Abstract
Enantioselective indicator displacement assays (eIDAs) were used for the determination of enantiomeric excess (ee) of α-amino acids as an alternative to the labor-intensive technique of chromatography. In this study, eIDAs were implemented by the use of two chiral receptors [(CuII(1)]2+, [CuII(2)]2+) in conjunction with the indicator chrome azurol S. The two receptors were able to enantioselectively discriminate 13 of the 17 analyzed α-amino acids. Enantiomeric excess calibration curves were made using both receptors and then used to analyze true test samples to check the system’s ability to determine ee accurately. The proposed method uses a conventional UV-vis spectrophotometer to monitor the colorimetric signal, which allows for a potential high-throughput screening (HTS) method for determining ee. The techniques created consistently produced results accurate enough for rapid preliminary determination of ee.
Introduction
The ability to determine enantiomeric excess (ee) is important for the scientific community, especially in the production of chiral drugs and in asymmetric catalyst discovery. Enantiomers may have different biological activities, which is why there are strict guidelines on the identification and quantification of chiral compounds by the U.S. Food and Drug Administration (FDA).1–4 Furthermore, it has become increasingly common to use asymmetric catalysts to obtain the desired enantiomer of a compound. Asymmetric catalysts are often used because they save money by increasing the yield of the desired enantiomer, thereby minimizing the wasteful discarding of the inactive enantiomer. Screening of asymmetric catalysts encompasses finding the best catalyst and optimizing the reaction to produce the highest ee. The use of combinatorial libraries has allowed for a more rapid method of screening for asymmetric catalysts.5 Via parallel syntheses, a large number of samples are produced that require analysis. Hence, one of the current limitations in the use of a combinatorial library of catalysts is the determination of ee in a high-throughput (HT) fashion.4,6
The current methods to determine ee are chiral high-performance liquid chromatography (HPLC) and gas chromatography (GC), as well as HPLC coupled with circular dichroism (CD).4,7,8 The instruments used are costly and cannot analyze thousands of samples a day, as would be required for a true high-throughput screening (HTS) method.5 A number of other methods have been developed for the determination of ee, including NMR spectroscopy,9,10 capillary array electrophoresis,11–13 liquid crystals,14,15 mass spectroscopy,16–18 infrared thermography monitoring,19,20 enzymatic processes,21–24 molecularly imprinted polymers,25 and the use of kinetic resolution.26 Some of these methods have been optimized to be used in microwell plates,4,18,19,22 allowing for rapid determination of ee. However, most of these methods require derivatization of the analytes, which increases the analysis time. Implementation of many of these techniques requires expensive specialized equipment to which not every laboratory has access. The methods that use enzymes or antibodies require a separate development for each analyte due to the high selectivity of such receptors.
The scientific community has also explored the use of chromogenic receptors to determine ee. Besides speed, this method offers several advantages, the most important being the use of less expensive and more commonly used instruments, such as a fluorimeter or a UV-vis spectrophotometer. The use of these instruments provides easy conversion to HTS assays through the incorporation of a microplate reader, which allows for rapid analysis of multiple samples.
Many research groups have developed supramolecular sensors with chromogenic units for the differentiation of enantiomers. Pu has created a bis-binaphthyl fluorescent sensor27 that shows biased fluorescence enhancement upon binding to chiral α-hydroxy acids. Others include Lin’s ReI-based luminescent chiral molecular squares,28 Ahn’s chiral tripodal oxazoline receptor,29 Wolf’s chiral scandium N,N′-dioxide complex,30,31 Hyun’s fluorescent anthracene thiourea derivative receptors,32 and Corradini’s modified β-cyclodextrins with an appended dansyl fluorophore.33 There are numerous other examples of receptors that can differentiate enantiomers.30,34–40 Only a handful of these sensors, however, were actually used to determine ee, and several of them required multiple synthetic steps to obtain the receptor. Furthermore, in these systems, the chromophoric unit is covalently attached to the receptor, which requires resynthesis of the scaffold when the chromophoric unit does not produce a satisfactory signal for the detection of the binding event. To overcome these disadvantages, our group introduced the use of indicator displacement assays (IDAs)41 because less covalent bond architecture is needed, allowing the use of a number of indicators with the same receptor system and enabling a secondary tuning of selectivity.42 Also, the receptors are obtained in a few synthetic steps from commercially available starting material.
