Abstract
Background/Aim: Influenza viruses, corona viruses and related pneumotropic viruses cause sickness and death partly by inducing cytokine storm, a hyper-proinflammatory host response by immune cells and cytokines in the host airway. Based on our in vivo experience with digitoxin as an inhibitor of TNFα-driven NFĸB signaling for cytokine expression in prostate cancer in rats and in cystic fibrosis in humans, we hypothesize that this drug will also block a virally-activated cytokine storm. Materials Methods: Digitoxin was administered intraperitoneally to cotton rats, followed by intranasal infection with 107TCID50/100 g of cotton rat with influenza strain A/Wuhan/H3N2/359/95. Daily digitoxin treatment continued until harvest on day 4 of the experiment. Results: The cardiac glycoside digitoxin significantly and differentially suppressed levels of the cytokines TNFα, GRO/KC, MIP2, MCP1, and IFNγ, in the cotton rat lung in the presence of influenza virus. Conclusion: Since cytokine storm is a host response, we suggest that digitoxin may have a therapeutic potential not only for influenza and but also for coronavirus infections.
Keywords: Virus, influenza, coronavirus, SARS, COVID-19, RNAvirus, cardiac glycosides, digitoxin, digoxin, ouabain, oleandrin, inflammation, cytokine storm, TNFα, GRO/KC, MIP-2, MCP1, IFNγ
Influenza, corona and related pneumotropic viruses cause sickness and death partly by inducing a hyper-proinflammatory immune response in the host airway. This immune overreaction, called a cytokine storm, can lead to multiorgan failure and death (1). For example, Influenza A (H5N1) has been shown to activate the TNFα-driven NFĸB signaling pathway in a mouse host during viral infection, generating a massive overproduction of cytokines, including interleukin 8 (IL-8) and monocyte chemoattractant protein 1 (MCP1), known as cytokine storm (2). As anticipated, inhibitors of NFĸB acutely suppress cytokine storm and increase survival in a mouse model of SARS-CoV infection (3). Recent data show that COVID-19 also activates NFĸB (4). Cytokine storm marks the airways of SARS-CoV-2-infected patients that were admitted to the Intensive Care Unit (ICU) with more severe disease (5). Since there are multiple strains of influenza as well as coronavirus, there might be an advantage to develop therapies that suppress host-induced cytokine storm, in addition to developing strain-specific vaccines.
The clinical problem is that there are limited options for treating respiratory cytokine storm, most of which are predicated on inhibiting NFĸB-activated cytokine expression (6-8). The absence of NFĸB inhibitory drugs from the human formulary is due to most candidate drugs being either neurotoxic or nephrotoxic when administered chronically (9). One drug that lacks these toxicities is the cardiac glycoside digitoxin. We have previously shown digitoxin to be amongst the most potent inhibitors of the proinflammatory TNFα/NFĸB pathway in the human airway and in other epithelial cells, both in vitro (10) and in vivo (11-13). Corroborating this discovery is a screen of 2800 drugs and bioactive compounds which found digitoxin to be one of the most potent inhibitors of TNFα-driven−NFĸB activity (14). Digitoxin is a drug that has been used to treat heart failure for decades, and a clinical trial (11) demonstrated that in addition to giving it to people with heart failure or heart arrythmias, it is also safe to give digitoxin for diseases like cystic fibrosis to children and adults with normal hearts who need to have reduction of lung inflammation (15). Digitoxin was also recently shown to block MERS-CoV infectivity in vitro (16). The digitoxin analogues digoxin and ouabain also block SARS-CoV-2 infectivity in vitro (17). In a clinical trial where digitoxin was administered to young adults with the proinflammatory lung disease cystic fibrosis, it proved safe. This clinical trial also showed that "the mRNAs encoding chemokine/cytokine or cell surface receptors in immune cells were decreased in nasal epithelial cells, leading to pathway-mediated reductions in IL-8 and IL-6 levels, lung epithelial inflammation, neutrophil recruitment and mucus hypersecretion" (11).
