Abstract
MicroRNAs (miRNAs) are known to take part in different biological mechanisms, including biotic as well as abiotic cellular stresses. The present investigation was aimed to identify comparative expression profile of differentially expressed miRNAs among Sahiwal (Bos indicus) and Frieswal (Bos indicus × Bos taurus) cattle breeds during summer stress. Stress responses in animals were characterized by recording various physiological parameters, biochemical assays and expression profiling of heat shock protein 70 (Hsp70) during elevated environmental temperature. Ion Torrent-based deep sequencing as well as CLC-genomic analysis identified 322 and 420 Bos taurus annotated miRNAs among Sahiwal and Frieswal, respectively. A total 69 common miRNAs were identified to be differentially expressed during summer among the breeds. Out of the 69, a total 14 differentially expressed miRNAs viz. bta-mir 6536-2, bta-mir-2898, bta-mir-let-7b, bta-mir-425, bta-mir-2332, bta-mir-2478, bta-mir-150, bta-mir142, bta-mir-16a, bta-mir-2311, bta-mir-1839, bta-mir-1248-1, bta-mir-103-2 and bta-mir-181b were randomly selected for qRT-PCR-based validation. bta-mir-2898, bta-mir-6536-1, bta-mir-let-7b, bta-mir-2478, bta-mir-150, bta-mir-16a, bta-mir-2311, bta-mir-1032-b and bta-mir-181-b were significantly (p < 0.01) upregulated during summer among Frieswal in comparison to Sahiwal while, bta-mir 6536-2, bta-mir-2332, bta-mir142, bta-mir-1839 and bta-mir-1248-1 was significantly (p < 0.01) expressed at higher level in Sahiwal in contrast to Frieswal correlation coefficient analysis revealed that bta-mir(s)-150, 16a and 181b are negatively correlated (p < 0.05) with Hsp70 expression. Thus, this study identified that miRNA expression during summer stress can vary between the breeds which may reflect their differential post-transcriptional regulation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-020-02608-4.
Keywords: miRNAs, Sahiwal, Frieswal, Cattle, HSP70.1, Differential gene expression
Introduction
Environmental heat stress is one of the important abiotic factors that influence the normal physiology of dairy cattle which directly or indirectly impact on their productivity in terms of growth rate, milk production, as well as reproductive efficiency (Belhadj et al. 2015). Apart from short-term managemental strategies to alleviate the heat stress impacts, understanding the molecular mechanisms of the stress response may assist for developing long-term goals for the selection of thermo-tolerant animals.
Indigenous cattle breeds are low producers as compared to European breeds and thus extensive crossbreeding program was undertaken to upsurge the milk production of these local cattle breeds (Hansen 2004). Selection of superior genotypes for higher productivity of dairy animals undertakes the presence of optimum environmental conditions. Nevertheless, with climate change scenario, thermos tolerance is also an important trait to be considered in addition to production trait of dairy animals. Owing to their long time in habitation in the tropical climate, Zebu (Bos indicus) cattle is better fortified against heat stress as compared to the European cattle (Bos taurus) (Najjar et al. 2010). The underlying genetic causes of the thermo-tolerance are the subject of intense study. Thermo-tolerance is a complex trait regulated by multiple stressors genes. Nonetheless, the gene sequence variations can only explain a portion of phenotypic traits and the left over part is considered to be influenced by the epigenetics patterns of animals.
microRNAs (miRNAs) are noncoding RNAs of about 22 nucleotide in size, reported to play significant role in regulation of target genes related to biotic as well as abiotic stresses (Leung and Sharp 2010). Most of the miRNAs are transcribed from DNA sequences into “primary miRNAs” which processed into “precursor miRNAs” followed by mature miRNAs (O’Brien et al. 2018). Series of studies revealed the function miRNAs during stress in different mammalian systems (Xu et al. 2003; van Rooij et al. 2007; Flynt et al. 2007; Leung and Sharp 2010; Tiwari et al. 2018; Correia et al. 2019; Rattanapan et al. 2020).
At present, few of the reports also suggested the role of miRNAs in stress response among livestock species (Zheng et al. 2014; Muroya et al. 2016; Sengar et al. 2018a, b). Our recent two studies independently identified and catalogued a list of miRNAs differentially expressed among Frieswal (Bos indicus × Bos taurus) (Sengar et al. 2018b) and Sahiwal (Bos indicus) (Sengar et al. 2018a) cattle breeds during summer stress. The present study identified the comparative miRNA expression pattern among Sahiwal and Frieswal cattle breeds.
Materials and methods
Animal experimental protocols described in the present study was approved by the Institutional Animal Ethics Committee (IAEC), ICAR-Central institute for Research on Cattle, Meerut, Uttar Pradesh, India.
Animals and sample collection
Ten each clinically healthy and non-pregnant heifers of Sahiwal (Bos indicus) and Frieswal (Bos taurus × Bos indicus) cattle breed maintained under similar pattern of managemental procedure were selected randomly. Blood samples were collected from all the animals by jugular vein puncture using sodium heparin (10 IU/ml) as anticoagulant substances. Temperature Humidity Index (THI) as heat stress indicator regardless of whether the experimental animals were stressed or not. Samples were obtained from all the experimental animals during the range of two environmental temperature zones viz. (a) January–February, when the temperature ranges in between 15 and 18 °C with THI rang in between 61 and 66 (designated as “Normal Zone”) and April–June, when the temperature ranges in between 42 and 45 °C with THI range in between 82 and 85 (designated as “Summer Zone”). In both the cases, we used samples were collected at 2:00 P.M. on four to five different days in each zone from each animal and at least for 1 h prior to collection of the blood samples, animals were exposed to the environmental temperatures (either normal or summer zone). Blood samples were subjected for plasma separation as per our earlier described protocols (2018a, b).