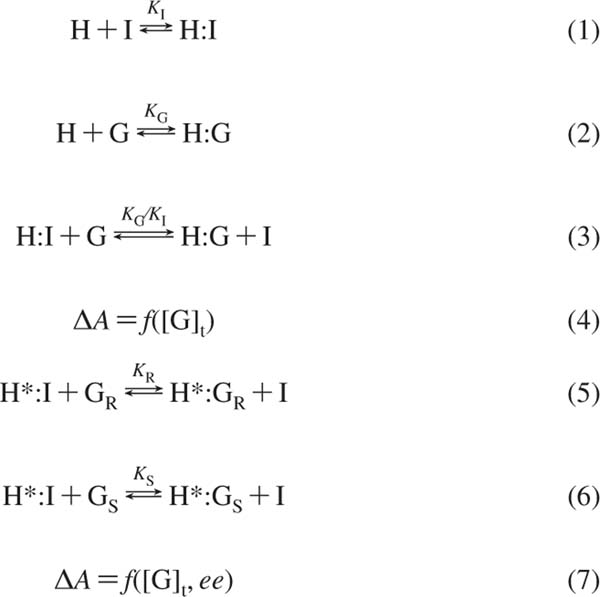
Indicator displacement assays43,44 have been widely explored as a method of sensing for a range of analytes (i.e., glucose,45 citrate,46 calcium,46 phosphoesters,47 tartrate48). The equilibria involved in an IDA are outlined in Scheme 1. For an IDA to be applicable, a suitable indicator (I) must be able to reversibly bind to the host (H) (Scheme 1, eq 1) and signal the binding through a change in a spectroscopic signal. The analyte (guest, G) must also be able to bind to the host (Scheme 1, eq 2), which leads to the indicator being displaced from the complex (Scheme 1, eq 3). The displacement produces a change in the indicator’s optical properties and thus signals the binding event. The resulting competition between indicator and guest for binding to the host can be monitored through the change in the indicator’s spectral properties, which are sensitive to the guest total concentration ([G]t) (Scheme 1, eq 4).41,44,49 As [G]t increases, more indicator is displaced and a larger change in absorbance is observed.
Scheme 1.
Equilibria of an IDA and eIDAa
a H = host/receptor; H* = chiral host/receptor; I = indicator; G, GR, GS = analyte/guest; [G]t = total guest concentration; K, KI, KG, KR, KS = binding constant; ΔA = change in absorbance.
An IDA can be used as a method of enantioselective discrimination if a chiral receptor (H*) is used.41 Upon association of the chiral receptor with an enantiomeric mixture of chiral analytes (GR, GS), multiple equilibria are simultaneously established in solution, leading to the formation of two diastereomeric host-guest complexes (H*:GR, H*:GS). The diastereomers have different stabilities, and therefore the two equilibria (Scheme 1, eqs 5 and 6) will have different equilibrium constants (KR, KS). This difference results in the differential displacement of indicator (I) and, consequently, a difference in the absorbance of the two solutions (Scheme 1, eq 7). Mixtures of the enantiomers will give a color that is between the extremes of the two pure enantiomers.
This work focuses on the detection of α-amino acids due to their importance as a class of biologically active compounds. Amino acids are important food components, and their optical purity can be modified as a consequence of fermentation, aging, heating, irradiation, or treatment at high pH.50 Further, amino acids are commonly used as chiral auxiliaries, catalysts, and chiral starting materials in organic synthesis, so the ability to obtain them enantiomerically pure is important.51,52 This is especially true for unnatural amino acids and for d-amino acids, which are rare in nature and therefore require asymmetric synthesis.53
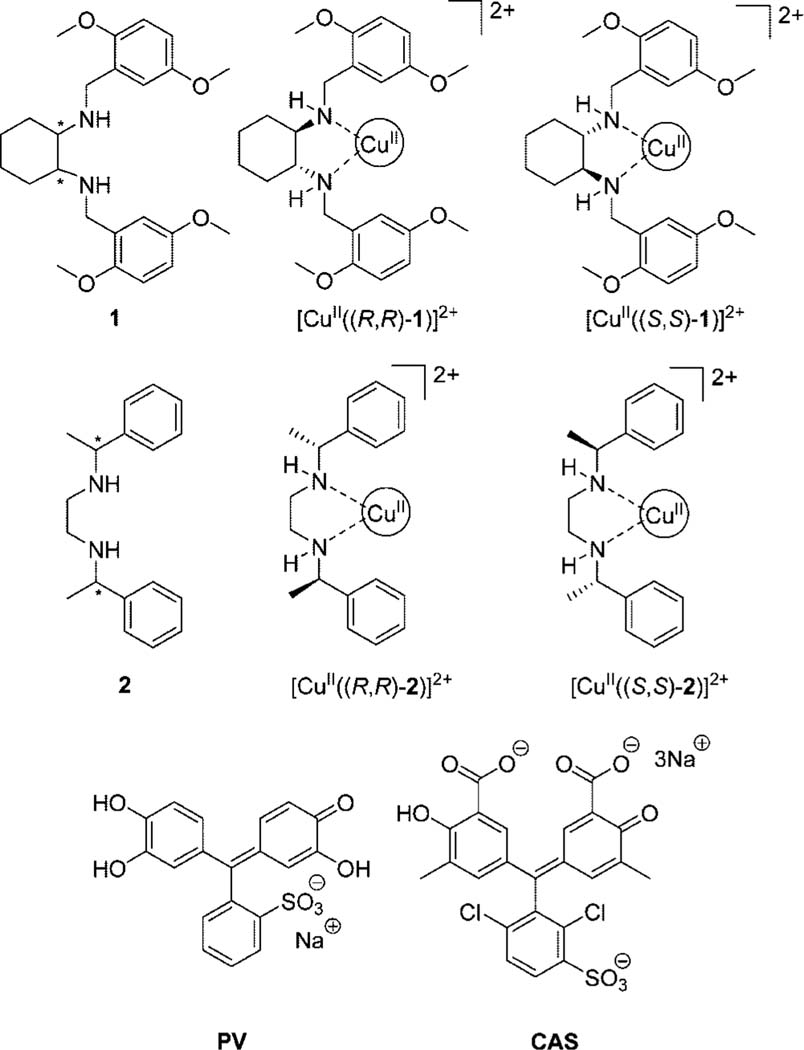
Previously published communications by our group have shown that hosts [CuII(1)]2+ 54 and [CuII(2)]2+ 55 (see Figure 1) can enantioselectively discriminate α-amino acids. The complex of 1 with Cu(OTf)2 (OTf = trifluoromethanesulfonate) and pyrocatechol violet (PV, Figure 1) showed enantioselectivity for four hydrophobic α-amino acids (valine, tryptophan, leucine, and phenylalanine).54 Further studies were conducted using an array of three hosts (1 and 2 included) and three indicators to enantioselectively discriminate l- and d-amino acids and differentiate the same four hydrophobic amino acids from one another using a pattern recognition technique.55 This study extends the scope of the previous study by encompassing amino acids other than just hydrophobic ones, and determining ee of true independent test samples rather than merely performing validation by jack-knife analysis, as in the prior work.