To further test the ability of digitoxin to inhibit cytokine storm in related pneumotropic viruses, we used the cotton rat model of influenza infection to investigate the effects of digitoxin in influenza-associated cytokine storm. The cotton rat model has the important advantage of susceptibility to influenza infection without engineered adaptation (18). Based on mRNA changes in an experiment from the same laboratory that we used, when the cotton rats were given only the influenza A H3N2 virus but no drug, the cytokine levels were shown to increase in the cotton rat lung from baseline 10-fold for tumor necrosis factor alpha (TNFα), 40-fold for interferon gamma (IFNγ), 10-fold for growth-regulated oncogene/ keratinocyte chemoattractant (GRO/KC), and 35-fold for macrophage inflammatory protein-2 beta (MIP1β) (19). Consistently, there is a close relationship between mRNA and protein changes for cytokine proteins (20). In addition, it has also been shown in vivo that in the presence of lipopolysaccharides (LPS), GRO/KC and MIP2 mRNAs increase 50-fold and 20-fold, respectively (21). Thus, cytokine levels in the absence of virus or other immune stimulant in the cotton rat lung are very low. Furthermore, it has been shown that the cytokine response of the cotton rat to this virus strain evokes a pattern of pulmonary cytokine changes that parallel the human response (19).
Materials and Methods
Animal protocol. Cotton rat experiments were performed as previously described (19). All experiments were performed using protocols that followed federal guidelines and were approved by the Institutional Animal Care and Use Committee. Animals were sacrificed by carbon dioxide inhalation.
Drugs and protocol for drug preparation. Digitoxin was obtained from Sigma-Aldrich (> 95% pure) (St. Louis, MO, USA). The drug was prepared as a stock solution in 95% ethanol. It was diluted into phosphate buffered saline (PBS) as the suspension solution from the stock solution. It was administered in a 200 μl volume. Digitoxin (0, 3, 10 and 30 μg per 100 g of body weight) was administered to 3 cotton rats in one dose intraperitoneally 6.25 h before intranasal infection with 107TCID50/100 g of cotton rat with influenza strain A/Wuhan/H3N2/359/95. This ensured drug availability at the time of virus infection, as distribution is complete within 4 to 6 h (22). Daily digitoxin treatment continued until harvest on day 4 of the experiment.
Tissue preparation and histological analysis. On day 4 post infection the animals were sacrificed by CO2 inhalation. The left lung was first tied off and reserved for cytokine analysis. These lung samples were then immediately frozen on dry ice, and kept at –80˚C until further processed. The remaining lung tissue was processed for histological analysis. The right lung was inflated with 70% formalin and fixed for histology, as previously described (19). The frozen left lungs were then transferred to author BSP.
Later, sections were stained by Hematoxylin and Eosin (H&E) and immune cell density was automatically analyzed in brightfield mode on a Zeiss Axio-Scan.Z1 digital slide scanner (Carl Zeiss AG, Oberkochen, Germany). Whole lung images (2 sections from each lung) were analyzed using NIH-ImageJ. Briefly, the program was trained to identify immune cells as dark stained objects in the section that were the size of neutrophils. The per cent total area occupied by such objects was measured.
Biochemical analysis. Frozen lung samples were weighed, thawed and minced with scissors in 10% (w/v) ice cold PBS, homogenized with 10 strokes of a Ten Broeck homogenizer (Vavantor, Radnor, PA, USA), and centrifuged at 20,000 × g for 30 min. The supernatant solutions were kept at –80˚C until assay. The supernatant solutions were brought to Bioassay Works (LLC, Ijamsville, MD, USA) by author BSP where they were tested for cytokines and chemokines by ELISA. The samples were then sent for corroboration to Pierce-Thermo (Waltham, MA, USA) for ELISA assay on the Searchlight® ELISA platform. Rat antibody reagents were used in both instances.
Statistics. All data are presented as means ± SEM from three animals for each digitoxin dose. The mean values for each cytokine were analyzed using one-way ANOVA followed by Dunnett’s post-hoc comparisons of each dose of digitoxin vs. vehicle (0 μg). Differences were considered significant when p<0.05. Complete statistical data are summarized in Table I. The same method was used to compute the significance of histology-based differences between immune cell density at each dose of digitoxin vs. vehicle (0 μg) in Table II.
Table I. Reduction of cytokine expression by digitoxin in cotton rat lungs.
