment regimens of human immunodeficiency virus (HIV) and hepatitis C virus (HCV) [80, 81]. Additionally, the use of antiviral agents with different mechanism and resistance profiles creates a higher barrier to genetic mutations, thereby hindering the emergence of resistance. Wang et al. [82] have demonstrated that the combination of two anti-EV drugs, rupintrivir and itraconazole, was shown to reduce the risk of generating drug-resistant EV-A71 mutants. Studies to investigate the in vivo combinatorial effects of anti-EV drugs such as disoxaril/guanidine/oxoglaucine and pleconaril/MDL-860/oxoglaucine in newborn mice infected with coxsackieviruses revealed that the combinations of these drugs prevented the development of drug resistance against the capsid binders [83–85].
Another strategy that has been explored to overcome antiviral resistance is by modifying the physical properties of existing antiviral agents. The study of structure–activity relationships has facilitated the selection of compound scaffolds that can facilitate the design of new inhibitors with limited antiviral resistance and unwanted side effects [74, 86]. For instance, the pleconaril scaffold has been used as the basis for the development of novel compounds such as the orally available compound 10g which could inhibit pleconaril-resistant EVs with IC50 values between 0.02 and 5.25 µM [74]. Additionally, compound 10g is a weaker inducer of CYP3A4 enzymes that pleconaril, lowering the risk of off-target effects [74].
Peptide-based anti-enteroviral development
Peptide-based strategy is another viable approach to develop anti-EV drugs especially with the success of the FDA-approved antiviral peptide drug enfuvirtide. Enfuvirtide is a peptide derived from a region within the human immunodeficiency virus (HIV-1) glycoprotein 41 (gp41) [87]. It inhibits HIV-1 infection by blocking the membrane fusion between HIV-1 and cellular membranes. The peptide-based strategy possesses several advantages over small-molecule compounds as they are easy to synthesize, exhibit a higher barrier to viral resistance and have a lower toxicity. In addition, peptides are better at targeting pockets that are too large to be occupied by small-molecule compounds. A study indicated that compounds that partially occupy the VP1 hydrophobic pocket exhibited weaker potency than larger compounds that have better occupancy [51]. However, larger chemical compounds are associated with difficulty to synthesize, high cost of production and poor bioavailability.
Multiple peptides have been identified to inhibit EV-A71 and other enteroviruses (Table 3). In a study by Tan et al. [88], four peptides SP40, SP45, SP55 and SP81 derived from the VP1 of EV-A71 were found to inhibit EV-A71 with the SP40 peptide exhibiting the highest antiviral activity with an IC50 of 6 µM. Synergistic antiviral activity assays revealed that SP40, SP45 and SP55 peptides might exert their activities against EV-A71 by inhibiting the viral attachment in the early phase of the infection. In contrast, SP81 exerted its activity at a later stage of EV-A71 infection viz. at post-viral entry [89]. Furthermore, SP40 peptide was shown to be active against other viruses in EV-A and EV-C groups such as CV-A16 and PV1, respectively [88].
Table 3.
Antiviral peptides against enteroviruses
| Peptide | Sequence | Potency (EC50 or IC50) | EV inhibited | Refs |
|---|---|---|---|---|
| SP40 | Ac-QMRRKVELFTYMRFD-NH2 | 6–9.3 µM | EV-A71, CV-A16, PV1 | [88] |
| SP45 | Ac-AEFTFVACTPTGEVV-NH2 | N/A | EV-A71 | [88] |
| SP55 | Ac-PESRESLAW-NH2 | N/A | EV-A71 | [88] |
| SP81 | Ac-SKSKYPLVVRIYMRMKHVRAW-NH2 | N/A | EV-A71 | [88] |
| LVLQTM | LVLQTM | nM–µM | EV-A71, Echo-6, RV-2 | [90, 91] |
| Tripeptide VAD | VAD | µM | RV-2 | [92] |
| Tetrapeptide AAPV | AAPV | µM | PV-1, CV-A21, RV-2 | [93] |
N/A not available
Another peptide (LVLQTM) that acted as a pseudosubstrate to the 2A protease was found to inhibit multiple EV infections [90]. This peptide binds to the active site of 2A protease to reduce its activity with an IC50 value of 0.3 μM [91]. Various other peptides have been shown to target 2A proteases from various EVs such as the tripeptide VAD and tetrapeptide AAPV with IC50 values of 5.6 μM and 20–65 μM, respectively [92, 93]. In general, the majority of the anti-EV peptides displayed lower potencies (> 0.3 μM) in comparison to the small-molecules with in vitro efficacies in the nM to pM range. Nevertheless, the development of anti-EV peptides is warranted as these peptides may be utilized in a combinatorial drug approach together with the small-molecules. Optimization of the peptides such as cholesterol tagging may improve the potency of these peptides, in particular the peptides targeting host proteins at the cellular membrane such as SP40 peptide.
Design of antiviral peptides targeting the canyon region on the surface of enteroviruses
Despite the wealth in structural information of the EV capsids, no direct-acting antiviral or host-targeting inhibitor has been designed using the structure-based drug design approach. There are several regions on the EV capsids that can be targeted by peptides. For instance, the atomic structure of EVs in complex with their receptors may guide the design of antiviral peptides derived from the complex interface on the receptors [18]. These peptides may act as direct-acting antivirals by interacting with the regions on the capsid and act as decoys to competitively inhibit EV attachment to host cells.
