Abstract
High-resolution structures of oligomers formed by the β-amyloid peptide, Aβ, are important for understanding the molecular basis of Alzheimer’s disease. Dimers of Aβ are linked to the pathogenesis and progression of Alzheimer’s disease, and tetramers of Aβ are neurotoxic. This paper reports the X-ray crystallographic structures of dimers and tetramers, as well as an octamer, formed by a peptide derived from the central and C-terminal regions of Aβ. In the crystal lattice, the peptide assembles to form two different dimers—an antiparallel β-sheet dimer and a parallel β-sheet dimer—that each further self-assemble to form two different tetramers—a sandwich-like tetramer and a twisted β-sheet tetramer. The structures of these dimers and tetramers derived from Aβ serve as potential models for dimers and tetramers of full-length Aβ that form in vitro and in Alzheimer’s disease brains.
Keywords: Amyloid, Aβ, Oligomer, Dimer, Tetramer, Crystal Structure, Alzheimer’s Disease
Graphical Abstract

INTRODUCTION
Interactions among β-sheets are ubiquitous in protein folding and protein-protein interactions. The self-assembly of β-sheets is particularly important in the aggregation of amyloidogenic peptides and proteins to form oligomers and fibrils. Understanding how β-sheets fold and assemble to form amyloid oligomers and fibrils is fundamental to peptide and protein science and is also important for understanding devastating diseases such as Alzheimer’s disease, Parkinson’s disease, and type II diabetes. Recent cryo-EM structural studies of amyloid fibrils have revealed a rich tapestry of β-sheet assemblies composed of continuous extended networks of parallel β-sheets that fold and intricately pack together.1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16 The structures of amyloid oligomers remain more elusive. X-ray crystallographic studies of fragments of amyloidogenic peptides and proteins, as well as NMR studies of full-length proteins, have provided clues about amyloid oligomer structures indicating that many amyloid oligomers are composed of antiparallel β-sheets and packed hydrophobic cores.17,18,19,20,21,22,23,24,25,26
The β-amyloid peptide, Aβ, assembles to form oligomers that vary in size and shape and are thought to be important in the pathogenesis and progression of Alzheimer’s disease.27,28 Elucidating the structures of these oligomers is central to understanding the molecular basis of Alzheimer’s disease. Recently, our laboratory has elucidated a wealth of X-ray crystallographic structures of oligomers formed by peptides designed to mimic Aβ β-hairpins. In studying these peptides, we have discovered new structural motifs for Aβ oligomers including triangular trimers, barrel-like and sandwich-like hexamers, ball-shaped dodecamers, and large annular pore-like structures.29,30,31,32,33,34 These structures have revealed the novel and unpredictable ways that β-hairpin peptides containing Aβ sequences can fit together to form oligomers, and may also help shed light on the structures of oligomers that full-length Aβ forms in Alzheimer’s disease.
In the current paper, we report the X-ray crystallographic structures of two different β-sheet dimers—one antiparallel β-sheet dimer and one parallel β-sheet dimer—formed by peptide 1, a β-hairpin derived from Aβ16–36. In the crystal lattice, these two dimers self-assemble to form different tetramers. The antiparallel dimer self-assembles with another antiparallel dimer to form a sandwich-like tetramer stabilized by a tightly packed hydrophobic core. In contrast, the parallel dimer self-assembles with another parallel dimer to form a tetramer composed of a highly twisted eight-stranded β-sheet. The parallel tetramer is stabilized by hydrophobic packing and edge-to-edge hydrogen bonding between the two parallel dimers. These oligomer structures further illustrate the remarkable ways in which β-hairpins derived from Aβ can interact to form oligomers, and add to the diversity of oligomers that β-hairpins can form.
RESULTS AND DISCUSSION
Design of peptide 1.
Peptide 1 is designed to mimic a β-hairpin formed by Aβ16–36 (Figures 1A and B). Peptide 1 contains Aβ residues 16–36 linked together at the N- and C-termini with a δ-linked ornithine turn unit and also contains a cross-strand disulfide bond in place of Ala21 and Ile31 to stabilize the β-hairpin structure.35 The disulfide bond at this position maintains the hydrophobic character of this region of the peptide. Peptide 1 also contains an N-methyl group on Gly33 to block uncontrolled aggregation of the peptide. These design features facilitate crystallization of the peptide. Variants of peptide 1 without the cross-strand disulfide bond or without the N-methyl group did not crystallize in any of the 864 different crystallization conditions tested. To facilitate determination of the X-ray crystallographic phases, peptide 1 also contains a para-iodo group on Phe19.36
Figure 1.

