Abstract
Background
Rapamycin is a promising agent for treating tumors, but clinical applications of rapamycin are limited due to its poor water solubility and low bioavailability. This paper constructs a liposome delivery system for rapamycin to improve the effect in treating colorectal cancer.
Methods
We prepared the rapamycin liposomes using the ethanol injection method. The cellular uptake and biodistribution were detected by LC-MS and in vivo imaging system. MTT assay, transwell migration experiment, flow cytometry, and Western blot analysis evaluated the antitumor effect of rapamycin liposomes in vitro. Furthermore, HCT-116 tumor-bearing mice were used to assess the therapeutic efficacy of rapamycin liposomes in vivo.
Results
The prepared rapamycin liposomes had a particle size of 100±5.5 nm and with a narrow size distribution. In vitro cellular uptake experiments showed that the uptake of rapamycin liposomes by colorectal cells was higher than that of free rapamycin. Subsequently, in vivo imaging experiments also demonstrated that rapamycin liposomes exhibited higher tumor accumulation. Therefore, the ability of rapamycin liposomes to inhibit tumor proliferation, migration and to induce tumor apoptosis is superior to that of free rapamycin. We also demonstrated in vivo good antitumor efficacy of the rapamycin liposomes in HCT-116 xenograft mice. In addition, rapamycin liposomes and 5-FU can synergistically improve the efficacy of colorectal cancer via the Akt/mTOR and P53 pathways.
Conclusion
Collectively, rapamycin liposomes are a potential treatment for colorectal cancer, as it not only improves rapamycin’s antitumor effect but also synergistically enhances 5-FU’s chemotherapy effect.
Keywords: rapamycin liposomes, 5-fluorouracil, Akt/mTOR, P53, colorectal cancer
Introduction
Colorectal cancer (CRC) is a type of epithelial cancer and has the third-highest mortality rate worldwide. 5-Fluorouracil (5-FU)-based chemotherapy drugs are the first line of treatment for CRC. However, development of multidrug resistance has limited the effects of 5-FU, and 5-FU has a toxicity to normal cells.1 Currently, combination treatments of 5-FU with other agents is becoming a promising strategy to improve CRC therapy.
Rapamycin (Rapa) are traditionally used as immunosuppressors, and recent researches have found that Rapa have been extensively applied in various types cancer, including osteosarcoma, bladder cancer, cervical cancer, and skin cancer. Rapa possesses antitumor activities through various mechanisms, including cell proliferation, invasion, migration inhibition, apoptosis promotion, and anti-angiogenesis effect.2–4 In addition, accumulating evidences have indicated that clinical chemotherapy drugs (eg, the cisplatin and paclitaxel) can be combined with Rapa in order to improve its efficacy.5,6 For example, Rapa synergizes with cisplatin in anti-endometrial cancer by improving the IL-27-stimulated cytotoxicity of natural killer (NK) cells.7 In another study, combined Rapa and doxorubicin-loaded cyclic octapeptide liposomes improve therapeutic outcomes of triple-negative breast cancer.8 Importantly, a previous study reported that Rapa combined with 5-FU to treat colorectal cancer, which suppresses cancer cell progression by inhibiting the proliferation and inducing apoptosis.9,10 However, the Rapa achieved tumor response in only 36%, while the tumor inhibition rate of the combined group is 55%.
The development of clinical applications of Rapa is limited due to its poor water solubility (2.6 μg/mL).11 Everolimus is a macrolide derivative of Rapa which ameliorates the problem of poor solubility, but its a second-line drug for advanced renal cell carcinoma.12,13 Thus, further improving bioavailability and local drug concentration is a key strategy for improving the therapeutic effect. Nowadays, nano-based drug delivery systems are developed to deliver antitumor drugs to treat CRC.14–16 Furthermore, several evidences indicated that nanoscale delivery vehicles, such as liposomes, micelles, and nanoparticles improve Rapa’s aqueous solubility, bioavailability and enhance the drug’s antitumor properties.17–19
Based on these reports, this study aimed to construct a liposome delivery system for Rapa (Rapa liposomes) and assesses whether Rapa liposomes’s antitumor effect is superior to that of free rapamycin and whether Rapa liposomes improve the effect of 5-FU in treating CRC. Finally, the potential molecular mechanisms were further verified in vitro and in vivo.
Materials and Methods
Materials, Cell Culture, and Animals
Rapamycin and 5-FU were obtained from meilunbio (Dalian, China). The anti Caspase3 antibody, Cleaved Caspase3 antibody, p-AKT (ser 473) antibody, p-AKT (Thr 308) antibody, p70 S6 Kinase antibody, and p-p70S6 Kinase (Thr 389) antibody were purchased from Cell Signaling Technology (Danvers, MA, USA). The ki-67 antibody was purchased from Abcam (Cambridge, UK). The BCL-2 antibody, Bax antibody, AKT antibody, p53 antibody, E-Cadherin antibody, N-Cadherin antibody, Vimentin antibody, MMP-9 antibody and PCNA antibody were purchased from Proteintech (Wuhan, China). The GAPDH rabbit polyclonal antibody was obtained from XianZhi Biotech (Hangzhou, China). The peroxidase-conjugated secondary antibodies were obtained from CWBIO (Taizhou, China).
The human CRC cell line HCT-116 and SW-480 cells were purchased from iCell Bioscience Inc (Shanghai, China). The cells were cultured in a DMEM medium containing 10% FBS in a 37 °C incubator containing 5% CO2.
Female athymic nude mice (6–8 weeks old) were purchased from Guangdong Medical Laboratory Animal Center. All animal studies were performed in accordance with the Guide for the Care and Use of Laboratory Animals. The Laboratory Animal Ethics Committee of the Third Affiliated Hospital of Guangzhou Medical University approved all experimental protocols.
Preparation and Characterization of the Rapa Liposomes
The Rapa liposomes were prepared by ethanol injection method. The composition based on soy phosphatidylcholine:cholesterol:PEG-DSPE were employed for Rapa liposomes. Rapa to total lipid ratios were 1:10. Firstly, lipids and Rapa were dissolved in absolute ethanol by magnetic stirring. Then, the ethanol solution was poured into PBS buffer under stirring, the stirring speed was 450 rpm, and the stirring time was 30 min. The obtained liposomal colostrum was homogenized five times with a high pressure homogenizer, the homogenization pressure was 600 bar, and the liquid supply flow rate was 20 mL/min. The liposome solution was filtered by a 0.22 μm microporous membrane, and was lyophilized and stored at 4 °C before use. The DiR fluorescent liposome was prepared in the same manner as the Rapa liposomes, and Rapa was replaced with DiR fluorescein.
The intensity particle size and zeta potential were evaluated using dynamic light scattering (DLS, Malvern, nano ZS, UK). The Rapa liposomes were stained with 2% phosphotungstic acid, and their morphology was examined by transmission electronic microscopy (TEM, JEM-1400plus). The drug loading and encapsulation efficiency of Rapa liposomes was measured by high performance liquid chromatography (Dikma, Diamonsil ODS C18 column, 250×4.6 mm, 5 µm, HPLC, Agilent).