Characterization of heat stress response
Various physiological parameters (rectal temperature, breathing rate, pulse rate as well as heat tolerance coefficient) and different biochemical assays viz. thiobarbituric acid reactive substances (TBARS), catalase (CAT) and glutathione peroxidase (GPx) were included for determination of stress responses in both the breeds. Detailed description of experimental designs for recording different physiological parameters and estimation of different biochemical parameters have been described in our two other studies that established the heat stress phenotypes (2018a, b).
Construction of small RNA libraries and deep sequencing
Our previous two independent studies (2018a, b) constructed a total four small RNA libraries from Frieswal and Sahiwal cattle breeds using total RNA-Seqkit v2 for small RNA libraries kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instruction. All the purified cDNA libraries were run on Agilent™2100 Bioanalyzer™instrument with the Agilent™DNA 1000 kit and their smear analysis was accomplished through 2100 expert software to determine the molar concentration of each cDNA libraries having the size range between 50 and 300 bp. Deep sequencing of the constructed cDNA libraries was performed using IonTorrent Personal Genome Machine (Life Technologies, Pleasanton, CA, USA).
Analysis of deep sequencing data
Detailed methodologies used for identification of miRNAs from the Ion Torrent-based deep sequencing results among Frieswal and Sahiwal can be obtained from our previous reports by Sengar et al. (2018a, b), respectively. Briefly, identified raw sequences were analyzed through CLC-Genomics work bench 8.0.2 software (Qiagen, Denmark). Obtained 15–26 nucleotide ranges of small RNA sequences were processed for mRNA, RFam and Repbase filter. The remaining sequences were subjected for comparison with miRBase21 (Kozomara et al. 2019) by online BLASTn search tool to identify the conserved miRNAs in bovine species (http://www.mirbase.org/). Finally, identified mature sequences of miRNAs were subjected for BLAST analysis against drafted bovine genome sequences (http://www.272ncbi.nlm.nih.gov/genome/82). Target Scan (Agarwal et al. 2015) software was utilized for prediction of their target mRNA related to heat stress.
Real-time PCR-based quantification
Isolated blood samples from natural and summer zones were subjected for total RNA isolation using PAX gene blood miRNA Kit (Qiagen, USA) as per the manufacturer’s recommendation. Extracted total RNA was quantified spectrophotometrically and the RNA integrity was determined by visualization of 18S as well as 28S ribosomal rRNA bands on agarose gel electrophoresis (Schroeder et al. 2006). Using M-MuLV reverse transcriptase and random primers, cDNAs were synthesized from isolated total RNAs using ProtoScript first-strand cDNA synthesis kit (New England Biolabs, Beverly, MA, USA). 1:10 dilution of cDNAs were used for quantification using real-time qPCR (Step One, Applied Biosystems, Foster City, CA, USA). A total 14 differentially expressed miRNAs (bta-mir 6536-2, bta-mir-2898, bta-mir-let-7b, bta-mir-425, bta-mir-2332, bta-mir-2478, bta-mir-150, bta-mir142, bta-mir-16a, bta-mir-2311, bta-mir-1839, bta-mir-1248-1, bta-mir-103-2 and bta-mir-181b) were selected randomly from the identified miRNA deep sequence databases of Frieswal (Sengar et al. 2018b) and Sahiwal (Sengar et al. 2018a) based on their fold changes (log2 values). miRNA primers were designed using miRprimer software (Busk 2014). Detailed primer sequences used for the validation of different miRNAs are presented in Table 1. Primers synthesized for the selected miRNAs had the analogous sequences as the Bos taurus miRNA with appropriate adjustments at their 5′ ends. bta-mir-3596 miRNA was used as an endogenous control (unpublished data) to validate the selected miRNAs. RT PCR was also used for quantification of HSP70.1 gene using beta-actin gene as an endogenous control (Deb et al. 2014; Bhanuprakash et al. 2016). SYBR Green® PCR master mix kit (Applied Biosystems, Foster City, CA, USA) was used to perform the qPCR reaction with a final reaction volume of 10 μl. ΔΔCt method was applied for the quantification of various candidates in different samples (Livak and Schmittgen 2001). The expression values obtained were normalized against the housekeeping candidates for allowing the assessment of samples independently and all the determinations were performed in triplicate.
Table 1.
