Abstract
The direct analysis of 3D Optical Coherence Tomography (OCT) volumes enables deep learning models (DL) to learn spatial structural information and discover new bio-markers that are relevant to glaucoma. Down-sampling 3D input volumes is the state-of-art solution to accommodate for the limited number of training volumes as well as the available computing resources. However, this limits the network’s ability to learn from small retinal structures in OCT volumes. In this paper, our goal is to improve the performance by providing guidance to DL model during training in order to learn from finer ocular structures in 3D OCT volumes. Therefore, we propose an end-to-end attention guided 3D DL model for glaucoma detection and estimating visual function from retinal structures. The model consists of three pathways with the same network architecture but different inputs. One input is the original 3D-OCT cube and the other two are computed during training guided by the 3D gradient class activation heatmaps. Each pathway outputs the class-label and the whole model is trained concurrently to minimize the sum of losses from three pathways. The final output is obtained by fusing the predictions of the three pathways. Also, to explore the robustness and generalizability of the proposed model, we apply the model on a classification task for glaucoma detection as well as a regression task to estimate visual field index (VFI) (a value between 0 and 100). A 5-fold cross-validation with a total of 3782 and 10,370 OCT scans is used to train and evaluate the classification and regression models, respectively. The glaucoma detection model achieved an area under the curve (AUC) of 93.8% compared with 86.8% for a baseline model without the attention-guided component. The model also outperformed six different feature based machine learning approaches that use scanner computed measurements for training. Further, we also assessed the contribution of different retinal layers that are relevant to glaucoma. The VFI estimation model achieved a Pearson correlation and median absolute error of 0.75 and 3.6%, respectively, for a test set of size 3100 cubes.
Keywords: 3D convolutional neural networks, optical coherence tomography, gradient-weighted class activation maps, glaucoma detection, visual field estimation, attention guided deep learning
I. Introduction
GLaucoma is the leading cause of irreversible blindness worldwide. The number of worldwide glaucoma patients, aged 40–80 years, is estimated to be approximately 80 million in 2020 with about 20 million increase since 2010 [1]. Glaucoma is associated with optic nerve damage, functional vision loss and death of retinal ganglion cells [2]. Structural and functional methods are utilized jointly to determine the severity of glaucoma and monitor its progression [3]. One of the functional tests utilized is called visual field test (VFT), and it is used to evaluate vision loss due to glaucoma and other optic nerve diseases [4]. VFT, however, is costly, time-consuming and shows poor repeatability as it is greatly affected by cataracts, visual acuity, glaucoma medications, severity of glaucoma, learning effect, distraction and other factors [5], [6].
On the other hand, structural measurements are objective and based on the imaging of the optic nerve head (ONH), macula and surrounding regions. It enables the quantification of retinal structures relevant to glaucoma such as the retinal nerve fiber layer (RNFL) and ganglion cell-inner plexiform layer (GCIPL) complex [3]. Many researchers have investigated the relationships between visual field test results and structural measures that are produced by optical coherence tomography (OCT) scanners [7]–[9]. For instance in [7], [8], RNFL thickness was found to be linearly related to visual field loss at advanced disease stage. However, finding such relationship is very challenging as sometimes the optic nerve changes before the visual field loss [10]–[13], and other times visual field loss occurs prior to structural damage at the optic nerve [14].
Deep learning (DL) approaches have been previously used with fundus 2D-colour images for ocular disease detection and diagnosis [15], [16]. This includes segmentation of retinal vessels [17]–[20], optic disc and optic cup segmentation [18], [21], [22], classification of glaucoma [23]–[28], and image registration [29]. A more recent 3D imaging modality is the spectral-domain OCT technology that provides clinicians with high-resolution images and quantified measurements of the retinal structures. In clinics, OCT scans are the standard for eye care and are employed for diagnosing and monitoring various retinal diseases, evaluating progression, and assessing response to therapy [30]. This technology enables the use of 3D DL techniques to learn new structural parameters useful for the diagnosis and management of glaucoma, quantify its relevant ocular structures (such as the individual retinal layers, optic nerve head, choroid, and lamina cribrosa) [31], and investigate whether functional measurements such as visual field index (VFI) or mean deviation (MD) can be inferred from structure (i.e. OCT volumes).
Further, the literature shows that most of DL initiatives in OCT glaucoma detection have primarily depended on scanner measurements of different retinal structures such as the thickness of RNFL and the ganglion cell complex (GCC), limiting the generalizability of DL models to measurements from different commercial scanners since they are calculated differently. This also limits the ability of DL models to discover new structural biomarkers which are not quantified by the scanners. For example, in [32], glaucoma was diagnosed by training DL model using thicknesses maps for both RNFL and GCIPL with an AUC of 93.7%. Also in [33], AlexNet pretrained model [34] was used for feature extraction using probability and thickness maps of RNFL and GCIPL layers, followed by random forest classifier [35] to discriminate between healthy and glaucomatous eyes. The best performance was achieved using RNFL probability map with an accuracy of 93.1%. In another study by An et al. [36], VGG19-based transfer learning model was performed to detect glaucoma using both thickness and deviation maps for each of RNFL and GCC layers. Then, a random forest classifier combining features from different inputs achieved an AUC of 96%. Further, Wang et al. [37] proposed S-D net that has two parts, S-net for segmentation of 6 retinal layers and D-net for the diagnosis of glaucoma according to RNFL thickness vector of length 1024 calculated from the segmentation results. The method achieved a dice coefficient of 0.959 for S-net and an accuracy of 85.4% for D-net.
The only end-to-end DL model that relied on the 3D scans as an input was presented by Maetschke et al. [38]. A 3D-CNN model composed of 5 convolutional layers with ReLU activation, batch-normalization using input volumes that were downsampled by a factor of nearly 80 (64×64×128 (b-scans×a-scans×depth) vs original size of 200×200×1024). The highest achieved AUC was 94% which outperformed classical machine learning (ML) techniques. In [39], we extended the 3D-CNN model proposed by Maetschke et al. [38] to investigate whether utilizing larger input volumes would improve the network performance or not. We used an input volumes of size 128×128×256 to train a network with 8 convolutional layers. We obtained an AUC of 97% using the same dataset used in [38]. Further, in [40], Maetschke et al. extended his work to assess structural-functional correlation using 3D-CNN model. Specifically, VFI and MD functional measures were estimated directly from 3D raw OCT scans. The highest achieved Pearson Correlation (ρ) was 0.88 compared with 0.74 for the best performing classical ML algorithms.
Another important aspect is the clinical interpretability and transparency [41] of the developed DL models. In this regard, class activation maps (CAMs) [42] and gradient-weighted class activation maps (grad-CAMs) [43] have been recently proposed to reveal insights into the decisions of deep learning models. Both of these techniques identify areas of the images that the network relied on heavily to generate the classification. However, CAM requires a specific network architecture, namely the use of a global average pooling layer prior to the output layer. Grad-CAM is a generalized form of CAM and can be used with any CNN-based architecture without any additional requirements.