In vitro Cellular Uptake
The HCT-116 cells were seeded in six-well plates, and the cells were grown to 80%. The cells were incubated with free Rapa and Rapa liposomes containing the same concentration of 8 ug/mL of Rapa for 30 minutes, 60 minutes, 90 minutes and 120 minutes. The protein was extracted, and the intracellular protein concentration of each well was measured by BCA method (Beyotime Biotechnology, Shanghai). The cellular uptake of Rapa in HCT-116 was measured by LC-MS (Agilent SB-C18 2.1×100 mm 3.5 μm, internal standard substances: Danazol).
Cell Viability
The HCT-116 and SW-480 cells were seeded at a density of 5×103 cells/well in 96-well plates. Afterwards, the cells were incubated with free Rapa, Rapa liposomes and 5-FU alone or in combination, respectively. After 48 hours, the formulations were changed with DMEM medium contained MTT (5 mg/mL) for 4 hours. Then, MTT was removed, and 100 µL DMSO was dropped in to dissolve the formazan crystals at 37 °C (dark, lasted for 4 hours). A microplate reader (Bio-Rad Model 680, UK) was used to measure the absorbance at 490 nm. The drug concentration at which the growth of 50% of the cells was inhibited (defined as IC50) was detected by curve fitting of the cell viability data in comparison with that of the control groups.
Colony Formation Unit Assay
The tumor cells were seeded in six-well plates with a density of 1×103 cells/well. After the cells were attached, the cells were treated with free Rapa, Rapa liposomes and 5-FU alone or in combination. Seven to ten days after the administration, the colonies were stained with crystal violet and photographed.
Cell Apoptosis Analysis
The apoptosis was detected by a flow apoptosis assay kit (KeyGEN BioTECH, China), and the cells were treated with Rapa, Rapa liposomes and 5-FU alone or in combination. After 48 hours of administration, the cells were collected by centrifugation, washed twice with PBS, and suspended in 400 uL of Annexin V binding solution. Then, 5 uL of Annexin V-FITC staining solution were added, and the solution was incubated for 15 minutes in the dark. Next, 10 uL of PI staining solution were added, and the solution was incubated for 5 minutes in the dark and immediately tested by flow cytometry.
Cell Migration Experiments
Cell migration experiments were performed using a transwell plate with a pore size of 8 µm. The density of cell suspension was 5 x 106/mL with a serum-free medium, which was added to the upper chamber of the transwell chamber, and a 10% FBS medium was added to the lower chamber. Meanwhile, the drug was added to the upper chamber for treatment with Rapa, Rapa liposomes and 5-FU alone or in combination. After 36 hours of treatment, the cells that were not migrated in the upper chamber were wiped with a cotton swab, fixed by crystal violet staining, and photographed.
Western Blot Analysis
The HCT-116 or SW-480 cells were seeded in six-well plates and divided into six groups for treatment with free Rapa, Rapa liposomes and 5-FU alone or in combination. The protein was extracted by cell RIPA lysis buffer containing a protease and phosphatase inhibitor cocktail and the protein concentration was measured using the BCA method. Separation was performed using SDS-PAGE gel. The protein samples were transferred onto the PVDF membranes, blocked with 5% skim milk (Solarbio, Beijing) or 5% BSA, washed twice with TBST, and then incubated with appropriate dilution of the primary antibody (BCL-2, diluted 1:1000; Bax, diluted 1:2000; Caspase3, diluted 1:1000; Cleaved Caspase3, diluted 1:1000; AKT, diluted 1:1000; p-AKT (ser 473), diluted 1:1000; p-AKT (Thr 308), diluted 1:1000; p70 S6 Kinase, diluted 1:1000; p-p70S6 Kinase, diluted 1:1000; P53, 1:2000; E-Cadherin, diluted 1:1000; N-Cadherin, diluted 1:1000; Vimentin, diluted 1:1000; MMP-9, diluted 1:1000; PCNA, diluted 1:5000; GAPAD, 1:1000) at 4 °C overnight. After washing with TBST three times, the samples were incubated with a secondary antibody for 1 hour at room temperature, and ECL illuminating solution (Millipore, USA) was used for development. Finally, the gray value of the band was analyzed by Image J software.
In vivo Imaging
Biodistribution assay was performed as described previously.20 After inoculation of HCT-116 cells in nude mice, the tumor volume was as long as ~250 mm3, and free DiR and DiR liposomes were injected into the tail vein. The concentration of DiR was 1.5 mg/kg, and the distribution of DiR in vivo was observed and photographed by an Xenogen IVIS100 imaging system (XenogenCorp., Hopkinton, MA, USA) at time points of 6 hours, 18 hours, and 24 hours after liposomes administration. Mice were sacrificed at 24 hours and the fluorescence of the major organs (heart, liver, spleen, lung, stomach, kidneys, tumor, blood) were also measured.
In vivo Antitumor Effects
About 100 μL 5×106 HCT-116 cells in PBS mixed with 100 μL matrigel (BD Biosciences) were implanted subcutaneously into the backs of mice. Of the injected mice, 95% developed a tumor with an average volume of about 50~100 mm3 after seven days. The mice were randomly divided into six groups (n = 6), which were intravenously injected every two days with saline, Rapa (2 mg/kg), Rapa liposomes (2 mg/kg), 5-FU (30 mg/kg), Rapa joint 5-FU group and Rapa liposomes joint 5-FU group, respectively. Next, a vernier caliper was used to survey the tumor size every other day, and the tumor volume (V) = length×width2/2. The antitumor efficacy was determined based on tumor growth and terminal tumor weight.
Hematoxylin and Eosin (H&E) Staining
After the mice were sacrificed, the tumor tissues were embedded in paraffin, deparaffinized, stained with hematoxylin nuclear stain semen and 0.5% eosin staining solution, and sealed with neutral gum. The pictures were detected with microscope (Nikon, Ni-u, Tokyo, Japan) in five random fields.
Immunohistochemistry
Paraffin sections were deparaffinized, and blocked with serum, primary antibody working solution (ki-67, diluted 1:200) was added at 4 °C overnight, washed with PBS twice for 5 minutes each time, and incubated with a secondary antibody at room temperature for 30 minutes. Then, 1–2 drops of DAB Plus Chromogen was mixed with 1 mL of DAB plus substrate was incubated with the sections for 5~15 minutes. The samples were detected with a microscope (Nikon, Ni-u, Tokyo, Japan) and the positive ki-67 cells were counted in five random fields.
TUNEL Assay
The apoptotic cells were investigated by TdT-mediated dUTP nick-end labeling assay (TUNEL, Promega, USA) in vivo. In brief, paraffin sections were first dehydrated and transparent, followed by permeation with DNase-free proteinase K, and washed three times with PBS. Add the prepared TUNEL detection solution to the section and incubate at 37 °C for 60 minutes. Finally, the section was mounted with an anti-fluorescence quenching mounting solution and observed under a confocal microscope (Nikon, A1R+N-STORM, Tokyo, Japan).