Details of the primers used for the present study
| miRNA | Sequence | Length (base pair) |
|---|---|---|
| bta-miR-103-2 |
F: GCAGAGCAGCATTGTACAG R: GGTCCAGTTTTTTTTTTTTTTTCATAG |
19 27 |
| bta-mir-2898 |
F: GCAGTGGTGGAGATGC R: GGTCCAGTTTTTTTTTTTTTTTCCC |
16 25 |
| bta-mir-150 |
F: CTCCCAACCCTTGTACCA R: GGTCCAGTTTTTTTTTTTTTTTACACT |
18 27 |
| bta-mir-2478 |
F: GCAGGTATCCCACTTCTGA R: TCCAGTTTTTTTTTTTTTTTGGTGT |
19 25 |
| bta-mir-181b-2 |
F: GCAGAACATTCATTGCTGTC R: TCCAGTTTTTTTTTTTTTTTAACCCA |
20 26 |
| bta-mir-6536-2 |
F: GCCTAAGTATACGATGACTAGC R:TCCAGTTTTTTTTTTTTTTTACGA |
22 24 |
| bta-mir-2311 |
F: GTACTGAAACTGTGCTCGT R: TCCAGTTTTTTTTTTTTTTTACACCA |
19 26 |
| bta-mir-142 |
F: CGCAGCATAAAGTAGAAAGCA R: GGTCCAGTTTTTTTTTTTTTTTGTAGT |
21 27 |
| bta-mir-1248 |
F: AGACCTTCTTGTATAAGCACTGT R: GGTCCAGTTTTTTTTTTTTTTTAGCA |
19 26 |
| bta-mir-2332 |
F: GCGGTTTAAGGTCTTGGAG R: TCCAGTTTTTTTTTTTTTTTCTTTGTC |
19 27 |
| bta-mir-1839 |
F: GCAGAAGGTAGATAGAACAGGTC R: GGTCCAGTTTTTTTTTTTTTTTAACAAG |
23 28 |
| bta-mir-16a |
F: CGCAGGTACATGATGACT R: CAGGTCCAGTTTTTTTTTTTTTTTAGA |
18 27 |
| bta-let-7b |
F: GCGCAGTTTAATTATACGATAA R: TCCAGTTTTTTTTTTTTTTTGGAGG |
19 25 |
| bta-mir-425 |
F: GCGCGATTTAAGTATACGATCA R: TCCAGTTTTTTTTTTTTTTTCCAGT |
19 25 |
Statistical analysis
The data presented in this study (mean ± SEM) were analyzed by using SPSS statistical program (SPSS 10.0 for Windows; SPSS, Inc., Chicago, IL, USA). Significant differences were obtained by one-way ANOVA using the same SPSS program. Pearson Correlation Sig. (2-tailed) was used to study the correlation coefficient between the differentially expressed miRNAs during thermal stress with overexpressed heat shock protein 70 (HSP70).
Results
Characterization of stress response
Different physiological parameters (rectal temperature, respiratory rate and pulse rate) were recorded among Sahiwal and Frieswal during normal as well as summer zone of temperatures illustrated in Table 2. All the parameters were increased during summer months in comparison to normal environmental temperatures. Similarly, estimation of different biochemical parameters revealed significant (p < 0.05) higher levels of plasma catalase activity (nmol/min/ml), GPx (nmol/min/ml) and MDA (μM) concentration during summer seasons in both the breeds (Fig. 1). Further, HSP70 expression profiling revealed that during the summer zone the level of expression significantly (p < 0.05) increased in comparison to normal temperature zone (Fig. 2).
Table 2.
Different physiological parameters documented during different environmental temperature zone
| Breed | Zone | RT | RR | PR |
|---|---|---|---|---|
| Frieswal | NZ | 38.09 ± 0.52 | 32.46 ± 0.41 | 62.25 ± 0.22 |
| SZ | 39.72 ± 0.52 | 106.22 ± 0.41 | 101.23 ± 0.22 | |
| Sahiwal | NZ | 38.07 ± 0.46 | 31.15 ± 0.31 | 63.11 ± 0.11 |
| SZ | 39.26 ± 0.46 | 102.24 ± 0.31 | 98.29 ± 0.11 |
NZ Normal Zone, SZ Summer Zone, RT Rectal Temperature (°C), RR Respiratory Rate (times/min), PR Pulse Rate (rate/min)
Fig. 1.
Different biochemical parameters assessed in plasma samples of Frieswal an Sahiwal breeds of cattle. Estimation of different biochemical parameters revealed significant (p < 0.05) higher levels of plasma catalase activity (nmol/min/ml), GPx (nmol/min/ml) and MDA (μM) concentration during summer seasons in both the breeds. Catalase (CAT) activity (nmol/min/ml); glutathione peroxidase (GPx) level (nmol/min/ml) and c malondialdehyde (MDA) concentration (μM). SZ Summer zone, NZ Normal Zone. *p < 0.05
Fig. 2.
Relative mRNA expression (mean ± SEM) of HSP70.1 among Sahiwal and Frieswal during normal vs summer zone. The results highlighted that, during summer, the relative mRNA expression of HSP70.1 was significantly (p < 0.05) higher in comparison to normal zone among the breeds. NZ Normal Zone, SZ Summer Zone. *p < 0.05
Deep sequencing
A total of 742 number of Bos taurus annotated miRNAs were identified among Frieswal (420) and Sahiwal (322) through RNA deep sequencing (Sengar et al. 2018a, b). Detail statistics of deep sequencing results including the number of raw reads, quality filter reads, raw bases, quality filter bases, filtered reads, total identified small RNAs and total annotated miRNAs showed in Table 3. Out of a total 742 number of miRNAs across the breeds, 69 were found to be common and differentially expressed during summer among Sahiwal and Frieswal (Fig. 3). The differential expression levels of common miRNAs in both the breeds are illustrated as a heat map which was generated using a function of heatmap.2 in gplots by R platform (Fig. 4). The rows are centred and clustered using vector scaling and Euclidean distance, respectively. The intensity of the different colours specifies the level of variation in the expression of particular miRNA.