Further, the visualization of DL models for glaucoma detection has been studied in three papers [36], [38], [39]. An et al. [36] identified pathologic regions in 2D thickness maps using grad-CAM, which have shown to be in agreement with the important decision making regions used by physicians. Also, Maetschke et al. [38] implemented 3D-CAM to identify the important regions for detecting glaucoma in 3D OCT volumes. The maps were however, in a coarse resolution that matched the downsampled input image. This method also employed specific architecture changes to accommodate the requirements of CAM generation. It is also noteworthy that neither of these approaches analyzed the CAMs in any systematic fashion, and merely used the heat maps to validate findings in a small number of images that were qualitatively assessed. Lastly, in our previous work [39], we used 3D grad-CAM to visualize the important decision regions in a higher resolution than was available before. One of the conducted experiments was to quantitatively validate grad-CAM results for 3D OCT volumes by occluding important decision regions identified in the heat maps and assessing the impact of this on the performance of the model. Occluding the most important decision regions in grad-CAM heatmaps dropped the performance by nearly 40% while occluding the least important areas only resutled in a 4% drop in the performance. The paper also included a quantitative comparison between CAM and grad-CAM heatmaps with the later significantly outperforming CAM heatmaps. This has motivated us to use grad-CAM heatmaps to provide guidance to DL model during training and improve the performance by learning the finer ocular structures in 3D OCT volumes associated with disease as well as visual function.
In this paper, we propose an end-to-end attention guided DL framework for glaucoma detection and estimating VFI. The model is trained directly on 3D volumes using three inputs, one is the original 3D-OCT cube and the other two are computed during training guided by 3D grad-CAM heatmaps [43]. The model consists of three pathways that have the same network architecture. First pathway uses the original volumes as an input after downsampling to size 256×64×64. Then grad-CAM heatmaps are generated to identify retinal structures in the original volumes, which the network relies on for detecting glaucoma. Occlusion of the less important retinal structures in original cubes is used as an input for the second pathway. The input for the third pathway is obtained by cropping the region with the most important structures. The contribution of this work can be summarized as follows:
The proposed approach continues to avoid the dependency on segmented structural thicknesses through direct analysis of raw OCT scans, and also improves on previously approached techniques by focusing on the important decision areas, identified by grad-CAM heatmaps, to learn more about fine ocular structures.
The performance of the model is evaluated for two different tasks: i) A classification task for glaucoma detection and ii) A regression task for VFI functional parameter estimation.
The proposed DL framework provides analysis of 3D attention maps in a higher resolution than was available before. This facilitates the understanding and interpretation of the network’s decision for glaucoma detection and diagnosis.
For the first time, we provide a quantitative clinical assessment for the contribution of different ocular structures that the network relied on when detecting glaucoma.
Intensive experiments are conducted to demonstrate the effectiveness of the proposed approach and it was compared with another 3D-CNN and classical ML approaches trained on scanner computed measurements.
The rest of the paper is organized as follows. Section II explains the proposed network architecture and DL framework. In Sections III and IV, we describe the dataset and experimental setup used for training and testing each of the glaucoma detection and VFI estimation models, respectively. Section V discusses the experimental results and the performed clinical assessment techniques. Finally, we conclude and outline future research directions in Section VI.
II. Attention-guided Network Architecture
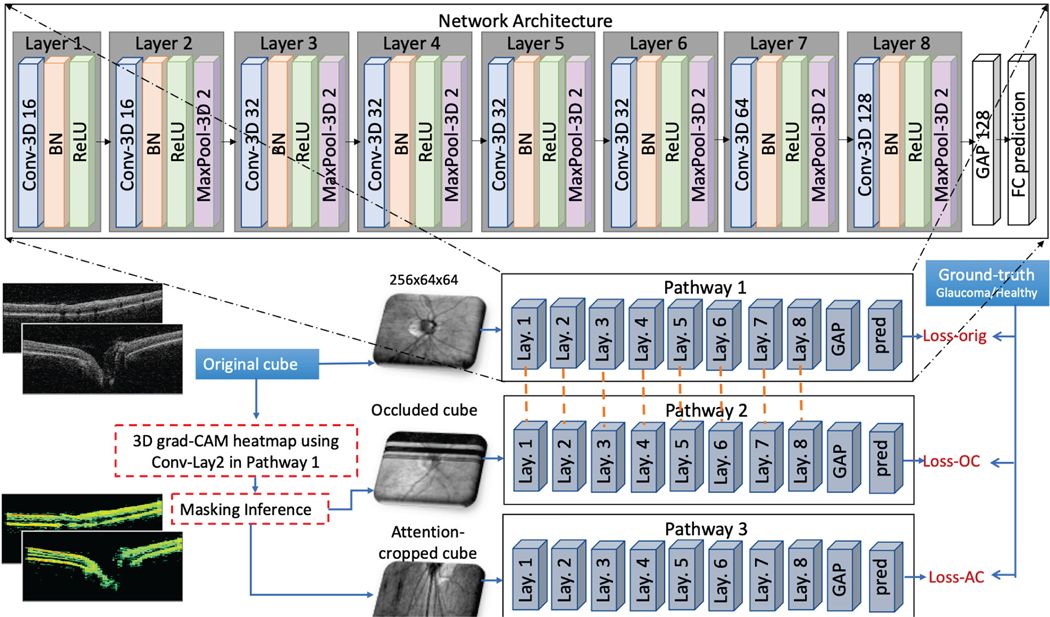
The framework of the proposed attention-guided DL model (AG-OCT) is presented in Figure 1. The model consists of three pathways called global, focused and local OCT structure pathways. They have same network architecture but different inputs with resolution of 256×64×64 (depth×b-scans×a-scans). Also, the first two pathways share same trainable weights, while the third one has its own learned weights. This is because the first two pathways have the same field of view, where the inputs are centered on the ONH and cover an area of 6×6×2 mm3. While the third pathway has a smaller field of view, i.e. different coverage area, as it focuses on a small region of the original area. The network architecture contains eight 3D-convolutional layers, each is followed by ReLU activation [44], batch-normalization [45] and max-pooling in order. The 3D convolutional layers have incremental number of filters of 16-16-32-32-32-32-64-128 with kernel size of 3, and stride of 1 for all layers. Also, 3D max-pooling layers have size of 2 and stride of 2. This is followed by global average pooling layer and a fully-connected output layer in order). Details about each pathway and the model loss are provided in the following sub-sections.
Fig. 1.
Framework of the proposed attention-guided DL model using 3D OCT volumes (AG-OCT)
A. Pathway#1: Global OCT Structure
This pathway learns the global OCT retinal structures that are relevant to the target task, i.e. glaucoma or VFI estimation. It receives its input by downsampling the original 3D-OCT cubes to size 256 ×64×64. We implement 3D grad-CAM to generate heatmaps that highlight the important decision areas in input volumes, following the explanation provided in [43]. In this context, grad-CAM heatmap is compute for conv#2 feature map that is 128×32×32 )(Lay. 2 in Figure 1). The generated heatmap is then used to derive the input of the other two pathways during training. We do not use CAM as it restricts the network architecture design. Further, CAM would generate heatmap visualization only for conv#8 feature map, which in our case has a size of 4×2×2. Hence, when resizing and overlaying on the original cube of size 1024×200×200 (depth×b-scans×a-scans) will not provide any meaningful results.