Statistical Analysis
GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA) was used to draw the picture. SPSS 17.0 was used for statistical analysis. One-way analysis of variance (ANOVA) was used to analyze the significance between the groups. All data were expressed as mean values ± SD, and P < 0.05 indicated significance.
Results and Discussion
Characterization of Rapa Liposomes
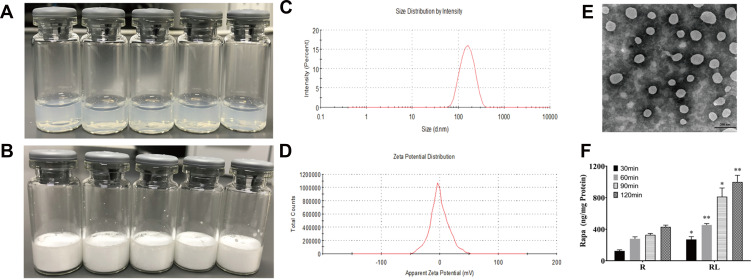
The delivery of drugs using nanocarriers is a new approach to the treatment of cancer.14–17 The appearance of Rapa liposomes is shown in Figure 1A, the clear solution of Rapa liposomes was observed, which indicated that delivery of Rapa by liposomes optimizes the solubility problems. Moreover, the Rapa liposomes could be lyophilized into a power form (Figure 1B). As can be noted from Figure 1C and D, the particle size of Rapa liposomes was about 100±5.5 nm, the polymer dispersity index (PDI) was 0.125–0.25, and the zeta potential of the Rapa liposomes was about −3.59±0.72 mV, which implied a nearly neutral surface charge. The results of the TEM show that the shape of Rapa liposomes was spherical (Figure 1E), and the particle size is similar to that measured by DLS. The Rapa liposomes had a drug loading and encapsulation efficiency of 9.0 ± 0.5% and 80 ± 2.5%, respectively. Meanwhile, the particle size and drug loading of re-dissolved Rapa liposomes was consistent with liposomes before lyophilization.
Figure 1.
Characterization of Rapamycin liposomes. (A) Photograph of rapamycin liposomes. (B) Photograph of freeze-dried powder of rapamycin liposomes. (C) The particle size distribution of rapamycin liposomes. (D) The Zeta potential of rapamycin liposomes. (E) Representative TEM image of rapamycin liposomes.Scale bar represents 200 nm. (F) Free rapamycin and rapamycin liposomes cellular uptake by HCT-116 cells. *p<0.05, and **p<0.01.
Abbreviations: R, rapamycin; RL, rapamycin liposomes.
We also used LC-MS to detect the cellular uptake of Rapa liposomes and free Rapa by the HCT-116 cells. The results showed that over a 30–120 min period, the accumulation Rapa concentration of the Rapa liposomes was found to 1.5–2 fold higher than that of free Rapa. These results indicated that Rapa liposomes are capable of obviously increasing the intracellular delivery of Rapa in a time dependent manner (*P<0.05,**P<0.01, Figure 1F). This phenomenon may be attributed to the liposomes present of endocytosis advantage.21,22 Besides, when the liposomes enter into HCT-116 cells, sustained intracellular release of Rapa also lead to a raised intracellular concentration of Rapa.23
Effects of Rapa Liposomes and 5-FU on Cell Proliferation
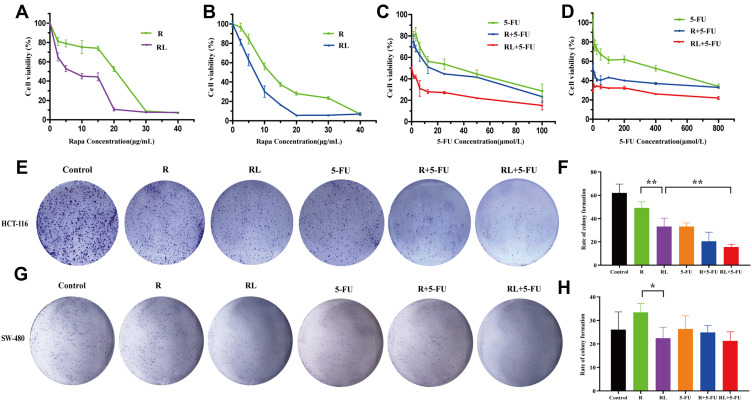
In order to verify whether the enhanced cellular uptake of Rapa liposomes increase the antitumor efficacy of rapamycin on CRC cells, the in vitro cytotoxicity effect of free Rapa and Rapa liposomes against HCT-116 and SW-480 cells was evaluated by the MTT assay. As shown in Figure 2A and B, free Rapa and Rapa liposomes inhibited the growth of HCT-116 and SW-480 cells in a dose-dependent manner, but the IC50 of the Rapa liposomes was lower than that of the free Rapa in the both cells (Table 1). These results clearly show that liposomes improved the cytotoxicity of Rapa to CRC cells.
Figure 2.
Cell proliferation and colony formation of colorectal cancer cells treated by different drug. The cell viability of (A) HCT-116 cells and (B) SW-480 cells after treatment rapamycin or rapamycin liposomes for 48 hours. The cell viability of (C) HCT116 cells and (D) SW-480 cells after treatment rapamycin liposomes or 5-Fu alone or in combination for 48 hours. The clone forming ability of HCT-116 cells was photographed (E) and their relative numbers were calculated (F). The clone forming ability of SW-480 cells was photographed (G) and their relative numbers were calculated (H). All data are expressed as the mean±SD (n=3).*p<0.05 and **p<0.01.
Abbreviations: R, rapamycin; RL, rapamycin liposomes.
Table 1.
Inhibitory Concentration (IC50 in μg/mL) of Free Rapa and Rapa Liposomes
| Cell Line | Free Rapa | Rapa Liposomes |
|---|---|---|
| HCT-116 | 15.34+1.18 | 5.61±0.75 |
| SW480 | 12.22+1.08 | 7.52±0.51 |
Next, we evaluated the cytotoxicity of 5-FU in the presence of free Rapa or Rapa liposomes of CRC cells. In contrast 5-FU alone and the combined treatment Rapa liposomes significantly increased the cytotoxicity in HCT-116 cells (Figure 2C). Similarly, the IC50 of 5-FU treatments was higher than that of combined treatments in SW-480 cells (Figure 2D). Overall, Rapa liposomes enhanced the 5-FU-induced cytotoxicity of CRC cells.
To further confirm the proliferation of CRC cells treatment Rapa liposomes or 5-Fu alone or in combination, colony formation was performed. The numbers of clones in the free Rapa group and Rapa liposomes group of HCT-116 cells were 49.2±5.2 and 33.1±7.3, respectively (**p<0.01, Figure 2E and F). At the same time, similar results were found in SW-480 cells which were shown in Figure 2G and H (*p<0.05). Moreover, the number of cell clones in the Rapa liposomes and 5-FU combination groups was obviously less than that of the single-drug in HCT-116 cell lines (**p<0.01).