Figure 1.
Structures of ligand 1, chiral receptors [CuII((R,R)-1)]2+ and [CuII((S,S)-1)]2+, ligand 2, chiral receptors [CuII((R,R)-2)]2+ and [CuII((S,S)-2)]2+, indicator pyrocatechol violet (PV), and chrome azurol S (CAS).
In this study, we show that an enantioselective indicator displacement assay (eIDA) with one of the two chiral receptors ([CuII(1)]2+ or [CuII(2)]2+) and chrome azurol S (CAS, Figure 1) as the indicator can enantioselectively discriminate 13 α-amino acids in aqueous medium buffered to pH 7.5 (1:1 MeOH:H2O solution of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)). Enantiomeric excess calibration curves were made to determine the ee of independent test samples on a UV-vis spectrophotometer, which demonstrated the ability of the eIDA to determine ee of unknown samples accurately enough for preliminary screening. Due to the use of colorimetric signaling, the system could inherently be converted to a HTS method (see the following paper).56
Results and Discussion
1. Design Criteria.
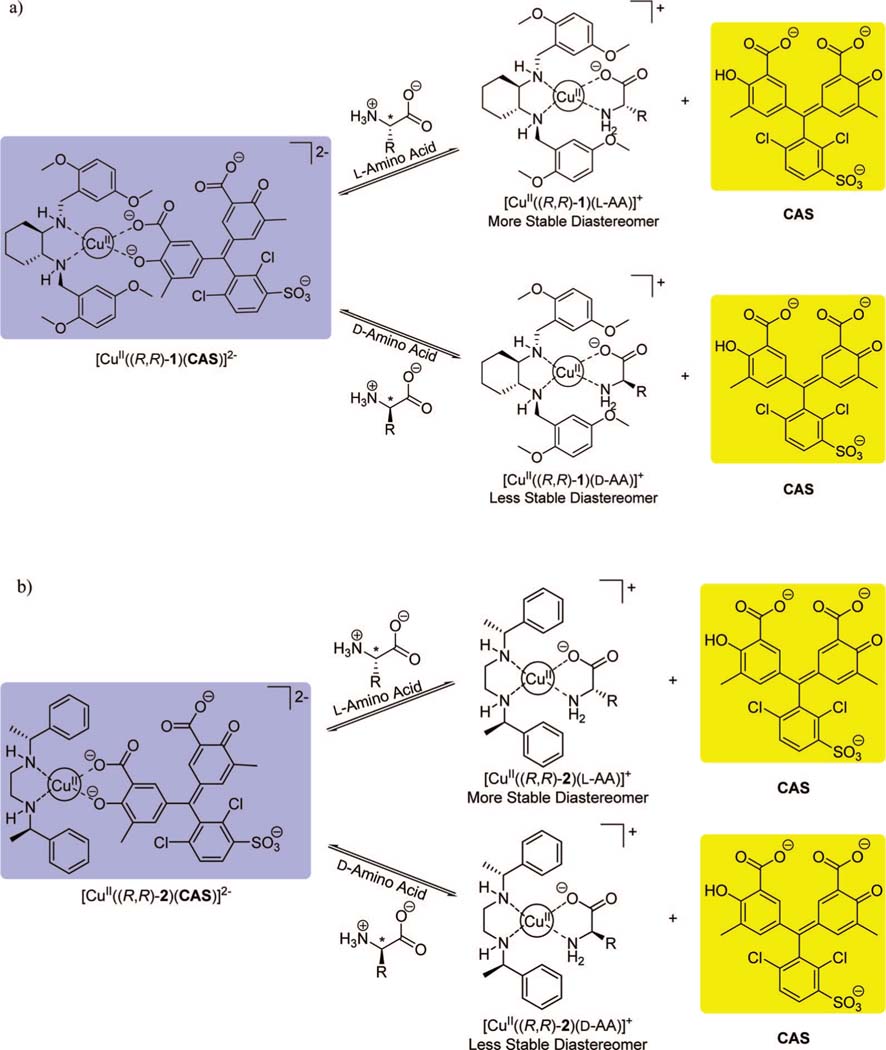
The coordination of chiral ligands 1 and 2 to a CuII metal center creates receptors with chiral coordination sites available for fast ligand exchange between an indicator and α-amino acids (Scheme 2).54 Ligands 1 and 2 were synthesized through procedures developed in the Anslyn and Floriani groups, respectively.54,57 Chrome azurol S was used as the indicator in this study instead of the previously used indicator, PV, to obtain a larger change in absorbance upon displacement of indicator. Chelation of CAS to the CuII metal center leads to a bathochromic absorbance shift from 429 to 602 nm, resulting in a colorimetric change from yellow to intense blue upon indicator coordination. The addition of α-amino acids leads to a reversal in the spectral change due to the displacement of the indicator from the receptor, allowing for the monitoring of the binding between the analyte and receptor (Scheme 2).