B. Pathway#2: Focused OCT Structure
The aim of the second pathway is to learn the correct output (e.g. glaucoma or healthy) using occluded cubes. The least important regions in the original cube are hidden, guiding the network to learn the location of the important decision areas. The rational is that if grad-CAM yields the correct decision areas, then hiding the least important decision areas should not have a great impact on the network performance results, since these areas are not important and most likely refer to noise and/or redundant information that are contained in the OCT volumes.
To do this, the input volumes are occluded by zeroing the rows and columns with the lowest heatmap weights. Specifically, we extract a set of indices with the lowest weights per each dimension using average pooling for spatial dimension reduction. For example, a heat map with size 1024×200×200 is reduced to a vector of size 1024×1×1 by averaging the values of each 200×200 map to get a rank of weights for the first dimension, i.e. depth. The indices of the lowest x values (i.e. weights) in the resultant vector represent the least important region for this dimension. We apply this process on the b-scans and depth dimensions with x values of 64 and 256 respectively (both values are chosen to match the desired input shape of the network), while we consider the 200 a-scan columns are all important. This means that a fixed region of size 256×64×200 is occluded for each volume in its original resolution. The occluded-cube is downsampled to sizefor the second pathway of the network.256×64×64 and is used as input
C. Pathway#3: Local OCT Structure
The third pathway enables the network to learn more about the local structures in the OCT volumes by retaining detail and image resolution in the important areas (i.e. a close up zoom into the important ocular structures). In this context, we use the generated grad-CAM heatmap to find the most important 3D sub-region in the input volume, which we call the attention-cropped cube. Specifically, we performed spatial dimension reduction method used in the second pathway to select the most important 64 b-scans from a total of 200 b-scans (i.e. rows with the highest weights along this dimension). This means that more than two-third of the voxels of input volumes are discarded. For example, given a cube of size 1024×200×200, the extracted attention-cropped cube size is 1024×64×200, while the size of the region which is taken away is 1024×136×200. The attention-cropped cube is also downsampled to size 256×64×64 and is used as an input for the third pathway.
D. Training Loss
The three pathways of the proposed model are trained concurrently, so that the attention maps are learned jointly. Each pathway has its prediction vector and loss as shown in Figure 1. The objective of the training is to minimize the total loss described in Equation 1, that is the sum of the three pathway losses, in addition to a regularization loss term to avoid overfitting during training.
| (1) |
Where are the predictions of pathway #1, 2, and 3 in order. Y is the ground truth vector and is the loss and is the regularization loss term for convolutional layers.
III. Glaucoma Detection
In this section, we explain how the proposed AG-OCT model is used for glaucoma detection. The model is trained to classify an OCT volume as healthy or glaucoma. The output has a value of [1,0] for healthy class and [0,1] for glaucoma class.
Dataset.
The dataset contains 3782 OCT scans from both eyes of 555 individuals, acquired on a Cirrus SD-OCT Scanner (Zeiss; Dublin, CA, USA) over multiple visits. The dataset has 427 healthy scans from 109 individuals and 3355 glaucoma scans from 446 individuals with primary open angle glaucoma (POAG). The clinical definition of healthy/glaucoma is made based on the visual field test results. The scans are centered on the ONH and has 200×200×1024 (a-scans×b-scans×depth) voxels per cube covering an area of 6×6×2 mm3. This study is an observational study that is conducted in accordance with the tenets of the Declaration of Helsinki and the Healthy Insurance Portability and Accountability Act. The Institutional Review Board of New York University and the University of Pittsburgh approved the study, and all subjects give written consent before participation.
Training and Testing.
We use a fully-connected softmax layer with 2 units for FC prediction layer (see Figure 1). The 3782 OCT volumes are split into a training, validation and testing subsets, containing 3031 (healthy: 325, POAG: 2706), 379 (healthy: 47, POAG: 332) and 372 (healthy: 55, POAG: 317) scans, respectively. OCT scans belonging to the same patient are included in only one of the three splits. The proposed model is trained using Adam optimizer with a learning rate of 1e−4. We also use weighted cross entropy loss [46] to avoid biased training due to the class size imbalance in the data. Training is performed with a batch of size 12 through 100 epochs. To avoid overfitting during training, we use L2 regularization loss with λ = 0.0001 and drop out layer with probability of 0.3. After each epoch, the area under the curve (AUC) was computed for the validation set, and the network is saved if an improvement in the AUC is observed.
Evaluation.
For the evaluation of the proposed model, five statistical performance measures are used, namely, AUC, accuracy, Matthews correlation coefficient (MCC), recall, precision and F1-score. Performance measures are computed using predictions from each pathway separately as well as the fusion of 3-pathway predictions using min, max and average operations. For reliable and stable results, we repeat the training 5 times and report the average performance measures for the five folds.
IV. Structural-Functional Correlation
The purpose of this section is to explore the generalizability and robustness of our attention-guided model (AG-OCT). To do this, we train the AG-OCT model to estimate the VFI parameter from structural data (i.e. OCT volumes), which is very important since clinicians use both structural and functional data to monitor glaucoma progression. The output is a value between 0 and 100.
Dataset.
We use a large dataset consisting of 10,370 ONH OCT volumes and their corresponding visual field test results. Structural OCT scans are captured from 1678 individuals across multiple visits using Cirrus SD-OCT scanners. Scans with signal strength less than 6 are discarded. The visual field test is performed using the Swedish interactive thresholding algorithm 24–2 perimetry (SITA standard; Humphrey Field Analyzer; Zeiss). The VFI can range from 0% (perimetrically blind field) to 100% (normal visual field). Similar to the previous cohort, all subjects give written consent before participation.
Training and Testing.
We trained and evaluated the regression model using the same architecture and experimental setup as the glaucoma detection experiment with three main changes. Firstly, we replace the last softmax layer with one unit fully connected layer with linear activation. Secondly, the mean squared error loss is used during training instead of weighted cross entropy. Lastly, we employ polynomial regression [47] for combining the 3-pathway predictions that is trained using same training data as the AG-OCT model (i.e. 70% of data) and the rest is used for testing (i.e. 30%, 3100 scans). Also, hyperparameter selection for polynomial degree is performed using Grid-search with 10-fold cross validation.
Evaluation.
For the evaluation, five evaluation metrics are computed namely, root mean squared error (RMSE), mean absolute error (MAE), median absolute error (MDAE), Pearson’s correlation coefficient (ρ) and Spearman’s rank correlation coefficient (r).
V. Experimental Results and Discussion
The proposed attention-guided DL model is implemented using Python and TensorFlow [48] on a single V100 GPU. We divide our results and experiments into five sections. In section V-A, we report the performance measures for glaucoma detection. In section V-B, we conduct different experiments to analyze the performance of our AG-OCT model. This followed by detailed clinical analysis of grad-CAM attention maps in section V-C. In section V-D, we provide comparative results with state-of-art approaches including classical ML techniques. Finally, in section V-E, we present the regression model results for VFI estimation.