Peptide 1. (A) Chemical structure of an Aβ16–36 β-hairpin. (B) Chemical structure of peptide 1. (C) X-ray crystallographic structure of a representative β-hairpin monomer formed by peptide 1 (PDB 6WXM). (D) Overlay of the 11 peptide 1 β-hairpins in the asymmetric unit.
X-ray crystallographic structure of peptide 1.
The X-ray crystallographic structure of peptide 1 reveals that the peptide folds to form a twisted β-hairpin and that the β-hairpins assemble to form oligomers. The asymmetric unit of the X-ray crystallographic structure contains 11 copies of peptide 1. Each copy folds to form a β-hairpin that is composed of an Aβ16–23 β-strand and an Aβ27–36 β-strand connected by an Aβ24–28 loop (Figure 1C). The peptide backbones of the β-strands share comparable geometries for all 11 copies of peptide 1 in the asymmetric unit, while the loop regions vary (Figure 1D).
In the crystal lattice, peptide 1 forms assemblies that may be thought of as oligomers. Peptide 1 forms two different β-sheet dimers—an antiparallel β-sheet dimer and a parallel β-sheet dimer. Both dimers comprise four-stranded β-sheets. The antiparallel dimer further assembles with another antiparallel dimer to form a sandwich-like tetramer. The parallel dimer further assembles with another parallel dimer to form a twisted β-sheet tetramer. Two of these twisted β-sheet tetramers further assemble to form an octamer. An additional antiparallel dimer, that is not part of a tetramer, rests upon the octamer in the crystal lattice. The following describes these oligomers in detail.
Antiparallel dimer.
The antiparallel dimer formed by peptide 1 consists of two peptide monomers arranged in an antiparallel orientation in which the Aβ16–23 β-strand of one monomer is across from the Aβ16–23 β-strand of the other monomer (Figure 2A). Five hydrogen-bonding interactions between the amide backbones of the two monomers stabilize the antiparallel dimer: Phe19I and Cys21 on one monomer pair with Cys21 and Phe19I on the other monomer; a water molecule bridges Asp23 on one monomer and Leu17 on the other monomer.
Figure 2.

Dimers formed by peptide 1. (A) Chemical structure (top) and X-ray crystallographic structure (bottom) of the antiparallel dimer formed by peptide 1. (B) Chemical structure (top) and X-ray crystallographic structure (bottom) of the parallel dimer formed by peptide 1.
Parallel dimer.
The parallel dimer formed by peptide 1 consists of two peptide monomers arranged in a parallel orientation in which the Aβ16–23 β-strand of one monomer is across from the Aβ16–23 β-strand of the other monomer (Figure 2B). Four hydrogen bonds between the amide backbones of the two monomers stabilize the parallel dimer: Phe20 and Cys21 on one monomer pair with Phe19I and Phe20 on the other monomer; a water molecule bridges Phe19I on one monomer and Leu17 on the other monomer.
The antiparallel dimer and the parallel dimer each self-assemble to form different tetramers. The antiparallel dimer assembles with another antiparallel dimer to form a sandwich-like tetramer, whereas the parallel dimer assembles with another parallel dimer to form a highly twisted β-sheet tetramer. The twisted β-sheet tetramer further assembles with another twisted β-sheet tetramer to form an octamer.
Sandwich-like tetramer formed by the antiparallel dimer.
Two antiparallel dimers further assemble to form a sandwich-like tetramer (Figure 3A). The sandwich-like tetramer is stabilized by packing between the two antiparallel dimers to create a hydrophobic core (Figure 3B). In the hydrophobic core, the side chains of Val18, Phe20, Ile32, Leu34, and Val36 from four copies of peptide 1 pack together, creating a dense core containing 20 hydrophobic amino acid side chains. The hydrophilic Aβ25–27 loops extend off of the sandwich-like tetramer and do not make any significant contacts.
Figure 3.

X-ray crystallographic structure of the sandwich-like tetramer (dimer of antiparallel dimers) formed by peptide 1. (A) Cartoon and stick model of the sandwich-like tetramer. (B) Cartoon and sphere model of the sandwich-like tetramer. The residues that comprise the hydrophobic core are shown as spheres.
Twisted β-sheet tetramer formed by the parallel dimer.