Effects of Rapa Liposomes and 5-FU on Cell Apoptosis
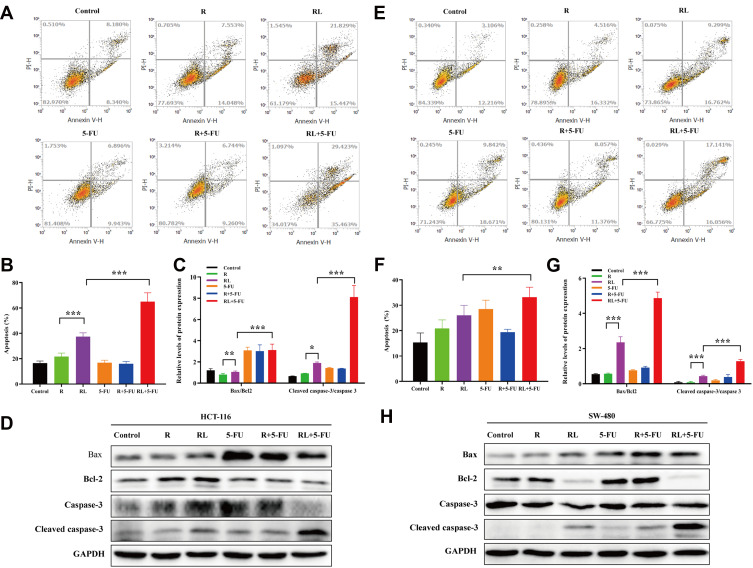
The apoptosis of HCT-116 and SW-480 cells was detected by Annexin V-PI double staining. The apoptotic rate of Rapa in HCT-116 cells was 21.6±2.8%, while the Rapa liposomes group markedly enhanced apoptosis rate, resulting in 37.2±3.1% (***P<0.001). Moreover, the combined treatment with Rapa liposomes and 5-FU resulted in 64.8±7.1% apoptosis rate (***P<0.001, Figure 3A and B). Figure 3E and F also show that the combination treatment induced cell apoptosis more effectively than the other treatments in SW-480 cell lines (**P<0.01). Subsequently, the corresponding apoptosis protein Bax/Bcl-2 ratio and cleaved caspase 3 were obviously up-regulated in the combination treatment group (Figure 3C, D, G and H).
Figure 3.
Cell apoptosis and the related signaling pathways of colorectal cancer cells treated by different drug. (A and B) Cell apoptosis analysis of HCT-116 cells via FITC Annexin V/PI double staining, and the percentage of apoptotic rates was calculated. (C and D) The Bax, Bcl-2, Caspase-3 and cleaved Caspase-3 protein expression in HCT-116 cells was examined by Western blot analysis, and the protein levels were quantified by densitometry. (E and F) Cell apoptosis analysis of SW-480 cells via FITC Annexin V/PI double staining, and the percentage of apoptotic rates was calculated. (G and H) The Bax, Bcl-2, Caspase-3 and cleaved Caspase-3 protein expression in SW-480 cells was examined by Western blot analysis, and the protein levels were quantified by densitometry. All data are expressed as the mean±SD (n=3). *p<0.05, **p<0.01 and ***p <0.001.
Abbreviations: R, rapamycin; RL, rapamycin liposomes.
Effects of Rapa Liposomes and 5-FU on Cell Migration
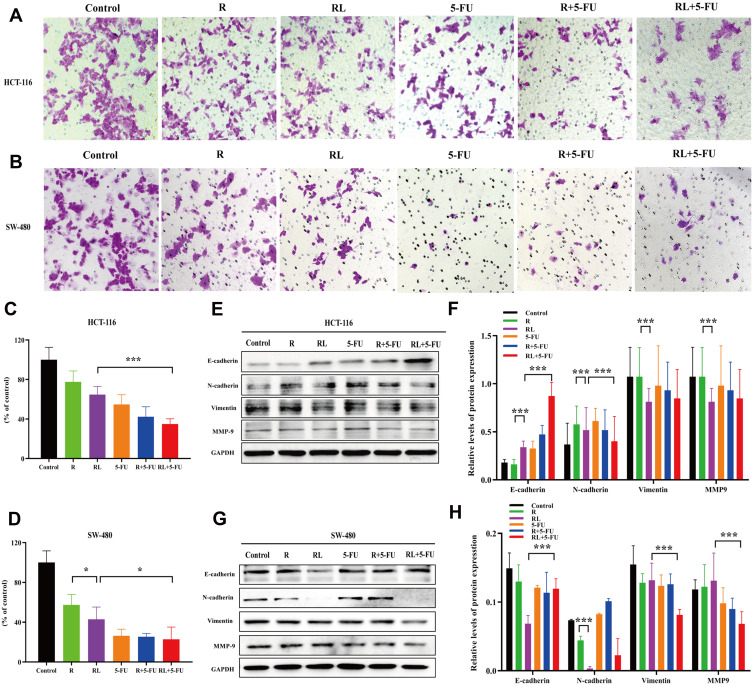
To determine whether Rapa liposomes or 5-Fu alone or in combination inhibited the metastasis of colorectal cells, we examined the cell migration by transwell assay. As shown in Figure 4B and D, compared with the free Rapa group, the migratory number of the Rapa liposomes group was markedly decreased in SW-480 cells (*P<0.05). Moreover, the migratory cell numbers of the Rapa liposomes and 5-FU combination group was slightly less than that of the single-drug group in HCT-116 (***P<0.01, Figure 4A and C) and SW-480 cells (*P<0.05). Taken together, these results demonstrate that a combination of Rapa liposomes and 5-FU shows a potential effect of inhibiting the metastasis of CRC.
Figure 4.
Cell migration and the related signaling pathways of colorectal cancer cells treated by different drug. Representative images (A) and quantitative data (C) of the transwell migration assay to assess the effect of drugs on HCT-116 cells. Representative images (B) and quantitative data (D) of the transwell migration assay to assess the effect of drugs on SW-480 cells. (E and F) The expression of the E-Cadherin, N-Cadherin, vimentin, and MMP-9 proteins in HCT-116 cells were analyzed by Western blot. (G and H) The expression of the E-Cadherin, N-Cadherin, vimentin, and MMP-9 proteins in SW-480 cells were analyzed by Western blot. All data are expressed as the mean±SD (n = 3) *p<0.05, and ***p<0.001. Scale bar = 50 μm.
Abbreviations: R, rapamycin; RL, rapamycin liposomes.
The occurrence of Epithelial-Mesenchymal Transition (EMT) in epithelial cancer cells is a key factor in tumor migration and invasion. Then, the expression of the proteins involved in EMT was detected. As shown in Figure 4E–H, compared with the free Rapa group, the expression of epithelial cell marker E-Cadherin was up-regulated in the Rapa liposomes, while N-Cadherin, vimentin, and MMP-9 were down-regulated. Additionally, we found the combination group further inhibited N-Cadherin, vimentin, and MMP-9 but promoted the E-cadherin expression in both CRC cells.