Scheme 2.
Enantioselective Indicator Displacement Assays for α-Amino Acids Based on Displacement of Chrome Azurol S (CAS) from Complex: (a) [CuII((R,R)-1)(CAS)]2– and (b) [CuII((R,R)-2)(CAS)]2-
[CuII((R,R)-1)]2+ selectively binds to l-amino acids, while [CuII((S,S)-1)]2+ selectively binds to d-amino acids due to the stability of the diastereomers formed. The relative stability of these diastereomers depends on the steric hindrance between the chiral receptor and the side chain of the α-amino acid, leading to a different degree of indicator displacement. This hypothesis is supported by the crystal structures obtained, which are discussed below. Ligand 2 was investigated in this study as well, due to the inability of receptor [CuII(1)]2+ to spectroscopically discriminate all of the 17 analyzed α-amino acids. The complexation and signaling pattern shown by receptor [CuII(2)]2+ is analogous to that of receptor [CuII(1)]2+: when bound to α-amino acids, diastereomers are formed ([CuII((R,R)-2)(l-amino acid)]+ vs [CuII((R,R)-2)(d-amino acid)]+, Scheme 2b).
2. Crystal Structures.
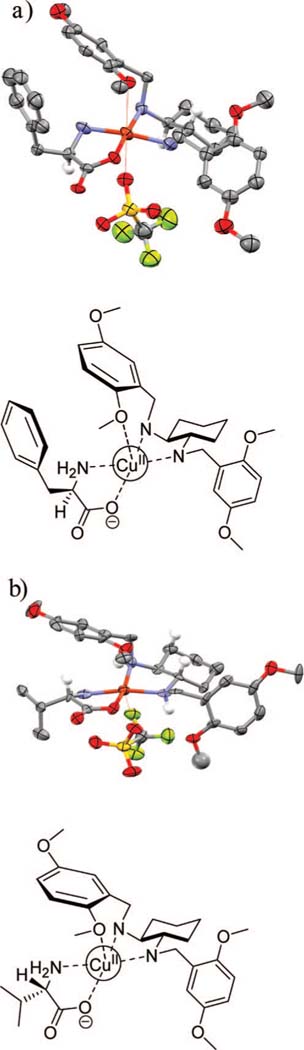
The previously published crystal structure of [CuII((S,S)-1))(d-Phe)]+ 54 is reported in Figure 2a. In the course of the present study, we have also obtained a crystal structure of [CuII((S,S)-1))(l-Val)]+ (Figure 2b). As shown in the crystal structures, the CuII metal center forms a square planar complex with the diamine ligand (S,S)-1 and the α-amino acid (d-Phe or l-Val). The counterion, trifluoromethane-sulfonate, is coordinated to the CuII metal center at a distant axial position. It is believed that the interaction between the counterion and the metal center is weak in solution and plays no role in the competitive binding to CuII. In the solid state, there is also a weak axial interaction between the oxygen from one of the methoxy group of the dimethoxybenzylic ring and the CuII metal center.
Figure 2.
X-ray crystal structures of (a) [CuII((S,S)-1)(d-Phe)](CF3SO3) and (b) [CuII((S,S)-1)(l-Val)](CF3SO3). Thermal ellipsoids are scaled to 30% probability. Most of the hydrogen atoms have been removed for clarity.
We propose two possible hypotheses for the observed enantioselectivity for l- and d-amino acid. Figure 2a shows binding of the preferred isomer, while Figure 2b shows binding of the less optimal stereoisomer. From the crystal structures the two dimethoxybenzylic rings are observed to be oriented on opposite sides of the CuII metal center (Figure 2, panels a and b), probably in order to minimize gauche interactions with the cyclohexane scaffold. Upon chelation of the amino acids to the CuII metal center, the side chain of the α-amino acid can be oriented either toward or away from the phenyl group of the dimethoxybenzylic ring with the coordinated methoxy (Figure 2, panels a and b, respectively). Preferential binding occurs with the stereochemistry of the amino acid such that the side chain of the amino acid is oriented toward this dimethoxybenzyl group. However, this interaction between the dimethoxybenzylic ring and the CuII metal center is likely to be absent in solution, where the axial position is probably occupied by a solvent molecule, so that the dimethoxybenzylic rings will be on opposite sides of the plane of the ligands, thus forming a C2-symmetric cavity, which could lead to steric interactions between the side chain of the amino acids and the dimethoxybenzylic ring that might be responsible for the observed enantioselectivity. With only these crystal structures in hand, the rationale for the observed enantioselectivity is still not definite.