Taking poliovirus in complex with PVR as an example, the PVR binds to the quasi-threefold axis region contacting with three capsid proteins VP1, VP2 and VP3 [12]. Structural analysis revealed that the canyon region of the PV capsid formed ionic and hydrophobic interactions with two regions spanning amino acids 60 to 99 and 126 to 130 within the apical domain of PVR (Fig. 2). Therefore, peptides derived from these two regions could potentially bind to the canyon and hinder the attachment of poliovirus to the PVR. There are several advantages associated with utilizing the peptide-based strategy to target the canyon region on the EV capsid surface. Firstly, targeting this region using peptides is advantageous as the peptides can engage the virus surface extracellularly. This removes the need to consider the permeability of the peptides. Secondly, peptides that act on the virus may display lower cytotoxicity than antivirals targeting host proteins that are prone to cause unwanted off-target effects. Lastly, capsid proteins are highly conserved among viral family, therefore they are promising to be developed as broad-spectrum antivirals against multiple EV infections.
Fig. 2.
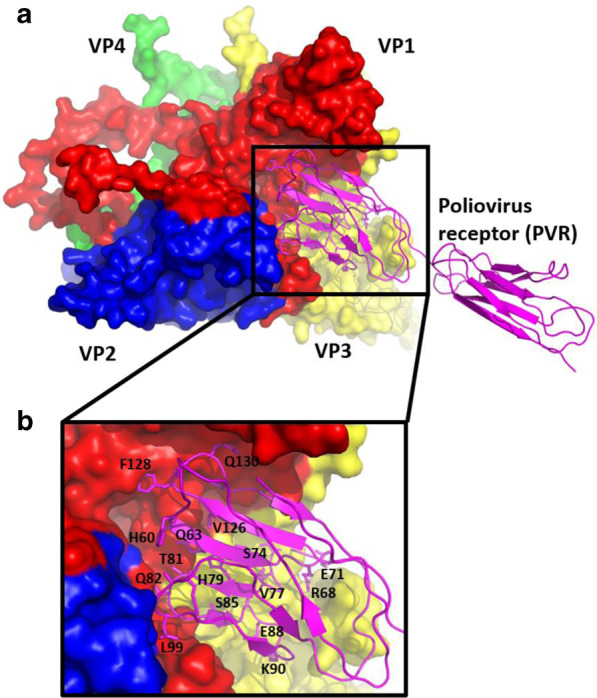
The structure of poliovirus in complex with PVR (PDB ID: 3EPD) [94]. a Overall view of the canonical picornavirus protomer with the capsid proteins VP1, VP2, VP3 and VP4 are shown in red, yellow, blue and green, respectively. The apical domain of PVR that binds to the canyon of the protomer is shown magenta. b The amino acids that make contacts with the canyon are shown in sticks and labeled
Design of antiviral peptides targeting the cellular receptors
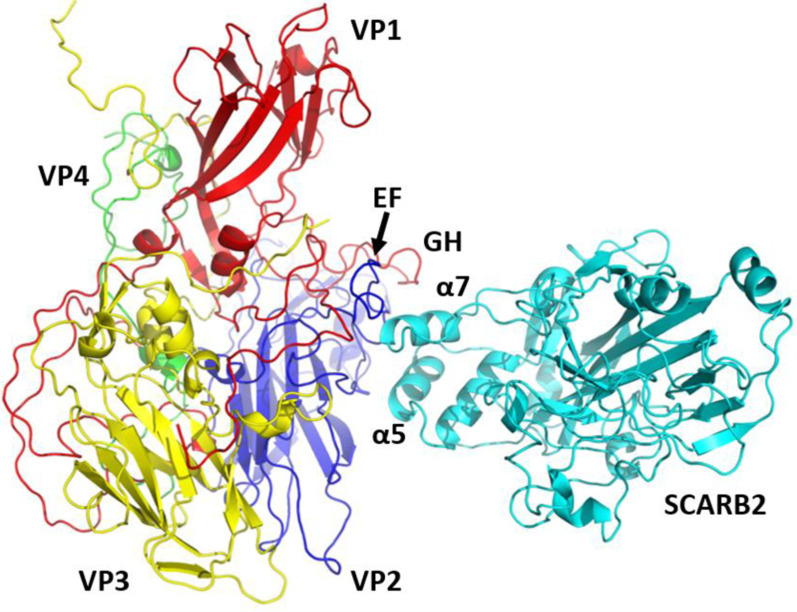
Apart from designing direct-acting antiviral peptides, the structural information of EV in complex with host receptors can be utilized to design peptides that can target host proteins, in particular the cellular receptors [81, 95]. Taking the structure of EV-A71 in complex with SCARB2 as an example, peptides derived from the capsid region can be evaluated for their ability to prevent viral attachment to host cells (Fig. 3) [18]. Structural analysis revealed that the interface of the EV-A71:SCARB2 complex is formed by the α5 (aa 152–163) and α7 (aa 183–193) helices of SCARB2 and VP1 GH and VP2 EF loops of EV-A71 [18]. Peptides mimicking the GH loop of VP1 and EF loop of VP2 could potentially bind to SCARB2, blocking the interactions between EV-A71 and SCARB2. There are several advantages of targeting the host cellular receptors [81]. Host-targeting antivirals generally possess a higher barrier to resistance than its virus-targeting counterparts [96]. In addition, antiviral peptides targeting the host receptor could provide a broad inhibition of multiple viruses from different genotypes and serotypes and possibly other viruses in the Picornaviridae family that utilize the same receptor for cellular attachment [97].
Fig. 3.