The Antitumor Mechanism of Rapa Liposomes and 5-FU In vitro
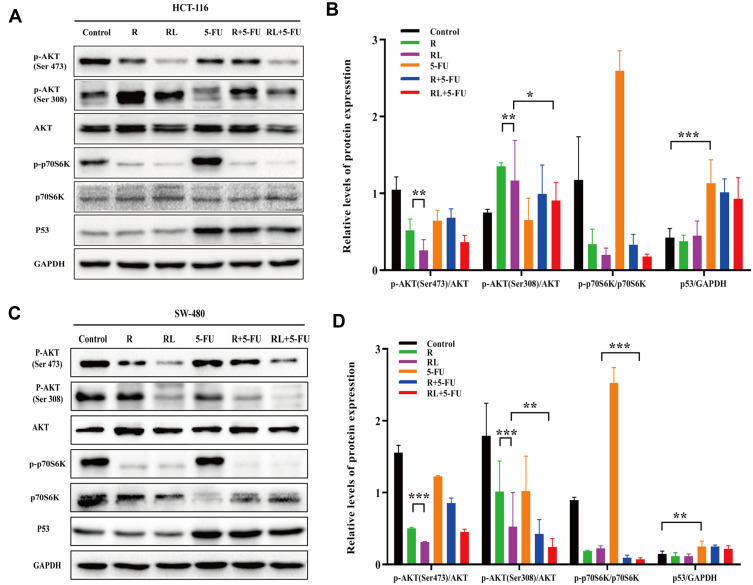
To analyze the possible mechanism of how Rapa liposomes and 5-FU inhibit proliferation and induce apoptosis in vitro, Western blot analysis was used to detect the expression of related proteins. The AKT/mTOR pathway regulates various cellular processes which include protein synthesis, cell proliferation, energy metabolism and autophagy.24–27 mTOR inhibitor Rapa inhibits the expression of p-p70S6K and regulates downstream pathways. Thus, pS6K is used as a marker for mTOR1 activity.28 As shown in Figure 5A–D, there was no change in total AKT and p70S6K protein expression in the HCT-116 and SW-480 cells, but the expression of p-AKT (Ser 473), p-AKT (Thr 308), and p-p70S6K (Thr 398) was decreased after treatment of tumors with free Rapa and Rapa liposomes. However, the protein levels were significantly down-regulated when treated with Rapa liposomes compared with free Rapa (***P<0.001).
Figure 5.
The antitumor mechanism of colorectal cancer cells treated by different drug. (A and B) The expression of the p-AKT (Ser 473), p-AKT (Thr 308), p-p70S6K (Thr 389) and P53 proteins in HCT-116 cells were analyzed by Western blot. (C and D) The expression of the p-AKT (Ser 473), p-AKT (Thr 308), p-p70S6K (Thr 389) and P53 proteins in SW-480 cells were analyzed by Western blot. All data are expressed as the mean±SD (n = 3) *p<0.05, **p<0.01 and ***p<0.001.
Abbreviations: R, rapamycin; RL, rapamycin liposomes.
On the other hand, 5-FU acting as a thymidylate synthase inhibitor and metabolite interferes with DNA and RNA, thus leading to inhibition of cell proliferation and inducing apoptosis.29 Our results show that 5-FU significantly increased P53 expression, thereby activating the downstream apoptotic pathway (Figure 5A–D). These results suggested that the Rapa liposomes synergized with the antitumor effect of 5-FU by targeting the AKT/mTOR and P53 signaling pathway.
Notably, excessive inhibition of mTOR negatively regulates the AKT pathway.30 However, Rapa combined with other drugs would attenuate this effect. For example, the combination of Rapa and melatonin blocked the negative feedback loop, which decreased head and neck squamous cell carcinoma cell viability, proliferation and clonogenic capacity.31 Meanwhile, autophagy activator Rapa combined 5-FU may overcome multidrug resistance in cancer.10,32 Collectively, Rapa is an ideal chemosensitizer for 5-FU treatment of CRC.
In vivo Biodistribution of DiR Liposomes
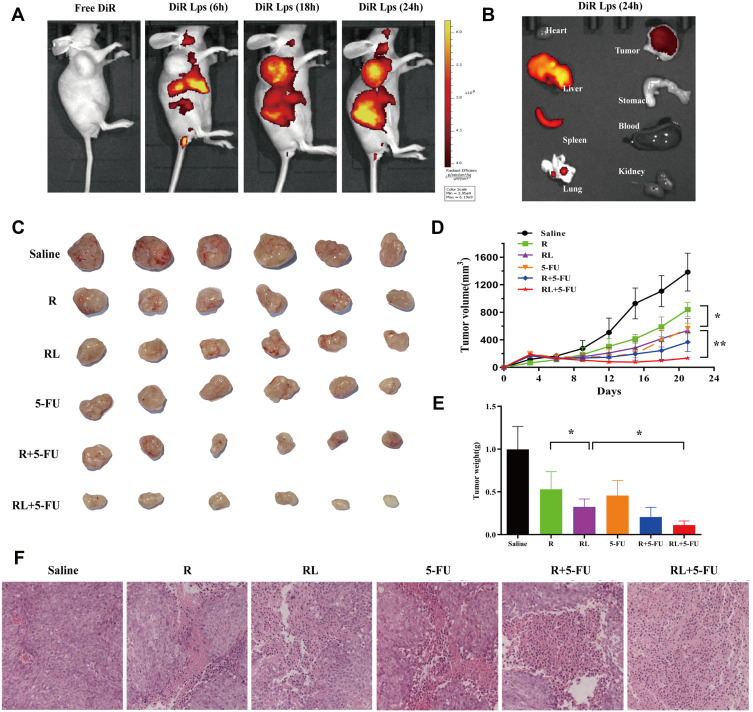
The distribution of DiR liposomes in nude mice was detected by in vivo imaging. The same doses of DiR and DiR liposomes were injected into the tail vein. As expected, DiR liposomes showed fluorescence in the transplanted tumor 6 h after administration, and obvious fluorescence enrichment occurred at 24 h (Figure 6A). In this study, the PEGylated liposomes were used to deliver Rapa to improve the anti-CRC effect. The PEG segments are located at the surface of the liposomes and formed a layer of hydration film, which reduces the interactions with serum proteins and improves the spatial stability.33,34 Therefore, the PEGylated liposomes extend the circulation time of liposomes in vivo and evade scavenging by the reticuloendothelial system (RES).35 Moreover, the liposomes with a small size (10–200 nm) passively accumulate in solid tumors due to the enhanced permeability and retention (EPR) effect.36–38 Figure 6B shows the fluorescent images of major organs and tumors after injection at 24h, which further suggests that liposomes exhibit high tumor accumulation. Some fluorescence intensity was also detected in the liver and spleen. The reason for this phenomenon is kupffer cells in liver participate in the uptaking and demoting of extra-phagocytosis.39
Figure 6.
The Biodistribution and antitumor efficacy of different treatment in HCT-116 xenograft mice. (A) Representative live animal fluorescence images of DiR-loaded liposomes in HCT-116 induced xenograft mice. (B) Representative fluorescence images of organs and tumor of mice at 24 h post injection. (C) Photo of subcutaneous tumors in each group. (D) Subcutaneous tumor volume development curve in each group. (E) Weight of subcutaneous tumors in each group. (F) Representative HE staining images of tumors in each group, Scale bar = 50 μm. All data are expressed as the mean±SD (n = 6). *p<0.05 and **p<0.01.