3. Structural Considerations.
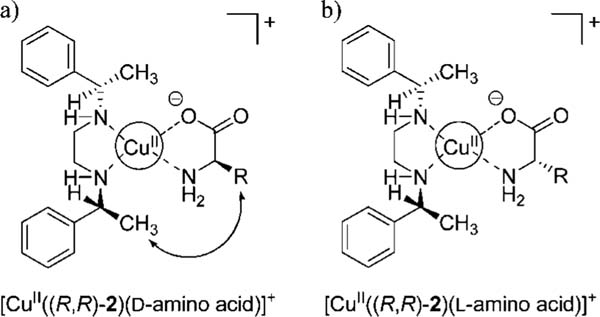
In the case of ligand 2, however, the inherent enantioselectivity is more readily postulated to arise from the avoidance of a steric interaction between the side chain of the amino acids and the ligand. Even though we did not obtain crystal structures as we did with ligand 1, examination of CPK models of the complex between [CuII-((R,R)-2)]2+ and amino acids suggests that there is a steric interaction between the side chain of d-amino acids and the methyl attached at the benzylic position of the (R,R)-2 ligand (Figure 3a). This interaction is absent in the case of l-amino acids (Figure 3b). This would explain the observed selectivity by [CuII((R,R)-2]2+ for l-amino acids shown in the UV-vis titrations, since the addition of l-amino acid to a solution of [CuII((R,R)-2)(CAS)]2- leads to more indicator being displaced than when d-amino acid is added.
Figure 3.
Enantioselective discrimination by ligand 2 explained through steric modeling of (a) [CuII((R,R)-2)(d-amino acid)]+ and (b) [CuII((R,R)-2)(l-amino acid)]+.
4. UV-Vis Displacement Isotherms.
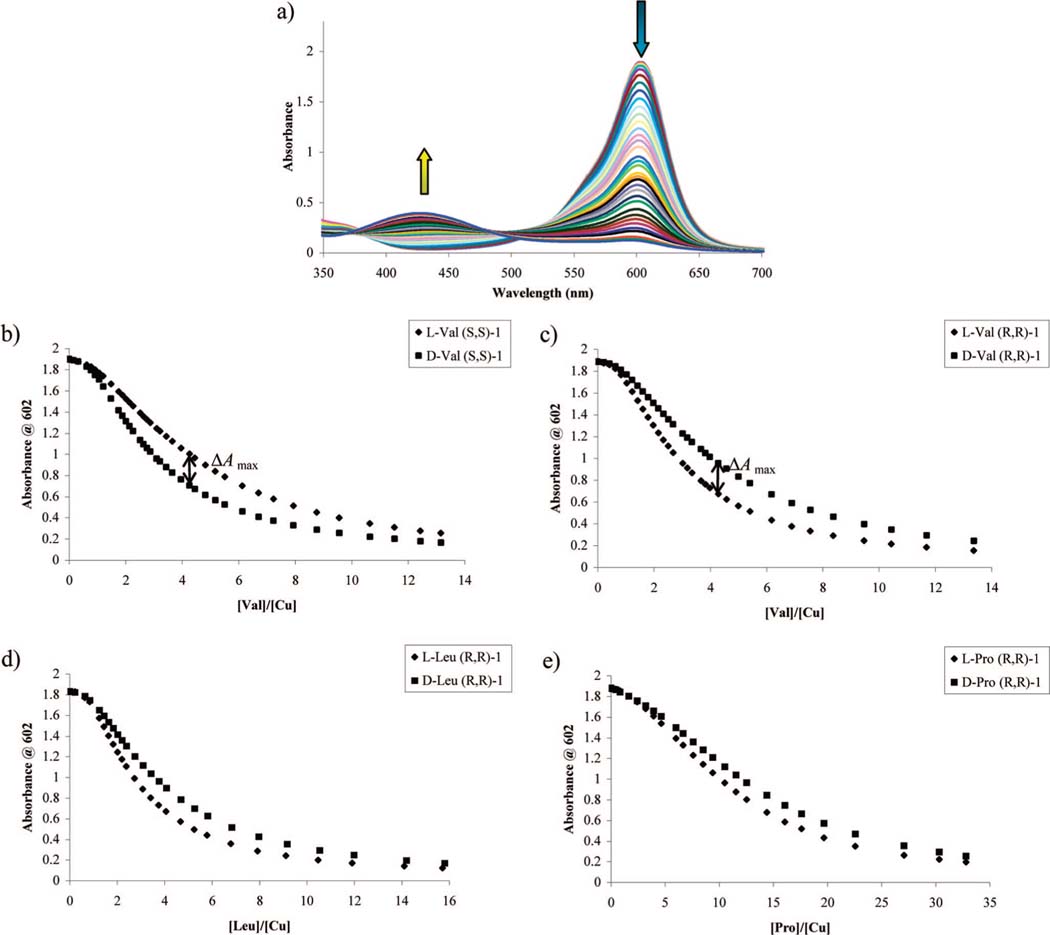
The present work was aimed to extend the scope of our eIDAs to as many naturally occurring α-amino acids as possible. To monitor the selectivity of the receptor [CuII(1)]2+ for l- and d-amino acids, UV-vis displacement isotherms were generated. The absorbance at 602 nm ([CuII(1)(CAS)]2-) was monitored on a UV-vis spectrophotometer as analyte (α-amino acid) was titrated into the CuII complex. Upon the addition of analyte, CAS is displaced, leading to a decrease in the absorbance measured at 602 nm ([CuII(1)(CAS)]2-) and an increase in absorbance at 429 nm (free CAS) (Figure 4a) that could be analyzed quantitatively (Scheme 1, eq 7). As expected, the decrease in absorbance at 602 nm ([CuII(1)(CAS)]2-) was different for the two enantiomers of a given amino acid, allowing enantioselective discrimination.
Figure 4.