Three-dimensional structure of EV-A71 in complex with SCARB2 (PDB: 6I2K) [18]. The EV-A71 capsid proteins VP1, VP2, VP3 and VP4 are shown in red, yellow, blue and green, respectively. The receptor SCARB2 is shown in cyan. VP2 EF loop and VP1 GH loop of EV-A71 interact with α5 and α7 helices of SCARB2 to form the interface of the complex
Limitations of peptide-based antiviral strategy
Despite the advantages of antiviral peptides, several limitations remain to be addressed. First, antiviral peptides generally exhibited a weaker potency than small-molecule compounds [81]. This issue could be resolved by modification strategies such as cholesterol tagging that have been proven to increase the potency of antiviral peptides. Cholesterol tagging was shown to enhance the local concentration of peptides at the membrane and improved membrane permeability [81, 98, 99]. Second, poor bioavailability and short half-life are common limitations for peptide-based strategy since peptides are susceptible to cleavage by peptidases and proteases [99]. The use of D-amino acids could decrease recognition and binding of peptides to proteolytic enzymes [100]. Terminal capping by post-translational modifications such as N-terminal acetylation and C-terminal amidation could enhance the ability of peptides to resist degradation by exopeptidases [101]. Furthermore, encapsulation into nanoparticles could increase the stability and bioavailability of antiviral peptides [102]. Lastly, there is an issue with the high production cost related to peptide synthesis and purification [99, 103]. Factors such as expensive reagents and low purity of final products coupled with challenging processes to introduce disulfide bridges in certain peptides significantly increased the production cost [99, 103]. Effective peptide production methods like the use of recombinant expression systems in hosts such as bacteria and yeast could significantly reduce the cost of peptide production [104, 105].
Conclusions
The eradication of EVs is challenging because these viruses are not easily inactivated and may survive well in water and sewage for long periods [106]. Therefore, the development of vaccines and antivirals should be pursued to mitigate EV epidemics. The ultimate goal of anti-EV research is to develop safe and effective antiviral agents without generating drug-resistant EVs. Many inhibitors targeting the surface capsid of EV have been identified with five of them being evaluated for their safety and efficacy in clinical trials. However, the majority of the inhibitors were found to cause unwanted side effects and failed to meet their clinical endpoints despite exhibiting potent in vitro activity in the nM to pM range. Furthermore, the inhibitors are prone to generate resistant variants, albeit the resistant variants exhibited reduced fitness in comparison to their wild-type counterparts. Nonetheless, researchers should explore the strategies such as drug combination therapy and drug optimization based on the structure–activity relationships to improve antiviral potency and increase the resistance barrier of the inhibitors. Lastly, the peptide-based antiviral strategy should be explored either as an alternative or to complement the anti-EV small-molecules.
Acknowledgements
Not applicable.
Authors’ contributions
MIA and FZ wrote the manuscript. CLP provided the critical revision. All authors read and approved the final manuscript.
Funding
This work was supported by the Ministry of Education, Malaysia (FRGS/1/2018/SKK11/SYUC/03/3) and Sunway University Research Centre Grant (STR-RCTR-CVVR-01-2020) to the Centre for Virus and Vaccine Research (CVVR).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Baggen J, Thibaut HJ, Strating JRPM, van Kuppeveld FJM. The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol. 2018;16:368–381. doi: 10.1038/s41579-018-0005-4. [DOI] [PubMed] [Google Scholar]
- 2.Anasir MI, Poh CL. Advances in antigenic peptide-based vaccine and neutralizing antibodies against viruses causing hand, foot, and mouth disease. Int J Mol Sci. 2019;20:1256. doi: 10.3390/ijms20061256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan J, Shen L, Wu J, Zou X, Gu J, Chen J, Mao L. Enterovirus A71 proteins: structure and function. Front Microbiol. 2018;9:286. doi: 10.3389/fmicb.2018.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benschop KSM, van der Avoort HGAM, Duizer E, Koopmans MPG. Antivirals against enteroviruses: a critical review from a public-health perspective. Antivir Ther. 2015;20:121–130. doi: 10.3851/IMP2939. [DOI] [PubMed] [Google Scholar]
- 5.Yang B, Liu F, Liao Q, Wu P, Chang Z, Huang J, Long L, Luo L, Li Y, Leung GM, et al. Epidemiology of hand, foot and mouth disease in China, 2008 to 2015 prior to the introduction of EV-A71 vaccine. Euro Surveill. 2017 doi: 10.2807/1560-7917.ES.2017.22.50.16-00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puenpa J, Wanlapakorn N, Vongpunsawad S, Poovorawan Y. The history of enterovirus A71 outbreaks and molecular epidemiology in the Asia-Pacific Region. J Biomed Sci. 2019;26:75. doi: 10.1186/s12929-019-0573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messacar K, Abzug MJ, Dominguez SR. 2014 outbreak of enterovirus D68 in North America. J Med Virol. 2016;88:739–745. doi: 10.1002/jmv.24410. [DOI] [PubMed] [Google Scholar]