Table 3.
Identified statistical parameters through deep sequencing
| Deep sequencing parameters | Frieswal (NZ) | Frieswal (SZ) | Sahiwal (NZ) | Sahiwal (SZ) |
|---|---|---|---|---|
| Raw Reads | 333,179 | 183,829 | 255,460 | 113,260 |
| Quality filter reads | 327,039 | 178,524 | 250,106 | 111,622 |
| Raw bases | 20,989,049 | 10,469,956 | 60,421,166 | 6,410,548 |
| quality filter bases | 20,696,590 | 10,241,331 | 60,162,444 | 6,338,186 |
| Filtered read | 6140 | 5305 | 5354 | 1638 |
| Total small RNAs | 30,227 | 44,873 | 24,064 | 26,835 |
| Total annotated miRNAs | 251 | 169 | 172 | 150 |
NZ Normal Zone, SZ Summer Zone
Fig. 3.

Venn diagram depicting the common differentially expressed miRNAs during elevated environmental temperature among Sahiwal and Frieswal. There was a total of 420 and 322 miRNAs were differentially expressed among Frieswal and Sahiwal breed of cattle, respectively. Among them, a total 69 miRNAs were identified to be common among the breeds
Fig. 4.
The heat map of differentially expressed common microRNA among Sahiwal and Frieswal. There are two specific groups. Columns 1 and 2 represent summer and normal zone, respectively in Frieswal while columns 2 and 4 represent summer and normal zone, respectively in Sahiwal. Heat map was generated using a function of heatmap.2 in gplots by R platform.The rows are centred and clustered using vector scaling and Euclidean distance, respectively. The intensity of the different colours specifies the level of variation in the expression of particular miRNA
Differential expression of miRNAs
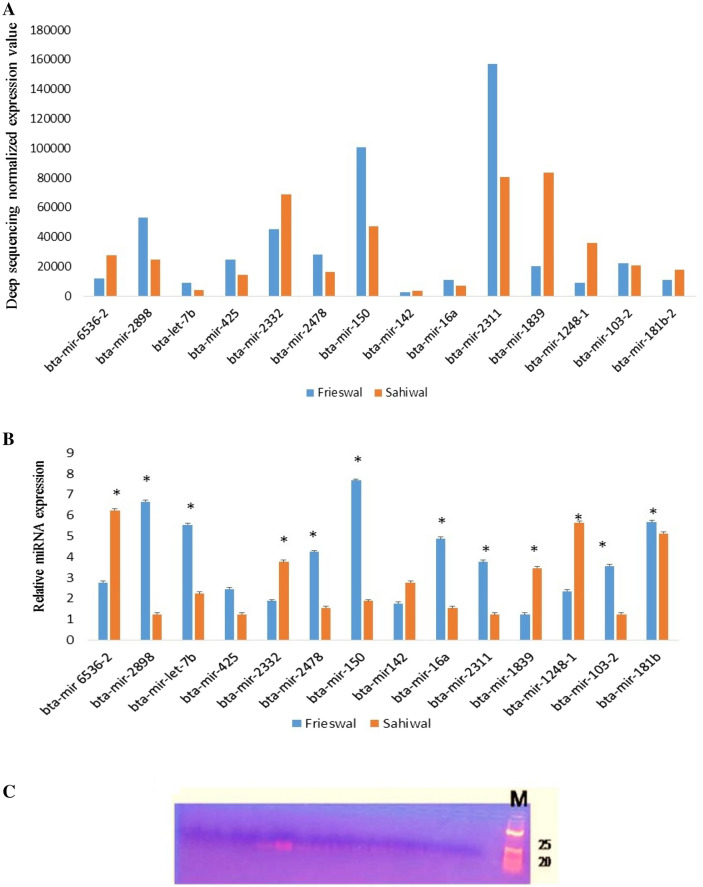
miRNAs fold change values were expressed as log2 values representing the variation in expression level of individual miRNA among Frieswal vs Sahiwal during summer stress (Table 4). Real-time PCR-based quantification was performed to validate the deep sequencing results (Fig. 5). Comparative expression profiling showed that bta-mir-2898, bta-mir-6536-1, bta-mir-let-7b, bta-mir-2478, bta-mir-150, bta-mir-16a, bta-mir-2311, bta-mir-1032-b and bta-mir-181-b having significantly (p < 0.01) higher expression during summer among Frieswal cattle in comparison to Sahiwal, while bta-mir 6536-2, bta-mir-2332, bta-mir142, bta-mir-1839 and bta-mir-1248-1 showing contrasting expression patterns. Target scan analysis revealed that identified miRNAs can target different heat shock family proteins.
Table 4.