Bold p-Values indicate statistical significance
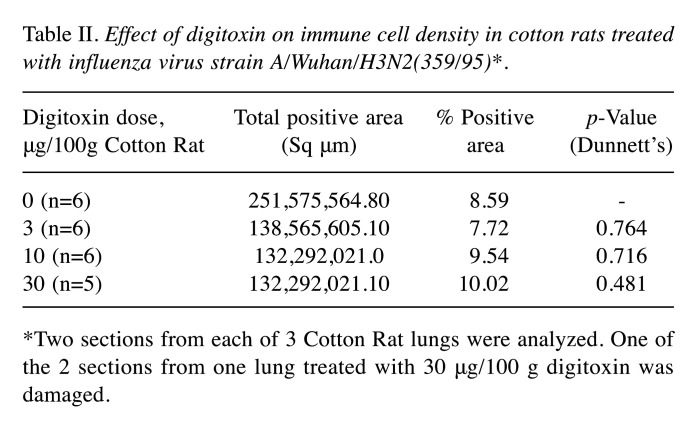
Table II. Effect of digitoxin on immune cell density in cotton rats treated with influenza virus strain A/Wuhan/H3N2(359/95)*.
*Two sections from each of 3 Cotton Rat lungs were analyzed. One of the 2 sections from one lung treated with 30 μg/100 g digitoxin was damaged
Results
Digitoxin blocks cytokine storm. As shown in Figure 1, protein data were collected for IFNγ, GRO/KC as the rodent equivalent of human IL-8, MIP2, chemokine (C-X-C motif) ligand 2 (CXCL2), TNFα, IL-1β, MCP1, and transforming growth factor beta (TGFβ). Figure 1 also shows the changes in cytokine protein in the lung due to digitoxin administration in the cotton rat following intranasal (IN) infection with a dose of 107TCID50/100 g of animal with influenza strain A/Wuhan/H3N2/359/95 virus. Animals were given three different doses of digitoxin, starting 6.25 h prior to virus administration and continuing with a daily dose until sacrifice on day 4. The dose range of digitoxin, 0-30 μg/100g cotton rat, was calculated (23) to approximate the human dose routinely used to treat heart failure (0.1 mg digitoxin, Merck KGaA, Darmstadt, Germany). As shown in Figure 1 and summarized in Table I, significant digitoxin-dependent reductions were found at >10 μg doses for 5 of the 7 cytokines. The digitoxin-dependent reductions were specific and saturating for each cytokine, but did not reduce any of them to zero.
Figure 1. Cytokine concentrations in lungs of cotton rats treated with digitoxin and influenza strain A/Wuhan/H3N2/359/95 virus. Animals were treated with different concentrations of digitoxin 6.25 hours before intranasal virus administration and daily thereafter for 4 days. Samples assayed were lung tissue. Digitoxin dose is in units of μg/100g. IFNγ: Interferon gamma; GRO/KC: growth-regulated oncogene/ keratinocyte chemoattractant; MIP2: macrophage inflammatory protein-2 ; CXCL2: chemokine (C-X-C motif) ligand 2; TNFα: tumor necrosis factor alpha; IL- 1β: interleukin 1 beta; MCP1: monocyte chemoattractant Protein 1 (or CCL2); TGFβ: transforming growth factor beta. *p<0.05, N=3.
Digitoxin differentially affects cytokine expression. Table I shows that the most significant digitoxin-dependent reductions of cytokine proteins were found in IFNγ (68.9%), GRO/KC (46.6%), and MCP1 (54.9%). Smaller but still significant reductions in cytokine proteins were found in MIP2 (32.2%) and TNFα (38.4%). In the cases of IL-1β and TGFβ changes were not significant. Taken together, digitoxin, dose-dependently and significantly lowers the individual concentrations in the lung of at least five cytokines that had been induced by viral infection.
Digitoxin leaves immune cell density intact in virus-infected lung. Figure 2a and Figure 2b show low-power views of cotton rat lung sections, taken 4 days after intranasal virus administration (no digitoxin and 10 μg digitoxin/100 g, respectively). Changes in cytokines appear to saturate at a dose of 10 μg/100 g cotton rat. Regions of heavy hematoxylin staining, representing infiltration foci of immune cells, are distributed in the lung. As previously described, pulmonary inflammatory changes can be seen in terms of peribronchitis (inflammatory cells clustered around the periphery of small airways), interstitial pneumonia (inflammatory cell infiltration and thickening of alveolar walls) and alveolitis (immune cells within the alveolar spaces) (19,24).
Figure 2. Histology of cotton rat lungs following infection with influenza strain A/Wuhan/H3N2/359/95 and treatment for 4 days with digitoxin. (a) Low power image of infected, control cotton rat lung (magnification 10×). (b) Low power image of infected cotton rat lung treated with 10 μg/100g digitoxin for 4 days. Analysis shows that the density of immune cells, marked by the NIH ImageJ program, is not statistically different when comparing all treatment conditions from control (also see Table II). Scale bar=1 mm.