- 8.Lyu K, Wang G-C, He Y-L, Han J-F, Ye Q, Qin C-F, Chen R. Crystal structures of enterovirus 71 (EV71) recombinant virus particles provide insights into vaccine design. J Biol Chem. 2015;290:3198–3208. doi: 10.1074/jbc.M114.624536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdelnabi R, Geraets JA, Ma Y, Mirabelli C, Flatt JW, Domanska A, Delang L, Jochmans D, Kumar TA, Jayaprakash V, et al. A novel druggable interprotomer pocket in the capsid of rhino- and enteroviruses. PLoS Biol. 2019;17:e3000281. doi: 10.1371/journal.pbio.3000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergelson JM, Coyne CB. Picornavirus entry. Adv Exp Med Biol. 2013;790:24–41. doi: 10.1007/978-1-4614-7651-1_2. [DOI] [PubMed] [Google Scholar]
- 11.Cifuente JO, Moratorio G. Evolutionary and structural overview of human picornavirus capsid antibody evasion. Front Cell Infect Microbiol. 2019;9:283. doi: 10.3389/fcimb.2019.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strauss M, Filman DJ, Belnap DM, Cheng N, Noel RT, Hogle JM. Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry. J Virol. 2015;89:4143–4157. doi: 10.1128/jvi.03101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shakeel S, Seitsonen JJ, Kajander T, Laurinmaki P, Hyypia T, Susi P, Butcher SJ. Structural and functional analysis of coxsackievirus A9 integrin alphavbeta6 binding and uncoating. J Virol. 2013;87:3943–3951. doi: 10.1128/jvi.02989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Organtini LJ, Makhov AM, Conway JF, Hafenstein S, Carson SD. Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J Virol. 2014;88:5755–5765. doi: 10.1128/jvi.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coyne CB, Bergelson JM. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell. 2006;124:119–131. doi: 10.1016/j.cell.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Ye X, Zhang X-Y, Zhu Z, Zhang X, Xu Z, Ding Z, Zou G, Liu Q, Kong L, et al. Coxsackievirus A10 atomic structure facilitating the discovery of a broad-spectrum inhibitor against human enteroviruses. Cell Discov. 2019;5:4. doi: 10.1038/s41421-018-0073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, Koike S. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat Med. 2009;15:798–801. doi: 10.1038/nm.1992. [DOI] [PubMed] [Google Scholar]
- 18.Zhou D, Zhao Y, Kotecha A, Fry EE, Kelly JT, Wang X, Rao Z, Rowlands DJ, Ren J, Stuart DI. Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2. Nat Microbiol. 2019;4:414–419. doi: 10.1038/s41564-018-0319-z. [DOI] [PubMed] [Google Scholar]
- 19.Yamayoshi S, Iizuka S, Yamashita T, Minagawa H, Mizuta K, Okamoto M, Nishimura H, Sanjoh K, Katsushima N, Itagaki T, et al. Human SCARB2-dependent infection by coxsackievirus A7, A14, and A16 and enterovirus 71. J Virol. 2012;86:5686–5696. doi: 10.1128/JVI.00020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchta D, Füzik T, Hrebík D, Levdansky Y, Sukeník L, Mukhamedova L, Moravcová J, Vácha R, Plevka P. Enterovirus particles expel capsid pentamers to enable genome release. Nat Commun. 2019;10:1138. doi: 10.1038/s41467-019-09132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyu K, Ding J, Han J-F, Zhang Y, Wu X-Y, He Y-L, Qin C-F, Chen R. Human enterovirus 71 uncoating captured at atomic resolution. J Virol. 2014;88:3114–3126. doi: 10.1128/JVI.03029-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth MS, Martin JH. Picornavirus uncoating. Mol Pathol. 2002;55:214–219. doi: 10.1136/mp.55.4.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer L, Lyoo H, van der Schaar HM, Strating JR, van Kuppeveld FJ. Direct-acting antivirals and host-targeting strategies to combat enterovirus infections. Curr Opin Virol. 2017;24:1–8. doi: 10.1016/j.coviro.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garriga D, Pickl-Herk A, Luque D, Wruss J, Castón JR, Blaas D, Verdaguer N. Insights into minor group rhinovirus uncoating: the X-ray structure of the HRV2 empty capsid. PLoS Pathog. 2012;8:e1002473. doi: 10.1371/journal.ppat.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dewindt B, van Eemeren K, Andries K. Antiviral capsid-binding compounds can inhibit the adsorption of minor receptor rhinoviruses. Antivir Res. 1994;25:67–72. doi: 10.1016/0166-3542(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 26.Zeichhardt H, Otto MJ, McKinlay MA, Willingmann P, Habermehl KO. Inhibition of poliovirus uncoating by disoxaril (WIN 51711) Virology. 1987;160:281–285. doi: 10.1016/0042-6822(87)90075-4. [DOI] [PubMed] [Google Scholar]
- 27.Otto MJ, Fox MP, Fancher MJ, Kuhrt MF, Diana GD, McKinlay MA. In vitro activity of WIN 51711, a new broad-spectrum antipicornavirus drug. Antimicrob Agents Chemother. 1985;27:883–886. doi: 10.1128/aac.27.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jubelt B, Wilson AK, Ropka SL, Guidinger PL, McKinlay MA. Clearance of a persistent human enterovirus infection of the mouse central nervous system by the antiviral agent disoxaril. J Infect Dis. 1989;159:866–871. doi: 10.1093/infdis/159.5.866. [DOI] [PubMed] [Google Scholar]
