Abstract
Data processing and learning has become a spearhead for the advancement of medicine, with pathology and laboratory medicine has no exception. The incorporation of scientific research through clinical informatics, including genomics, proteomics, bioinformatics, and biostatistics, into clinical practice unlocks innovative approaches for patient care. Computational pathology is burgeoning subspecialty in pathology that promises a better-integrated solution to whole-slide images, multi-omics data, and clinical informatics. However, computational pathology faces several challenges, including the ability to integrate raw data from different sources, limitation of hardware processing capacity, and a lack of specific training programs, as well as issues on ethics and larger societal acceptable practices that are still solidifying. The establishment of the entire industry of computational pathology requires far-reaching changes of the three essential elements connecting patients and doctors: the local laboratory, the scan center, and the central cloud hub/portal for data processing and retrieval. Computational pathology, unlocked through information integration and advanced digital communication networks, has the potential to improve clinical workflow efficiency, diagnostic quality, and ultimately create personalized diagnosis and treatment plans for patients. This review describes clinical perspectives and discusses the statistical methods, clinical applications, potential obstacles, and future directions of computational pathology.
Subject terms: Preventive medicine, Bioinformatics
Data processing and learning has become a spearhead for the advancement of medicine. Computational pathology is burgeoning subspecialty that promises a better-integrated solution to whole-slide images, multi-omics data and clinical informatics as innovative approach for patient care. This review describes clinical perspectives and discusses the statistical methods, clinical applications, potential obstacles, and future directions of computational pathology.
Introduction
Artificial intelligence (AI) refers to the simulation of the human mind in computer systems that are programmed to think like humans and mimic their actions such as learning and problem-solving. AI should be able to perform tasks that normally require human intelligence, such as visual perception, decision-making, and communication. AI-based computational pathology as an emerging discipline has recently shown great promise to increase both the accuracy and availability of high-quality health care to patients in many medical fields. The primary forces and limitations in this field are: (1) a shortage of experienced pathologists and the limitation of global health care resources [1]; (2) the ever increasing amount of health data available, including digital images, omics, clinical records, and patient demographic information, being generated through the process of patient care [2]; (3) the increased complexity that is created in managing and integrating the data across different sources in order to maximize patient care; and (4) machine learning-based algorithms need to be efficiently harnessed in order to process and understand the big data [3]. AI technologies have the ability to handle the gigantic quantity of data created throughout the patient care lifecycle to improve pathologic diagnosis, classification, prediction, and prognostication of diseases.
The most important advantage of the computational pathology is to reduce errors in diagnosis and classification. The Camelyon Grand Challenge 2016 (CAMELYON16 challenge), is a worldwide machine learning-based program to evaluate new algorithms for the automated detection of cancer in hematoxylin and eosin (H&E)-stained whole-slide imaging (WSI), has achieved encouraging results with a 92.4% sensitivity in tumor detection rate. In contrast, a pathologist could only achieve 73.2% sensitivity [4]. Computational pathology has the potential to transform the traditional core functions of pathology and not just growing sub-segments such as digital pathology, molecular pathology, and pathology informatics [5, 6]. Computational pathology aims to improve diagnostic accuracy, optimize patient care, and reduce costs by bringing global collaboration. As the rapid technological advancement drives individualized precision medicine [7], computational pathology is a critical factor in achieving this goal.
Digital pathology, machine learning, and computational pathology
The development of brightfield and fluorescent slide scanners made possible the virtualizing and digitalizing the whole glass slides [8]. Digital pathology includes the process of digitizing histopathology, immunohistochemistry or cytology slides using whole-slide scanners as well as the interpretation, management, and analysis of these digitized whole-slide images using computational approaches. The digital data of the slides can be stored in a central cloud-based space allowing for remote access to the information for manual review by a pathologist or automated review by a data algorithm. It makes AI, a branch of computational science which generates the data algorithms, to be applied in pathology possible [9]. Based on the degree of intelligence, AI can currently be divided into two major categories: weak AI and strong AI (Table 1). Weak AI, also known as artificial narrow intelligence, refers to the classification of data based on a well-established statistic model that has already been trained to perform specific tasks [10]. In contrast, strong AI, also known as artificial general intelligence (AGI), can create a system, which can function intelligently and independently by executing machine learning from any available normalized data.
Table 1.
List of terms and abbreviations appearing in this paper.
| Term | Abbreviation | Explanation |
|---|---|---|
| Artificial intelligence | AI | A branch of computer science dealing with tasks that normally require human intelligence. |
| Machine learning | ML | A branch of AI in which statistical algorithms establish their own patterns by being exposed to representative data to interpret and act on new data. |
| Support vector machine | SVM |
A statistical method to distinguish between classes of data with the widest possible margin by training. The number of trainable parameters: 10–100. |
| Random forest | RF | A statistical method using a network of decision trees to classify the data. |
| k-nearest neighbor | k-NN | A statistical method for data classification and regression based on the number of k neighbors. |
| Deep neural networks | DNNs | Also called deep learning (DL), which is a subset of machine learning using complex multilayered architectures including multiple hidden layers and a large number of nodal connections. |
| Artificial neural networks | ANNs |
A set of layered, interconnected artificial neurons based on deep neural networks to explore higher level features, mimicking biological brain. The number of trainable parameters ≥100,000. |
| Recurrent neural networks | RNNs | A type of ANNs but the connections between nodes within a hidden layer are cycled. |
| Convolutional neural networks | CNNs | A type of ANNs particularly designed for machine vision field. CNNs have been most commonly applied to analyze images such as image recognition and classification. |
| Computer vision | CV | Quickly and accurately parsed trends and patterns from digital images, mimicking biological vision. |
| Machine vision | MV | Shares similar benefits in terms of speed and accuracy of CV, but the applications are mainly focused on efficiency such as automatic inspection, robotic guidance process. |
| GoogLeNet | – | A CNNs model that was created by Google for computer vision and classification. |
| FaceNet | – | A CNNs model for face recognition and classification. |
| AlexNet | – | A CNNs model was designed by Alex Krizhevsky and achieved a top 5 of the ImageNet Large Scale Visual Recognition Challenge on 2012. |
| VGGNet | – | A CNNs model was created by Visual Geometry Group (VGG) at University of Oxford and achieved first runner-up of the ImageNet Large Scale Visual Recognition Competition on 2014. |
| Area under receiver operating curve | AUC | Performance measured by the area under the receiver operating characteristic curve (from 0.5 (lowest) to 1 (highest)). |
Generally, machine learning is an AI process to allow a computer system to automatically learn and improve from the data set by itself and to solve problems without being programmed during the process. Machine learning is an advanced branch of AGI using a large amount of initial data, training set, to build statistic algorithms to interpret and act on new data later on [11]. At present, various machine learning-based approaches have been developed and tested in pathology to assist pathologic diagnosis using the basic morphology pattern such as cancer cells, cell nuclei, cell divisions, ducts, blood vessels, etc. [12]. Deep learning (also known as deep structured learning) is a subfield of machine learning based on artificial neural networks (ANNs) in which the statistic models are established from input training data [13]. Deep neural networks provide architectures for deep learning. The ANNs can perform its own determination as to whether its interpretation or prediction is correct, resembling a biological complex neural network of the human brain [14]. ANNs are comprised of three functional layers of artificial neurons, known as “nodes”, which include an input layer, multiple hidden layers, and an output layer. The artificial neurons are connected to each other in the ANNs and the strength of their connections is known as “weights”. The connections between artificial neurons in the ANNs are assessed using statistic methods, including clustering algorithms, K-nearest neighbor, support vector machines (SVM), and logistic regressions [15]. The involved artificial neurons, which are related to the output event, and their associated connections, which bear different “weight”, need to be trained by qualified big data set to achieve an optimized algorithm for specific tasks (Fig. 1). The convolutional neural networks are a type of deep multilayer neural networks particularly designed for visual image. It employs convolutional kernels, a set of learnable filters, to build up a pooling layer that can effectively reduce the dimensions of the image data while still retaining its characteristics (Fig. 2). By flattening an image, removing or reducing the dimensions, convolutional kernels act as a preprocess treatment that then allows for computer vision and machine vision models to process, analyze, and classify the digital images, or parts of the image, into known categories.