A. Glaucoma Detection
Table I has the performance measures for glaucoma detection using the proposed attention-guided model. The table shows that the average of the 3-pathway predictions has the best performance with an AUC of 93.8%. Interestingly, the second best performance comes from pathway #2, i.e. the occluded cube with an AUC of 93.0%. This means that hiding some of the least important regions in 3D cubes improves the performance, because those regions might refer to noisy or redundant areas in 3D volume. This is followed by predictions from max-fusion, min-fusion, pathway #3, and pathway #1 with AUCs of 92.8, 92.2, 91.2, and 86.7% in order. Further, to examine the computational complexity of the proposed framework, we compute the average and standard deviation for execution time of the validation and test sets with 23.4± 2.2 and 22.7±3.8 (in seconds), respectively, for a batch of size 12. This means that the network takes less than 2 seconds for one cube to be processed.
TABLE I.
5-FOLD AVERAGE PERFORMANCE MEASURES FOR THE ATTENTION-GUIDED AND BASELINE MODELS
| Accuracy | MCC | Recall | Precision | F1-score | AUC | |
|---|---|---|---|---|---|---|
| Pathway#1 | 85.184 | 0.373 | 89.425 | 93.576 | 91.371 | 86.741 |
| Pathway#2 | 91.413 | 0.553 | 94.838 | 95.418 | 95.102 | 93.007 |
| Pathway#3 | 90.300 | 0.513 | 94.826 | 94.115 | 94.426 | 91.189 |
| Fusion - average | 91.073 | 0.557 | 95.119 | 94.730 | 94.882 | 93.769 |
| Fusion - min | 90.452 | 0.430 | 98.690 | 91.182 | 94.736 | 92.243 |
| Fusion - max | 85.319 | 0.528 | 85.216 | 97.873 | 91.056 | 92.785 |
| Baseline DL model | 86.315 | 0.399 | 90.928 | 93.409 | 92.088 | 86.803 |
B. Quantitative Analysis Results
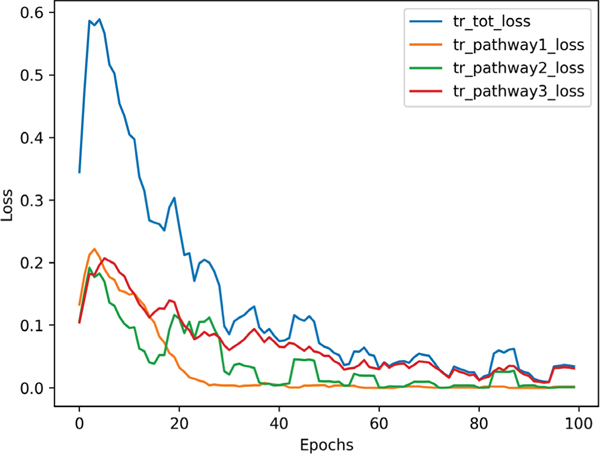
To demonstrate the influence of attention maps on the performance of the AG-OCT model, we trained one branch of our proposed framework, where the input is the downsampled original volumes and the output is the prediction label (i.e. glaucoma or healthy). This means that we trained pathway #1 only without the attention-guided branches, which we used as our baseline model. Table I reported an AUC of 86.8% for the baseline model that is very close to the performance of pathway #1 when it is learned jointly with Pathway #2 and 3. This confirms that training the network without the guidance of grad-CAM heatmaps has approximately 7% and 3% drop in the AUC and F1-score measures respectively.
Further, Figure 2 displays the training loss curves for each pathway separately as well as the total training loss. Loss of pathway#1 reached its minimum at epoch 20 while other losses decreased further until epoch 40. Also, the total loss was highly influenced by pathway#2 loss. This also clarifies why pathway#2 predictions has the best performance for glaucoma detection. This also suggests that weighting losses of 3-pathways might guide the network to pay more attention to the branch with slow convergence.
Fig. 2.
Training losses for glaucoma detection using AG-OCT model
We also examined the impact of sharing weights across branches/pathways by running 3 experiments that use same data split and different settings for sharing weights, colorbluenamely: i) full sharing: the 3-pathways share the same weights, ii) no sharing: each pathway learned its own weights, and iii) partial sharing: only pathway#1 and pathway#2 share the same weights, while pathway #3 had its own weights. Table II reports the performance measures for the 3 experiments, where separate weights for each pathway has the lowest performance, while partial sharing recorded the highest AUCs. The results confirm our hypothesis for sharing weights between first two pathways since they share the same field of view, where the inputs are centered on the ONH and cover an area of 6×6×2 mm3. While the third pathway has a smaller field of view, i.e. different coverage area, as it focuses on a small region of the originally scanned area.
TABLE II.
SHARING WEIGHTS IMPACT ON THE PERFORMANCE THE PROPOSED ATTENTION-GUIDED MODEL
| Accuracy | MCC | Recall | Precision | F1-score | AUC | |
|---|---|---|---|---|---|---|
| No weight sharing: each pathway learns its weights | ||||||
| Pathway#1 | 90.349 | 0.496 | 94.311 | 94.880 | 94.595 | 90.198 |
| Pathway#2 | 91.413 | 0.553 | 94.838 | 95.418 | 95.102 | 93.007 |
| Pathway#3 | 92.225 | 0.580 | 95.808 | 95.522 | 95.665 | 88.579 |
| Fusion - average | 91.421 | 0.521 | 95.808 | 94.675 | 95.238 | 91.392 |
| Fusion - min | 92.225 | 0.484 | 99.701 | 92.244 | 95.827 | 90.095 |
| Fusion - max | 89.008 | 0.533 | 91.018 | 96.508 | 93.683 | 90.922 |
| Full weight sharing: Layers of all pathways share weights | ||||||
| Pathway#1 | 90.349 | 0.585 | 91.916 | 97.152 | 94.462 | 90.527 |
| Pathway#2 | 91.153 | 0.586 | 93.413 | 96.594 | 94.977 | 91.528 |
| Pathway#3 | 92.493 | 0.599 | 95.808 | 95.808 | 95.808 | 91.475 |
| Fusion - average | 91.689 | 0.611 | 93.713 | 96.904 | 95.282 | 92.210 |
| Fusion - min | 93.298 | 0.601 | 97.904 | 94.783 | 96.318 | 91.481 |
| Fusion - max | 89.008 | 0.588 | 89.521 | 98.033 | 93.584 | 91.137 |
| Partial sharing: Only pathway 1 and 2 share weights | ||||||
| Pathway#1 | 89.544 | 0.520 | 92.216 | 95.950 | 94.046 | 90.970 |
| Pathway#2 | 90.080 | 0.534 | 92.814 | 95.975 | 94.368 | 93.054 |
| Pathway#3 | 92.225 | 0.590 | 95.509 | 95.796 | 95.652 | 93.432 |
| Fusion - average | 90.349 | 0.553 | 92.814 | 96.273 | 94.512 | 94.405 |
| Fusion - min | 93.298 | 0.584 | 98.802 | 94.017 | 96.350 | 92.647 |
| Fusion - max | 88.204 | 0.559 | 88.922 | 97.697 | 93.103 | 94.505 |
| FusionAvgPath.1&2 | 89.812 | 0.539 | 92.216 | 96.250 | 94.190 | 92.930 |
| FusionAvgPath.1&3 | 92.493 | 0.627 | 94.910 | 96.646 | 95.770 | 94.528 |
| FusionAvgPath.2&3 | 94.102 | 0.699 | 96.108 | 97.273 | 96.687 | 94.271 |
Table II also shows the average fusion results for all possible pairs of pathways namely, pathways# 1&2, 1&3 and 2&3. From the table, there is a very slight performance difference for average fusion of 3-pathways versus 2-pathways. More importantly, dropping the attention map pathway from the fusion (i.e. pathway# 3) resulted in approximately 2% decrease in the recorded performance measures. While, fusion of pathway# 2&3 had the highest performance measures.