- 29.Pevear DC, Tull TM, Seipel ME, Groarke JM. Activity of pleconaril against enteroviruses. Antimicrob Agents Chemother. 1999;43:2109–2115. doi: 10.1128/AAC.43.9.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andries K, Dewindt B, Snoeks J, Willebrords R, van Eemeren K, Stokbroekx R, Janssen PA. In vitro activity of pirodavir (R 77975), a substituted phenoxy-pyridazinamine with broad-spectrum antipicornaviral activity. Antimicrob Agents Chemother. 1992;36:100–107. doi: 10.1128/aac.36.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buontempo PJ, Cox S, Wright-Minogue J, DeMartino JL, Skelton AM, Ferrari E, Albin R, Rozhon EJ, Girijavallabhan V, Modlin JF, et al. SCH 48973: a potent, broad-spectrum, antienterovirus compound. Antimicrob Agents Chemother. 1997;41:1220–1225. doi: 10.1128/AAC.41.6.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oberste MS, Moore D, Anderson B, Pallansch MA, Pevear DC, Collett MS. In vitro antiviral activity of V-073 against polioviruses. Antimicrob Agents Chemother. 2009;53:4501–4503. doi: 10.1128/AAC.00671-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rotbart HA. Antiviral therapy for enteroviruses and rhinoviruses. Antivir Chem Chemother. 2000;11:261–271. doi: 10.1177/095632020001100402. [DOI] [PubMed] [Google Scholar]
- 34.Hayden FG, Herrington DT, Coats TL, Kim K, Cooper EC, Villano SA, Liu S, Hudson S, Pevear DC, Collett M, et al. Efficacy and safety of oral pleconaril for treatment of colds due to picornaviruses in adults: results of 2 double-blind, randomized, placebo-controlled trials. Clin Infect Dis. 2003;36:1523–1532. doi: 10.1086/375069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayden FG, Hipskind GJ, Woerner DH, Eisen GF, Janssens M, Janssen PA, Andries K. Intranasal pirodavir (R77,975) treatment of rhinovirus colds. Antimicrob Agents Chemother. 1995;39:290–294. doi: 10.1128/aac.39.2.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirabelli C, Scheers E, Neyts J. Novel therapeutic approaches to simultaneously target rhinovirus infection and asthma/COPD pathogenesis. F1000Research. 1860;2017:6. doi: 10.12688/f1000research.11978.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feil SC, Hamilton S, Krippner GY, Lin B, Luttick A, McConnell DB, Nearn R, Parker MW, Ryan J, Stanislawski PC, et al. An orally available 3-ethoxybenzisoxazole capsid binder with clinical activity against human rhinovirus. ACS Med Chem Lett. 2012;3:303–307. doi: 10.1021/ml2002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collett MS, Hincks JR, Benschop K, Duizer E, van der Avoort H, Rhoden E, Liu H, Oberste MS, McKinlay MA, Hartford M. Antiviral activity of pocapavir in a randomized, blinded, placebo-controlled human oral poliovirus vaccine challenge model. J Infect Dis. 2017;215:335–343. doi: 10.1093/infdis/jiw542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayden FG, Turner RB, Gwaltney JM, Chi-Burris K, Gersten M, Hsyu P, Patick AK, Smith GJ, Zalman LS. Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob Agents Chemother. 2003;47:3907–3916. doi: 10.1128/aac.47.12.3907-3916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patick AK, Brothers MA, Maldonado F, Binford S, Maldonado O, Fuhrman S, Petersen A, Smith GJ, Zalman LS, Burns-Naas LA, et al. In vitro antiviral activity and single-dose pharmacokinetics in humans of a novel, orally bioavailable inhibitor of human rhinovirus 3C protease. Antimicrob Agents Chemother. 2005;49:2267–2275. doi: 10.1128/AAC.49.6.2267-2275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller FD, Monto AS, DeLong DC, Exelby A, Bryan ER, Srivastava S. Controlled trial of enviroxime against natural rhinovirus infections in a community. Antimicrob Agents Chemother. 1985;27:102–106. doi: 10.1128/aac.27.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinz BA, Vance LM. The antiviral compound enviroxime targets the 3A coding region of rhinovirus and poliovirus. J Virol. 1995;69:4189–4197. doi: 10.1128/JVI.69.7.4189-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Palma AM, Thibaut HJ, van der Linden L, Lanke K, Heggermont W, Ireland S, Andrews R, Arimilli M, Al-Tel TH, De Clercq E, et al. Mutations in the nonstructural protein 3A confer resistance to the novel enterovirus replication inhibitor TTP-8307. Antimicrob Agents Chemother. 2009;53:1850–1857. doi: 10.1128/AAC.00934-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nikolaeva L, Galabov AS. Synergistic inhibitory effect of enviroxime and disoxaril on poliovirus type 1 replication. Acta Virol. 1995;39:235–241. [PubMed] [Google Scholar]
- 45.Barnard DL, Hubbard VD, Smee DF, Sidwell RW, Watson KGW, Tucker SPT, Reece PAR. In vitro activity of expanded-spectrum pyridazinyl oxime ethers related to pirodavir: novel capsid-binding inhibitors with potent antipicornavirus activity. Antimicrob Agents Chemother. 2004;48:1766–1772. doi: 10.1128/aac.48.5.1766-1772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tijsma A, Franco D, Tucker S, Hilgenfeld R, Froeyen M, Leyssen P, Neyts J. The capsid binder Vapendavir and the novel protease inhibitor SG85 inhibit enterovirus 71 replication. Antimicrob Agents Chemother. 2014;58:6990–6992. doi: 10.1128/AAC.03328-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patick AK, Binford SL, Brothers MA, Jackson RL, Ford CE, Diem MD, Maldonado F, Dragovich PS, Zhou R, Prins TJ, et al. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob Agents Chemother. 1999;43:2444–2450. doi: 10.1128/AAC.43.10.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dragovich PS, Webber SE, Babine RE, Fuhrman SA, Patick AK, Matthews DA, Lee CA, Reich SH, Prins TJ, Marakovits JT, et al. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 1. Michael acceptor structure–activity studies. J Med Chem. 1998;41:2806–2818. doi: 10.1021/jm980068d. [DOI] [PubMed] [Google Scholar]