Fig. 1. The basic structure of a deep neural network.
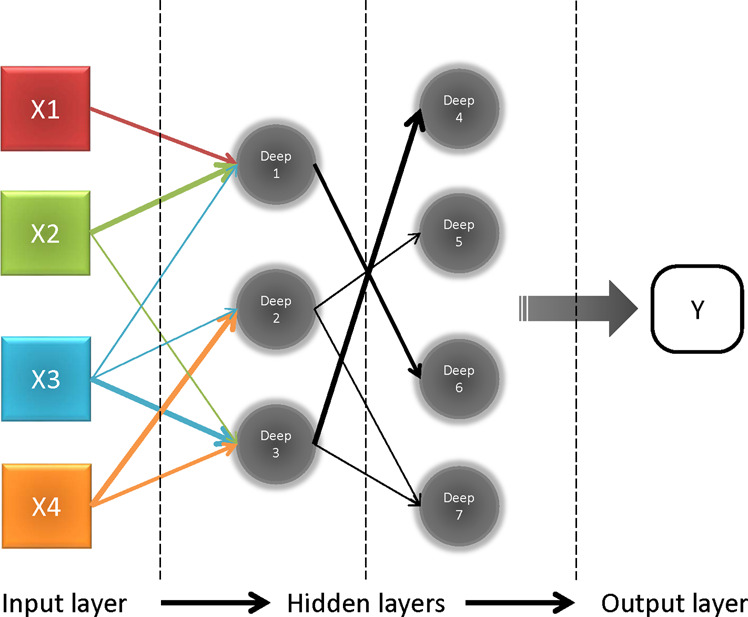
Each input value is assigned to each node from “X1–3” to “Xn” in the input layer (left). Nodes labeled “deep 1–7” are hidden layers behind the input layer. The values of hidden layers are not directly visible and are sent to the output layer after processed. Arrows connecting each node represent the direction and weight from previous layers. Both weights and nodes impact the network in generating an output (Y).
Fig. 2. The principle of convolutional neural networks.
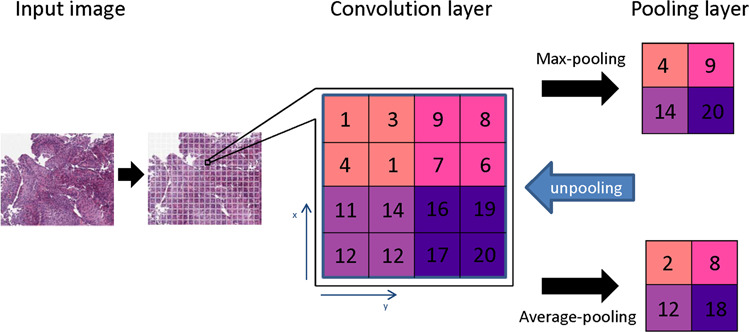
The input image is converted to numerical data (1–20) as the convolution layer. The convolutional neural network generates a pooling layer to reduce the dimensions of the image data as well as retain its characteristics for the statistic modeling. Several types of pooling methods including max pooling, which returns the maximum value from the portion of the image, and average pooling, which returns the average of all the values from the portion of the image. In addition, max pooling also performs de-noising along with dimensionality reduction, which improved analysis and accuracy.
With slide scanning technology getting faster and more reliable, a larger volume of WSI data becomes available to train and validate convolutional neural network models. In combination with clinical information, biomarkers, and multi-omics data, computational pathology will become part of the new standard of care [16]. Computational pathology not only facilitates a more efficient pathology workflow, but also provides a more comprehensive and personalized view, enabling pathologists to address the progress of complex diseases for better patient care [17].
Algorithm training progress
Case selection
Patients’ selection is the initial step to train the algorithm (Fig. 3). Both training set and validation set must include all sample types or variants, which are related to the subject of diseases including stages, grades, histologic classification, complication, etc. to eliminate false-negative and false-positive scenarios. Still very much a machine driven process, algorithms have no way to recognize the variants that has not been included in the training set. The criteria for the samples and subsequent slide selection for the learning set need to be established by experienced pathologists alongside a computational team. Confounding variables have to be isolated and removed. For example, the patients with other medical conditions who may interfere with the outcome. In addition, inadequate slide preparation including blurred vision, over- or under-staining, air-bubbles, and folded tissue can produce inaccurate resulting algorithms. The comprehensive initial and follow-up clinical information, as well as laboratory results, should be collected and included. The more relevant the information included, the more accurate the resulting algorithm.
Fig. 3. Flow chart of algorithm training.
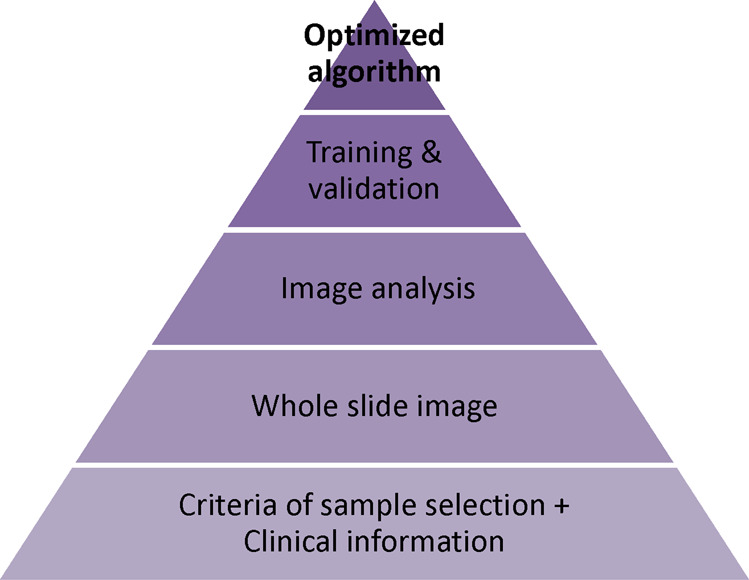
The process of creating an algorithm is divided into four necessary phases. The initial phase is to collect applicable samples along with clinical information. Next phase is to create whole-slide images with annotation. Based on the image analysis data, an algorithm was developed and trained by both the training set and the independent validation set.