C. Clinical Analysis of Attention Maps for Glaucoma
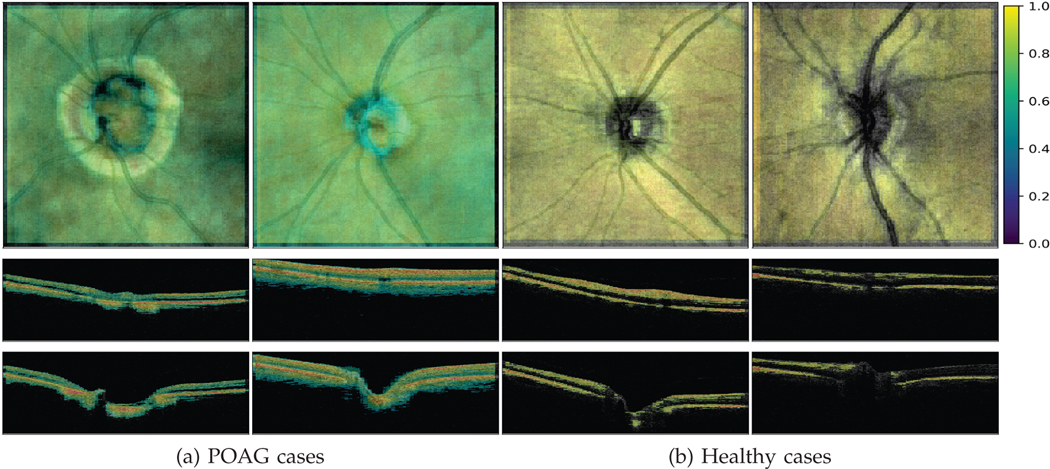
In this section, we present detailed analysis of grad-CAM heatmaps to give insights about the clinical biomarkers that our AG-OCT model relies on for glaucoma detection. This is very essential not only to understand the network decision, but also to increase reliability of DL approaches for direct analysis of 3D-OCT scans by showing agreement of decision making process between DL approaches and clinicians. Figure 3 visualizes the important retinal structures for both healthy and glaucoma cases by overlaying grad-CAM heatmap on the original volumes. The figure displays the overlaid heatmaps for both the enface/top view as well as the b-scans/side view. It is clear from the figure that the AG-OCT model depends on the OCT retinal layers region for detecting glaucoma.
Fig. 3.
Grad-CAM attention maps. First row shows overlaid grad-CAM heatmap for enface view while second and third rows show b-scan slices# 50 and 100 in order
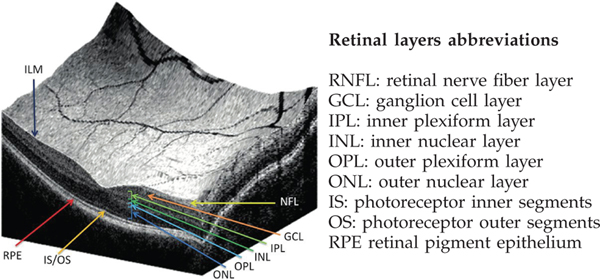
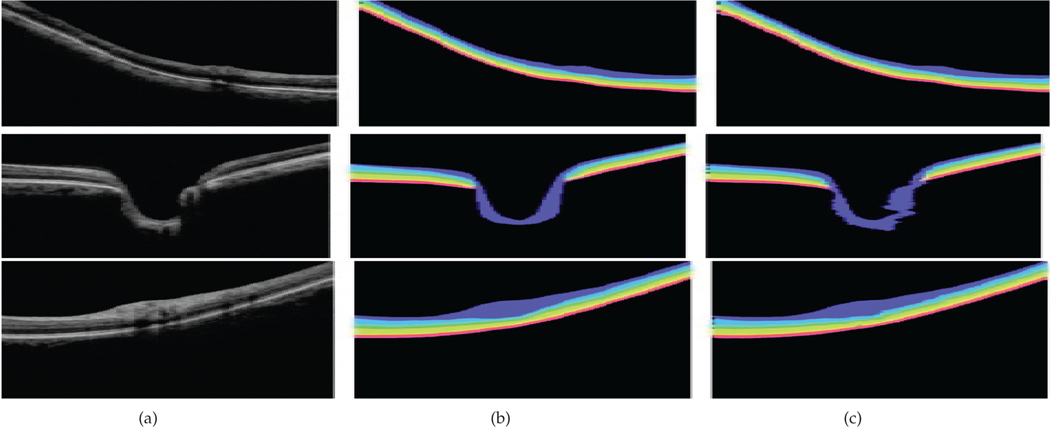
Further, to show which retinal structure/layer has the greatest impact, we quantify the presence of each retinal layer in the generated grad-CAM heatmaps. If the presence of a specific retinal layer, i.e bio-marker, is high, then this means that our model depends on this retinal structure for detecting glaucoma. Visualization and abbreviation of retinal layers are presented in Figure 4. In this regard, we adopt the OCT retinal layers segmentation method described in [49], to classify each 2D b-scan slice into 9 classes, namely: background and 8 different retinal layers, namely RNFL, GCL+IPL, INL, OPL, ONL, IS, OS, and RPE as shown in Figure 5.
Fig. 4.
Visualization and abbreviation of different retinal layers in OCT scan. The left image is taken from this paper [50]
Fig. 5.