(a) Hypsochromic absorbance shift upon the addition of l-valine (5.01 mM) into a solution of CAS (10 μM), Cu(OTf)2 (200 μM), and (R,R)-1 (2.5 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5. (b) Displacement isotherms at 602 nm upon the addition of l- or d-valine (5.01 mM) into a solution containing CAS (10 μM), Cu(OTf)2 (200 μM), and (S,S)-1 (2.5 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5. (c-e) Displacement isotherms at 602 nm obtained for a solution containing CAS (10 μM), Cu(OTf)2 (200 μM), and (R,R)-1 (2.5 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5, upon the addition of (c) l- or d-valine (5.01 mM), (d) l- or d-leucine (5.11 mM), or (e) l- or d-proline (9.96 mM). ΔAmax is defined as the largest difference in absorbance observed in separate titrations of a [CuII((R,R)-1)(CAS)]2– solution at the same concentration of d- and l-amino acid.
As expected, the two enantiomeric ligands (S,S)-1 and (R,R)-1 should be cross-reactive. This is indeed the case, as shown in Figure 4b,c. In a UV-vis titration, d-valine displaced the indicator from the [CuII((S,S)-1)(CAS)]2- complex more readily than l-valine, as evidenced by steeper decrease in absorbance at 602 nm (Figure 4b), allowing enantioselective discrimination between l- and d-valine. Receptor [CuII((R,R)-1)]2+ showed opposite selectivity compared to [CuII((S,S)-1)]2+: the absorbance at 602 nm decreased more readily upon addition of l-valine (Figure 4c). The cross-reactivity pattern shown by [CuII((R,R)-1)]2+ and [CuII((S,S)-1)]2+ was applicable for all α-amino acids, so only results from [CuII((R,R)-1)]2+ are shown from here on. Receptor [CuII((R,R)-1)]2+ was used to analyze 17 of the 20 naturally occurring α-amino acids.58 Cysteine and tyrosine were not completely soluble in the 1:1 MeOH:H2O, 50 mM HEPES buffer solution used in these studies and therefore could not be analyzed. Glycine was not analyzed in this study because it has no stereocenter. A selection of the resulting UV-vis displacement isotherms is shown in Figure 4c-e.
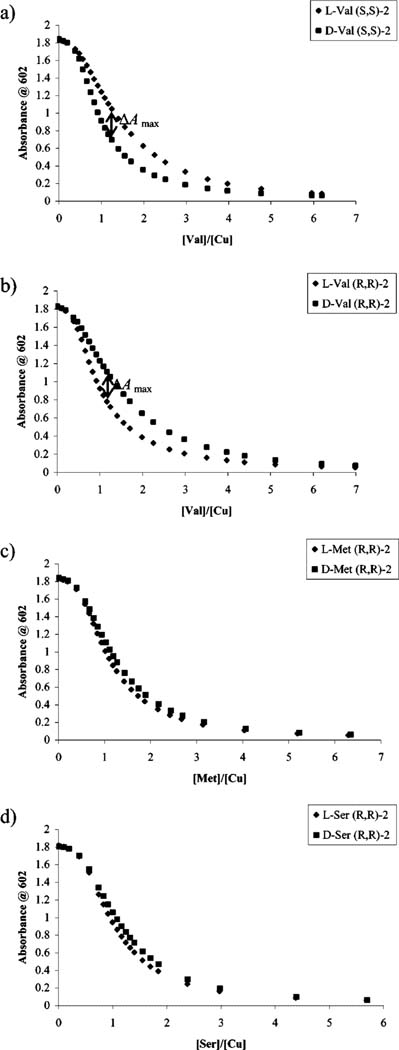
In analogy to receptor [CuII(1)]2+, a similar enantioselectivity and cross-reactivity pattern was observed on addition of enantiomerically pure valine to solutions of [CuII((S,S)-2)(CAS)]2- and [CuII((R,R)-2)(CAS)]2-, with preference for the d- and l-valine, respectively (Figure 5a,b). This cross-reactivity pattern was applicable to all α-amino acids, so in analogy to receptor [CuII(1)]2+, only results obtained with [CuII((R,R)-2)]2+ are reported from here on. A selection of these UV-vis displacement isotherms is shown in Figure 5b-d.59
Figure 5.
(a) Displacement isotherm at 602 nm upon the addition of l- or d-valine (1.25 mM) into a solution containing CAS (10 μM), Cu(OTf)2 (105 μM), and (S,S)-2 (8.8 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5. (b-d) Displacement isotherms at 602 nm obtained for a solution containing CAS (10 μM), Cu(OTf)2 (105 μM), and (R,R)-2 (8.8 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5, upon the addition of (b) l- or d-valine (1.25 mM), (c) l- or d-methionine (1.27 mM), or (d) l- or d-serine (1.25 mM).