Whole-slide imaging (WSI)
Several slide scanning systems for whole-slide imaging have been approved by the US Food and Drug Administration (FDA) to be used in clinical settings (Table 2) [18]. The first FDA-approved Ultra-Fast Scanner, the Philips IntelliSite Pathology Solution (PIPS), has a resolution of 0.25 μm/pixel, scanning speed of 60 s for a 15 × 15-mm scan area and scanning capacity of 300 slides in one load [19]. The Aperio AT2 DX System from Leica Biosystems has 400 slide capacity for brightfield and fluorescent slides [20]. File sizes of digital images at applicable resolution vary depending on the scan area on the glass slides. In general, pathology images are tremendously large, in the range of 1–3 GB per image. Therefore, it requires a high-capacity and fast digital working computer. Furthermore, the number of slides needed to achieve a clinically accepted algorithm may vary by tissue type and diagnosis. Campanella et al. showed that at least 10,000 slides are necessary for training to reach a good performance. The authors also observed the discrepancy of the prediction between Leica Aperio and PIPS and found that brightness, contrast, and sharpness affect the prediction performance [21].
Table 2.
Whole-slide images systems.
| Device | Brightfield scanning technology | Fluorescence scanning technology | Company | Slide loading capacity | Brightfield scanning speed | Fluorescence scanning speed | Highest throughput slides/h | Resolution (μm/pixel) ×20/×40 objective |
|---|---|---|---|---|---|---|---|---|
| VS200 | 2/3-inch CMOS, 3.45 × 3.45 μm pixel size | 1-inch CMOS, 3.45 × 3.45 μm pixel size | Olympus | 210 | 80 s | – | – | 0.274/0.137 |
| Aperio AT2 | TDI line scan | NO | Leica | 400 | 72 s (20×)/3 min (40×) | NO | 50 | 0.50/0.25 |
| HLM8 | – | – | Histo-Line Laboratories | – | – | – | – | – |
| PathFusion | 5 MP CMOS Color | ASI | 99 | – | – | – | – | |
| TissueScope LE120 | 0.75 | NO | HURON Digital Pathology | 120 | <1 min per slide | NO | – | 0.4/0.2 |
| Axiocam 208 color | NO | 1/2.1-inch Sony CMOS, 1.85 × 1.85 μm pixel size | ZEISS | – | – | – | – | – |
| Axiocam 202 mDavidono | 1/2.3-inch Sony CMOS, 5.86 × 5.86 μm pixel size | NO | ZEISS | – | – | – | – | – |
| VENTANA iScan HT | – | Integrated LED | ROCHE | 160 | <2 min per slide | – | – | – |
| ProScanner APro 5 | – | – | AMOS | – | – | – | – | – |
| 3DHISTECH Pannoramic 250 FLASH III | 12 MP 12-bit camera with Xenon Flash illumination | Additional 4.2 MP 16-bit camera with 6-channel LED | Epredia | 250 | 35 s (20×)/1 min 35 s (40×) |
5 min @ 31× 15 min @ 62× |
60 | 0.242/0.121 |
| Philips IntelliSite Pathology Solution | TDI line scanning | – | Philips | 300 | 60 s | – | – | 0.25 |
CMOS complementary metal oxide semiconductor, TDI time delay and integration.
Image analysis and automation
For digital slide analysis, Senaras et al. [22] described a novel deep-learning framework, called DeepFocus, which enables the automatic identification of blurry regions in digital slides for immediate re-scan in order to improve image quality for pathologists and image analysis algorithms. Janowczyk et al. [23] presented an open-source tool called HistoQC to assess color histograms, brightness, and contrast of each slide and to identify cohort-level outliers (e.g., darker or lighter stain than other slides in the cohort). These methods play an essential role in the quality control of whole-slide images to standardize the quality of images in computational pathology. Due to improvements in various smart image-recognition algorithmic discriminators, based on high-capacity deep neural network models [24], the pathologist can be released from extensive manual annotations for each whole-slide images at the pixel level so that they can focus other parts of the clinical workflow. The patch-based whole-slide images (224 × 224 to 256 × 256) have been widely used in many machine learning domains to train classifiers for diagnostic or prognostic tasks. For example, Campanella et al. [21] employed multiple instance learning (MIL) approaches with “bag” and “instance” based on convolutional neural networks and recurrent neural networks to classify the prostate cancer images of H&E slides. Kapil et al. [25] applied deep semi-supervised architecture and auxiliary classifier generative adversarial networks, including one generator network and one discriminator network, to automatically analyze the PD-L1 expression in immunohistochemistry slide of late stage non-small cell lung cancer needle biopsies. Barker et al. [26] revealed an elastic net linear regression model and weighted voting system to differentiate glioblastoma multiforme and lower-grade glioma with an accuracy of 93.1%.
Pathologist-centered medical system
Pathologist-centered medical system
Although most AI research is still focused on the detection and grading of tumors in digital pathology and radiology, computational pathology is not limited to the detection of a morphological pattern. It can also contribute to the complex process of analysis and judgment using demographic information, digital pathology, -omics, and laboratory results [27]. Therefore, AI has the potential to contribute to nearly all aspects of the clinical workflow, from more accurate diagnosis to prognosis, and individualized treatment. Multiple sources of clinical data [28] are incorporated into mathematic models to generate diagnostic inferences and predictions, to enable physicians, patients, and laboratory personnel to make the best possible medical decisions [29]. For example, deep neural networks have been applied to automated biomarker assessment of breast tumor images, such as HER2, ER, and Ki67 [30]. Hamidinekoo et al. [31] created a novel convolutional neural network-based mammography–histology–phenotype–linking–model to connect and map the features and phenotypes between mammographic abnormalities and their histopathological representation. Mobadersany et al. [32] developed a genomic survival convolutional neural network model to integrate information from both histology images and genomic data to predict time-to-event outcomes and demonstrated the prediction accuracy surpassed the current clinical paradigm for predicting the overall survival of patients diagnosed with glioma.
As electronic health record (EHR) systems enable us to collect medical data such as age, race, gender, social history, and clinic history, applying these data as independent factors of a particular disease to an appropriate mathematic algorithm becomes feasible [33, 34]. These integrated data allow pathologists to gain deeper insights and to switch between different algorithms of treatment at different stages of the disease and/or for different statuses of the patient. As the health-related apps on mobile devices and smart personal trackers become popular, direct access to continuous real-time health information, such as temperature, heart rate, respiratory rate, electrocardiogram, body mass index, blood glucose, and blood oxygen content, can be recorded into individual health data. These data can then be incorporated into the EHR and laboratory information systems (LIS) [35] to reintegrate into a virtualized and digitalized person, which was not possible previously and was beyond what the human brain alone can accomplish [36] (Fig. 4). This new system of data-driven care requires the pathology, as a cornerstone of modern medicine, to integrate data, algorithms, and analytics to deliver high-quality and efficient care. The combination of computational pathology and big data mining offers the potential to create a revolutionary way of practicing evidence-based, personalized medicine.
