Abstract
Background/Aim: Histone modification is associated with tumorigenesis and cancer progression. Recent studies have revealed the prognostic value of histone modification; however, its prognostic role in distal bile duct cancer remains unclear. Patients and Methods: We analyzed the expression of H3K9me3, H4K20me3, and H3K36me3 and its correlation with survival outcomes in resected samples from 88 patients with distal bile duct cancer. Results: Low expression rates of H3K9me3, H4K20me3, and H3K36me3 were significantly associated with poor overall survival (p=0.003, 0.008, and 0.047, respectively) and event-free survival (p=0.03 for H3K9m3). Additionally, low-expression of H3K9me3 was an independent poor prognostic indicator (p<0.001; HR=7.85; 95% CI=2.693-22.883). Conclusion: H3K9me3 was an independent poor prognostic factor in distal common bile duct cancer. Our results suggest that histone markers are potential prognostic markers and provide better management for patients at risk for an aggressive course of disease.
Keywords: Distal bile duct cancer, histone, methylation
Cholangiocarcinoma (CCA) is a heterogeneous group of epithelial malignancies divided into intrahepatic (I), perihilar (P), and distal (D) CCA based on the anatomical location of the tumors. DCCA is defined as a tumor arising from the epithelial cells of the bile duct between the origin of the cystic duct and the ampulla of Vater and accounts for 20-30% of all CCA (1). Most DCCA cases are diagnosed at an advanced, unresectable stage, and prognosis is generally poor (2,3).
Epigenetic changes such as DNA methylation and histone modification play a crucial role in tumorigenesis and cancer progression. Histone modification is one of the important epigenetic regulatory mechanisms and includes methylation, acetylation, ubiquitylation, sumoylation, and phosphorylation. Histone methylation is defined as the addition of methyl groups to lysine or arginine residues of histone proteins (4). These modifications can affect the overall chromatin structure and thus regulate downstream gene expression as well as DNA repair, replication, and recombination (4,5). Functional consequences of methylation depend on the histone protein, the methylated residue, and the number of methyl groups. Trimethylation of lysine 9 on histone H3 (H3K9me3) and lysine 20 on histone H4 (H4K20me3) is associated with the suppression of gene expression. In contrast, trimethylation of lysine 36 on histone H3 (H3K36me3) is associated with the activation of gene expression (5,6).
The histone methylation status and its prognostic significance have been investigated in various cancers. Previous studies have revealed that histone methylation was related to the prognosis of various cancers, including esophagus (7,8), liver (9,10), colorectal (11,12), and lung cancer (13,14). Recently, a pioneering study demonstrated that mutations in gene promoters with H3K27 trimethylation (H3K27me3) were frequently found in fluke-positive ICCA, while they were rarely detected in fluke-negative DCCA (15). To the best of our knowledge, the methylation status of other major histone proteins in DCCA has not been reported yet.
Histone trimethylation antibodies are widely used surrogate markers for detecting the trimethylation status (16). Diffuse and strong nuclear immunoreactivity of these antibodies is regarded as indicative of trimethylation, whereas negative expression of these antibodies indicates the lack of trimethylation. To investigate the methylation status of the major histone proteins in DCCA, we determined the expression rates of H3K9me3, H4K20me3, and H3K36me3 in resected samples from patients with DCCA using immunohistochemistry and analyzed their relationship with various prognostic factors.
Patients and Methods
Patients and tissue samples. Patients with DCCA were identified from the surgical pathology database of the Department of Pathology at the Kyung Hee University Hospital (Seoul, Republic of Korea). Between 1982 and 2017, 100 patients who underwent surgical resection for DCCA were included in the study. Follow-up data after surgery were available for 88 patients (88%). Tumors were histologically diagnosed based on the World Health Organization Classification (17). Two experienced pathologists (H.G.K. and S.W.K.) reviewed all available hematoxylin-eosin slides and selected the most representative slide of each patient for immunohistochemical staining. We also reviewed the medical records and pathology reports to record clinicopathological features, including age at diagnosis, sex, histological grade, tumor size, pathological tumor stage, nodal metastasis (pN), and stage group. Local recurrences and distant metastases were identified in imaging studies, including computed tomography and magnetic resonance imaging. To analyze event-free survival (EFS), we defined the primary end point as the time of relapse in the form of local recurrence or distant metastasis This study (2020-01-039) was reviewed and approved by the Institutional Review Board of the Kyung Hee University Hospital (Seoul, Republic of Korea). Review Board and the investigation conformed with the principles outlined in the Declaration of Helsinki.