Densities of immune cells were not affected by digitoxin. Table II shows that the positive area (%) occupied by immune cells in treated animals was not statistically different from the area occupied by immune cells in untreated, virus-infected animals. Thus, digitoxin appears to inhibit the cytokine storm host response to influenza A infection but does not significantly affect the density and distribution of immune cells as seen in the microscope on the 4th-day after infection.
Discussion
Our experimental results in Figure 1 and Table 1 show that administration of digitoxin to the cotton rat inhibits expression of five cytokines in the lung in the presence of influenza strain A/Wuhan/H3N2/359/95. These cytokines include TNFα, the key activator of the TNFα−driven NFĸB inflammation pathway. The data also show that digitoxin inhibits cytokine storm, but does not appear to significantly affect the density of immune cells in the lung four days after viral infection. The data further suggest that digitoxin acts on multiple cell types (25). For example, IFNγ is secreted only from activated T lymphocytes and NK cells of the immune system 5). The remainder of the cytokines are secreted by epithelial cells in the airway, as well as by endothelial cells, immune cells and others (26,27). GRO/KC [CXCL1, the rodent equivalent of human IL-8 (28)], and MIP2 are a key target of NFĸB signaling and are major chemoattractants for neutrophils (29). MIP1 induces entry and accumulation of monocytes and macrophages into the lung, and are targets of NFĸB (29). TGFβ is indirectly dependent on NFĸB-signaling and indirectly drives NFĸB (12). IL-1β also drives NFĸB, albeit not through TNFα. The data from Figure 1 and Table I support the interpretation that digitoxin-dependent reduction in TNFα-driven NFĸB signaling may be sufficient to suppress influenza A-associated cytokine storm.
The reduction of influenza A-driven TNFα expression by digitoxin is specifically relevant to what is known regarding the mechanism of influenza A virus RNA production and propagation (30). For example, TNFα drives NFĸB activation and signaling by host NFĸB, which has been shown to be a prerequisite for influenza virus infection (30-32). Knockdown of host NFĸB_p65 has also been found to reduce influenza virus replication and vRNA synthesis (30). This relationship appears to be dependent on viral genes because mouse-adapted descendants of the avian Influenza A strain H7N7 can be genetically engineered to function independently of NFĸB, using CRISPR-Cas9 editing (33). More recently, it was shown that cirsimaritin, a pure flavonoid from Chinese medicine, blocks NFĸB signaling induced in MDCK and THP-1 cells by influenza A strains H1N1 and H3N2, and suppresses virally activated expression of TNFα, IL-8 and other cytokines (34). In prospect, our present work shows that digitoxin, a specific blocker of TNFα-driven NFĸB signaling, achieves an analogous result in vivo with influenza A strain H3N2.
The decision to analyze the response to digitoxin on the 4th day after infection was based on the observation that in the cotton rat lung, mRNA expression for many cytokines reaches maximal level on that day in response to A/Wuhan/H3N2/359/95 infection (19). Our data show that digitoxin treatment causes the most profound reduction in INFγ expression relative to the other cytokines. The potential importance of digitoxin-dependent INFγ reduction may be manifest by a recent report, where simply neutralizing INFγ in a mouse model of infection with influenza A virus strain A/California/07/2009 (H1N1v; "Swine Flu") was sufficient not only to alleviate acute lung injury but also to increase weight and survival rate (35). Why digitoxin is so powerful a suppressor of INFγ is not immediately obvious. However, it is known that IFNγ expression is driven by a combination of both NFĸB and NFAT acting on the IFNγ promoter (36). We have previously reported that digitoxin not only reduces NFĸB, but also reduces NFAT expression (37). It is, therefore, possible that the suppression of both of these transcription factors may contribute to digitoxin’s potent suppression of virally-induced INFγ expression.