- 49.Kim J, Jung YK, Kim C, Shin JS, Scheers E, Lee J-Y, Han SB, Lee C-K, Neyts J, Ha J-D, et al. A novel series of highly potent small molecule inhibitors of rhinovirus replication. J Med Chem. 2017;60:5472–5492. doi: 10.1021/acs.jmedchem.7b00175. [DOI] [PubMed] [Google Scholar]
- 50.Ho J-Y, Chern J-H, Hsieh C-F, Liu S-T, Liu C-J, Wang Y-S, Kuo T-W, Hsu S-J, Yeh T-K, Shih S-R, et al. In vitro and in vivo studies of a potent capsid-binding inhibitor of enterovirus 71. J Antimicrob Chemother. 2016;71:1922–1932. doi: 10.1093/jac/dkw101. [DOI] [PubMed] [Google Scholar]
- 51.De Colibus L, Wang X, Spyrou JAB, Kelly J, Ren J, Grimes J, Puerstinger G, Stonehouse N, Walter TS, Hu Z, et al. More-powerful virus inhibitors from structure-based analysis of HEV71 capsid-binding molecules. Nat Struct Mol Biol. 2014;21:282–288. doi: 10.1038/nsmb.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Colibus L, Wang X, Tijsma A, Neyts J, Spyrou JAB, Ren J, Grimes JM, Puerstinger G, Leyssen P, Fry EE, et al. Structure elucidation of coxsackievirus A16 in complex with GPP3 informs a systematic review of highly potent capsid binders to enteroviruses. PLoS Pathog. 2015;11:e1005165. doi: 10.1371/journal.ppat.1005165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishimura Y, Lee H, Hafenstein S, Kataoka C, Wakita T, Bergelson JM, Shimizu H. Enterovirus 71 binding to PSGL-1 on leukocytes: VP1-145 acts as a molecular switch to control receptor interaction. PLoS Pathog. 2013;9:e1003511. doi: 10.1371/journal.ppat.1003511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan CW, Poh CL, Sam I-C, Chan YF. Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J Virol. 2013;87:611–620. doi: 10.1128/JVI.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McLeish NJ, Williams ÇH, Kaloudas D, Roivainen MM, Stanway G. Symmetry-related clustering of positive charges is a common mechanism for heparan sulfate binding in enteroviruses. J Virol. 2012;86:11163–11170. doi: 10.1128/JVI.00640-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X, Shi J, Ye X, Ku Z, Zhang C, Liu Q, Huang Z. Coxsackievirus A16 utilizes cell surface heparan sulfate glycosaminoglycans as its attachment receptor. Emerg Microbes Infect. 2017;6:e65. doi: 10.1038/emi.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Israelsson S, Gullberg M, Jonsson N, Roivainen M, Edman K, Lindberg AM. Studies of Echovirus 5 interactions with the cell surface: heparan sulfate mediates attachment to the host cell. Virus Res. 2010;151:170–176. doi: 10.1016/j.virusres.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi K, Koike S. Cellular receptors for enterovirus A71. J Biomed Sci. 2020;27:23. doi: 10.1186/s12929-020-0615-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiedemar N, Hauser DA, Mäser P. 100 years of suramin. Antimicrob Agents Chemother. 2020;64:e01168–e1219. doi: 10.1128/AAC.01168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren P, Zheng Y, Wang W, Hong L, Delpeyroux F, Arenzana-Seisdedos F, Altmeyer R. Suramin interacts with the positively charged region surrounding the 5-fold axis of the EV-A71 capsid and inhibits multiple enterovirus A. Sci Rep. 2017;7:42902. doi: 10.1038/srep42902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishimura Y, McLaughlin NP, Pan J, Goldstein S, Hafenstein S, Shimizu H, Winkler JD, Bergelson JM. The suramin derivative NF449 interacts with the 5-fold vertex of the Enterovirus A71 capsid to prevent virus attachment to PSGL-1 and heparan sulfate. PLoS Pathog. 2015;11:e1005184. doi: 10.1371/journal.ppat.1005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ren P, Zou G, Bailly B, Xu S, Zeng M, Chen X, Shen L, Zhang Y, Guillon P, Arenzana-Seisdedos F, et al. The approved pediatric drug suramin identified as a clinical candidate for the treatment of EV71 infection-suramin inhibits EV71 infection in vitro and in vivo. Emerg Microbes Infect. 2014;3:e62. doi: 10.1038/emi.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meng T, Jia Q, Wong S-M, Chua K-B. In vitro and in vivo inhibition of the infectivity of human enterovirus 71 by a sulfonated food azo dye, brilliant black BN. J Virol. 2019;93:e00061-19. doi: 10.1128/JVI.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rivero-Buceta E, Sun L, Martínez-Gualda B, Doyagüez EG, Donckers K, Quesada E, Camarasa M-J, Delang L, San-Félix A, Neyts J, et al. Optimization of a class of tryptophan dendrimers that inhibit HIV replication leads to a selective, specific, and low-nanomolar inhibitor of clinical isolates of enterovirus A71. Antimicrob Agents Chemother. 2016;60:5064–5067. doi: 10.1128/AAC.00626-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun L, Lee H, Thibaut HJ, Lanko K, Rivero-Buceta E, Bator C, Martinez-Gualda B, Dallmeier K, Delang L, Leyssen P, et al. Viral engagement with host receptors blocked by a novel class of tryptophan dendrimers that targets the 5-fold-axis of the enterovirus-A71 capsid. PLoS Pathog. 2019;15:e1007760. doi: 10.1371/journal.ppat.1007760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kobayashi K, Sudaka Y, Takashino A, Imura A, Fujii K, Koike S. Amino acid variation at VP1–145 of enterovirus 71 determines attachment receptor usage and neurovirulence in human scavenger receptor B2 transgenic mice. J Virol. 