Fig. 4. Pathologist-centered medical system.
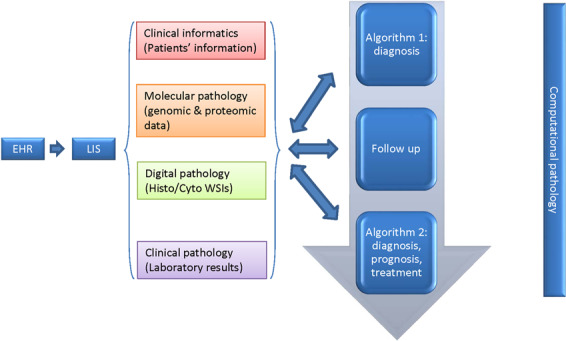
Clinical information from EHR, -omics data from molecular pathology, WSIs from digital pathology, and results from clinical laboratories aggregated into LIS to create “algorithm 1” for diagnosis. The updated disease-related data during follow-up are integrated into the previous data to build the “algorithm 2” over time for improved patient care.
Global pathology service model
Three essential advancements happened in recent years: the possibility to store a great amount of data from network-attached storage to cloud storage, the growing speed of network from WIFI-6 to 5 G, and high-performance central processing unit (CPU) and graphics processing unit. These technological improvements not only enhance people’s daily life, but also have a great impact on medicine, especially digital and computational pathology [37] (Fig. 5). Together with the surging development of network and information technology, these technologic improvements allow for the centralization of medical and computing resources—with the benefit of larger sample data volume for optimization of algorithms. Furthermore, the central cloud-based AI laboratory and data bank of digital and computational pathology make the global network of computational pathology possible. In local laboratories or centralized scanning centers, histology slides can be converted to whole-slide images and numerical data. These data can then be transferred to the central laboratory together with EHR data and multi-omics data for further analysis [38] (Fig. 6). Patients in different geographic areas around the world can benefit from more efficient and effective diagnosis, treatment, and follow-up. In the meantime, pathologists are able to access the information they need to care for patients or to collaborate with specialists anytime and anywhere. Deep-learning platforms have the potential to facilitate the discovery of more complicated or subtle connections and to help pathologists make the best clinical decisions to meet every patient’s needs.
Fig. 5. The relation between different levels of artificial intelligence and the four bottlenecks that are facing currently.
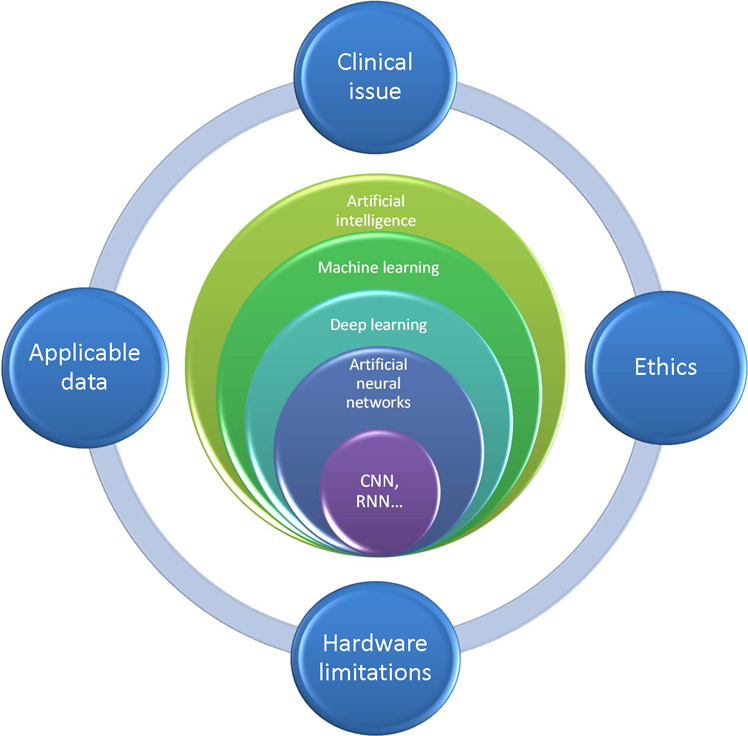
The four challenges are experienced computational clinicians who are capable of developing algorithms of particular clinical issues, hardware limitations (i.e., cloud storage, computational capacity, network speed), qualified applicable data, and ethical issues.
Fig. 6. Global pathology service model.
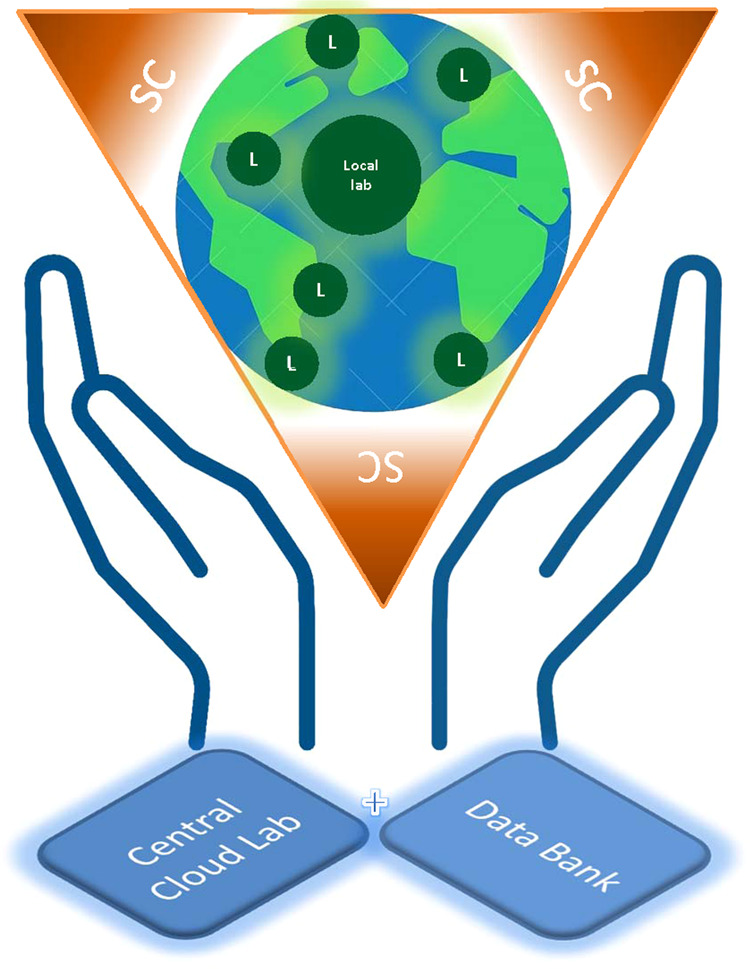
Each small green circle labeled “L” represents a local laboratory where the slides are scanned. Alternatively, slides can be scanned at a centralized scanning center (SC) where many local laboratories can send their slides for scanning. A central cloud laboratory with a large data storage and high capacity of computation will integrate, analyze, and store WSI data together with other medically related data.