Immunohistochemistry (IHC). Blocks of tumor specimens were used for IHC staining. An antigen retrieval procedure was performed using the Bond ER2 Solution for 30 min at 100˚C. Endogenous peroxidases were quenched by incubation with hydrogen peroxide for 5 min. The sections were incubated for 15 min at ambient temperature with primary antibodies against H3K9me3 (1:200, ab8898, Abcam, Cambridge, MA, USA), H4K20me3 (1:200, ab9053, Abcam), and H3K36me3 (1:500, ab9050, Abcam). The Bond Polymer Intense Detection System (Vision Biosystems, Waverley Victoria, Australia) was used for immunohistochemical staining according to the manufacturer’s instructions with minor modifications. Nuclei were counterstained with hematoxylin. Normal intestinal mucosa and hepatocytes were used as positive controls. The negative control was prepared by substituting non-immune serum for primary antibodies.
Statistical analysis. The relationship between clinicopathological parameters and the expression of antibodies was determined by the Chi-squared test. The Kaplan-Meier method with the log-rank test was performed to evaluate the survival curves. A Cox proportional regression model was used for univariate and multivariate analyses of the impact of histone methylation and other prognostic factors. Statistical analyses were performed using the SPSS software (version 22.0, SPSS Inc., Chicago, IL, USA). A p-value less than 0.05 was considered statistically significant.
Results
Demographics. This study included a total of 88 patients consisting of 62 males and 26 females. The median age of patients was 61 years (range=33-83 years). The pathological T stages according to the American Joint Committee on Cancer (AJCC, 8th edition) staging system were as follows: T1, 25% (n=22); T2, 52% (n=46); and T3, 23% (n=20). Twenty-eight patients (32%) had nodal metastases. The overall stage distribution was as follows: I, 19% (n=17); IIA, 43% (n=38); IIB, 36% (n=32); and IIIA, 1% (n=1). None of the patients underwent neoadjuvant chemotherapy or neoadjuvant concurrent chemoradiation therapy.
Expression pattern of histone markers in DCCA. Seventeen samples (19%) contained normal biliary epithelial cells. The normal biliary epithelial cells of most patients showed at least weak expression of the antibodies against H3K9me3 (76%), H4K20me3 (88%), and H3K36me3 (88%), but we also observed moderate to strong expression of these antibodies in normal cells (71%, 71%, and 76%, respectively). Figure 1 shows the expression of histone markers in normal biliary epithelial and tumor cells. Compared to normal biliary epithelial cells, tumor cells expressed considerably less histone markers. The tumor cells of only 28-41% of the patients showed homogenous and strong expression of the histone markers (H3K9me3, H4K20me3, and H3K36me3; 41%, 30%, and 28%, respectively), whereas those of 35-54% of the patients showed heterogeneous expression (H3K9me3, H4K20me3, and H3K36me3; 35%, 54%, and 53%, respectively).
Figure 1. Representative images of immunohistochemical staining of histone trimethylation antibodies in distal cholangiocarcinoma and normal biliary epithelial cells. (A-B) H3K9me3, (C-D) H4K20me3, and (E-F) H3K36me3. Original magnification, ×200.
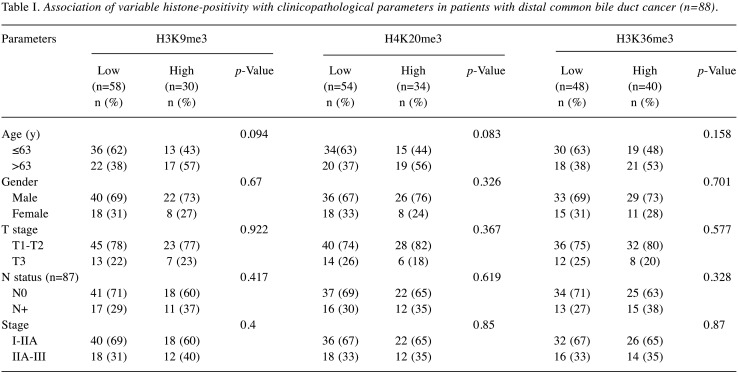
Association between histone methylation and clinicopathological parameters. For statistical analysis, the cases were divided into two groups according to the proportion of tumor cells with nuclear positivity to each antibody (as indicated by immunostaining): low-expression (0-49%) and high-expression groups (≥50%). Two independent pathologists (H.G.K. and K.N) evaluated the immunostaining scores. The degree of agreement between the two observers was calculated with a kappa value of 0.94. Two cases of discrepancy were re-reviewed, and their final scores were determined by consensus. Fifty-eight cases (66%) of H3K9me3 (H3K9me3-low), 54 cases (61%) of H4K20me3 (H4K20me3-low), and 48 cases (55%) of H3K36me3 (H3K36me3-low) were in the low-expression group. Next, we analyzed the relationship between the expression level and clinicopathological parameters, including patient age, sex, pT, pN, and stage (Table I). None of the histone methylation markers was related to any clinicopathological parameters.