Two parallel dimers further assemble in a parallel orientation to form a highly twisted β-sheet tetramer comprising an eight-stranded β-sheet (Figures 4A and B). In the twisted β-sheet tetramer, the Aβ27–36 β-strand of one dimer is across from the Aβ27–36 β-strand of the other dimer. Four hydrogen bonds between the amide backbones of the two parallel dimers stabilize the twisted β-sheet tetramer: Gly29 and Cys31 on one parallel dimer pair with Lys28 and Ala30 on the other parallel dimer. The twisted β-sheet tetramer is further stabilized by hydrophobic packing between the two parallel dimers to create a hydrophobic core (Figure 4C). In the hydrophobic core, the side chains of Phe19I, Phe20, Cys21, Val24, Ala30, Cys31, Ile32, Leu34, and Val36 of each parallel dimer pack together, creating a dense core containing 18 hydrophobic amino acid side chains.
Figure 4.

The twisted β-sheet tetramer formed by the parallel dimer. (A) Chemical structure. (B) X-ray crystallographic structure of the twisted β-sheet tetramer (cartoon and stick model; the side chains are omitted for clarity). (C) X-ray crystallographic structure of the twisted β-sheet tetramer (cartoon and sphere model; the residues that comprise the hydrophobic core are shown as spheres).
Octamer.
Two twisted β-sheet tetramers further assemble to form an octamer. The inner four β-hairpin monomers of the octamer create a continuous hydrogen-bonding network containing 12 intermolecular hydrogen bonds between the peptide backbones (Figure 5A). Packing between hydrophobic residues on the inner four β-hairpin monomers further stabilizes the octamer. The side chains of Leu17, Phe19I, Phe20, Ile32, Leu34, Met35, and Val36 pack together, creating a core containing 22 hydrophobic amino acid side chains.
Figure 5.

X-ray crystallographic structure of the octamer (dimer of twisted β-sheet tetramers) formed by peptide 1. (A) Cartoon and stick model of the octamer (side chains are omitted for clarity). (B) Cartoon and sphere model of the octamer illustrating the hydrophobic packing between the two twisted β-sheet tetramers. The side chains of the hydrophobic core are shown as spheres.
The dimers, tetramers, and octamer are all part of the same crystal lattice.
The assembly of peptide 1 into a crystal lattice reveals the ways in which the peptide can self-assemble with other copies of the peptide to form oligomers. The oligomers described above are all part of the same crystal lattice. The asymmetric unit of the X-ray crystallographic structure of peptide 1 contains 11 copies of the peptide (Figure 6A). The asymmetric unit contains two crystallographically unique twisted β-sheet tetramers that comprise the octamer (Figure 6B, cyan strands). The octamer is flanked by three additional copies of peptide 1: Two copies — by a symmetry operation — comprise the sandwich-like tetramer (Figure 6B, magenta strands). One copy — by a symmetry operation — forms an antiparallel dimer that rests upon the octamer in the crystal lattice (Figure 6B, yellow strands).
Figure 6.

(A) The asymmetric unit of the X-ray crystallographic structure of peptide 1. (B) The crystal lattice of peptide 1, illustrating the relationship between the sandwich-like tetramer (magenta) and the twisted β-sheet tetramers that comprise the octamer (cyan). An antiparallel dimer that rests on the octamer is shown in yellow.
Oligomers of Aβ40 and Aβ42 in SDS-PAGE.
Dimers and tetramers of synthetic or expressed Aβ have been observed to form in vitro. In SDS-PAGE, Aβ40 predominantly forms two oligomers that migrate at molecular weights consistent with a dimer and tetramer, in addition to the monomer (Figure 7). Aβ42 predominantly forms two oligomers that migrate at molecular weights consistent with a trimer and tetramer, in addition to the monomer (Figure 7). The structures of these oligomers are unknown. The parallel and antiparallel dimers formed by peptide 1 provide two potential structural models for the dimer formed by Aβ40 in SDS-PAGE. Furthermore, the sandwich-like tetramer and twisted β-sheet tetramer formed by peptide 1 provide potential structural models for the tetramers formed by Aβ40 and Aβ42 in SDS-PAGE.
Figure 7.

Silver-stained SDS-PAGE of recombinantly expressed Aβ40 and Aβ42 illustrating the oligomers that the peptides form in vitro. A 5-μL aliquot of each peptide concentration in a serial dilution was run on the gel.