Examples of pathology AI
Increasingly, AI detection is being applied to different subspecialties with various sample types [39]. Early reports on accuracy have shown to be promising and that the AI-assisted systems have the potential to classify accurately at an unprecedented scale [40] and lay the foundation for the deployment of computational pathology in nearly all subspecialties [41].
Prostate cancer
Campanella et al. validated a high-capacity deep neural network-based algorithm to analyze image classification and categorization of 44,732 whole-slide images across three different cancer types, including prostate cancer, basal cell carcinoma, and breast cancer metastases to axillary lymph nodes. In terms of whole-slide images, they found that ×5 magnification has higher accuracy. They trained a statistic model with MIL-based tile classifier for each tissue type and achieved area under receiver operating curve (AUC) above 0.98 for all cancer types. Its clinical application would allow pathologists to exclude 65–75% of slides while retaining 100% sensitivity [21]. Wildeboer et al. discussed deep-learning techniques based on different imaging sources including magnetic resonance imaging, echogenicity in ultrasound imaging, and radio density in computed tomography as computer-aided diagnostic tools for prostate cancer. They found that the algorithm of convolutional neural network architecture performed equal or better than SVM or random forest classifiers in machine learning [42].
Colorectal cancer
Korbar et al. developed multiple deep-learning algorithms, modified version of a residual network architecture, which can accurately classify whole-slide images of five types of colorectal polyps, including hyperplastic, sessile serrated, traditional serrated, tubular, and tubulovillous/villous polyps. Among 2074 of images, 90% of them were used for model training and the remaining 10% of images were assigned to the validation set. The overall accuracy for classification of colorectal polyps was 93% (confidence interval (CI) 95%, 89.0–95.9%) [43]. Bychkov et al. combined convolutional neural networks and recurrent neural network architectures to predict colorectal cancer outcomes based on tissue microarray (TMA) samples from 420 colorectal cancer patients. Their results show that the AUC of deep neural network-based outcome prediction was 0.69 (hazard ratio, 2.3; CI 95%, 1.79–3.03). For comparison, pathology experts performed inferiorly on both TMA samples (HR, 1.67; CI 95%, 1.28–2.19; AUC, 0.58) and whole-slide level (HR, 1.65; CI 95%, 1.30–2.15; AUC, 0.57), which implied that deep neural networks could extract more prognostic information from the tissue morphology of colorectal cancer than an experienced pathologist [44].
Breast cancer
Wang et al., the team of winner of competitions in the CAMELYON16 challenge, used input 256 × 256 pixel patches from positive and negative regions of the whole-slide images of breast sentinel lymph nodes to train various classification models including GoogLeNet Patch, AlexNet, VGG16, and FaceNet. The patch classification accuracy is 98.4, 92.1, 97.9, and 96.8% separately. Among the algorithms, GoogLeNet has the best performance and is generally faster and more stable, which achieved AUC of 0.925 for whole-slide images classification. With the assistance of deep-learning system, the accuracy of pathologist’s diagnoses improved significantly as the AUC increased from 0.966 to 0.995, representing ~85% reduction of human error rate [45]. Furthermore, the open resource of a data set of annotated whole-slide images for CAMELYON16 and CAMELYON17 challenges enable testing of new machine learning and image analysis strategies for digital pathology [46].
Cytology
Martin et al. applied convolutional neural networks for classifying cervical cytology images into five diagnostic categories, including negative for intraepithelial lesion or malignancy, atypical squamous cells of undetermined significance, low-grade squamous intraepithelial lesion, atypical squamous cells cannot exclude how-grade squamous intraepithelial lesion and high-grade squamous intraepithelial lesion, and achieved accuracies of 56, 36, 72, 17, and 86% separately, which implies convolutional neural networks are able to learn cytological features [47]. In another cytopathology study, the authors used morphometric algorithm and semantic segmentation network based on VGG-19 to classify urine cytology whole-slide images according to Paris System for Urine Cytopathology and achieved a sensitivity of 77%, false-positive rate of 30% and AUC of 0.8 [48].
COVID-19
During the outbreak of COVID-19, telemedicine and computer-aided medicine are rapidly entering the market in many countries. Highly contagious nature, systemic risks, and social isolation brought unexpected challenges to traditional medicine. Applying AI-based computer-aided medicine along with clinical data from EHR, including individuals’ clinical risk factors of human-to-human interactions and a variety of diverse social data, may provide a quick control of this public health emergency with a better quality and safety [49].
Several AI companies have been working on products to address the COVID-19 pandemic. For example, JLK Inspection, Korea (http://www.jlk-inspection.com/#/medical/main) is integrating the reverse transcription-polymerase chain reaction (RT-PCR) results, imaging tests, and their universal AI platform, AIHuB, to provide COVID-19 diagnosis. Persivia, Massachusetts (https://persivia.com/covid-19-detection/) announced a new surveillance module based on their Soliton AI engine to identify and alert patients who are presumed positive for COVID-19. Biofourmis, Massachusetts (https://www.biofourmis.com/) developed an analytic platform called Biovitals Sentinel, which provides 24/7 remote monitor to identify early clinical deterioration and enable earlier interventions. Schaar et al. described that machine learning could significantly enhance both the efficiency and effectiveness of randomized clinical trials for COVID-19. It has the capability to speed up recruiting subjects from identifiable subgroups and assigning subjects to treatment or control groups as well as significantly reducing error and requiring many fewer patients [50].
Challenges
Although machine learning has produced promising results and provided many benefits in computational pathology, the following limitations need to be addressed before deep machine learning can be implemented in the clinical setting.
Standardization and normalization
The successful adaptation of whole-slide images in digital pathology heavily depends on each step of high-quality pathology slide preparation, including embedding, cutting, staining, and scanning. Folded tissue section during cutting, staining variation and the presence of air bubble during covering slide as well as different settings of brightness, intensity disparity, average color, and boundary intensity during scanning can cause unreliable raw data and produce inaccurate results [8, 51]. The protocols and systemic quality controls need to be standardized to reduce the system errors and random errors resulted from different instruments since a single noise in big data can cause misclassification and change the slide prediction, possibly resulting in a large number of false positives or negatives. The data used to generate algorithms are analyzed by different models by different developers. The larger the accumulated data, the more accurate algorithms, especially for rear diseases and specific small populations. A standard data format and normalization method of data analysis should be engaged to merge consecutive data sets from different resources and train them into one algorithm since different data resources may cause variation in classification accuracy in practice. The digital imaging and communications in medicine (DICOM) developed standards for medical images including radiology (www.dicomstandard.org). It defines the formats for medical images that can be exchanged with the data and quality necessary for clinical use. DICOM Standard now provides support for WSI, by incorporating a way to handle tiled large images as multi-frame images and multiple images at varying resolutions (http://dicom.nema.org/Dicom/DICOMWSI/).