Comparative miRNA fold changes among Frieswal and Sahiwal during summer
| miRNAs | Frieswal | Sahiwal | Fold changesa |
|---|---|---|---|
| bta-mir-181b-2 | 10,767.85 | 17,744.717 | − 0.216939919 |
| bta-mir-30e | 72,915.15 | 24,032.519 | 0.482018467 |
| bta-mir-103-2 | 22,066.43 | 20,779.998 | 0.026086497 |
| bta-mir-26c | 23,957.83 | 14,038.043 | 0.232140983 |
| bta-mir-361 | 20,246.06 | 40,038.081 | − 0.296132799 |
| bta-mir-1248-1 | 9126.794 | 36,097.825 | − 0.597162787 |
| bta-mir-342 | 945,569.8 | 534,267.013 | 0.247935251 |
| bta-mir-1839 | 19,964.86 | 83,350.881 | − 0.620643882 |
| bta-mir-20a | 16,871.71 | 4448.676 | 0.578928428 |
| bta-mir-103-1 | 6654.954 | 13,160.665 | − 0.296132777 |
| bta-mir-26a-2 | 28,521.23 | 7520.38 | 0.57892848 |
| bta-mir-138-2 | 51,956.75 | 7610.987 | 0.834200989 |
| bta-mir-93 | 24,891.26 | 8204.051 | 0.48201846 |
| bta-mir-2889 | 58,912.71 | 10,355.933 | 0.755019747 |
| bta-mir-671 | 4060.65 | 10,706.982 | − 0.421071515 |
| bta-mir-140 | 28,035.76 | 13,440.68 | 0.319291139 |
| bta-mir-210 | 29,336.12 | 16,115.1 | 0.260169728 |
| bta-mir-221 | 4355.97 | 5742.836 | − 0.120041534 |
| bta-mir-2311 | 156,814.9 | 80,399.701 | 0.290132933 |
| bta-mir-15a | 25,978.37 | 11,416.481 | 0.357079707 |
| bta-mir-2404-2 | 14,303.19 | 4714.268 | 0.482018488 |
| bta-mir-320a-1 | 8765.061 | 11,555.706 | − 0.120041541 |
| bta-mir-2892 | 13,821.83 | 33,407.843 | − 0.383282983 |
| bta-mir-222 | 4355.97 | 17,228.507 | − 0.597162764 |
| bta-mir-378-2 | 25,218.77 | 8311.999 | 0.48201847 |
| bta-mir-16a | 10,767.57 | 7097.887 | 0.180988461 |
| bta-mir-2484 | 390,891 | 369,883.964 | 0.023990145 |
| bta-mir-193b | 5772.972 | 3805.494 | 0.18098841 |
| bta-mir-142 | 2753.774 | 3630.528 | − 0.120041495 |
| bta-mir-150 | 100,622.9 | 47,378.395 | 0.327116503 |
| bta-mir-2904-3 | 65,028.41 | 45,122.281 | 0.158712069 |
| bta-mir-2478 | 28,002.66 | 16,408.102 | 0.232140987 |
| bta-mir-677 | 497,306.6 | 282,356.094 | 0.245827022 |
| bta-mir-2891 | 977,370.7 | 585,051.398 | 0.22286531 |
| bta-mir-2332 | 45,137.95 | 68,664.341 | − 0.182189435 |
| bta-mir-2904-1 | 88,986.24 | 85,732.335 | 0.016178209 |
| bta-mir-1246 | 32,818.95 | 34,614.353 | − 0.023131523 |
| bta-mir-23b | 19,964.86 | 10,528.532 | 0.277898491 |
| bta-mir-2440 | 22,066.43 | 24,935.998 | − 0.053094756 |
| bta-mir-30d | 106,099 | 63,171.194 | 0.225192123 |
| bta-mir-23a | 16,409.48 | 12,980.382 | 0.101807213 |
| bta-mir-423 | 30,584.47 | 23,521.189 | 0.114041673 |
| bta-mir-425 | 24,783.97 | 14,522.114 | 0.232140963 |
| bta-mir-21 | 33,274.77 | 17,547.554 | 0.277898475 |
| bta-let-7b | 8873.272 | 3899.456 | 0.35707977 |
| bta-mir-1434 | 7,019,974 | 9,185,783.89 | − 0.116780744 |
| bta-mir-92a-1 | 6143.034 | 8098.871 | − 0.120041562 |
| bta-mir-17 | 17,112.74 | 22,561.141 | − 0.120041533 |
| bta-mir-3432b | 2469.88 | 13,024.988 | − 0.722101478 |
| bta-let-7g | 5772.972 | 11,416.481 | − 0.296132807 |
| bta-mir-574 | 10,727.39 | 14,142.805 | − 0.120041564 |
| bta-mir-320a-2 | 5843.374 | 23,111.412 | − 0.597162796 |
| bta-mir-484 | 11,408.49 | 5013.587 | 0.357079687 |
| bta-mir-155 | 34,225.48 | 10,027.174 | 0.533170996 |
| bta-mir-223 | 46,584.68 | 14,622.962 | 0.503207747 |
| bta-mir-378b | 2444.677 | 3223.02 | − 0.120041514 |
| bta-mir-2411 | 37,336.88 | 24,612.153 | 0.180988464 |
| bta-mir-2898 | 52,893.92 | 24,612.153 | 0.332256148 |
| bta-mir-2904-2 | 116,366.6 | 49,634.51 | 0.370044692 |
| bta-mir-30c | 59,324.16 | 54,146.738 | 0.039659307 |
| bta-mir-138-1 | 212,409.7 | 48,843.707 | 0.638365655 |
| bta-mir-539 | 3071.517 | 4049.436 | − 0.120041616 |
| bta-mir-26a-1 | 10,647.93 | 3509.511 | 0.482018416 |
| bta-let-7a-3 | 9712.635 | 4268.324 | 0.35707969 |
| bta-mir-2424 | 8984.188 | 3948.2 | 0.357079687 |
| bta-mir-4286-2 | 83,662.28 | 140,380.431 | − 0.224776886 |
| bta-mir-6536-1 | 26,619.82 | 46,793.477 | − 0.244980267 |
| bta-mir-6536-2 | 11,978.92 | 27,637.397 | − 0.363079581 |
alog2 (Frieswal/Sahiwal)
Fig. 5.