2018 doi: 10.1128/JVI.00681-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujii K, Sudaka Y, Takashino A, Kobayashi K, Kataoka C, Suzuki T, Iwata-Yoshikawa N, Kotani O, Ami Y, Shimizu H, et al. VP1 amino acid residue 145 of enterovirus 71 is a key residue for its receptor attachment and resistance to neutralizing antibody during cynomolgus monkey infection. J Virol. 2018 doi: 10.1128/JVI.00682-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pourianfar HR, Poh CL, Fecondo J, Grollo L. In vitro evaluation of the antiviral activity of heparan sulfate mimetic compounds against Enterovirus 71. Virus Res. 2012;169:22–29. doi: 10.1016/j.virusres.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 69.Earley DF, Bailly B, Maggioni A, Kundur AR, Thomson RJ, Chang C-W, von Itzstein M. Efficient blocking of enterovirus 71 infection by heparan sulfate analogues acting as decoy receptors. ACS Infect Dis. 2019;5:1708–1717. doi: 10.1021/acsinfecdis.9b00070. [DOI] [PubMed] [Google Scholar]
- 70.Hsieh C-F, Jheng J-R, Lin G-H, Chen Y-L, Ho J-Y, Liu C-J, Hsu K-Y, Chen Y-S, Chan YF, Yu H-M, et al. Rosmarinic acid exhibits broad anti-enterovirus A71 activity by inhibiting the interaction between the five-fold axis of capsid VP1 and cognate sulfated receptors. Emerg Microbes Infect. 2020;9:1194–1205. doi: 10.1080/22221751.2020.1767512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin W-Y, Yu Y-J, Jinn T-R. Evaluation of the virucidal effects of rosmarinic acid against enterovirus 71 infection via in vitro and in vivo study. Virol J. 2019;16:94. doi: 10.1186/s12985-019-1203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma Y, Abdelnabi R, Delang L, Froeyen M, Luyten W, Neyts J, Mirabelli C. New class of early-stage enterovirus inhibitors with a novel mechanism of action. Antivir Res. 2017;147:67–74. doi: 10.1016/j.antiviral.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 73.Duyvesteyn HME, Ren J, Walter TS, Fry EE, Stuart DI. Glutathione facilitates enterovirus assembly by binding at a druggable pocket. Commun Biol. 2020;3:9. doi: 10.1038/s42003-019-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Egorova A, Kazakova E, Jahn B, Ekins S, Makarov V, Schmidtke M. Novel pleconaril derivatives: influence of substituents in the isoxazole and phenyl rings on the antiviral activity against enteroviruses. Eur J Med Chem. 2020;188:112007. doi: 10.1016/j.ejmech.2019.112007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmidtke M, Hammerschmidt E, Schüler S, Zell R, Birch-Hirschfeld E, Makarov VA, Riabova OB, Wutzler P. Susceptibility of coxsackievirus B3 laboratory strains and clinical isolates to the capsid function inhibitor pleconaril: antiviral studies with virus chimeras demonstrate the crucial role of amino acid 1092 in treatment. J Antimicrob Chemother. 2005;56:648–656. doi: 10.1093/jac/dki263. [DOI] [PubMed] [Google Scholar]
- 76.Groarke JM, Pevear DC. Attenuated virulence of pleconaril-resistant coxsackievirus B3 variants. J Infect Dis. 1999;179:1538–1541. doi: 10.1086/314758. [DOI] [PubMed] [Google Scholar]
- 77.Liu H-M, Roberts JA, Moore D, Anderson B, Pallansch MA, Pevear DC, Collett MS, Oberste MS. Characterization of poliovirus variants selected for resistance to the antiviral compound V-073. Antimicrob Agents Chemother. 2012;56:5568–5574. doi: 10.1128/AAC.00539-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kelly JT, De Colibus L, Elliott L, Fry EE, Stuart DI, Rowlands DJ, Stonehouse NJ. Potent antiviral agents fail to elicit genetically-stable resistance mutations in either enterovirus 71 or Coxsackievirus A16. Antivir Res. 2015;124:77–82. doi: 10.1016/j.antiviral.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pevear DC, Hayden FG, Demenczuk TM, Barone LR, McKinlay MA, Collett MS. Relationship of pleconaril susceptibility and clinical outcomes in treatment of common colds caused by rhinoviruses. Antimicrob Agents Chemother. 2005;49:4492–4499. doi: 10.1128/AAC.49.11.4492-4499.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peterson CW, Younan P, Jerome KR, Kiem H-P. Combinatorial anti-HIV gene therapy: using a multipronged approach to reach beyond HAART. Gene Ther. 2013;20:695–702. doi: 10.1038/gt.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anasir MI, Ramanathan B, Poh CL. Structure-based design of antivirals against envelope glycoprotein of dengue virus. Viruses. 2020;12:367. doi: 10.3390/v12040367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y, Li G, Yuan S, Gao Q, Lan K, Altmeyer R, Zou G. In vitro assessment of combinations of enterovirus inhibitors against Enterovirus 71. Antimicrob Agents Chemother. 2016;60:5357–5367. doi: 10.1128/AAC.01073-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stoyanova A, Nikolova I, Galabov AS. Effect of consecutive alternating administration (CAA) of a triple anti-enteroviral combination on Coxsackievirus B1 neuroinfection in mice. Antivir Res. 2015;121:138–144. doi: 10.1016/j.antiviral.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 84.Vassileva-Pencheva R, Galabov AS. Effectiveness of the consecutive alternating administration course of a triple antiviral combination in Coxsackievirus B3 infections in mice. Drug Res. 2016;66:639–643. doi: 10.1055/s-0042-112970. [DOI] [PubMed] [Google Scholar]