Influenza A is known to drive activation of IL-1β and TGFβ, but in these experiments digitoxin did not significantly change the expression of these cytokines, as it did to the others. The importance of IL-1β for influenza A is that it is synthesized by alveolar macrophages and dendritic cells in response to viral infection (38). Its role is to drive neutrophilic inflammation in a manner unrelated to the levels of GRO/KC or MIP-2α in the virus-infected mouse lung (39). We conclude that further understanding of this complexity will depend on additional investigation. TGFβ expression is also driven by influenza A in response to viral infection (40); however, the regulation of TGFβ expression itself remains poorly understood, and is also not directly dependent on NFĸB (12). It is possible that the difference may lie in the fact that digitoxin acts directly on the TNFα-driven NFĸB pathway, but that IL-1β and TGFβ act on NFĸB indirectly or by alternative pathways. For example, digitoxin acts directly to suppress TNFα-driven NFĸB signaling by blocking the binding of the TNFα/TNFR1 to TRADD (41,42). Tumor necrosis factor receptor type 1-associated death domain (TRADD) is the first intracellular adaptor for the TNFα/TNFR1 complex, and the resulting ternary complex directly drives the downstream activation of IKKα, β, & γ, phosphorylation of IĸBα, and, thus, activation of NFĸB. Increased cytokine expression follows NFĸB activation.
Finally, it is a limitation of this study that there may be antiviral effects of digitoxin that may contribute to reduction in host-driven cytokine storm, and may also have implications for COVID-19 therapy. This is because digitoxin and the other approved cardiac glycosides, digoxin and ouabain have been shown to have inhibitory properties for coronaviruses and other viruses (43). With respect to COVID-19, digoxin and ouabain have been shown to block cell penetration and infectivity when tested against SARS-CoV-2 (17). Furthermore, digitoxin itself has been shown to block Middle East respiratory syndrome, MERS CoV penetration into target cells and subsequent infectivity (16). Previously, digoxin was shown to block MERS-CoV penetration and infectivity (44). In silico molecular docking analysis based on CRYO-EM structures has shown that digitoxin, out of 15,000 molecular candidates, binds best to the receptor binding domain (RBD) of the SARS-CoV-2 Spike (45). A similar study by others has come to the same conclusion (46). Both the latter authors suggested that digitoxin may block the interaction of SARS-CoV-2 with the receptor ACE2. Using the same in silico screening approach, but also followed by an in-vitro test, ouabain was shown to dock optimally to the SARS-CoV-2 Main protease (Mpro), and to block viral penetration and infectivity (47). Consistently, digitoxigenin, a digitoxin without the three sugars at the 3’OH position, has also been shown to dock optimally to the Mpro (48). With respect to the activation of the cytokine storm, we have already noted that TNFα-driven NFĸB activation by the host drives cytokine storm for Influenza A (2), for SARS-CoV (3), and for SARS-CoV-2 (4). Consistently, digitoxin potently blocks this host-response process, independent of viral activation, at low nM concentrations (10,11,13,37,41). Since the cardiac glycosides digitoxin, digoxin and ouabain are approved drugs, we conjecture that the suppressive effects of digitoxin on influenza A cytokine storm shown here could be quite relevant to future tests of cardiac glycoside-based therapies for COVID-19.
In conclusion, these data show that digitoxin blocks the host over-production of cytokines raised by influenza strain A/Wuhan/H3N2/359/95 in the cotton rat lung. Since digitoxin has already been shown to be safe in an FDA Phase II clinical trial with cystic fibrosis patients with pulmonary disease and a normal heart, and has been shown to cause a similar reduction in NFĸB driven cytokine expression, this drug may be a good candidate for further investigation as a therapy for influenza and potentially for COVID-19.
Conflicts of Interest
Bette Pollard has a patent on anti-inflammatory and immune properties of cardiac glycosides, such as digitoxin and its use in treatment of diseases.
Authors’ Contributions
BSP requested the experiment, conceived and designed the experiment, analyzed the data, and realized that additional analysis was necessary after histology did not show an effect, and wrote the manuscript. GAP was the pathologist and analyzed the data. JCB designed the experiment, provided the virus, and performed the experiment. JRP analyzed the data and wrote the manuscript. All authors approved the manuscript for publication.
Acknowledgements
The Authors would like to thank Val Hemming, Harvey B. Pollard, and Dennis McDaniel for technical advice, and Cara Olsen and Max Tran for providing statistical advice.