Abbreviations: R, rapamycin; RL, rapamycin liposomes.
The Antitumor Effects of Rapa Liposomes and 5-FU In vivo
We investigated the antitumor effects of Rapa liposomes and 5-FU in HCT-116 cell xenograft mice. A representative tumor from each group was shown in Figure 6C, the volume of the saline group, Rapa group, Rapa liposomes group, and Rapa liposomes and 5-FU combination group were 1383.3±276.1 mm3, 839.8±103.9 mm3, 536.8±174.1 mm3 (*P<0.05), and 134.2±31.1 mm3 (**P<0.01), respectively. The tumor inhibition rate of the combination group resulted in 90% when compared with the saline group (Figure 6D). Those indicating that the combination of Rapa liposomes and 5-FU is a potentially effective treatment for CRC. This is consistent with previous studies that the delivery of antitumor drugs by micelles or nanoparticles can improve the therapeutic effect of the drug.14,19,40,41 Importantly, the antitumor effect of Rapa has been greatly improved compared to what was previously reported in the literature.9
The removed tumor tissues were weighed, the group treated with Rapa liposomes showed reduced tumor weight in comparison with free Rapa (*P<0.05). Moreover, the tumor weight of the combination of Rapa liposomes and 5-FU group was lower than that of a single-drug administration group (*P<0.05, Figure 6E). Noteworthy, HE staining in the tumor tissue found that the combined administration group exhibited a larger death area (Figure 6F).
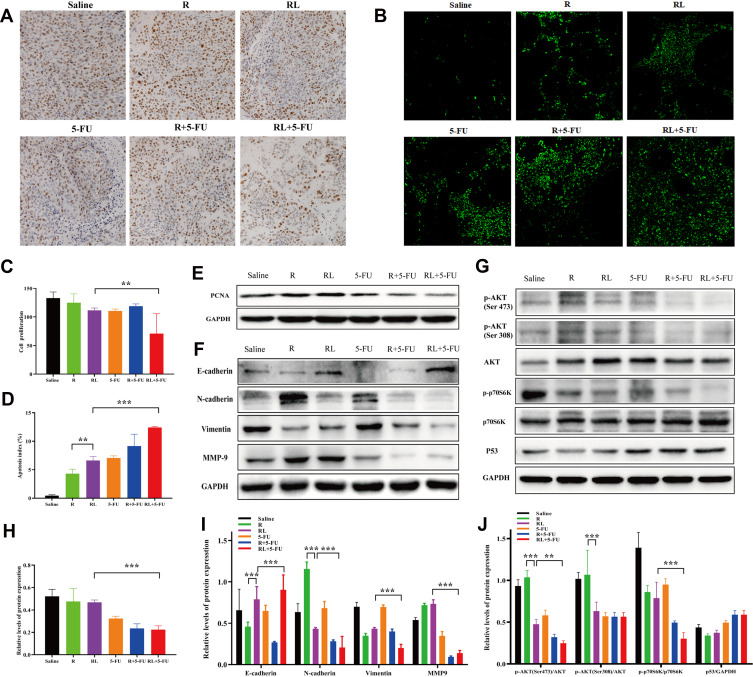
Simultaneously, the proliferation of tumor cells in vivo was detected by ki-67 staining. As shown in Figure 7A and C, the ki-67 positive cells were significantly decreased in the combination group compared with that in other groups (**P<0.01). Furthermore, we also examined the expression of PCNA protein in tumor tissues. The PCNA protein expression in the combination group was lower than that of the single-drug group (***P<0.001, Figure 7E and H). These findings imply that inhibiting the proliferation of tumor cells may be one of its antitumor mechanisms.
Figure 7.
Analysis proliferation, apoptosis and metastasis in vivo and detection of related signaling pathways. Representative Ki67 immunohistochemistry images (A) and quantitative data (C) of tumors in each group. Representative TUNEL immunofluorescent images (B) and quantitative data (D) of tumors in each group. (E and H) The expression of the PCNA protein in tumors were analyzed by Western blot. (F and I) The expression of the E-Cadherin, N-Cadherin, vimentin, and MMP-9 proteins in tumors were analyzed by Western blot. (G and J) The expression of the p-AKT (Ser 473), p-AKT (Thr 308), p-p70S6K (Thr 389) and P53 proteins in tumors were analyzed by Western blot. All data are expressed as the mean±SD (n = 3) Scale bar = 50 μm. **p<0.01 and ***p <0.001.
Abbreviations: R, rapamycin; RL, rapamycin liposomes.
In addition, the TUNEL assay was applied to discover the apoptosis in vivo. As shown in Figure 7B, the greenish fluorescence was observed in colorectal cancer tumor tissue from the group treated with the drug. The apoptosis index was in the combination of Rapa liposomes and 5-FU group (12.42%±0.15%, ***P<0.001) significantly higher than that in groups treated with free Rapa (4.32%±0.75%), Rapa liposomes (6.62%±0.70%, **P<0.01), and 5-FU (7.07%±0.36%, Figure 7D), respectively. These results suggested that the induction of tumor cell apoptosis is another antitumor mechanism in vivo.
The Antitumor Mechanism of Rapa Liposomes and 5-FU In vivo
To further confirm the antitumor mechanism in vivo, we examined the expression of proteins in a tissue specimen of HCT-116 xenograft tumor. Firstly, the EMT associated protein levels of N-Cadherin, vimentin and MMP-9 expression was down-regulation, but promoted E-cadherin expression after treatment of tumors with Rapa liposomes and 5-FU (Figure 7F and I). Our results demonstrated that combination treatments can inhibit tumor cell EMT consistent with previous studies.42
In addition, the AKT/mTOR and P53 pathway were detected in tumor tissues. As shown in Figure 7G and J, Rapa and Rapa liposomes inhibit the AKT/mTOR pathway by down-regulating the expression of p-AKT (Ser 473), p-AKT (Ser308), and p-p70S6K (Thr 389) proteins. Meanwhile, 5-FU increased the expression of the P53 protein. These results showed that combination of Rapa liposomes and 5-FU affected the expression of Akt/mTOR and P53 signaling pathway and achieved synergistic antitumor effects in vivo.
Conclusion
In general, we developed Rapa liposomes for treatment CRC. The cellular uptake, cytotoxicity and cell apoptosis of Rapa liposomes were increased in vitro compared with free Rapa. Meanwhile, Rapa liposomes were more effective in suppressing tumor growth in vivo. Furthermore, Rapa liposomes combined with 5-FU achieves a synergistic antitumor effect via the Akt/mTOR and P53 pathways in vitro and in vivo. In the future, multiple researches indicate that the liposomes or nanoparticles-based co-delivery of rapamycin and clinical chemotherapy drugs system achieve the synergistic/combined effect, while decreasing the systemic toxicity. Besides, co-delivery of chemotherapeutic agents by liposomes can overcome tumor multidrug resistance.43,44 Of note, further modification of the liposome surface, such as, folic acid or hyaluronic acid, enhance the active targeting effect and improve the antitumor effect.45,46
Acknowledgments
This work was supported by the Science and Technology Major Project Foundation of Guangzhou of Guangdong (Grant-in-Aid 201604020166) and Guangzhou Science and Technology Program key projects (No. 201903010014).