Segmentation method results adopted from [49] for clinical assessment of retinal structures relevant to glaucoma. (a) original b-scan slice, (b) ground truth, (c) segmentation results based on the method described in [49]
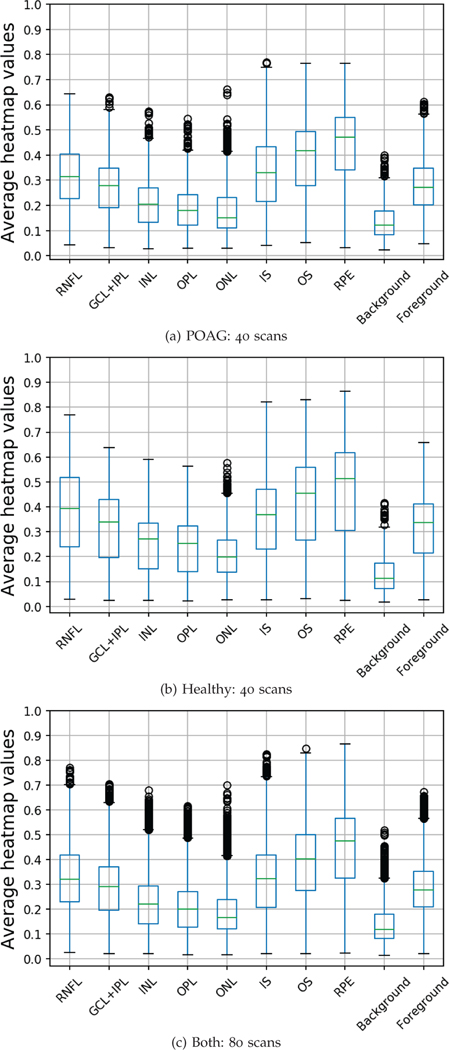
To run this experiment, we perform the following steps. We select 80 3D OCT scans from the test set (40: healthy and 40: POAG) and apply the segmentation method on each b-scan separately to generate 9 binary masks: one for each of the eight retinal layers in addition to the background mask. We also extracted the foreground mask to assess the influence of the whole retina area. Then, we computed the average heatmap weights for each b-scan in each generated mask. In total, this resulted in 16000 average heatmap values for each binary mask (200 b-scans × 80 cubes). The validation process was done for each set of healthy and POAG cases, separately.
To report the contribution of each retinal layer, we use box-plots to represent the average computed heatmap values for each layer separately, as shown in the first row of Figure 6. From the figure, RPE, OS, IS, RNFL and GCL+IPL have the highest correlation with grad-CAM heatmaps, in order, where the median heatmap value lies between 0.3 and 0.5 for those layers. While ONL, INL, and ONL have shown less influence on the network decision where median value lies between 0.1 and 0.3. These findings are non-intuitive for clinicians because these layers are not traditionally associated with glaucoma. However, these findings show that DL approaches might depend on information from unknown features in the tissue such as thinning of the inner retina layers. Also, the figure demonstrates that RNFL has higher heatmap values in healthy cases versus POAG cases with median values of 0.3 and 0.4 respectively. As expected, background area has the least influence on the network decision.
Fig. 6.
Contribution of different retinal layers for glaucoma detection using the proposed AG-OCT model
D. Comparative Results
For comparative study, we follow Maetschke et al. [38] and compare our AG-OCT model against feature based ML approaches, where we use Cirrus OCT scanner computed measurements for training classical ML algorithms. Specifically, we use 22 measurements including peripapillary RNFL thickness at 12 clock-hours, peripapillary RNFL thickness in the four quadrants, average RNFL thickness, rim area, disc area, average cup-to-disc ratio, vertical cup-to-disc ratio and cup volume. We normalize all features by subtracting the mean and scaling to unit variance.
Six ML classifiers are trained, namely, Support Vector Machine (SVM), Logistic Regression, Naïve Bayes, Gradient Boosting, Extra Trees and Random Forest. We used the same dataset used for training AG-OCT model and same split (i.e. train, validation and test). We used the validation set to select the best hyper-parameters for each classifier using grid-search. We also computed the same performance measures. For reliability, we perform 5-fold cross-validation and report the average performance measures for the test set as shown in Table III.
TABLE III.
5-FOLD AVERAGE PERFORMANCE MEASURES FOR GLAUCOMA DETECTION USING FEATURE-BASED MACHINE LEARNING METHODS
| Accuracy | MCC | Recall | Precision | F1-score | AUC | |
|---|---|---|---|---|---|---|
| Extra Trees | 85.563 | 0.436 | 87.367 | 96.160 | 91.498 | 89.556 |
| Gradient Boosting | 90.318 | 0.395 | 95.660 | 93.692 | 94.586 | 90.380 |
| Logistic Regression | 82.908 | 0.475 | 83.223 | 97.464 | 89.660 | 91.499 |
| Naive Bayes | 81.583 | 0.448 | 82.370 | 96.806 | 88.776 | 89.689 |
| Random Forest | 88.128 | 0.469 | 91.026 | 95.588 | 93.176 | 89.098 |
| SVM (Linear) | 81.120 | 0.454 | 81.202 | 97.477 | 88.450 | 91.550 |
| SVM (Poly) | 88.008 | 0.447 | 91.166 | 95.244 | 93.117 | 91.204 |
| SVM (RBF) | 80.749 | 0.442 | 81.126 | 97.140 | 88.254 | 90.602 |
| Proposed AG-OCT | 91.073 | 0.557 | 95.119 | 94.730 | 94.882 | 93.769 |
From the table, the best feature based ML model that has the highest AUC and a good balance between recall and precision is gradient boosting with an AUC of 91.5%, that is 2.3% less than our AUC (i.e. AG-OCT). This is followed by SVM with polynomial kernel with AUC of 91.2%, with higher measure for precision than recall. All other feature based ML classifiers either showed an AUC less than 89% or strongly biased towards one class (i.e. significant difference between recall and precision). In a nutshell, not only does this experiment confirm the effectiveness of our proposed approach but it also shows that DL approaches have the potential to learn directly from raw volumes with better performance than the one achieved by relying on scanner extracted features.
E. Structural-Functional Correlation
Table IV reports the performance measures for the attention-guided regression model. It is revealed from the table that polynomial regression using 3-pathway predictions has significantly outperformed the other predictions with Pearson correlation (ρ) of 0.75 and MAE of 8 for a test set of size 3100 cubes. The table also shows that predictions from pathway#2 has slightly better performance measures than the other two pathways with Pearson correlation (ρ) of 0.65 and MAE of 11.6. Also, refining the predictions from the individual pathways had decreased the MAE by at least a values of 2.5 (i.e. MAE = 9.1).
TABLE IV.
PERFORMANCE MEASURES FOR VISUAL FIELD INDEX ESTIMATION EXPERIMENT
| Predictions | RMSE | MAE | MDAE | r | ρ |
|---|---|---|---|---|---|
| Pathway#1 | 17.139 | 12.362 | 9.071 | 0.497 | 0.648 |
| Pathway#2 | 16.783 | 11.628 | 8.000 | 0.491 | 0.649 |
| Pathway#3 | 19.371 | 14.060 | 10.701 | 0.459 | 0.582 |
| Fusion-regression (3-pathways) | 13.403 | 7.954 | 3.615 | 0.582 | 0.750 |
| Regression (pathway#1) | 14.806 | 9.103 | 4.441 | 0.497 | 0.680 |
| Regression (pathway#2) | 14.834 | 9.035 | 3.729 | 0.451 | 0.681 |
| Regression (pathway#3) | 15.874 | 10.078 | 4.602 | 0.451 | 0.620 |
VI. CONCLUSION AND FUTURE WORK
We present an end-to-end 3D attention-guided model that can be used for multiple tasks including classification and regression through direct analysis of 3D raw volumes that outperformed the scanner computed measurements. The model leverages the rich structural information embedded in the high resolution 3D OCT cubes by the guidance of grad-CAM attention map, which resulted in better performance compared with baseline models and feature based ML approaches. Importantly, we showed that using the attention-guided framework we can identify the important regions in the OCT volumes, whereby redundant regions of the scan can be excluded from the analysis. Also, grad-CAM allowed for a qualitative clinical analysis and understanding of the DL network. In particular, we quantitatively measured the importance of different retinal layers in 3D OCT cubes which the network relied on for detecting glaucoma. Further, the glaucoma detection and VFI estimation experiments confirmed the effectiveness, robustness and generalization of the proposed model that is able to learn from high resolution 3D volumes. Both tasks showed that the fusion of predictions from the three pathways (i.e. attention-guided) had the best performance. In the future, we will apply this approach for estimating other functional parameters and detecting other ocular diseases. We also plan to improve the performance of the model by enhancing the attention map.