References
- 1.Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76(1):16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmolke M, Viemann D, Roth J, Ludwig S. Essential impact of NF-kappaB signaling on the H5N1 influenza A virus-induced transcriptome. J Immunol. 2009;183(8):5180–5189. doi: 10.4049/jimmunol.0804198. [DOI] [PubMed] [Google Scholar]
- 3.DeDiego ML, Nieto-Torres JL, Regla-Nava JA, Jimenez-Guardeño JM, Fernandez-Delgado R, Fett C, Castano-Rodriguez C, Perlman S, Enjuanes L. Inhibition of NF-ĸB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J Virol. 2014;88(2):913–924. doi: 10.1128/JVI.02576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Tan K-S, Wang D-Y, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7(1):11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang M, Cao L, Xie M, Yu Y, Kang R, Yang L, Zhao M, Tang D. Chloroquine inhibits HMGB1 inflammatory signaling and protects mice from lethal sepsis. Biochem Pharmacol. 2013;86(3):410–418. doi: 10.1016/l.bcp.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teijaro JR, Walsh KB, Long JP, Tordoff KP, Stark GV, Eisfeld AJ, Kawaoka Y, Rosen H, Oldstone M. Protection of ferrets from pulmonary injury due to H1N1 2009 influenza virus infection: immunopathology tractable by sphingosine-1-phosphate 1 receptor agonist therapy. Virology. 2014;452-453:152–157. doi: 10.1016/j.virol.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang S, Qiang L, Sample A, Shah P, He YY. NF-ĸB Signaling Activation Induced by Chloroquine Requires Autophagosome, p62 Protein, and c-Jun N-terminal Kinase (JNK) Signaling and Promotes Tumor Cell Resistance. J Biol Chem. 2017;292(8):3379–3388. doi: 10.1074/jbc.M116.756536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, Lenardo MJ, Baltimore D. 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell. 2017;168(1-2):37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srivastava M, Eidelman O, Zhang J, Paweletz C, Caohuy H, Yang Q, Jacobson KA, Heldman E, Huang W, Jozwik C, Pollard BS, Pollard HB. Digitoxin mimics gene therapy with CFTR and suppresses hypersecretion of IL-8 from cystic fibrosis lung epithelial cells. Proc Nat Acad Sci (USA) 2004;101(20):7693–7698. doi: 10.1073/pnas.0402030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeitlin PL, Diener-West M, Callahan KA, Lee S, Talbot CC Jr., Pollard B, Boyle MP, Lechtzin N. Digitoxin for airway inflammation in cystic fibrosis: preliminary assessment of safety, pharmacokinetics, and dose finding. Ann Amer Thor Soc. 2017;14(2):220–229. doi: 10.1513/AnnalsATS.201608-649OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollard BS, Suckow MA, Wolter WR, Starr JM, Eidelman O, Dalgard CL, Kumar P, Cattacharyya S, Srivastava M, Biswas R, Wilkerson MD, Xhang X, Yang Q, Pollard HB. Digitoxin inhibits epithelial-to-mesenchymal-transition in hereditary castration resistant prostate cancer. Front Oncol. 2019;9:630. doi: 10.3389/fonc.2019.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Q, Soltis AR, Sukumar G, Zhang X, Caohuy H, Freedy J, Dalgard CL, Wilkerson MD, Pollard HB, Pollard BS. Gene therapy-emulating small molecule treatments in cystic fibrosis airway epithelial cells and patients. Resp Res. 2019;20(1):290. doi: 10.1186/s12931-019-1214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller SC, Huang R, Sakamuru S, Shukla SJ, Attene-Ramos MS, Shinn P, Van Leer D, Leister W, Austin CP, Xia M. Identification of known drugs that act as inhibitors of NF-kappaB signaling and their mechanism of action. Biochem Pharmacol. 2010;79(9):1272–1280. doi: 10.1016/j.bcp.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman BJ, Bigger J, Gilman AG, Rall TW. Digitalis and Allied Cardiac Glycosides in Goodman and Gilman’s The Pharmacological Basis of Therapeutics. New York: Permagon Press. 1990;Eighth Edition [Google Scholar]