Comparative miRNAs expression among Frieswal and Sahiwal cattle breeds during summer stress. A total 14 miRNAs were selected for qRT- PCR assay based on their fold changes. Comparative expression profiling showed that bta-mir-2898, bta-mir-6536-1, bta-mir-let-7b, bta-mir-2478, bta-mir-150, bta-mir-16a, bta-mir-2311, bta-mir-1032-b and bta-mir-181-b having significantly (p < 0.01) higher expression during summer among Frieswal cattle in comparison to Sahiwal, while bta-mir 6536-2, bta-mir-2332, bta-mir142, bta-mir-1839 and bta-mir-1248-1 showing contrasting expression patterns. a Normalized expression values through deep sequencing platform and b relative miRNA expression through RT-PCR *p < 0.01. c RT-PCR products (20–25 bp size) of different miRNAs run in agarose gel electrophoresis. M DNA molecular weight marker
Correlation between miRNAs with HSP70.1
Correlation coefficients between the differentially expressed miRNAs during thermal stress with overexpressed HSP70.1 gene was studied taken Frieswal as a model. The results revealed that there is a negative correlation (p < 0.05) between the HSP70.1 expression with bta-mir(s)-150, 16a and 181b (Table 5).
Table 5.
Correlation coefficients between the differentially expressed miRNAs during thermal stress in crossbred cattle with over-expressed heat shock protein 70 (HSP70)
| HSP70 | bta-mir-378 | bta-mir-142 | bta-mir-23a | bta-mir-425 | bta-mir-4286 | |
|---|---|---|---|---|---|---|
| HSP70 | 1 | |||||
| bta-mir-150a | − 0.52391 | 1 | ||||
| bta-mir-2898 | − 0.202814 | − 0.39607 | 1 | |||
| bta-mir-16aa | − 0.455747 | − 0.34938 | − 0.64304 | 1 | ||
| bta-mir-103-2 | 0.217698 | − 0.4739 | − 0.31172 | 0.454756 | 1 | |
| bta-mir-181ba | − 0.628902 | − 0.54069 | − 0.45395 | 0.904063 | 0.726681 | 1 |
aCorrelation is significant at the 0.05 level (2-tailed)
Discussion
Being endogenous post-transcriptional regulators of gene expression, miRNAs are known to play a crucial role in various biological processes. The expression profile of various miRNAs is strongly reliant on different physiological stimuli which reflects the functional state of a cell and; thus, making the miRNA signature as an exciting biomarker candidate. miRNAs are also known to be an important regulators for thermal stress response in mammalian systems (Islam et al. 2013; Place and Noonan 2014). Scanty of reports is available on the role of miRNAs in bovine thermoregulation (Zhang et al. 2014). Recently, our two independent studies identified differentially expressed miRNAs during thermal stress among Sahiwal (Bos indicus) and Frieswal (Bos taurus × Bos indicus) cattle breeds (Sengar et al. 2018a, b). The present study identified the comparative expression profile of certain miRNAs during elevated natural environmental temperatures among Sahiwal and Frieswal cattle breeds of India. Frieswal is a national milch crossbred cattle (Kumar et al. 2018), developed by the crossing of Holstein Friesian with Sahiwal aiming to improve milk production. However, the drawback of crossbreds is that they are highly susceptible to changing environmental temperatures. Our earlier reports also suggested that Sahiwal having the better thermo-tolerance ability in comparison to Frieswal due to their certain superior inherent capacities like better innate immunity as well as a higher level of heat shock proteins (Deb et al. 2014; Bhanuprakash et al. 2016, 2017). Thus, summer stress can vary between the breeds, which may reflects their differential post-transcriptional regulation.
In the present study, the stress response of animals was characterized by recoding various physiological parameters. Bianaca (1961) reported that body temperature, respiration rate and heart rate can give an instant response to the climatic stress in animals. Thus, these parameters can be used as an important heat stress indicator for dairy cattle (Charoensook et al. 2012). In the present study, we observed that all the physiological parameters viz. rectal temperature, pulse rate and respiratory rate was comparatively higher during summer in comparison to normal environmental temperatures which corroborated our earlier findings (Deb et al. 2013, 2014; Bhanuprakash et al. 2017; Sengar et al. 2018a, b).
Thermal stress is one of the prime factors which leads to oxidative stress in animals (Ganaie et al. 2013). Variations in the antioxidant enzyme activity among different breeds may lead to their inherent capacity to curtail the adverse effect of oxidative stress during the extreme environmental temperatures (Sengar et al. 2018b). It was observed that during summer months both the breeds can produce significantly (p < 0.05) higher levels of antioxidants viz. catalase (CAT) and glutathione peroxidase (GPX) in comparison to normal temperature zone. It was also noticed that during the summer concentration of lipid peroxidation by-product, i.e. thiobarbituric acid (TBARS)/malondialdehyde (MDA) was much (p < 0.05) higher among both the breeds.