The role in the computational pathology
Computational pathology is not only important in medical research, but also needed to address clinical questions in practice [52]. To achieve this goal, a team of experts in different fields is needed to participate in computational pathology projects, including data scientists such as statisticians and bio-informaticians for algorithm design and architect as well as engineer for the construction of physical environments and maintaining hardware (Fig. 7). Among them, the pathologists play a critical role to introduce medical questions and clinical applications to the developer team and to trigger the downstream industry development [53]. The new role of the pathologist in computational pathology not only requires solid clinical knowledge and experience, but also requires the knowledge of statistical analysis and data mining to bridge the gap between clinical medicine and AI so that when a disease suddenly happens, or a new biomarker is discovered, the pathologist can quickly react to the opportunity to either create a new algorithm or optimize an existing algorithm to assist the clinician [54]. Furthermore, pathologists who understand the potential problems that may occur during the data analysis can address the clinical problems clearly with computational thinking. Good communication among the team members can help to design a more efficient algorithm because the algorithms with different coding approaches will consume different amount of computing resources and time, especially for big data, although they generate the same end result. Furthermore, AI pathology provides excellent tools for experimental pathology by the integration of morphology at organ, histology, cell, and organelle levels with molecular details of omics data.
Fig. 7. Computational pathology team.
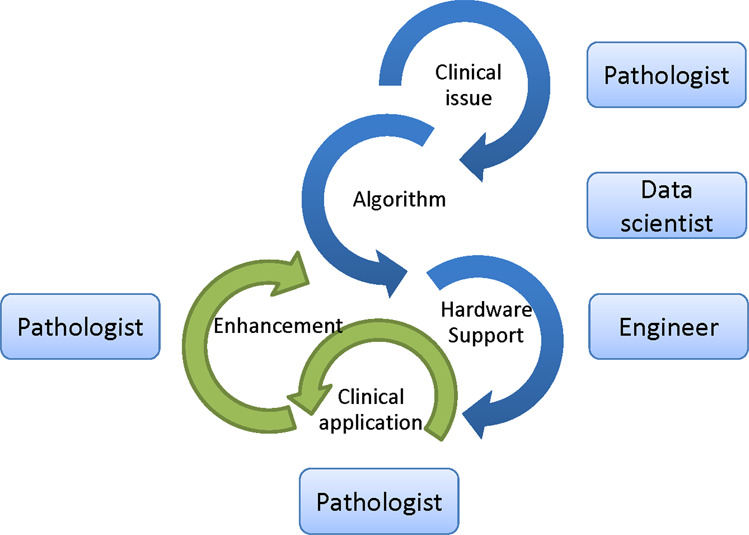
A computational pathology developer team includes pathologists to establish a clinically relevant issue, data scientists to develop and train the algorithm, and engineer to support the operating environment. During actual clinical practice, pathologists play a pivotal role in applying and monitoring the algorithm and relay feedback to the team to keep optimizing it.
Hardware limitations
The accuracy of applications in the computational pathology heavily depends on large amounts of data, reliable hardware and software, and a supportive network environment. Large image file size (around 3 GB per slide scan) requires significant big storage space with backup capability in both local and cloud. In addition, deep machine learning solutions, especially when applying analysis of pathology images, heavily depend on graphics processing unit, which is a chip on the computer’s graphics card for rapidly manipulating graphics and processing images [55]. A powerful graphics processing unit can provide significant performance enhancement alongside the CPU to boost computing capacity and reduce turn-around time. For implementation of either data transmission or cloud-based image processing, data bandwidth of both intranet and internet becomes a bottleneck, which limits the speed of upload and download [56]. Only if all these related elements in a network have been developed and evolved into a robust system (Fig. 5), computational pathology can move forward to assist resolving more complex and multifaceted medical and clinical questions and research tasks. Finally, the clearance as medical devices (510k) by FDA is critical to ensure the clinical reliability and the acceptance by pathology community.
Ethics
In the new era of computation-driven decision-making processes based on AI and machine learning, computational pathology must involve in more complicated interactions of massive information from clinical history, omics data, living environment to social habits [57]. It is very likely that the experts involved in these decision-making processes will no longer be exclusively pathologists. Instead, the decision-making panel will include other experts such as data statisticians and bio-informaticians, which may raise ethical concerns [58]. A continuous massive, sensitive health data transfer among clinics, laboratories, and data banks can enable higher precision medicine, but at the same time increases the security vulnerability. Policies around the strict protection of patient privacy and personal data creates an obstacle for computational pathology to access the health databases need to create more comprehensive training data sets. General Data Protection Regulation was enacted in May 2018 in Europe to impose new responsibilities on organizations who process the data of European Union citizens for scientific research [59]. This concept highlights the proportionate approach to regulate computational pathology-related security and ethical issues while not limiting innovation unduly, which is difficult but critical.
Future prospects
Technological innovation in health care is growing at an increasingly fast pace and has been integrated into both our daily lives, such as smart healthy tracker, and diagnostic algorithm in medical practice [60]. With the rapid development of digital pathology, molecular pathology, and informatics pathology, computational pathology is increasingly involved in many subspecialties such as pulmonary, renal, gastrointestinal, neurology, and gynecology pathology. We believe the initial phase of AI will start with specific tasks such as the diagnosis of a particular cancer and classification of tissue types, which require limited and simple criteria [61]. For example, the common subtypes and variants of benign and malignant neoplasm in prostate should be included in the training and validation to ensure the feasibility of daily pathology practice. As a result of more data collection and more powerful computing capacity over time, the clinical applications of AI will be broader and the number of nonspecific cases in the gray zone or with red flags classified by AI for manual review will be decreased.
The growing medical data, including genomics, proteomics, informatics, and whole-slide images [62], is expected to integrate together to become a data-rich pathomics and lead to rapid development and prosperity of an AI-assisted computational pathology. Although many challenges remain, the computational pathology with the deployment of digital pathology technology and statistic algorithm will continue to improve clinical workflows and collaboration among pathologist and other members in the patient care team. The improved infrastructure of the network environment, the enhanced computing capacity, and broad integration of informatics have ushered in new horizons for both computational pathology and collaborative pattern, which make data travel and cloud-based central laboratory and data bank to deliver better care for patients at lower costs possible.
Conclusion
In the new era of deep learning-assisted pathology, the data banking, integration, and cloud laboratory are becoming an essential part of daily practice of pathology. Furthermore, the pathologists, data scientists, and industry are starting to incorporate the genomics, proteomics, bioinformatics, and computer algorithms into a large amount of complex clinical information. Through this process, the computational pathology can contribute valuable insights to the diagnosis, prognosis, and treatment of disease ultimately. Although many technical and ethical challenges need to be addressed, computational pathology as a synergistic system will lead to a boosting workflow, enabling clinical teams to share and analyze image data in a broader platform.