VII. Acknowledgments
This work is supported by a grant from the National Institutes of Health National Eye Institute R01EY013178.
Contributor Information
Yasmeen George, AI/Medical Image Analytics Researcher with IBM Research Australia. She received her B.Sc. and M.Sc. degrees in computer science from Faculty of Computer and Information Sciences, Ain Shams University (Egypt), in 2008 and 2013, respectively. Yasmeen also received her Ph.D. in 2018 at the University of Melbourne (UoM), Department of Electrical and Electronic Engineering.
Bhavna J. Antony, IBM Research Australia
Hiroshi Ishikawa, Department of Ophthalmology, NYU Langone Health, New York, United States.
Gadi Wollstein, Department of Ophthalmology, NYU Langone Health, New York, United States.
Joel S. Schuman, Department of Ophthalmology, NYU Langone Health, New York, United States
Rahil Garnavi, IBM Research Australia.
References
- [1].Flaxman SR, Bourne RR, Resnikoff S, Ackland P, Braithwaite T, Cicinelli MV, Das A, Jonas JB, Keeffe J, Kempen JH et al. , “Global causes of blindness and distance vision impairment 1990–2020: a systematic review and meta-analysis,” The Lancet Global Health, vol. 5, no. 12, pp. e1221–e1234, 2017. [DOI] [PubMed] [Google Scholar]
- [2].Davis BM, Crawley L, Pahlitzsch M, Javaid F, and Cordeiro MF, “Glaucoma: the retina and beyond,” Acta neuropathologica, vol. 132, no. 6, pp. 807–826, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lucy KA and Wollstein G, “Structural and functional evaluations for the early detection of glaucoma,” Expert review of ophthalmology, vol. 11, no. 5, pp. 367–376, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Broadway DC, “Visual field testing for glaucoma–a practical guide,” Community eye health, vol. 25, no. 79–80, p. 66, 2012. [PMC free article] [PubMed] [Google Scholar]
- [5].Peracha M, Hughes B, Tannir J, Momi R, Goyal A, Juzych M, Kim C, McQueen M, Eby A, and Fatima F, “Assessing the reliability of humphrey visual field testing in an urban population,” Investigative Ophthalmology & Visual Science, vol. 54, no. 15, pp. 3920–3920, 2013. [Google Scholar]
- [6].Ho J, Ameen S, Crawley L, Normando E, Cordeiro MF, Bloom P, and Ahmed F, “Key predictors of visual field test reliability,” Investigative Ophthalmology & Visual Science, vol. 54, no. 15, pp. 2632–2632, 2013. [Google Scholar]
- [7].Leite MT, Zangwill LM, Weinreb RN, Rao HL, Alencar LM, and Medeiros FA, “Structure-function relationships using the cirrus spectral domain optical coherence tomograph and standard automated perimetry,” Journal of glaucoma, vol. 21, no. 1, p. 49, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nilforushan N, Nassiri N, Moghimi S, Law SK, Giaconi J, Coleman AL, Caprioli J, and Nouri-Mahdavi K, “Structure–function relationships between spectral-domain oct and standard achromatic perimetry,” Investigative ophthalmology & visual science, vol. 53, no. 6, pp. 2740–2748, 2012. [DOI] [PubMed] [Google Scholar]
- [9].Malik R, Swanson WH, and Garway-Heath DF, “Structure– function relationship in glaucoma: past thinking and current concepts,” Clinical & experimental ophthalmology, vol. 40, no. 4, pp. 369–380, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kuang TM, Zhang C, Zangwill LM, Weinreb RN, and Medeiros FA, “Estimating lead time gained by optical coherence tomography in detecting glaucoma before development of visual field defects,” Ophthalmology, vol. 122, no. 10, pp. 2002–2009, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sung KR, Kim S, Lee Y, Yun S-C, and Na JH, “Retinal nerve fiber layer normative classification by optical coherence tomography for prediction of future visual field loss,” Investigative ophthalmology & visual science, vol. 52, no. 5, pp. 2634–2639, 2011. [DOI] [PubMed] [Google Scholar]
- [12].Medeiros FA, Zangwill LM, Bowd C, Mansouri K, and Weinreb RN, “The structure and function relationship in glaucoma: implications for detection of progression and measurement of rates of change,” Investigative ophthalmology & visual science, vol. 53, no. 11, pp. 6939–6946, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Norouzifard M, Nemati A, Abdul-Rahman A, GholamHosseini H, and Klette R, “A comparison of transfer learning techniques, deep convolutional neural network and multilayer neural network methods for the diagnosis of glaucomatous optic neuropathy,” in International Computer Symposium. Springer, 2018, pp. 627–635. [Google Scholar]
- [14].Group EGPSE et al. , “Results of the european glaucoma prevention study,” Ophthalmology, vol. 112, no. 3, pp. 366–375, 2005. [DOI] [PubMed] [Google Scholar]
- [15].Panwar N, Huang P, Lee J, Keane PA, Chuan TS, Richhariya A, Teoh S, Lim TH, and Agrawal R, “Fundus photography in the 21st century—a review of recent technological advances and their implications for worldwide healthcare,” Telemedicine and e-Health, vol. 22, no. 3, pp. 198–208, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Saine PJ and Tyler ME, Ophthalmic photography: retinal photography, angiography, and electronic imaging. Butterworth-Heinemann Boston, 2002, vol. 132. [Google Scholar]
- [17].Li Q, Feng B, Xie L, Liang P, Zhang H, and Wang T, “A cross-modality learning approach for vessel segmentation in retinal images,” IEEE transactions on medical imaging, vol. 35, no. 1, pp. 109–118, 2016. [DOI] [PubMed] [Google Scholar]
- [18].Maninis K-K, Pont-Tuset J, Arbelaez P, and Van Gool L, “Deep´ retinal image understanding,” in International conference on medical image computing and computer-assisted intervention Springer, 2016, pp. 140–148. [Google Scholar]
- [19].Dasgupta A. and Singh S, “A fully convolutional neural network based structured prediction approach towards the retinal vessel segmentation,” in 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). IEEE, 2017, pp. 248–251. [Google Scholar]
- [20].Fu H, Xu Y, Wong DWK, and Liu J, “Retinal vessel segmentation via deep learning network and fully-connected conditional random fields,” in 2016 IEEE 13th international symposium on biomedical imaging (ISBI). IEEE, 2016, pp. 698–701. [Google Scholar]
- [21].Fu H, Cheng J, Xu Y, Wong DWK, Liu J, and Cao X, “Joint optic disc and cup segmentation based on multi-label deep network and polar transformation,” IEEE transactions on medical imaging, vol. 37, no. 7, pp. 1597–1605, 2018. [DOI] [PubMed] [Google Scholar]