- 16.Ko M, Chang SY, Byun SY, Choi I, Hung A-LP, d’Orengiani D’A, Shum D, Min J-Y, Windisch MP. Screening of FDAapproved drugs using a MERS-CoV clinical isolate from South Korea identifies potential therapeutic options for COVID-19. bioRxiv. 2020 doi: 10.1101/2020.02.25.965582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho J, Lee YJ, Kim J-H, Kim SI, Kim SS, Choi B-S, Choi J-H. Antiviral activity of digoxin and ouabain against SARS-CoV-2 infection and its implication for COVID-19. Sci Rep, 2020;10(1):16200. doi: 10.1038/s41598-020-72879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mifsud EJ, Tal CMK, Hurt AC. Animal models to assess influenza antivirals. Expert Opin Drug Discov. 2018;13:12,1131–1139. doi: 10.1080/17460441.2018.1540586. [DOI] [PubMed] [Google Scholar]
- 19.Ottolini MG, Blanco JCG, Eichelberger MC, Porter DD, Pletneva L, Richardson JY, Prince GA. The cotton rat provides a useful small-animal model for the study of influenza virus pathogenesis. J Gen Virol. 2005;86(Pt 10):2823–2830. doi: 10.1099/vir.0.81145-0. [DOI] [PubMed] [Google Scholar]
- 20.Kalb DM, Adikari SH, Hong-Geller E, Werner JH. Single-cell correlations of mRNA and protein content in a human monocytic cell line after LPS stimulation. PLoS One. 2019;14(4):e0215602. doi: 10.1371/journal.ponme.0215602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Filippo K, Henderson RB, Laschinger M, Hogg N. Neutrophil chemokines KC and Macrophage-Inflammatory Protein-2 are newly synthesized by tissue macrophages using distinct TLR signaling pathways. J Immunol. 2008;180:4308–4315. doi: 10.4049/jimmunol.180.6.4308. [DOI] [PubMed] [Google Scholar]
- 22.Perrier D, Mayersohn M, Marcus FI. Clinical pharmacology of digitoxin. Clin Pharmacokinet. 1977;2:292–311. doi: 10.2165/00003088-197702040-00005. [DOI] [PubMed] [Google Scholar]
- 23.Nair AB, Jacob S. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharmacy. 2016;7:27–31. doi: 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prince GA, Curtis SJ, Yim KC, Porter DD. Vaccine-enhanced respiratory syncytial virus disease in cotton rats following immunization with Lot 100 or a newly prepared reference vaccine. J Gen Virol. 2001;82(Pt 12):2881–2888. doi: 10.1099/0022-1317-82-12-2881. [DOI] [PubMed] [Google Scholar]
- 25.Mah AY, Cooper MA. Metabolic regulation of natural killer cell IFN-γ production. Crit Rev Immunol. 2016;36(2):131–147. doi: 10.1615/CritRevImmunol.2016017387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills PR, Davies RJ, Devalia JL. Airway epithelial cells, cytokines, and pollutants. Am J Respir Crit Care Med. 1999;160(5 Pt 2):S38–S43. doi: 10.1164/ajrccm.160.supplement_1.11. [DOI] [PubMed] [Google Scholar]
- 27.Liu T, Zhang L, Joo D, Sun SC. NF-ĸB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hol J, Wilhelmsen L, Haraldsen G. The murine Il-8 homologues KC, MIP-2 and LIX are found in endothelial cytoplasmic granules but not Weibel-palade bodies. J Leuk Biol. 2009;87(3):501–508. doi: 10.1189/jlb.0809532. [DOI] [PubMed] [Google Scholar]
- 29.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyote chemoattracrant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29(6) doi: 10.1089/jir.2008.0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar N, Xin ZT, Liang Y, Ly H, Liang Y. NF-kappaB signaling differentially regulates influenza virus RNA synthesis. J Virol. 2008;82(20):9880–9889. doi: 10.1128/JVI.00909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nimmerjahn F, Dudziak D, Dirmeier U, Hobom G, Riedel A, Schlee M, Staudt LM, Rosenwald A, Behrends U, Bornkamm GW, Mautner J. Active NF-kappaB signalling is a prerequisite for influenza virus infection. J Gen Virol. 2004;85(Pt 8):2347–2356. doi: 10.1099/vir.0.79958-0. [DOI] [PubMed] [Google Scholar]
- 32.Wurzer WJ, Ehrhardt C, Pleschka S, Berberich-Siebelt F, Wolff T, Walczak H, Planz O, Ludwig S. NF-kappaB-dependent induction of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Fas/FasL is crucial for efficient influenza virus propagation. The J Biol Chem. 2004;279(30):30931–30937. doi: 10.1074/jbc.M403258200. [DOI] [PubMed] [Google Scholar]