Genetic configuration of animals is accountable for their phenotypic traits such as milk production as well as stress resilience. However, the DNA sequence variations can only explain a portion of phenotypic traits. The remaining part is considered to be influenced by certain epigenetic patterns of the animals. Studies identified that, miRNAs are one of the important regulators of mammalian thermal stress responses (Islam et al. 2013; Place and Noonan 2014). Recently few reports also suggested that miRNAs are also an important key player towards thermoregulation mechanisms in livestock (Zheng et al. 2014; Sengar et al. 2018a, b).
In the present study, we identified that few of the miRNAs are differentially expressed among Frieswal and Sahiwal cattle breeds during summer seasons in contrast to normal environmental temperature zone. Our earlier studies identified that, bta-mir (s)-103-2, 2898, 150, 181-b-2, 2311 and 142 and bta-mir (s)-1248-1, 2332, 2478 and 1839 were upregulated in Frieswal and Sahiwal, respectively, during summer stress. Although bta-mir-6536-2 and bta-mir(s) 16a, let-7b,142 and 425 were downregulated in Frieswal and Sahiwal, respectively (Sengar et al. 2018a, b). The present studies revealed that Frieswal can express higher levels of bta-mir-2898, bta-mir-let-7b, bta-mir-425, bta-mir-2478, bta-mir-150, bta-mir-16a, bta-mir-2311, bta-mir-103-2 and bta-mir-181b in comparison to Sahiwal breed during summer, while bta-mir 6536-2, bta-mir-2332, bta-mir142, bta-mir-1839 and bta-mir-1248-1 were highly expressed during summer in Sahiwal in comparison to Frieswal. Braud et al. (2017) identified the genome-wide miRNA binding site variation among cattle breeds, nonetheless the study provides the first note on differential expression of miRNAs among the cattle breeds. Thus, these studies may speculate the differential expression pattern of miRNAs among different breeds.
It was stated that concurrently expression of both miRNA and it is target mRNA could be informative for getting an insight on functional miRNA-mRNA relationships (Wang and Li 2019). In the present study, the coefficient correlation analysis revealed that the expression of few miRNAs like bta-mir(s)-150, 16a and 181b having significant negative correlation (p < 0.005) with HSP70 expression in Frieswal, which may show their post-translational regulation of the bovine Hsp70 gene during thermal stress. However, further studies are necessary to draw concise statements about the inhibitory/modulatory effect of miRNAs on major stressor genes during thermal stress in cattle.
Overall, the present study identified differentially expressed miRNA signatures during summer stress among native vs crossbred cattle. This study may highlight the differential thermoregulatory mechanisms among the breeds for their climatic adaptation.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are thankful to the Director, ICAR-CIRC for providing necessary facilities to carry out the present research. We are also thankful to Military Farm, Meerut, India for providing experimental animals; Ome Research Laboratory, Anand Agricultural University, Gujrat, India for NGS analysis
Funding
The authors acknowledge the Science and Engineering Research Board, Government of India for providing financial support under the project YSS/2014/000279 to RD.
Compliance with ethical standards
Conflict of interests
The authors declared that they have no conflict of interest.
References
- Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. elife. 2015;12:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhadj Slimen I, Najar T, Ghram A, Abdrrabba M. Heat stress effects on livestock: molecular, cellular and metabolic aspects, a review. J Anim Physiol Anim Nutr. 2015;100:401–412. doi: 10.1111/jpn.12379. [DOI] [PubMed] [Google Scholar]
- Bhanuprakash V, Singh U, Sengar G, Sajjanar B, Bhusan B, Raja TV, Alex R, Kumar S, Singh R, Ashish ARR, Kumar S, Deb R. Differential effect of thermal stress on HSP70 expression, nitric oxide production and cell proliferation among native and crossbred dairy cattle. J Therm Biol. 2016;59:18–25. doi: 10.1016/j.jtherbio.2016.04.012. [DOI] [PubMed] [Google Scholar]
- Bhanuprakash V, Singh U, Sengar GS, Raja TV, Sajjanar B, Alex R, Kumar S, Alyethodi RR, Kumar A, Kumar S, Bhusan B, Deb R. Comparative expression profile of NOD1/2 and certain acute inflammatory cytokines in thermal stressed cell culture model of native and crossbred cattle. Int J Biometeorol. 2017;61:931–941. doi: 10.1007/s00484-016-1273-1. [DOI] [PubMed] [Google Scholar]
- Bianca W. Heat tolerance in cattle-its concept, measurement and dependence on modifying factors. Int J Biometeorol. 1961;5:5–30. doi: 10.1007/BF02186917. [DOI] [Google Scholar]
- Braud M, Magee DA, Park SD, Sonstegard TS, Waters SM, MacHugh DE, Spillane C (2017) Genome-wide microRNA binding site variation between extinct wild aurochs and modern cattle identifies candidate microRNA-regulated domestication genes. Front Genet 8:3 [DOI] [PMC free article] [PubMed]