Crystallographically based models of two Aβ12–40 tetramers.
We envision that the full-length Aβ peptide can assemble in the same fashion as peptide 1 to form sandwich-like tetramers and twisted β-sheet tetramers. To better understand what a sandwich-like tetramer (dimer of antiparallel dimers) and a twisted β-sheet tetramer (dimer of parallel dimers) containing full-length Aβ might look like, we modeled Aβ12–40 into the crystallographic coordinates of each tetramer.37 We first appended the N- and C-terminal regions 12–15 (VHHQ) and 37–40 (GGVV) onto the crystallographic coordinates of the four peptide 1 monomers that comprise each tetramer, and mutated all modified residues back to the native residues. We then performed replica-exchange molecular dynamics (REMD) to generate realistic conformations the N- and C-terminal regions of the β-hairpins (Figures 8A and B).38,39 In these models, the β-hairpins constitute the cores of the tetramers, and the N- and C-termini surround the cores. The REMD simulations shows that both tetramers can accommodate the N- and C-terminal residues without steric clashes and suggests that full-length Aβ could form a sandwich-like tetramer or a twisted β-sheet tetramer.
Figure 8.
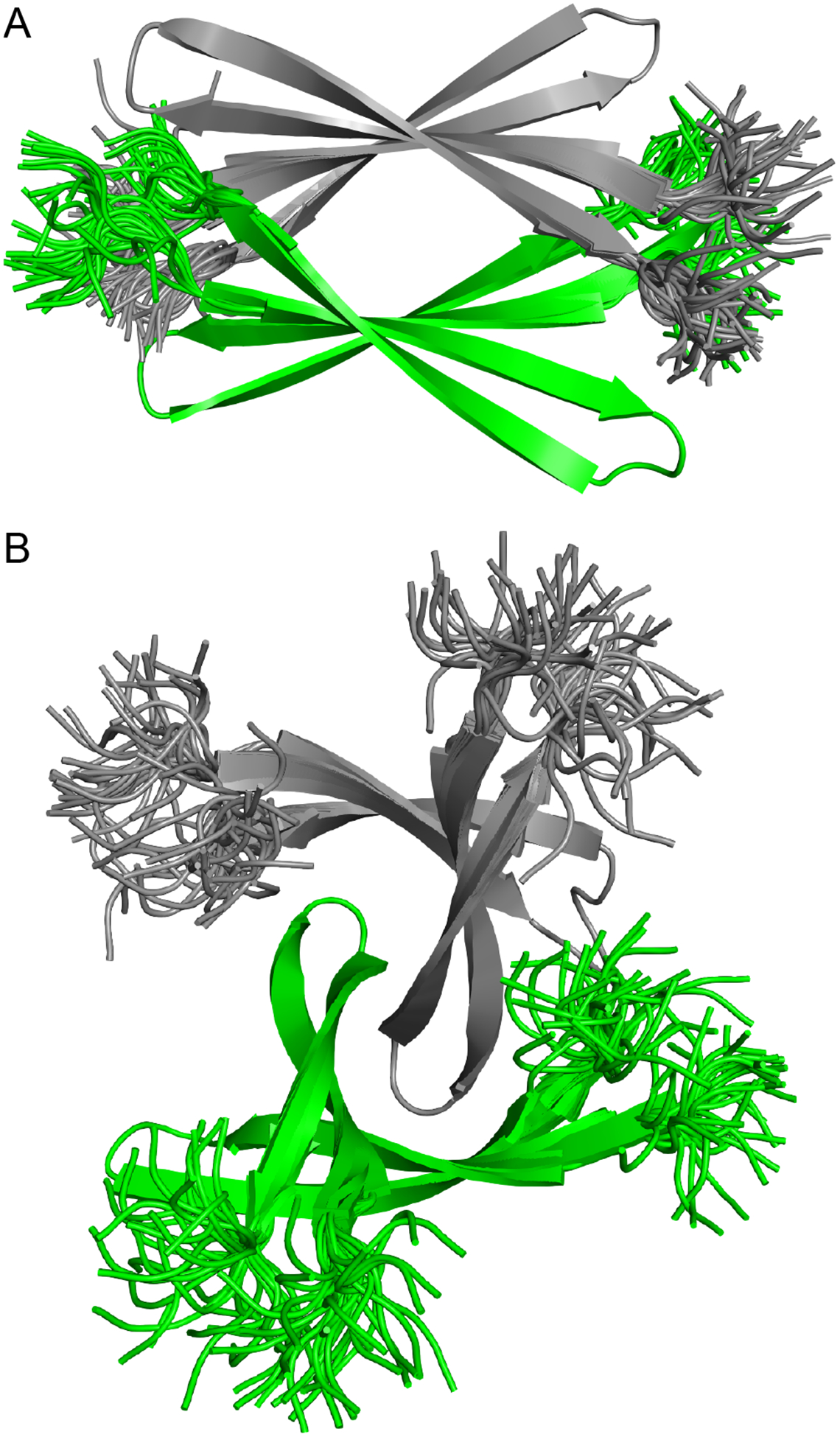
Crystallographically based models of an Aβ12–40 sandwich-like tetramer (A) and an Aβ12–40 twisted β-sheet tetramer (B). Superpositions of 32 structures generated by replica-exchange molecular dynamics.