Disclosure
The authors declare no conflicts of interests.
References
- 1.Dekker E, Tanis PJ, Vleugels J, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. doi: 10.1016/S0140-6736(19)32319-0 [DOI] [PubMed] [Google Scholar]
- 2.Tian T, Li X, Zhang J. mTOR signaling in cancer and mTOR inhibitors in solid tumor targeting therapy. Int J Mol Sci. 2019;20(3):755. doi: 10.3390/ijms20030755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marquard FE, Jucker M. PI3K/AKT/mTOR signaling as a molecular target in head and neck cancer. Biochem Pharmacol. 2020;172:113729. doi: 10.1016/j.bcp.2019.113729 [DOI] [PubMed] [Google Scholar]
- 4.Gopalakrishnan K, Venkatesan S, Low E, Hande MP. Effects of rapamycin on the mechanistic target of rapamycin (mTOR) pathway and telomerase in breast cancer cells. Mutat Res Genet Toxicol Environ Mutagen. 2018;836(Pt B):103–113. doi: 10.1016/j.mrgentox.2018.03.008 [DOI] [PubMed] [Google Scholar]
- 5.Iorio AL, Da RM, Pisano C, de Martino M, Genitori L, Sardi I. Combined treatment with doxorubicin and rapamycin is effective against in vitro and in vivo models of human glioblastoma. J Clin Med. 2019;8(3). doi: 10.3390/jcm8030331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B, Yang J, Lu Z, Liu B, Liu F. A study on the mechanism of rapamycin mediating the sensitivity of pancreatic cancer cells to cisplatin through PI3K/AKT/mTOR signaling pathway. J BUON. 2019;24(2):739–745. [PubMed] [Google Scholar]
- 7.Zhou WJ, Chang KK, Wu K, et al. Rapamycin synergizes with cisplatin in antiendometrial cancer activation by improving IL-27-stimulated cytotoxicity of NK cells. Neoplasia. 2018;20(1):69–79. doi: 10.1016/j.neo.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai W, Yang F, Ma L, et al. Combined mTOR inhibitor rapamycin and doxorubicin-loaded cyclic octapeptide modified liposomes for targeting integrin alpha3 in triple-negative breast cancer. Biomaterials. 2014;35(20):5347–5358. doi: 10.1016/j.biomaterials.2014.03.036 [DOI] [PubMed] [Google Scholar]
- 9.Chao TH, Chang GR, Chen WY, Chen PL, Mao FC. The synergistic effect of rapamycin combined with 5-fluorouracil in BALB/cByJNarl mice bearing CT-26 tumor cells. Anticancer Res. 2014;34(7):3329–3335. [PubMed] [Google Scholar]
- 10.Yang JW, Zhang QH, Liu T. Autophagy facilitates anticancer effect of 5-fluorouracil in HCT-116 cells. J Cancer Res Ther. 2018;14(Supplement):S1141–S1147. doi: 10.4103/0973-1482.204898 [DOI] [PubMed] [Google Scholar]
- 11.Haeri A, Osouli M, Bayat F, Alavi S, Dadashzadeh S. Nanomedicine approaches for sirolimus delivery: a review of pharmaceutical properties and preclinical studies. Artif Cells Nanomed Biotechnol. 2018;46(sup1):1–14. doi: 10.1080/21691401.2017.1408123 [DOI] [PubMed] [Google Scholar]
- 12.Falkowski S, Woillard JB. Therapeutic drug monitoring of everolimus in oncology: evidences and perspectives. Ther Drug Monit. 2019;41(5):568–574. doi: 10.1097/FTD.0000000000000628 [DOI] [PubMed] [Google Scholar]
- 13.Flaifel A, Xie W, Braun DA, et al. PD-L1 expression and clinical outcomes to cabozantinib, everolimus, and sunitinib in patients with metastatic renal cell carcinoma: analysis of the randomized clinical trials METEOR and CABOSUN. Clin Cancer Res. 2019;25(20):6080–6088. doi: 10.1158/1078-0432.CCR-19-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan R, Tong A, Li X, et al. Enhanced antitumor effects by docetaxel/LL37-loaded thermosensitive hydrogel nanoparticles in peritoneal carcinomatosis of colorectal cancer. Int J Nanomedicine. 2015;10:7291–7305. doi: 10.2147/IJN.S89066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Duo Y, Bi J, et al. Targeted delivery of anti-miR-155 by functionalized mesoporous silica nanoparticles for colorectal cancer therapy. Int J Nanomedicine. 2018;13:1241–1256. doi: 10.2147/IJN.S158290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashour AE, Badran M, Kumar A, Hussain T, Alsarra IA, Yassin A. Physical PEGylation enhances the cytotoxicity of 5-fluorouracil-loaded PLGA and PCL nanoparticles. Int J Nanomedicine. 2019;14:9259–9273. doi: 10.2147/IJN.S223368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabary DM, Helmy MW, Elkhodairy KA, Fang JY, Elzoghby AO. Hyaluronate/lactoferrin layer-by-layer-coated lipid nanocarriers for targeted co-delivery of rapamycin and berberine to lung carcinoma. Colloids Surf B Biointerfaces. 2018;169:183–194. doi: 10.1016/j.colsurfb.2018.05.008 [DOI] [PubMed] [Google Scholar]
- 18.Yoon HY, Chang IH, Goo YT, et al. Intravesical delivery of rapamycin via folate-modified liposomes dispersed in thermo-reversible hydrogel. Int J Nanomedicine. 2019;14:6249–6268. doi: 10.2147/IJN.S216432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YC, Lo CL, Lin YF, Hsiue GH. Rapamycin encapsulated in dual-responsive micelles for cancer therapy. Biomaterials. 2013;34(4):1115–1127. doi: 10.1016/j.biomaterials.2012.10.034 [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Wu Y, Chen J, et al. A simple glutathione-responsive turn-on theranostic nanoparticle for dual-modal imaging and chemo-photothermal combination therapy. Nano Lett. 2019;19(8):5806–5817. doi: 10.1021/acs.nanolett.9b02769 [DOI] [PubMed] [Google Scholar]
- 21.Gilabert-Oriol R, Chernov L, Anantha M, Dragowska WH, Bally MB. In vitro assay for measuring real time topotecan release from liposomes: release kinetics and cellular internalization. Drug Deliv Transl Res. 2017;7(4):544–557. doi: 10.1007/s13346-017-0380-9 [DOI] [PubMed] [Google Scholar]
- 22.Pu Y, Zhang H, Peng Y, et al. Dual-targeting liposomes with active recognition of GLUT(5) and α(v)β(3) for triple-negative breast cancer. Eur J Med Chem. 2019;183:111720. doi: 10.1016/j.ejmech.2019.111720 [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Chen R, Morott J, Repka MA, Wang Y, Chen M. mPEG-b-PCL/TPGS mixed micelles for delivery of resveratrol in overcoming resistant breast cancer. Expert Opin Drug Deliv. 2015;12(3):361–373. doi: 10.1517/17425247.2014.951634 [DOI] [PubMed] [Google Scholar]