- 85.Stoyanova A, Nikolova I, Pürstinger G, Dobrikov G, Dimitrov V, Philipov S, Galabov AS. Anti-enteroviral triple combination of viral replication inhibitors: activity against coxsackievirus B1 neuroinfection in mice. Antivir Chem Chemother. 2015;24:136–147. doi: 10.1177/2040206616671571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Egorova A, Ekins S, Schmidtke M, Makarov V. Back to the future: advances in development of broad-spectrum capsid-binding inhibitors of enteroviruses. Eur J Med Chem. 2019;178:606–622. doi: 10.1016/j.ejmech.2019.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wild CT, Shugars DC, Greenwell TK, McDanal CB, Matthews TJ. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tan CW, Chan YF, Sim KM, Tan EL, Poh CL. Inhibition of enterovirus 71 (EV-71) infections by a novel antiviral peptide derived from EV-71 capsid protein VP1. PLoS ONE. 2012;7:e34589. doi: 10.1371/journal.pone.0034589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yap MS, Tang YQ, Yeo Y, Lim WL, Lim LW, Tan KO, Richards M, Othman I, Poh CL, Heng BC. Pluripotent human embryonic stem cell derived neural lineages for in vitro modelling of enterovirus 71 infection and therapy. Virol J. 2016;13:5. doi: 10.1186/s12985-015-0454-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Falah N, Montserret R, Lelogeais V, Schuffenecker I, Lina B, Cortay J-C, Violot S. Blocking human enterovirus 71 replication by targeting viral 2A protease. J Antimicrob Chemother. 2012;67:2865–2869. doi: 10.1093/jac/dks304. [DOI] [PubMed] [Google Scholar]
- 91.Falah N, Violot S, Décimo D, Berri F, Foucault-Grunenwald M-L, Ohlmann T, Schuffenecker I, Morfin F, Lina B, Riteau B, et al. Ex vivo and in vivo inhibition of human rhinovirus replication by a new pseudosubstrate of viral 2A protease. J Virol. 2012;86:691–704. doi: 10.1128/JVI.05263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deszcz L, Seipelt J, Vassilieva E, Roetzer A, Kuechler E. Antiviral activity of caspase inhibitors: effect on picornaviral 2A proteinase. FEBS Lett. 2004;560:51–55. doi: 10.1016/S0014-5793(04)00069-9. [DOI] [PubMed] [Google Scholar]
- 93.Molla A, Hellen CU, Wimmer E. Inhibition of proteolytic activity of poliovirus and rhinovirus 2A proteinases by elastase-specific inhibitors. J Virol. 1993;67:4688–4695. doi: 10.1128/JVI.67.8.4688-4695.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang P, Mueller S, Morais MC, Bator CM, Bowman VD, Hafenstein S, Wimmer E, Rossmann MG. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc Natl Acad Sci USA. 2008;105:18284–18289. doi: 10.1073/pnas.0807848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Anasir MI, Poh CL. Structural vaccinology for viral vaccine design. Front Microbiol. 2019;10:738. doi: 10.3389/fmicb.2019.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ji X, Li Z. Medicinal chemistry strategies toward host targeting antiviral agents. Med Res Rev. 2020;40:1519–1557. doi: 10.1002/med.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin K, Gallay P. Curing a viral infection by targeting the host: the example of cyclophilin inhibitors. Antivir Res. 2013;99:68–77. doi: 10.1016/j.antiviral.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pessi A, Langella A, Capitò E, Ghezzi S, Vicenzi E, Poli G, Ketas T, Mathieu C, Cortese R, Horvat B, et al. A general strategy to endow natural fusion-protein-derived peptides with potent antiviral activity. PLoS ONE. 2012;7:e36833. doi: 10.1371/journal.pone.0036833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vilas Boas LCP, Campos ML, Berlanda RLA, de Carvalho Neves N, Franco OL. Antiviral peptides as promising therapeutic drugs. Cell Mol Life Sci. 2019;76:3525–3542. doi: 10.1007/s00018-019-03138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feng Z, Xu B. Inspiration from the mirror: D-amino acid containing peptides in biomedical approaches. Biomol Concepts. 2016;7:179–187. doi: 10.1515/bmc-2015-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brinckerhoff LH, Kalashnikov VV, Thompson LW, Yamshchikov GV, Pierce RA, Galavotti HS, Engelhard VH, Slingluff CLJ. Terminal modifications inhibit proteolytic degradation of an immunogenic MART-1(27–35) peptide: implications for peptide vaccines. Int J Cancer. 1999;83:326–334. doi: 10.1002/(sici)1097-0215(19991029)83:3<326::aid-ijc7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 102.Lembo D, Donalisio M, Civra A, Argenziano M, Cavalli R. Nanomedicine formulations for the delivery of antiviral drugs: a promising solution for the treatment of viral infections. Expert Opin Drug Deliv. 2018;15:93–114. doi: 10.1080/17425247.2017.1360863. [DOI] [PubMed] [Google Scholar]
- 103.Andersson L, Blomberg L, Flegel M, Lepsa L, Nilsson B, Verlander M. Large-scale synthesis of peptides. Biopolymers. 2000;55:227–250. doi: 10.1002/1097-0282(2000)55:3<227::AID-BIP50>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 104.Gaglione R, Pane K, Dell’Olmo E, Cafaro V, Pizzo E, Olivieri G, Notomista E, Arciello A. Cost-effective production of recombinant peptides in Escherichiacoli. N Biotechnol. 2019;51:39–48. doi: 10.1016/j.nbt.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 105.Cao J, de la Fuente-Nunez C, Ou RW, Torres MDT, Pande SG, Sinskey AJ, Lu TK. Yeast-based synthetic biology platform for antimicrobial peptide production. ACS Synth Biol. 2018;7:896–902. doi: 10.1021/acssynbio.7b00396. [DOI] [PubMed] [Google Scholar]
- 106.Tiwari S, Dhole TN. Assessment of enteroviruses from sewage water and clinical samples during eradication phase of polio in North India. Virol J. 2018;15:157. doi: 10.1186/s12985-018-1075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.