- Busk PK. A tool for design of primers for microRNA-specific quantitative RT-qPCR. BMC Bioinform. 2014;15:1–9. doi: 10.1186/1471-2105-15-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoensook R, Gatphayak K, Sharifi AR, Chaisongkram C, Brenig B, Knorr C. Polymorphisms in the bovine HSP90AB1 gene are associated with heat tolerance in Thai indigenous cattle. Trop Anim Health Prod. 2012;44:921–928. doi: 10.1007/s11250-011-9989-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia DS, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs’ action through miRNA editing. Int J Mol Sci. 2019;20:6249. doi: 10.3390/ijms20246249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb R, Sajjanar B, Singh U, Kumar S, Brahmane MP, Singh R, Sengar G, Sharma A (2013) Promoter variants at AP2 box region of Hsp70. 1 affect thermal stress response and milk production traits in Frieswal cross bred cattle. Gene 532(2):230–235 [DOI] [PubMed]
- Deb R, Sajjanar B, Singh U, Kumar S, Singh R, Sengar G, Sharma A. Effect of heat stress on the expression profile of Hsp90 among Sahiwal (Bos indicus) and Frieswal (Bos indicus × Bos taurus) breed of cattle: a comparative study. Gene. 2014;536:435–440. doi: 10.1016/j.gene.2013.11.086. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganaie AH, Shanker G, Bumla NA, Ghasura RS, Mir NA, Wani SA, Dudhatra GB. Biochemical and physiological changes during thermal stress in bovines. J Vet Sci Technol. 2013;4:126–132. [Google Scholar]
- Hansen PJ. Physiological and cellular adaptations of zebu cattle to thermal stress. Anim Reprod Sci. 2004;82–83:349–360. doi: 10.1016/j.anireprosci.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Islam A, Deuster PA, Devaney JM, Ghimbovschi S, Chen Y. An exploration of heat tolerance in mice utilizing mRNA and microRNA expression analysis. PLoS ONE. 2013;8:72258. doi: 10.1371/journal.pone.0072258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M, Datta J, Lang JC, Teknos TN. Down regulation of RhoC by microRNA-138 results in de-activation of FAK, Src and Erk ½ signaling pathway in head and neck squamous cell carcinoma. Oral Oncol. 2014;50:448–456. doi: 10.1016/j.oraloncology.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;8:155–162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Alex R, Gaur GK, Mukherjee SS, Mandal DK, Singh U, Tyagi S, Kumar A, Das AK, Deb R, Kumar M, Sirohi AS, Chand N, Prasad R, Bhasin V, Prakash B, Kashyap S. Evolution of Frieswal cattle: a crossbred dairy animal of India. Indian J Anim Sci. 2018;88:265–275. [Google Scholar]
- Leung AKL, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Muroya S, Hagi T, Kimura A, Aso H, Matsuzaki M, Nomura M. Lactogenic hormones alter cellular and extracellular microRNA expression in bovine mammary epithelial cell culture. J Anim Sci Biotechnol. 2016;7:8. doi: 10.1186/s40104-016-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar RG, Pyke CR, Adams MB, Breitburg D, Hershner C, Kemp M, Howarth R, Mulholland MR, Paolisso M, Secor D, Sellner K. Potential climate-change impacts on the Chesapeake Bay. Estuar Coast Shelf Sci. 2010;86:1–20. doi: 10.1016/j.ecss.2009.09.026. [DOI] [Google Scholar]
- O'Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Noonan EJ. Non-coding RNAs turn up the heat: an emerging layer of novel regulators in the mammalian heat shock response. Cell Stress Chaperones. 2014;19:159–172. doi: 10.1007/s12192-013-0456-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattanapan Y, Korkiatsakul V, Kongruang A, Siriboonpiputtana T, Rerkamnuaychoke B, Chareonsirisuthigul T. MicroRNA expression profiling of epithelial ovarian cancer identifies new markers of tumor subtype. Microrna (Shariqah, United Arab Emirates) 2020 doi: 10.2174/2211536609666200722125737. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:1–4. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengar GS, Deb R, Singh U, Junghare V, Hazra S, Raja TV, Alex R, Kumar A, Alyethodi RR, Kant R, Jakshara S, Joshi CG. Identification of differentially expressed microRNAs in Sahiwal (Bos indicus) breed of cattle during thermal stress. Cell Stress Chaperones. 2018;23:1019–1032. doi: 10.1007/s12192-018-0911-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengar GS, Deb R, Singh U, Raja TV, Kant R, Sajjanar B, Alex R, Alyethodi RR, Kumar A, Kumar S, Singh R. Differential expression of microRNAs associated with thermal stress in Frieswal (Bos taurus × Bos indicus) crossbred dairy cattle. Cell Stress Chaperones. 2018;23:155–170. doi: 10.1007/s12192-017-0833-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A, Mukherjee B, Dixit M. MicroRNA key to angiogenesis regulation: miRNA biology and therapy. Curr Cancer Drug Targets. 2018;18:266–277. doi: 10.2174/1568009617666170630142725. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Wang Y-P, Li K-B. Correlation of expression profiles between microRNAs and mRNA targets using NCI-60 data. BMC Genom. 2019;10:218. doi: 10.1186/1471-2164-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:90–795. doi: 10.1016/S0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Chen K, Zheng X, Li H, Wang G. Identification and bioinformatics analysis of microRNAs associated with stress and immune response in serum of heat-stressed and normal Holstein cows. Cell Stress Chaperones. 2014;19:973–981. doi: 10.1007/s12192-014-0521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.