ΔAmax was taken as an empirical measurement of enantioselectivity for the analyzed amino acids by each receptor, [CuII((R,R)-1)]2+ and [CuII((R,R)-2)]2+. ΔAmax is defined as the largest difference in absorbance observed in separate titrations of a [CuII((R,R)-1)(CAS)]2- solution with the same concentration of d- or l-amino acid, respectively. The observed ΔAmax values for titrations using receptor [CuII((R,R)-1)]2+ are shown in Table 1a. We arbitrarily chose to consider systems with ΔAmax > 0.1 as being enantioselectively discriminated. Only nine of the 17 α-amino acids analyzed met our arbitrary criterion with [CuII((R,R)-1)]2+, and therefore receptor [CuII((R,R)-2)]2+ was used in an attempt to enantioselectively discriminate the eight remaining amino acids. Since valine showed good enantioselectivity using receptor [CuII((R,R)-1)]2+, it was also analyzed using [CuII((R,R)-2)]2+ as a reference/benchmark in the process of finding optimum experimental conditions for analysis with [CuII((R,R)-2)]2+. The ΔAmax values obtained using receptor [CuII((R,R)-2)]2+ showed appreciable enantioselectivity for four of the eight remaining amino acids (Table 1b). It is interesting to note that the other four amino acids that were not considered enantioselectively discriminated by our arbitrary definition had ΔAmax values above 0.07, which is not very far from our arbitrary threshold of 0.1, but we wanted to explore the use of an eIDA to determine ee of unknown samples accurately. In case the arbitrary threshold were not sufficient or too strict, it would be easy to adjust this criterion when necessary, so even though these amino acids were not far from the arbitrary threshold, they were not considered to be enantioselectively discriminated. Thus, by combining the results obtained with ligands 1 and 2 (Table 1), 13 of the 17 α-amino acids were considered to be enantioselectively detectable (ΔAmax > 0.1) using only two simple synthetic receptors and the commercially available indicator chrome azurol S.
Table 1.
ΔAmax Values Obtained from Displacement Isotherms with Two Receptors
| (a) [CuII((R,R)-1)]2+a |
(b) [CuII((R,R)-2)]2+b |
||
|---|---|---|---|
| Amino Acid | ΔAmax | Amino Acid | ΔAmax |
| Ala | 0.025 | Ala | 0.103 |
| Arg | 0.119 | Gln | 0.087 |
| Asn | −0.262 | Glu | 0.078 |
| Asp | −0.169 | Lys | 0.079 |
| Gln | −0.075 | Met | 0.105 |
| Glu | 0.043 | Phe | 0.074 |
| His | −1.125 | Ser | 0.120 |
| Ile | 0.392 | Trp | 0.117 |
| Leu | 0.235 | Val | 0.331 |
| Lys | 0.070 | ||
| Met | 0.023 | ||
| Phe | 0.020 | ||
| Pro | 0.167 | ||
| Ser | 0.070 | ||
| Thr | 0.118 | ||
| Trp | 0.096 | ||
| Val | 0.292 | ||
ΔAmax at 602 nm with the addition of l- or d-α-amino acids (various concentrations of amino acids were used, refer to Table S-1 in the Supporting Information) into a solution containing CAS (10 μM), Cu(OTf)2 (200 μM), and (R,R)-1 (2.5 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5. All displacement isotherms are available in Figure S-1 in the Supporting Information.
ΔAmax at 602 nm with the addition of l- or d-α-amino acids (various concentrations of amino acids were used, refer to Table S-2 in the Supporting Information) into a solution containing CAS (10 μM), Cu(OTf)2 (105 μM), and (R,R)-2 (8.8 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5. All displacement isotherms are available in Figure S-2 in the Supporting Information.
5. Unexpected Enantioselectivity.
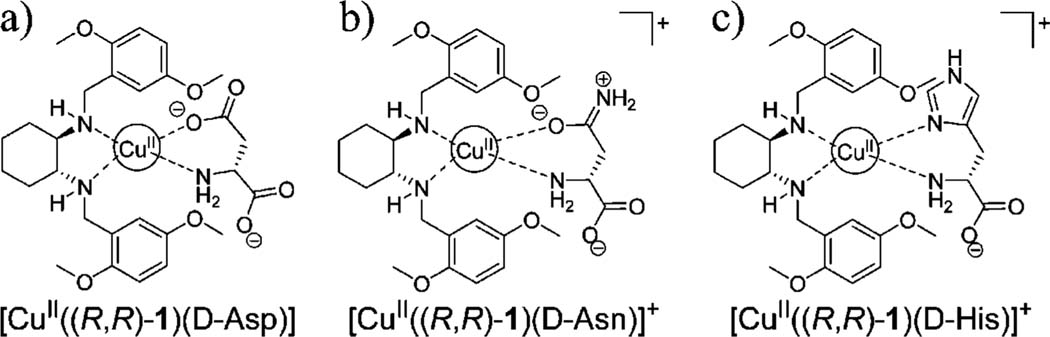
As discussed above, receptor [CuII((R,R)-1)]2+ was generally found to bind more strongly to l-α-amino acids. However, in the case of aspartate, asparagine, and histidine, the d-enantiomer was preferred by receptor [CuII((R,R)-1)]2+, leading to the observed negative ΔAmax values shown in Table 1a. This behavior can be explained by postulating a different coordination mode for these amino acids, involving binding motifs present in their side chains, as shown in Figure 6. However, direct experimental evidence of these structures has not yet been obtained. With these same amino acids, analysis with receptor [CuII((S,S)-1)]2+ has shown a preference for the l-form, thus conserving the required cross-reactivity by the chiral ligand 1.
Figure 6.
Possible binding modes to explain opposite enantioselectivity by [CuII((R,R)-1)]2+ for (a) d-aspartate, (b) d-asparagine, and (c) d-histidine.