The structures of the dimers and tetramers formed by peptide 1 provide new models for Aβ oligomers. Aβ dimers are thought to have special significance in the pathogenesis and progression of Alzheimer’s disease. Aβ plaques from Alzheimer’s disease patients contain cross-linked Aβ dimers that are composed of different Aβ alloforms.40 Aβ dimers appear to be the building blocks of large, mildly cytotoxic oligomers with molecular weights ranging from 150–650 kDa.41 Aβ dimers also promote phosphorylation and aggregation of the microtubule-associated protein tau, which is also involved in Alzheimer’s disease progression.42 The structures of these dimers are unknown. The parallel and antiparallel β-sheet dimers formed by peptide 1 provide two potential structural models for the dimers observed in Alzheimer’s disease brains.
Aβ tetramers have been observed in protein extracts from Alzheimer’s disease brains, but their exact roles in the pathogenesis and progression of the disease is less clear than that of dimers.43,44 Aβ tetramers prepared in vitro are toxic toward both neuroblastoma cells and cultured hippocampal neurons.45,46 Furthermore, covalently stabilized Aβ tetramers prepared using photo-induced cross-linking of unmodified proteins (PICUP) interact with the cell membranes of hippocampal neurons.47 The structures of the Aβ tetramers in these studies are unknown. The sandwich-like tetramer and twisted β-sheet tetramer formed by peptide 1 provide two potential structural models for the tetramers observed in Alzheimer’s disease brains, as well as the tetramers prepared in vitro.
CONCLUSIONS
The structures of the dimers, tetramers, and octamer formed by peptide 1 contribute to the rich structural landscape of amyloidogenic peptides and proteins. CryoEM structures of fibrils formed by amyloidogenic peptides and proteins such as Aβ, islet amyloid polypeptide, tau, α-synuclein, and human prion protein have revealed that the peptides and proteins adopt highly convoluted shapes and pack together to form twisted filaments.8,10,12,13,14,15,16 The twisted shapes of the oligomers formed by peptide 1 are distinct from the twists of fibrils and filaments. The structure of the sandwich-like tetramer formed by peptide 1 (PDB 6WXM) is reminiscent of the Aβ tetramer reported by Streltsov et al. (PDB 3MOQ), as well as the tetramer formed by transthyretin (e.g., PDB 1TTC).21,48 It is also evocative of the Aβ tetramer and octamer structural models from Ciudad et al. (PDB 6RHY).26 Prominent features of these structures include antiparallel β-hairpins that pack together in a sandwich-like fashion to form a hydrophobic core, much like the sandwich-like tetramer formed by peptide 1.
The parallel and antiparallel β-sheet dimers, the sandwich-like and twisted β-sheet tetramers, and the octamer formed by peptide 1 add to the diversity of Aβ-derived oligomers observed by our laboratory. In studying, β-hairpin peptides that contain the central and C-terminal regions of Aβ, we have discovered a variety of different structures (Figure 9). These structures reveal the intricate ways that Aβ β-hairpins can fit together to form compact oligomers that are stabilized by edge-to-edge hydrogen bonding interactions and hydrophobic cores. We believe the variety of structures we have observed exemplifies the heterogeneity of oligomers formed by Aβ in vitro and in the brain. Investigating the exact relationship between the oligomer structures we have observed crystallographically and the structures of Aβ oligomers in Alzheimer’s disease is an active area of research in our laboratory, and we will report our findings from these studies in due course.
Figure 9.

Representative oligomers of Aβ-derived peptides observed in our laboratory by X-ray crystallography.