Table I. Association of variable histone-positivity with clinicopathological parameters in patients with distal common bile duct cancer (n=88).
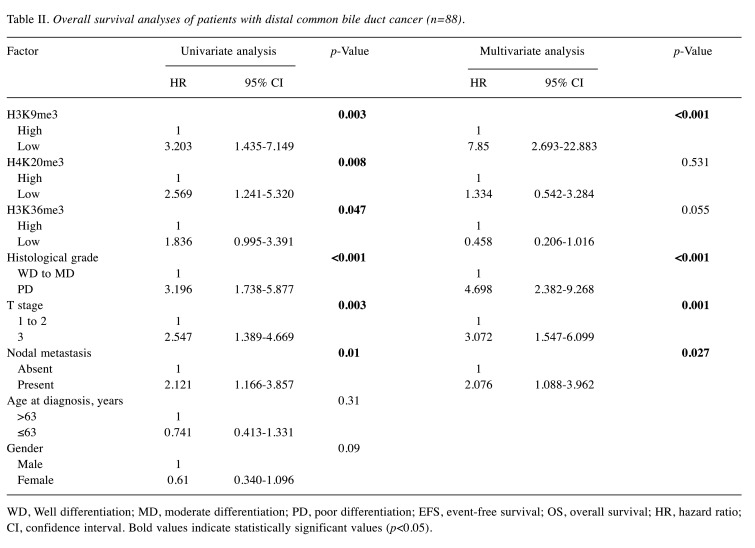
Survival analysis based on histone methylation. The median follow-up was 40 months, and 50 of the 88 patients (57%) died during the follow-up. Low-expression of the histone markers was significantly associated with poor overall survival (OS) [Table II and Figure 2A, C and E, H3K9me3: p=0.003, hazard ratio (HR)=3.203, 95% confidence interval (CI)=1.435-7.149; H4K20me3: p=0.008, HR=2.569, 95% CI=1.241-5.320; H3K36me3: p=0.047, HR=1.836, 95% CI=0.995-3.391] compared to the OS in the high-expression group. In addition, H3K9me3-low patients had a significantly shorter EFS than H3K9me3-high patients (Figure 2B, p=0.03, HR=1.978, 95% CI=1.042-3.757). The status of the other histone markers was not related to significant differences in EFS (Figure 2D and F).
Table II. Overall survival analyses of patients with distal common bile duct cancer (n=88).
WD, Well differentiation; MD, moderate differentiation; PD, poor differentiation; EFS, event-free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval. Bold values indicate statistically significant values (p<0.05)
Figure 2. Kaplan-Meier curves for event-free (EFS) and overall survival (OS). (A-B) H3K9me3, (C-D) H4K20me3, and (E-F) H3K36me3 expression. Low expression of (A) H3K9me3, (C) H4K20me3, and (E) H3K36me3 is significantly associated with poor OS (p=0.003, 0.008, and 0.047, respectively). In addition, low expression of (B) H3K9me3 is significantly associated with poor EFS (p=0.03).
In order to determine whether histone methylation was an independent prognostic factor, Cox regression analysis was performed with other clinicopathological factors (Table II). Univariate analysis showed that low-expression of histone markers (H3K9me3, H4K20me3, and H3K36me3), histologic grade, pT stage, and nodal metastasis were poor prognostic factors for OS. Factors found to be significant in the univariate analysis were included in the multivariate analysis. After adjusting for these variables, expression of H3K9me3, histologic grade, pT, and nodal metastasis were identified as independent prognostic factors for OS and EFS.