- [22].Sadhukhan S, Ghorai GK, Maiti S, Karale VA, Sarkar G, and Dhara AK, “Optic disc segmentation in retinal fundus images using fully convolutional network and removal of false-positives based on shape features,” in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer, 2018, pp. 369–376. [Google Scholar]
- [23].Sevastopolsky A, “Optic disc and cup segmentation methods for glaucoma detection with modification of u-net convolutional neural network,” Pattern Recognition and Image Analysis, vol. 27, no. 3, pp. 618–624, 2017. [Google Scholar]
- [24].Li Z, He Y, Keel S, Meng W, Chang RT, and He M, “Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs,” Ophthalmology, vol. 125, no. 8, pp. 1199–1206, 2018. [DOI] [PubMed] [Google Scholar]
- [25].Raghavendra U, Fujita H, Bhandary SV, Gudigar A, Tan JH, and Acharya UR, “Deep convolution neural network for accurate diagnosis of glaucoma using digital fundus images,” Information Sciences, vol. 441, pp. 41–49, 2018. [Google Scholar]
- [26].Phene S, Dunn RC, Hammel N, Liu Y, Krause J, Kitade N, Schaekermann M, Sayres R, Wu DJ, Bora A. et al. , “Deep learning to assess glaucoma risk and associated features in fundus images,” arXiv preprint arXiv:181208911, 2018. [Google Scholar]
- [27].Kim M, Janssens O, Park H.-m, Zuallaert J, Van Hoecke S, and De Neve W, “Web applicable computer-aided diagnosis of glaucoma using deep learning,” arXiv preprint arXiv:181202405, 2018. [Google Scholar]
- [28].Ahn JM, Kim S, Ahn K-S, Cho S-H, Lee KB, and Kim US, “A deep learning model for the detection of both advanced and early glaucoma using fundus photography,” PloS one, vol. 13, no. 11, p. e0207982, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mahapatra D, Antony B, Sedai S, and Garnavi R, “Deformable medical image registration using generative adversarial networks,” in 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE, 2018, pp. 1449–1453. [Google Scholar]
- [30].Fujimoto J. and Swanson E, “The development, commercialization, and impact of optical coherence tomography,” Investigative ophthalmology & visual science, vol. 57, no. 9, pp. OCT1–OCT13, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lavinsky F, Wollstein G, Tauber J, and Schuman JS, “The future of imaging in detecting glaucoma progression,” Ophthalmology, vol. 124, no. 12, pp. S76–S82, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Asaoka R, Murata H, Hirasawa K, Fujino Y, Matsuura M, Miki A, Kanamoto T, Ikeda Y, Mori K, Iwase A. et al. , “Using deep learning and transfer learning to accurately diagnose early-onset glaucoma from macular optical coherence tomography images,” American journal of ophthalmology, vol. 198, pp. 136–145, 2019. [DOI] [PubMed] [Google Scholar]
- [33].Muhammad H, Fuchs TJ, De NC, De CM, Blumberg DM, Liebmann JM, Ritch R, and Hood DC, “Hybrid deep learning on single wide-field optical coherence tomography scans accurately classifies glaucoma suspects.” Journal of glaucoma, vol. 26, no. 12, pp. 1086–1094, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Krizhevsky A, Sutskever I, and Hinton GE, “Imagenet classification with deep convolutional neural networks,” in Advances in neural information processing systems, 2012, pp. 1097–1105. [Google Scholar]
- [35].Breiman L, “Random forests,” Machine learning, vol. 45, no. 1, pp. 5–32, 2001. [Google Scholar]
- [36].An G, Omodaka K, Hashimoto K, Tsuda S, Shiga Y, Takada N, Kikawa T, Yokota H, Akiba M, and Nakazawa T, “Glaucoma diagnosis with machine learning based on optical coherence tomography and color fundus images,” Journal of Healthcare Engineering, vol. 2019, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang J, Chen C, Li F, Wang Z, Qu G, Qiao Y, Lv H, and Zhang X, “Sd net: Joint segmentation and diagnosis revealing the diagnostic significance of using entire rnfl thickness in glaucoma,” in Conference on Medical Imaging with Deep Learning (MIDL), 2018. [Google Scholar]
- [38].Maetschke S, Antony B, Ishikawa H, and Garvani R, “A feature agnostic approach for glaucoma detection in oct volumes,” arXiv preprint arXiv:180704855, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].George Y, Antony B, Ishikawa H, Wollstein G, Schuman J, and Garnavi R, “3d-cnn for glaucoma detection using optical coherence tomography,” in International Workshop on Ophthalmic Medical Image Analysis. Springer, 2019, pp. 52–59. [Google Scholar]
- [40].Maetschke S, Antony B, Ishikawa H, Wollstein G, Schuman J, and Garnav R, “Inference of visual field test performance from oct volumes using deep learning,” arXiv preprint arXiv:190801428, 2019. [Google Scholar]
- [41].Razzak MI, Naz S, and Zaib A, “Deep learning for medical image processing: Overview, challenges and the future,” in Classification in BioApps. Springer, 2018, pp. 323–350. [Google Scholar]
- [42].Zhou B, Khosla A, Lapedriza A, Oliva A, and Torralba A, “Learning deep features for discriminative localization,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 2921–2929. [Google Scholar]
- [43].Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, and Batra D, “Grad-cam: Visual explanations from deep networks via gradient-based localization,” in Proceedings of the IEEE International Conference on Computer Vision, 2017, pp. 618–626. [Google Scholar]
- [44].Glorot X, Bordes A, and Bengio Y, “Deep sparse rectifier neural networks,” in Proceedings of the fourteenth international conference on artificial intelligence and statistics, 2011, pp. 315–323. [Google Scholar]
- [45].Ioffe S. and Szegedy C, “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” arXiv preprint arXiv:1502.03167, 2015. [Google Scholar]
- [46].Goodfellow I, Bengio Y, and Courville A, Deep learning. MIT press, 2016. [Google Scholar]
- [47].Bini SA, “Artificial intelligence, machine learning, deep learning, and cognitive computing: what do these terms mean and how will they impact health care?” The Journal of arthroplasty, vol. 33, no. 8, pp. 2358–2361, 2018. [DOI] [PubMed] [Google Scholar]
- [48].Gulli A. and Pal S, Deep Learning with Keras. Packt Publishing Ltd, 2017. [Google Scholar]
- [49].Sedai S, Antony B, Mahapatra D, and Garnavi R, “Joint segmentation and uncertainty visualization of retinal layers in optical coherence tomography images using bayesian deep learning,” in Computational Pathology and Ophthalmic Medical Image Analysis. Springer, 2018, pp. 219–227. [Google Scholar]
- [50].Stankiewicz A, Marciniak T, Dabrowski A, Stopa M, Rakowicz P, and Marciniak E, “Denoising methods for improving automatic segmentation in oct images of human eye,” Bulletin of the Polish Academy of Sciences Technical Sciences, vol. 65, no. 1, pp. 71–78, 2017. [Google Scholar]