METHODS
Synthesis of peptide 1, X-ray crystallographic procedures, Aβ oligomer preparation, SDS-PAGE and silver staining, and replica-exchange molecular dynamics were performed as described previously.29,30,31,32,33 These procedures are restated in detail in the Materials and Methods section in the Supporting Information.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) for funding (Grant GM097562).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at http://pubs.acs.org/.
(1) Procedures for the synthesis of peptide 1, crystallization of peptide 1, preparation of Aβ oligomers, SDS-PAGE and silver staining, and replica-exchange molecular dynamics; (2) details of X-ray crystallographic data collection, processing, and refinement; (3) characterization data for peptide 1 (PDF). Crystallographic data for peptide 1 (cif file).
Crystallographic coordinates of peptide 1 were deposited into the Protein Data Bank (PDB) with code 6WXM.
The authors declare no competing financial interest.
REFERENCES AND NOTES
- 1.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci U S A. 2008. Nov 25;105(47):18349–54. Epub 2008 Nov 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006. Jan 17;45(2):498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013. Sep 12;154(6):1257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiang W, Yau WM, Luo Y, Mattson MP, Tycko R. Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils. Proc Natl Acad Sci U S A. 2012. Mar 20;109(12):4443–8. Epub 2012 Mar 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wälti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Böckmann A, Güntert P, Meier BH, Riek R. Atomic-resolution structure of a disease-relevant Aβ(1–42) amyloid fibril. Proc Natl Acad Sci U S A. 2016. Aug 23;113(34):E4976–84. Epub 2016 Jul 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, Griffin RG. Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils. J Am Chem Soc. 2016. Aug 3;138(30):9663–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Y, Ma B, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y. Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015. Jun;22(6):499–505. Epub 2015 May 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gremer L, Schölzel D, Schenk C, Reinartz E, Labahn J, Ravelli RBG, Tusche M, Lopez-Iglesias C, Hoyer W, Heise H, Willbold D, Schröder GF. Fibril structure of amyloid-β(1–42) by cryo-electron microscopy. Science. 2017. Oct 6;358(6359):116–119. Epub 2017 Sep 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, Wan W, Stubbs G, Schwieters CD, Lee VM, George JM, Rienstra CM. Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat Struct Mol Biol. 2016. May;23(5):409–15. Epub 2016 Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, Crowther RA, Ghetti B, Goedert M, Scheres SHW. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature. 2017. Jul 13;547(7662):185–190. Epub 2017 Jul 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dregni AJ, Mandala VS, Wu H, Elkins MR, Wang HK, Hung I, DeGrado WF, Hong M. In vitro 0N4R tau fibrils contain a monomorphic β-sheet core enclosed by dynamically heterogeneous fuzzy coat segments. Proc Natl Acad Sci U S A. 2019. Aug 13;116(33):16357–16366. Epub 2019 Jul 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falcon B, Zhang W, Murzin AG, Murshudov G, Garringer HJ, Vidal R, Crowther RA, Ghetti B, Scheres SHW, Goedert M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature. 2018. Sep;561(7721):137–140. Epub 2018 Aug 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kollmer M, Close W, Funk L, Rasmussen J, Bsoul A, Schierhorn A, Schmidt M, Sigurdson CJ, Jucker M, Fändrich M. Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue. Nat Commun. 2019. Oct 29;10(1):4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glynn C, Sawaya MR, Ge P, Gallagher-Jones M, Short CW, Bowman R, Apostol M, Zhou ZH, Eisenberg DS, Rodriguez JA. Cryo-EM structure of a human prion fibril with a hydrophobic, protease-resistant core. Nat Struct Mol Biol. 2020. May;27(5):417–423. Epub 2020 Apr 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schweighauser M, Shi Y, Tarutani A, Kametani F, Murzin AG, Ghetti B, Matsubara T, Tomita T, Ando T, Hasegawa K, Murayama S, Yoshida M, Hasegawa M, Scheres SHW, Goedert M. Structures of α-synuclein filaments from multiple system atrophy. Nature. 2020. May 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Röder C, Kupreichyk T, Gremer L, Schäfer LU, Pothula KR, Ravelli RBG, Willbold D, Hoyer W, Schröder GF. Cryo-EM structure of islet amyloid polypeptide fibrils reveals similarities with amyloid-β fibrils. Nat Struct Mol Biol. 2020. Jun 15. [DOI] [PubMed] [Google Scholar]