Discussion
In this study, we observed that low-expression of H3K9me3, H4K20me3, and H3K36me3 was significantly associated with poor OS in patients with DCCA. Furthermore, H3K9me3 was found to be an independent poor prognostic factor for OS and EFS by multivariate Cox regression analysis. Due to the late diagnosis of DCCA in an advanced stage and uncertainty about the eligibility of the patient for adjuvant therapy, prognosis is poor. Furthermore, the survival of patients remains low even after potentially curative surgery because of tumor recurrence. Our findings suggest that the status of histone methylation can predict patient survival outcomes and allows an accurate prognostic stratification of patients with DCCA.
Epigenetic alterations such as DNA methylation and histone modification are crucial in carcinogenesis. Histone methylation influences the chromatin structure and can affect the accessibility of transcription factors to nearby genes (5,18). This important modification can be detected by immunohistochemical histone markers. SETDB1, a histone methyltransferase, is responsible for histone methylation at H3K9 and promotes silencing of tumor suppressor genes (19). The expression of SETDB1 is up-regulated in most cancers and thus closely related to carcinogenesis of various cancers, such as colorectal cancer (20), hepatocellular carcinoma (21), and lung cancer (22). In a colorectal cancer study, SETDB1 was recruited to the promoter region of TP53 and caused promoter methylation to inhibit apoptosis, thereby promoting colorectal carcinogenesis (20). SUV420H2 is involved in the repression of gene expression by histone methylation at H4K20 (23). This enzyme silences specific genes and promotes mesenchymal-epithelial transition in pancreatic cancer (24). Expression of H4K20me3 is detected not only in lung squamous cell carcinoma but also in early precursor lesions in which the histone level tends to decrease with disease progression (14).
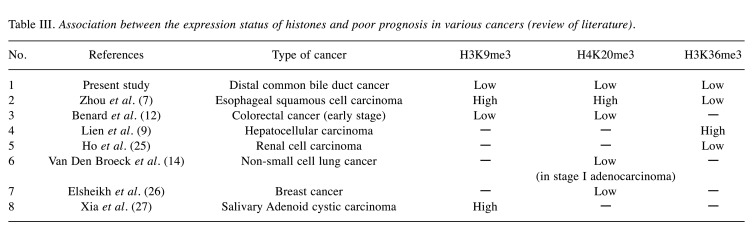
The association between histone methylation and prognosis is controversial. To date, research on the prognostic value of the histone status in various cancers has been conducted. Table III summarizes some of the findings concerning the relationship between the histone status and the survival rate for various cancers, including esophageal squamous cell carcinoma (7), colorectal cancer (12), hepatocellular carcinoma (9), renal cell carcinoma (25), non-small cell lung cancer (14), breast cancer (26), and salivary adenoid cystic carcinoma (27). Loss of H3K36me3 expression correlates with poor prognosis of most cancers, such as renal cell carcinoma (25) and esophageal squamous cell carcinoma (7). Sequencing of renal cell carcinoma revealed a loss-of-function mutation of the tumor suppressor SETD2, which is related to the loss of H3K36me3 (25). This SETD2 gene mutation is also found in other cancers (28); however, it does not necessarily lead to loss of H3K36me3 expression. H3K36me3 induces CDH1 exon 8 skipping by a mechanism that regulates gene expression through gene splicing, thereby causing cell discohesion and affecting cancer progression (29).
Table III. Association between the expression status of histones and poor prognosis in various cancers (review of literature).
Of note, no study has been conducted to evaluate the association between the global histone methylation status and clinical outcomes in DCCA. In this study, we investigated the expression of histone markers, including H3K9me3, H4K20me3, and H3K36me3, and revealed their poor prognostic value in DCCA using immunohistochemistry. The current study has certain limitations, including its retrospective single-center design and small case numbers. In addition, the mechanisms underlying histone modification in DCCA carcinogenesis were not examined. Therefore, future studies on DCCA should include larger patient cohorts with external validation, and further experimental studies are needed.
In conclusion, our results indicate that low-expression of H3K9me3 in distal bile duct cancer is independently and strongly related to poor prognosis. These results imply that histone modification markers can be used as prognostic markers in patients who are at risk for an aggressive course of disease.
Conflicts of Interest
The Authors have no conflicts of interest to declare regarding this study.
Authors’ Contributions
All Authors made substantial contributions to the conception and design of the study, acquisition of data, analysis and interpretation of the data, as well as drafting the manuscript and revising it critically for important intellectual content, and providing final approval of the version to be published.