- 24.Alzahrani AS. PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin Cancer Biol. 2019;59:125–132. doi: 10.1016/j.semcancer.2019.07.009 [DOI] [PubMed] [Google Scholar]
- 25.Costa R, Han HS, Gradishar WJ. Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: a review. Breast Cancer Res Treat. 2018;169(3):397–406. doi: 10.1007/s10549-018-4697-y [DOI] [PubMed] [Google Scholar]
- 26.Xu Z, Han X, Ou D, et al. Targeting PI3K/AKT/mTOR-mediated autophagy for tumor therapy. Appl Microbiol Biotechnol. 2020;104(2):575–587. doi: 10.1007/s00253-019-10257-8 [DOI] [PubMed] [Google Scholar]
- 27.Mirza-Aghazadeh-Attari M, Ekrami EM, Aghdas S, et al. Targeting PI3K/Akt/mTOR signaling pathway by polyphenols: implication for cancer therapy. Life Sci. 2020:117481. doi: 10.1016/j.lfs.2020.117481. [DOI] [PubMed] [Google Scholar]
- 28.Jhanwar-Uniyal M, Wainwright JV, Mohan AL, et al. Diverse signaling mechanisms of mTOR complexes: mTORC1 and mTORC2 in forming a formidable relationship. Adv Biol Regul. 2019;72:51–62. doi: 10.1016/j.jbior.2019.03.003 [DOI] [PubMed] [Google Scholar]
- 29.Mazrouei R, Raeisi E, Lemoigne Y, Heidarian E. Activation of p53 gene expression and synergistic antiproliferative effects of 5-fluorouracil and β-escin on MCF7 cells. J Med Signals Sens. 2019;9(3):196–203. doi: 10.4103/jmss.JMSS_44_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Huang F, Zhang Z, et al. Feedback activation of SGK3 and AKT contributes to rapamycin resistance by reactivating mTORC1/4EBP1 axis via TSC2 in breast cancer. Int J Biol Sci. 2019;15(5):929–941. doi: 10.7150/ijbs.32489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen YQ, Guerra-Librero A, Fernandez-Gil BI, et al. Combination of melatonin and rapamycin for head and neck cancer therapy: suppression of AKT/mTOR pathway activation, and activation of mitophagy and apoptosis via mitochondrial function regulation. J Pineal Res. 2018;64(3). doi: 10.1111/jpi.12461 [DOI] [PubMed] [Google Scholar]
- 32.Murugan AK. mTOR: role in cancer, metastasis and drug resistance. Semin Cancer Biol. 2019;59:92–111. doi: 10.1016/j.semcancer.2019.07.003 [DOI] [PubMed] [Google Scholar]
- 33.Yan X, Scherphof GL, Kamps JA. Liposome opsonization. J Liposome Res. 2005;15(1–2):109–139. doi: 10.1081/lpr-64971 [DOI] [PubMed] [Google Scholar]
- 34.Zhao T, Liu Y, Gao Z, et al. Self-assembly and cytotoxicity study of PEG-modified ursolic acid liposomes. Mater Sci Eng C Mater Biol Appl. 2015;53:196–203. doi: 10.1016/j.msec.2015.04.022 [DOI] [PubMed] [Google Scholar]
- 35.Gao X, Wang S, Wang B, et al. Improving the anti-ovarian cancer activity of docetaxel with biodegradable self-assembly micelles through various evaluations. Biomaterials. 2015;53:646–658. doi: 10.1016/j.biomaterials.2015.02.108 [DOI] [PubMed] [Google Scholar]
- 36.Kalyane D, Raval N, Maheshwari R, Tambe V, Kalia K, Tekade RK. Employment of enhanced permeability and retention effect (EPR): nanoparticle-based precision tools for targeting of therapeutic and diagnostic agent in cancer. Mater Sci Eng C Mater Biol Appl. 2019;98:1252–1276. doi: 10.1016/j.msec.2019.01.066 [DOI] [PubMed] [Google Scholar]
- 37.Kang H, Rho S, Stiles WR, et al. Size-dependent EPR effect of polymeric nanoparticles on tumor targeting. Adv Healthc Mater. 2020;9(1):e1901223. doi: 10.1002/adhm.201901223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry JL, Reuter KG, Luft JC, Pecot CV, Zamboni W, DeSimone JM. Mediating passive tumor accumulation through particle size, tumor type, and location. Nano Lett. 2017;17(5):2879–2886. doi: 10.1021/acs.nanolett.7b00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao W, Zeng X, Wu J, et al. Polydopamine-based surface modification of novel nanoparticle-aptamer bioconjugates for in vivo breast cancer targeting and enhanced therapeutic effects. Theranostics. 2016;6(4):470–484. doi: 10.7150/thno.14184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan L, Yang L, Jin J, et al. Micro/nano-bubble-assisted ultrasound to enhance the EPR effect and potential theranostic applications. Theranostics. 2020;10(2):462–483. doi: 10.7150/thno.37593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang J, Zhao X, Wang X, et al. Targeted co-delivery of the iron chelator deferoxamine and a HIF1alpha inhibitor impairs pancreatic tumor growth. ACS Nano. 2019;13(2):2176–2189. doi: 10.1021/acsnano.8b08823 [DOI] [PubMed] [Google Scholar]
- 42.Lei HW, Cai J, Li CM, et al. Rapamycin combine with TAE on the growth, metastasis, and prognosis of hepatocellular carcinoma in rat models. Ann Hepatol. 2018;17(4):645–654. doi: 10.5604/01.3001.0012.0948 [DOI] [PubMed] [Google Scholar]
- 43.Eftekhari RB, Maghsoudnia N, Samimi S, Zamzami A, Dorkoosh FA. Co-delivery nanosystems for cancer treatment: a review. Pharm Nanotechnol. 2019;7(2):90–112. doi: 10.2174/2211738507666190321112237 [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Wang X, Yan J, et al. Nanoparticle ferritin-bound erastin and rapamycin: a nanodrug combining autophagy and ferroptosis for anticancer therapy. Biomater Sci. 2019;7(9):3779–3787. doi: 10.1039/c9bm00653b [DOI] [PubMed] [Google Scholar]
- 45.Zhao Z, Ukidve A, Kim J, Mitragotri S. Targeting strategies for tissue-specific drug delivery. Cell. 2020;181(1):151–167. doi: 10.1016/j.cell.2020.02.001 [DOI] [PubMed] [Google Scholar]
- 46.Huang G, Huang H. Hyaluronic acid-based biopharmaceutical delivery and tumor-targeted drug delivery system. J Control Release. 2018;278:122–126. doi: 10.1016/j.jconrel.2018.04.015 [DOI] [PubMed] [Google Scholar]