Currently, deep learning has been applied to solve more and more specialized tasks in medicine. Several studies discussed above showed that algorithm assistance has the potential to not only improve the sensitivity and accuracy of the diagnoses but also improve turn-around time. Moreover, according to Sarwar’s et al. [63] study, around 75% of pathologists across 59 countries in the world are interested and excited about using AI as a diagnostic tool. Finally, despite the challenges and obstacles, the potential of computational pathology which will change and improve the current health care system is promising and exciting.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.George MR, Johnson KA, Gratzinger DA, Brissette MD, McCloskey CB, Conran RM, et al. Will I need to move to get my first job?: geographic relocation and other trends in the pathology job market. Arch Pathol Lab Med. 2020;144:427–34. doi: 10.5858/arpa.2019-0150-CP. [DOI] [PubMed] [Google Scholar]
- 2.Goldenberg SL, Nir G, Salcudean SE. A new era: artificial intelligence and machine learning in prostate cancer. Nat Rev Urol. 2019;16:391–403. doi: 10.1038/s41585-019-0193-3. [DOI] [PubMed] [Google Scholar]
- 3.Parwani AV. Next generation diagnostic pathology: use of digital pathology and artificial intelligence tools to augment a pathological diagnosis. Diagn Pathol. 2019;14:138. doi: 10.1186/s13000-019-0921-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Gadepalli K, Norouzi M, Dahl GH, Kohlberger T, Boyko A, et al. Detecting cancer metastases on gigapixel pathology images. arXiv. 2017. https://arxiv.org/abs/1703.02442.
- 5.McAlpine ED, Michelow P. The cytopathologist’s role in developing and evaluating artificial intelligence in cytopathology practice. Cytopathology. 2020;31:385–92. doi: 10.1111/cyt.12799. [DOI] [PubMed] [Google Scholar]
- 6.Huss R, Coupland SE. Software-assisted decision support in digital histopathology. J Pathol. 2020;250:685–92. doi: 10.1002/path.5388. [DOI] [PubMed] [Google Scholar]
- 7.Serag A, Ion-Margineanu A, Qureshi H, McMillan R, Saint Martin MJ, Diamond J, et al. Translational AI and deep learning in diagnostic pathology. Front Med. 2019;1:185. doi: 10.3389/fmed.2019.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, Saltz JH. Patch-based convolutional neural network for whole slide tissue image classification. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2016;2016:2424–33. doi: 10.1109/CVPR.2016.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang HY, Jung CK, Woo JI, Lee S, Cho J, Kim SW, et al. Artificial intelligence in pathology. J Pathol Transl Med. 2019;53:1–12. doi: 10.4132/jptm.2018.12.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tizhoosh HR, Pantanowitz L. Artificial intelligence and digital pathology: challenges and opportunities. J Pathol Inform. 2018;14:38. doi: 10.4103/jpi.jpi_53_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komura D, Ishikawa S. Machine learning methods for histopathological image analysis. Comput Struct Biotechnol J. 2018;9:34–42. doi: 10.1016/j.csbj.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komura D, Ishikawa S. Machine learning approaches for pathologic diagnosis. Virchows Arch. 2019;475:131–8. doi: 10.1007/s00428-019-02594-w. [DOI] [PubMed] [Google Scholar]
- 13.Levine AB, Schlosser C, Grewal J, Coope R, Jones SJM, Yip S. Rise of the machines: advances in deep learning for cancer diagnosis. Trends Cancer. 2019;5:157–69. doi: 10.1016/j.trecan.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018;15:20170387. doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rashidi HH, Tran NK, Betts EV, Howell LP, Green R. Artificial intelligence and machine learning in pathology: the present landscape of supervised methods. Acad Pathol. 2019;6:2374289519873088. doi: 10.1177/2374289519873088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krempel R, Kulkarni P, Yim A, Lang U, Habermann B, Frommolt P. Integrative analysis and machine learning on cancer genomics data using the Cancer Systems Biology Database (CancerSysDB) BMC Bioinform. 2018;24:156. doi: 10.1186/s12859-018-2157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cong L, Feng W, Yao Z, Zhou X, Xiao W. Deep learning model as a new trend in computer-aided diagnosis of tumor pathology for lung cancer. J Cancer. 2020;26:3615–22. doi: 10.7150/jca.43268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans AJ, Bauer TW, Bui MM, Cornish TC, Duncan H, Glassy EF, et al. US Food and Drug Administration approval of whole slide imaging for primary diagnosis: a key milestone is reached and new questions are raised. Arch Pathol Lab Med. 2018;142:1383–7. doi: 10.5858/arpa.2017-0496-CP. [DOI] [PubMed] [Google Scholar]
- 19.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703–15. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang DY. An outline of office-based bladder and prostate biopsy pathology. Nova Science Publishers Inc.; 2019.
- 21.Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25:1301–9. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senaras C, Niazi MKK, Lozanski G, Gurcan MN. DeepFocus: detection of out-of-focus regions in whole slide digital images using deep learning. PLoS ONE. 2018;25:e0205387. doi: 10.1371/journal.pone.0205387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janowczyk A, Zuo R, Gilmore H, Feldman M, Madabhushi A. HistoQC: an open-source quality control tool for digital pathology slides. JCO Clin Cancer Inform. 2019;3:1–7. doi: 10.1200/CCI.18.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakata N. Recent technical development of artificial intelligence for diagnostic medical imaging. Jpn J Radiol. 2019;37:103–8. doi: 10.1007/s11604-018-0804-6. [DOI] [PubMed] [Google Scholar]
- 25.Kapil A, Meier A, Zuraw A, Steele KE, Rebelatto MC, Schmidt G, et al. Deep semi supervised generative learning for automated tumor proportion scoring on NSCLC tissue needle biopsies. Sci Rep. 2018;26:17343. doi: 10.1038/s41598-018-35501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barker J, Hoogi A, Depeursinge A, Rubin DL. Automated classification of brain tumor type in whole-slide digital pathology images using local representative tiles. Med Image Anal. 2016;30:60–71. doi: 10.1016/j.media.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;3:181–93.e7. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harmon SA, Tuncer S, Sanford T, Choyke PL, Turkbey B. Artificial intelligence at the intersection of pathology and radiology in prostate cancer. Diagn Interv Radiol. 2019;25:183–8. doi: 10.5152/dir.2019.19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moscatelli M, Manconi A, Pessina M, Fellegara G, Rampoldi S, Milanesi L, et al. An infrastructure for precision medicine through analysis of big data. BMC Bioinform. 2018;15(Suppl 10):351. doi: 10.1186/s12859-018-2300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson S, Azizpour H, Smith K, Hartman J. Digital image analysis in breast pathology-from image processing techniques to artificial intelligence. Transl Res. 2018;194:19–35. doi: 10.1016/j.trsl.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Hamidinekoo A, Denton E, Rampun A, Honnor K, Zwiggelaar R. Deep learning in mammography and breast histology, an overview and future trends. Med Image Anal. 2018;47:45–67. doi: 10.1016/j.media.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velazquez Vega JE, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci USA. 2018;27:E2970–9. doi: 10.1073/pnas.1717139115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aeffner F, Zarella MD, Buchbinder N, Bui MM, Goodman MR, Hartman DJ, et al. Introduction to digital image analysis in whole-slide imaging: a white paper from the digital pathology association. J Pathol Inform. 2019;8:9. doi: 10.4103/jpi.jpi_82_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller DD, Brown EW. Artificial intelligence in medical practice: the question to the answer? Am J Med. 2018;131:129–33. doi: 10.1016/j.amjmed.2017.10.035. [DOI] [PubMed] [Google Scholar]