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, Information and Communications Technology (ICT), and Future Planning (NRF-2020R1G1A1003692).
References
- 1.DeOliveira ML, Cunningham SC, Cameron JL, Kamangar F, Winter JM, Lillemoe KD, Choti MA, Yeo CJ, Schulick RD. Cholangiocarcinoma: thirty-one-year experience with 564 patients at a single institution. Ann Surg. 2007;245:755–762. doi: 10.1097/01.sla.0000251366.62632.d3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrling J, Andersson R, Sasor A, Lindell G, Ansari D, Nilsson J, Andersson B. Outcome and evaluation of prognostic factors after pancreaticoduodenectomy for distal cholangio-carcinoma. Ann Gastroenterol. 2017;30:571–577. doi: 10.20524/aog.2017.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogenberger JM, DeLeon TT, Arora M, Ahn DH, Borad MJ. Emerging role of precision medicine in biliary tract cancers. NPJ Precis Oncol. 2018;2:21. doi: 10.1038/s41698-018-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limpaiboon T. Epigenetic aberrations in cholangiocarcinoma: potential biomarkers and promising target for novel therapeutic strategies. Asian Pac J Cancer Prev. 2012;13 Suppl:41–45. [PubMed] [Google Scholar]
- 6.Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta. 2014;1839:1362–1372. doi: 10.1016/j.bbagrm.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou M, Li Y, Lin S, Chen Y, Qian Y, Zhao Z, Fan H. H3K9me3, H3K36me3, and H4K20me3 expression correlates with patient outcome in esophageal squamous cell carcinoma as epigenetic markers. Dig Dis Sci. 2019;64:2147–2157. doi: 10.1007/s10620-019-05529-2. [DOI] [PubMed] [Google Scholar]
- 8.Chen YW, Kao SY, Wang HJ, Yang MH. Histone modification patterns correlate with patient outcome in oral squamous cell carcinoma. Cancer. 2013;119:4259–4267. doi: 10.1002/cncr.28356. [DOI] [PubMed] [Google Scholar]
- 9.Lien HC, Jeng YM, Jhuang YL, Yuan RH. Increased trimethylation of histone H3K36 associates with biliary differentiation and predicts poor prognosis in resectable hepatocellular carcinoma. PLoS One. 2018;13:e0206261. doi: 10.1371/journal.pone.0206261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He C, Xu J, Zhang J, Xie D, Ye H, Xiao Z, Cai M, Xu K, Zeng Y, Li H, Wang J. High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Hum Pathol. 2012;43:1425–1435. doi: 10.1016/j.humpath.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Li Y, Li J, Liu Y, Cui B. H3K4me3 and Wdr82 are associated with tumor progression and a favorable prognosis in human colorectal cancer. Oncol Lett. 2018;16:2125–2134. doi: 10.3892/ol.2018.8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benard A, Goossens-Beumer IJ, van Hoesel AQ, de Graaf W, Horati H, Putter H, Zeestraten EC, van de Velde CJ, Kuppen PJ. Histone trimethylation at H3K4, H3K9 and H4K20 correlates with patient survival and tumor recurrence in early-stage colon cancer. BMC Cancer. 2014;14:531. doi: 10.1186/1471-2407-14-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avila-Moreno F, Armas-Lopez L, Alvarez-Moran AM, Lopez-Bujanda Z, Ortiz-Quintero B, Hidalgo-Miranda A, Urrea-Ramirez F, Rivera-Rosales RM, Vazquez-Manriquez E, Pena-Mirabal E, Morales-Gomez J, Vazquez-Minero JC, Tellez-Becerra JL, Ramirez-Mendoza R, Avalos-Bracho A, de Alba EG, Vazquez-Santillan K, Maldonado-Lagunas V, Santillan-Doherty P, Pina-Sanchez P, Zuniga-Ramos J. Overexpression of MEOX2 and TWIST1 is associated with H3K27me3 levels and determines lung cancer chemoresistance and prognosis. PLoS One. 2014;9:e114104. doi: 10.1371/journal.pone.0114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Den Broeck A, Brambilla E, Moro-Sibilot D, Lantuejoul S, Brambilla C, Eymin B, Khochbin S, Gazzeri S. Loss of histone H4K20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin Cancer Res. 2008;14:7237–7245. doi: 10.1158/1078-0432.Ccr-08-0869. [DOI] [PubMed] [Google Scholar]