- 33.Dam S, Kracht M, Pleschka S, Schmitz ML. The influenza A virus genotype determines the antiviral function of NF-ĸB. J Virol. 2016;90(17):7980–7990. doi: 10.1128/JVI.00946-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan H, Wang H, Ma L, Ma X, Yin J, Wu S, Huang H, Li Y. Cirsimaritin inhibits influenza A virus replication by downregulating the NF-ĸB signal transduction pathway. Virol J. 2018;15(1):88. doi: 10.1186/s12985-018-0995-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu B, Bao L, Wang L, Li F, Wen M, Li H, Deng W, Zhang X, Cao B. Anti-IFN-γ therapy alleviates acute lung injury induced by severe influenza A (H1N1) pdm09 infection in mice. J Microbiol Immunol Infect. 2019;12:S1684-11829(18)30438-9. doi: 10.1016/j.jmii.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Sica A, Dorman L, Viggiano V, Cippitelli M, Ghosh P, Rice N, Young HA. Interaction of NF-kappaB and NFAT with the interferon-gamma promoter. J Biol Chem. 1997;272(48):30412–30420. doi: 10.1074/jbc.272.48.30412. [DOI] [PubMed] [Google Scholar]
- 37.Yang QF, Dalgard CL, Eidelman O, Jozwik C, Pollard BS, Srivastava M, Pollard HB. Digitoxin induces apoptosis in cancer cells by inhibiting nuclear factor of activated T-cells-driven c-MYC expression. J Carcinogenesis. 2013;12:8. doi: 10.4103/1477-3163.112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206(1):79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sichelstiel A, Yadava K, Trompette A, Salami O, Iwakura Y, Nicod LP, Marsland BJ. Targeting IL-1β and IL-17A driven inflammation during influenza-induced exacerbations of chronic lung inflammation. PloS one. 2014;9(2):e98440. doi: 10.1371/journal.pone.0098440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jolly L, Stavrou A, Vanderstoken G, Meliopoulos VA, Habgood A, Tatler AL, Porte J, Knox A, Weinreb P, Violette S, Hussell T, Kolb M, Stampfli MR, Schultz-Cherry S, Jenkins G. Influenza promotes collagen deposition via αvβ6 integrin-mediated transforming growth factor β activation. J Biol Chem. 2014;289(51):35246–35263. doi: 10.1074/jbc.M114.582262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Q, Huang W, Jozwik C, Lin Y, Glasman M, Caohuy H, Sruvastava M, Esposito D, Gillette W, Hartley J, Pollard HB. Cardiac glycosides inhibit TNF-alpha/NF-kappaB signaling by blocking recruitment of TNF receptor-associated death domain to the TNF receptor. Proc Nat Acad Sci (USA) 2005;102(27):9631–9636. doi: 10.1073/pnas.0504097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Cebotaru L, Lee HW, Yang Q, Pollard BS, Pollard HB, Guggino WB. CFTR controls the activity of NF-kappaB by enhancing the degradation of TRADD. Cell Physiol and Biochem. 2016;40(5):1063–1078. doi: 10.1159/000453162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amarelle L, Lecuona E. The antiviral effects of Na,KATPase inhibition: A minireview. Int J Mol Sci. 2018;19(8) doi: 10.3390/ijms19072154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burkard C, Verheije MH, Haagmans BL, van Kuppeveld FJ, Rottier PJ, Bosch BJ, de Haan CAM. ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells. J Virol. 2015;89(8):4434–4448. doi: 10.1128/JVI.03274-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei T, Wang H, Wu X, Lu Y, Guan S, Dong F, Dong C, Zhu G, Bao Y, Zhang J, Wang G, Li H. in silico screening of potential spike glycoprotein inhibitors of SARS-CoV-2 with drug repurposing strategy. Chin J Integr Med. 2020;26(9):663–669. doi: 10.1007/s11655-020-3427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senathilake KS, Samarakoon S, Tennekoon K. Virtual screening of inhibitors against spike glycoprotein of SARS-CoV- 2: a drug repurposing approach. Preprints. 2020 doi: 10.20944/preprints202003.0042.v2. [DOI] [Google Scholar]
- 47.Farag A, Wang P, Boys IN, Eitson JL, Ohlson MB, Fan W, McDougal MB, Ahmed M, Schoggins JW, Sadek H. Identification of atovaquone, ouabain and mebendazole as FDA approved drugs targeting SARS-CoV-2. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12003930. [DOI] [Google Scholar]
- 48.Aanouz I, Belhassan A, El-Khatabi K, Lakhlifi T, El-Ldrissi M, Bouachrine M. Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. J Biomol Struct Dyn. 2020;6:1–9. doi: 10.1080/07391102.2020.1758790. [DOI] [PMC free article] [PubMed] [Google Scholar]