- 35.Farnell DA, Huntsman D, Bashashati A. The coming 15 years in gynaecological pathology: digitisation, artificial intelligence, and new technologies. Histopathology. 2020;76:171–7. doi: 10.1111/his.13991. [DOI] [PubMed] [Google Scholar]
- 36.Granter SR, Beck AH, Papke DJ., Jr AlphaGo, deep learning, and the future of the human microscopist. Arch Pathol Lab Med. 2017;141:619–21. doi: 10.5858/arpa.2016-0471-ED. [DOI] [PubMed] [Google Scholar]
- 37.Mazzanti M, Shirka E, Gjergo H, Hasimi E. Imaging, health record, and artificial intelligence: hype or hope? Curr Cardiol Rep. 2018;10:48. doi: 10.1007/s11886-018-0990-y. [DOI] [PubMed] [Google Scholar]
- 38.Smail-Tabbone M, Rance B. Contributions from the 2018 literature on bioinformatics and translational informatics. Yearb Med Inform. 2019;28:190–3. doi: 10.1055/s-0039-1677945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landau MS, Pantanowitz L. Artificial intelligence in cytopathology: a review of the literature and overview of commercial landscape. J Am Soc Cytopathol. 2019;8:230–41. doi: 10.1016/j.jasc.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Houssami N, Kirkpatrick-Jones G, Noguchi N, Lee CI. Artificial Intelligence (AI) for the early detection of breast cancer: a scoping review to assess AI’s potential in breast screening practice. Expert Rev Med Devices. 2019;16:351–62. doi: 10.1080/17434440.2019.1610387. [DOI] [PubMed] [Google Scholar]
- 41.Jang HJ, Cho KO. Applications of deep learning for the analysis of medical data. Arch Pharm Res. 2019;42:492–504. doi: 10.1007/s12272-019-01162-9. [DOI] [PubMed] [Google Scholar]
- 42.Wildeboer RR, van Sloun RJG, Wijkstra H, Mischi M. Artificial intelligence in multiparametric prostate cancer imaging with focus on deep-learning methods. Comput Methods Programs Biomed. 2020;189:105316. doi: 10.1016/j.cmpb.2020.105316. [DOI] [PubMed] [Google Scholar]
- 43.Korbar B, Olofson AM, Miraflor AP, Nicka CM, Suriawinata MA, Torresani L, et al. Deep learning for classification of colorectal polyps on whole-slide images. J Pathol Inform. 2017;8:30. doi: 10.4103/jpi.jpi_34_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bychkov D, Linder N, Turkki R, Nordling S, Kovanen PE, Verrill C, et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci Rep. 2018;8:3395. doi: 10.1038/s41598-018-21758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang DY, Khosla A, Gargeya R, Irshad H, Beck AH. Deep learning for identifying metastatic breast cancer. arXiv. 2016. https://arxiv.org/abs/1606.05718.
- 46.Litjens G, Bandi P, Ehteshami Bejnordi B, Geessink O, Balkenhol M, Bult P, et al. 1399 H&E-stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset. Gigascience. 2018;7:giy065. doi: 10.1093/gigascience/giy065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin V, Kim TH, Kwon M, Kuko M, Pourhomayoun M, Martin S. A more comprehensive cervical cell classification using convolutional neural network. J Am Soc Cytopathol. 2018. 10.1016/j.jasc.2018.06.156.
- 48.Vaickus LJ, Suriawinata AA, Wei JW, Liu X. Automating the Paris System for urine cytopathology-A hybrid deep-learning and morphometric approach. Cancer Cytopathol. 2019;127:98–115. doi: 10.1002/cncy.22099. [DOI] [PubMed] [Google Scholar]
- 49.Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20:e253–61. doi: 10.1016/S1470-2045(19)30154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van der Schaar M, Alaa A. How artificial intelligence and machine learning can help healthcare systems respond to COVID-19. Mach Learn. 2020. 10.1007/s10994-020-05928-x. [DOI] [PMC free article] [PubMed]
- 51.Conceicao T, Braga C, Rosado L, Vasconcelos MJM. A review of computational methods for cervical cells segmentation and abnormality classification. Int J Mol Sci. 2019;20:5114. doi: 10.3390/ijms20205114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulkarni S, Seneviratne N, Baig MS, Khan AHA. Artificial intelligence in medicine: where are we now? Acad Radiol. 2020;27:62–70. doi: 10.1016/j.acra.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Jha S, Topol EJ. Adapting to artificial intelligence: radiologists and pathologists as information specialists. JAMA. 2016;316:2353–4. doi: 10.1001/jama.2016.17438. [DOI] [PubMed] [Google Scholar]
- 54.Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: challenges and opportunities. Med Image Anal. 2016;33:170–5. doi: 10.1016/j.media.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chan HP, Samala RK, Hadjiiski LM, Zhou C. Deep learning in medical image analysis. Adv Exp Med Biol. 2020;1213:3–21. doi: 10.1007/978-3-030-33128-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abels E, Pantanowitz L, Aeffner F, Zarella MD, van der Laak J, Bui MM, et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J Pathol. 2019;249:286–94. doi: 10.1002/path.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin JC, Fan CT, Liao CC, Chen YS. Taiwan Biobank: making cross-database convergence possible in the Big Data era. Gigascience. 2018;7:1–4. doi: 10.1093/gigascience/gix110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stoekle HC, Mamzer-Bruneel MF, Frouart CH, Le Tourneau C, Laurent-Puig P, Vogt G, et al. Molecular tumor boards: ethical issues in the new era of data medicine. Sci Eng Ethics. 2018;24:307–22. doi: 10.1007/s11948-017-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chico V. The impact of the General Data Protection Regulation on health research. Br Med Bull. 2018;128:109–18. doi: 10.1093/bmb/ldy038. [DOI] [PubMed] [Google Scholar]
- 60.Mintz Y, Brodie R. Introduction to artificial intelligence in medicine. Minim Invasive Ther Allied Technol. 2019;28:73–81. doi: 10.1080/13645706.2019.1575882. [DOI] [PubMed] [Google Scholar]
- 61.Allen TC. Regulating artificial intelligence for a successful pathology future. Arch Pathol Lab Med. 2019;143:1175–9. doi: 10.5858/arpa.2019-0229-ED. [DOI] [PubMed] [Google Scholar]
- 62.Wang S, Yang DM, Rong R, Zhan X, Fujimoto J, Liu H, et al. Artificial intelligence in lung cancer pathology image analysis. Cancers. 2019;11:1673. doi: 10.3390/cancers11111673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarwar S, Dent A, Faust K, Richer M, Djuric U, Van Ommeren R, et al. Physician perspectives on integration of artificial intelligence into diagnostic pathology. NPJ Digit Med. 2019;2:28. doi: 10.1038/s41746-019-0106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