- 15.Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, Choo SP, Myint SS, Thanan R, Nagarajan S, Lim WK, Ng CCY, Boot A, Liu M, Ong CK, Rajasegaran V, Lie S, Lim AST, Lim TH, Tan J, Loh JL, McPherson JR, Khuntikeo N, Bhudhisawasdi V, Yongvanit P, Wongkham S, Totoki Y, Nakamura H, Arai Y, Yamasaki S, Chow PK, Chung AYF, Ooi L, Lim KH, Dima S, Duda DG, Popescu I, Broet P, Hsieh SY, Yu MC, Scarpa A, Lai J, Luo DX, Carvalho AL, Vettore AL, Rhee H, Park YN, Alexandrov LB, Gordan R, Rozen SG, Shibata T, Pairojkul C, Teh BT, Tan P. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 2017;7:1116–1135. doi: 10.1158/2159-8290.Cd-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurdistani SK. Histone modifications as markers of cancer prognosis: a cellular view. Brit J Cancer. 2007;97:1. doi: 10.1038/sj.bjc.6603844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumours of the digestive system. World Health Organization. 2010 [Google Scholar]
- 18.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 19.Yang L, Xia L, Wu DY, Wang H, Chansky HA, Schubach WH, Hickstein DD, Zhang Y. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene. 2002;21:148–152. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- 20.Chen K, Zhang F, Ding J, Liang Y, Zhan Z, Zhan Y, Chen LH, Ding Y. Histone methyltransferase SETDB1 promotes the progression of colorectal cancer by inhibiting the expression of TP53. J Cancer. 2017;8:3318–3330. doi: 10.7150/jca.20482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong CM, Wei L, Law CT, Ho DW, Tsang FH, Au SL, Sze KM, Lee JM, Wong CC, Ng IO. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63:474–487. doi: 10.1002/hep.28304. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Paredes M, Martinez de Paz A, Simó-Riudalbas L, Sayols S, Moutinho C, Moran S, Villanueva A, Vázquez-Cedeira M, Lazo PA, Carneiro F, Moura CS, Vieira J, Teixeira MR, Esteller M. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene. 2014;33:2807–2813. doi: 10.1038/onc.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viotti M, Wilson C, McCleland M, Koeppen H, Haley B, Jhunjhunwala S, Klijn C, Modrusan Z, Arnott D, Classon M, Stephan JP, Mellman I. SUV420H2 is an epigenetic regulator of epithelial/mesenchymal states in pancreatic cancer. J Cell Biol. 2018;217:763–777. doi: 10.1083/jcb.201705031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho TH, Kapur P, Joseph RW, Serie DJ, Eckel-Passow JE, Tong P, Wang J, Castle EP, Stanton ML, Cheville JC, Jonasch E, Brugarolas J, Parker AS. Loss of histone H3 lysine 36 trimethylation is associated with an increased risk of renal cell carcinoma-specific death. Mod Pathol. 2016;29:34–42. doi: 10.1038/modpathol.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elsheikh SE, Green AR, Rakha EA, Powe DG, Ahmed RA, Collins HM, Soria D, Garibaldi JM, Paish CE, Ammar AA, Grainge MJ, Ball GR, Abdelghany MK, Martinez-Pomares L, Heery DM, Ellis IO. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69:3802–3809. doi: 10.1158/0008-5472.Can-08-3907. [DOI] [PubMed] [Google Scholar]
- 27.Xia R, Zhou R, Tian Z, Zhang C, Wang L, Hu Y, Han J, Li J. High expression of H3K9me3 is a strong predictor of poor survival in patients with salivary adenoid cystic carcinoma. Arch Pathol Lab Med. 2013;137:1761–1769. doi: 10.5858/arpa.2012-0704-OA. [DOI] [PubMed] [Google Scholar]
- 28.Kudithipudi S, Jeltsch A. Role of somatic cancer mutations in human protein lysine methyltransferases. Biochim Biophys Acta. 2014;1846:366–379. doi: 10.1016/j.bbcan.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Li XW, Shi BY, Yang QL, Wu J, Wu HM, Wang YF, Wu ZJ, Fan YM, Wang YP. Epigenetic regulation of CDH1 exon 8 alternative splicing in gastric cancer. BMC Cancer. 2015;15:954. doi: 10.1186/s12885-015-1983-5. [DOI] [PMC free article] [PubMed] [Google Scholar]