- 17.Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao M, Pensalfini A, Soriaga AB, Landau M, Teng PK, Cascio D, Glabe C, Eisenberg D. Atomic view of a toxic amyloid small oligomer. Science. 2012. Mar 9;335(6073):1228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sangwan S, Zhao A, Adams KL, Jayson CK, Sawaya MR, Guenther EL, Pan AC, Ngo J, Moore DM, Soriaga AB, Do TD, Goldschmidt L, Nelson R, Bowers MT, Koehler CM, Shaw DE, Novitch BG, Eisenberg DS. Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS). Proc Natl Acad Sci U S A. 2017. Aug 15;114(33):8770–8775. Epub 2017 Jul 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apostol MI, Perry K, Surewicz WK. Crystal structure of a human prion protein fragment reveals a motif for oligomer formation. J Am Chem Soc. 2013. Jul 17;135(28):10202–5. Epub 2013 Jul 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu L, Edalji R, Harlan JE, Holzman TF, Lopez AP, Labkovsky B, Hillen H, Barghorn S, Ebert U, Richardson PL, Miesbauer L, Solomon L, Bartley D, Walter K, Johnson RW, Hajduk PJ, Olejniczak ET. Structural characterization of a soluble amyloid beta-peptide oligomer. Biochemistry. 2009. Mar 10;48(9):1870–7. [DOI] [PubMed] [Google Scholar]
- 21.Streltsov VA, Varghese JN, Masters CL, Nuttall SD. Crystal structure of the amyloid-β p3 fragment provides a model for oligomer formation in Alzheimer’s disease. J Neurosci. 2011. Jan 26;31(4):1419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. Evidence of fibril-like β-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s β-amyloid. Nat Struct Mol Biol. 2007. Dec;14(12):1157–64. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed M, Davis J, Aucoin D, Sato T, Ahuja S, Aimoto S, Elliott JI, Van Nostrand WE, Smith SO. Structural conversion of neurotoxic amyloid-beta(1–42) oligomers to fibrils. Nat Struct Mol Biol. 2010. May;17(5):561–7. Epub 2010 Apr 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang D, Zimmerman MI, Martin PK, Nix AJ, Rosenberry TL, Paravastu AK. Antiparallel β-Sheet Structure within the C-Terminal Region of 42-Residue Alzheimer’s Amyloid-β Peptides When They Form 150-kDa Oligomers. J Mol Biol. 2015. Jul 3;427(13):2319–28. Epub 2015 Apr 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lendel C, Bjerring M, Dubnovitsky A, Kelly RT, Filippov A, Antzutkin ON, Nielsen NC, Härd T. A hexameric peptide barrel as building block of amyloid-β protofibrils. Angew Chem Int Ed Engl. 2014. Nov 17;53(47):12756–60. Epub 2014 Sep 26. [DOI] [PubMed] [Google Scholar]
- 26.Ciudad S, Puig E, Botzanowski T, Meigooni M, Arango AS, Do J, Mayzel M, Bayoumi M, Chaignepain S, Maglia G, Cianferani S, Orekhov V, Tajkhorshid E, Bardiaux B, Carulla N. Aβ(1–42) tetramer and octamer structures reveal edge conductivity pores as a mechanism for membrane damage. Nat Commun. 2020. Jun 15;11(1):3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benilova I, Karran E, De Strooper B. The toxic Aβ oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat Neurosci. 2012. Jan 29;15(3):349–57. Review. [DOI] [PubMed] [Google Scholar]
- 28.Larson ME, Lesné SE. Soluble Aβ oligomer production and toxicity. J Neurochem. 2012. Jan;120 Suppl 1:125–39. Epub 2011 Nov 28. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spencer RK, Li H, Nowick JS. X-ray crystallographic structures of trimers and higher-order oligomeric assemblies of a peptide derived from Aβ(17–36). J Am Chem Soc. 2014. Apr 16;136(15):5595–8. Epub 2014 Apr 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreutzer AG, Yoo S, Spencer RK, Nowick JS. Stabilization, Assembly, and Toxicity of Trimers Derived from Aβ. J Am Chem Soc. 2017. Jan 18;139(2):966–975. Epub 2017 Jan 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kreutzer AG, Hamza IL, Spencer RK, Nowick JS. X-ray Crystallographic Structures of a Trimer, Dodecamer, and Annular Pore Formed by an Aβ17–36 β-Hairpin. J Am Chem Soc. 2016. Apr 6;138(13):4634–42. Epub 2016 Mar 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salveson PJ, Spencer RK, Kreutzer AG, Nowick JS. X-ray Crystallographic Structure of a Compact Dodecamer from a Peptide Derived from Aβ(16–36). Org Lett. 2017. Jul 7;19(13):3462–3465. Epub 2017 Jun 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreutzer AG, Spencer RK, McKnelly KJ, Yoo S, Hamza IL, Salveson PJ, Nowick JS. A Hexamer of a Peptide Derived from Aβ(16–36). Biochemistry. 2017. Nov 14;56(45):6061–6071. Epub 2017 Oct 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kreutzer AG, Nowick JS. Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer’s Disease and Other Amyloid Diseases. Acc Chem Res. 2018. Mar 20;51(3):706–718. Epub 2018 Mar 6. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowick JS, Brower JO. A new turn structure for the formation of beta-hairpins in peptides. J Am Chem Soc. 2003. Jan 29;125(4):876–7. [DOI] [PubMed] [Google Scholar]
- 36.Mutation of Phe19 to serine has previously been shown to impact the assembly of Aβ42. (Marshall KE, Vadukul DM, Dahal L, Theisen A, Fowler MW, Al-Hilaly Y, Ford L, Kemenes G, Day IJ, Staras K, Serpell LC. A critical role for the self-assembly of Amyloid-β1–42 in neurodegeneration. Sci Rep. 2016. July 22;6:30182.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.To simplify the REMD simulation and reduce computing time, we truncated the N-terminus to Val12.
- 38.Zhou R Replica exchange molecular dynamics method for protein folding simulation. Methods Mol Biol. 2007;350:205–23. [DOI] [PubMed] [Google Scholar]
- 39.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem 2005;26;1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brinkmalm G, Hong W, Wang Z, Liu W, O’Malley TT, Sun X, Frosch MP, Selkoe DJ, Portelius E, Zetterberg H, Blennow K, Walsh DM. Identification of neurotoxic cross-linked amyloid-β dimers in the Alzheimer’s brain. Brain. 2019. May 1;142(5):1441–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang T, Li S, Xu H, Walsh DM, Selkoe DJ. Large Soluble Oligomers of Amyloid β-Protein from Alzheimer Brain Are Far Less Neuroactive Than the Smaller Oligomers to Which They Dissociate. J Neurosci. 2017. Jan 4;37(1):152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin M, Shepardson N, Yang T, Chen G, Walsh D, Selkoe DJ. Soluble amyloid beta-protein dimers isolated from Alzheimer cortex directly induce Tau hyperphosphorylation and neuritic degeneration. Proc Natl Acad Sci U S A. 2011. Apr 5;108(14):5819–24. Epub 2011 Mar 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLean CA, Cherny RA, Fraser FW, Fuller SJ, Smith MJ, Beyreuther K, Bush AI, Masters CL. Soluble pool of Abeta amyloid as a determinant of severity of neurodegeneration in Alzheimer’s disease. Ann Neurol. 1999. Dec;46(6):860–6. [DOI] [PubMed] [Google Scholar]
- 44.Sokolow S, Henkins KM, Bilousova T, Miller CA, Vinters HV, Poon W, Cole GM, Gylys KH. AD synapses contain abundant Aβ monomer and multiple soluble oligomers, including a 56-kDa assembly. Neurobiol Aging. 2012. Aug;33(8):1545–55. Epub 2011 Jul 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea JE, Ruotolo BT, Robinson CV, Bowers MT. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat Chem. 2009. Jul;1(4):326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ono K, Condron MM, Teplow DB. Structure-neurotoxicity relationships of amyloid beta-protein oligomers. Proc Natl Acad Sci USA. 2009. Sep 1;106(35):14745–50. Epub 2009 Aug 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jana MK, Cappai R, Pham CL, Ciccotosto GD. Membrane-bound tetramer and trimer Aβ oligomeric species correlate with toxicity towards cultured neurons. J Neurochem. 2016. Feb;136(3):594–608. [DOI] [PubMed] [Google Scholar]
- 48.Hamilton JA, Steinrauf LK, Braden BC, Liepnieks J, Benson MD, Holmgren G, Sandgren O, Steen L. The x-ray crystal structure refinements of normal human transthyretin and the amyloidogenic Val-30-->Met variant to 1.7-A resolution. J Biol Chem. 1993. Feb 5;268(4):2416–24. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