6. Equilibria in Solution.
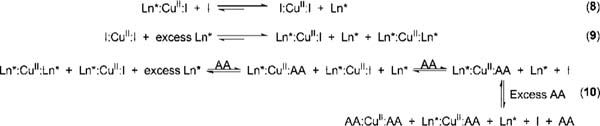
The data obtained could not be fit to a displacement model based on the two simple 1:1 equilibria (Scheme 1, eqs 1 and 2). The equilibria in solution that presumably led to the observed UV-vis isotherms in this study are shown in Scheme 3. Chrome azurol S binds more strongly to the CuII metal center than the chiral ligands 1 and 2 (Ln*, Scheme 3, eq 8), which is why an excess of chiral ligand (Ln*, Scheme 3, eq 9) was used in the UV-vis titrations. This favors the formation of [CuII(1)(CAS)]2- and [CuII(2)(CAS)]2- (Ln*: CuII:I, Scheme 3, eq 9). Due to the excess of chiral ligands 1 and 2 in solution, [CuII(1)2]2+ and [CuII(2)2]2+ (Ln*:CuII:Ln*) also form (Scheme 3, eq 9). Upon the addition of amino acid (AA), one chiral ligand from the Ln*:CuII:Ln* complex is displaced from the CuII metal center (Scheme 3, eq 10) preferentially over displacing an indicator or chiral ligand from Ln*:CuII:I, since there is an excess of Ln*:CuII:Ln* complex in solution compared to Ln*:CuII:I complex, due to a significantly lower concentration of indicator used compared to CuII and Ln*. Also, the amino acid would preferentially displace the chiral ligand instead of CAS due to the ligand’s weaker binding to the metal center, which leads to the observed plateau at the start of the UV-vis titrations. Upon the addition of more amino acid, CAS will be displaced from the metal center, leading to the observed increase in absorbance at 429 nm and a decrease in absorbance at 602 nm (hypsochromic shift). Because the indicator binds to the CuII metal center more strongly than the amino acid, excess AA is required to displace all the indicator, and upon doing so, saturation is reached. The concentration of amino acids used in the titrations was such that complete displacement of CAS could be observed. The receptor’s (Ln*:CuII) different affinities toward the α-amino acids affected the amount of amino acid required to displace all the CAS from the CuII complex (refer to Tables S-1 and S-2 in the Supporting Information).
Scheme 3.
Equilibria in Solutiona
a Ln* = chiral ligand 1 or 2; I )CAS; AA = α-amino acid.
7. “Naked Eye” Detection.
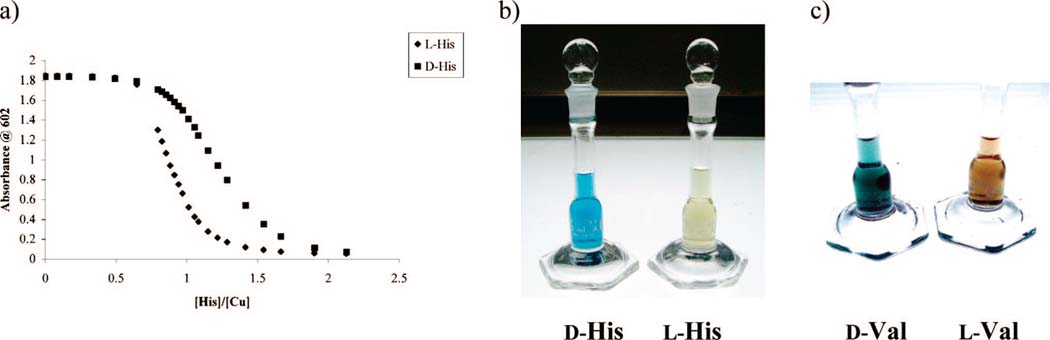
As shown in Table 1a and by the displacement isotherm (Figure 7a), histidine had the largest ΔAmax. Hence, there is a large spectroscopic difference between the additions of l-histidine and d-histidine to a solution of [CuII((S,S)-1)(CAS)]2-. The intervening color change is so striking that it can be easily observed by the “naked eye” (Figure 7b). Among the other considered amino acids, valine also induced a very noticeable change in color when added to a solution containing [CuII((R,R)-2)(CAS)]2- (Figure 7c). The color differences are shown in Figure 7b,c. The solution containing [CuII((S,S)-1)(CAS)]2- and l-histidine was light yellow, indicating displacement of CAS, while the d-histidine-containing solution was blue, indicating that most of the CAS was still bound to the metal center (Figure 7b). Solutions of valine (Figure 7c) showed similar colorimetric differences with receptor [CuII((R,R)-2)]2+, with the solution of l-valine being brown and the one containing d-valine being blue. The ability to discriminate between l- and d-amino acids by “naked eye” detection is not intended to substitute for a precise instrumental method, but it certainly allows for a very rapid and simple method for roughly gauging the enantioselectivity of reactions, showing the advantage of using a colorimetric method such as an eIDA.
Figure 7.
(a) Displacement isotherms at 602 nm obtained upon the addition of l- or d-histidine (2.02 mM) into a solution containing CAS (10 μM), Cu(OTf)2 (200 μM), and (S,S)-1 (2.5 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5. (b) Histidine (480 μM) was added to a solution containing CAS (21 μM), Cu(OTf)2 (375 μM), and (S,S)-1 (4.7 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5. (c) Valine (124 μM) was added to a solution containing CAS (30 μM), Cu(OTf)2 (120 μM), and (R,R)-2 (50 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.0.
8. Determining the Enantiomeric Excess of Test Sampleson a UV-Vis Spectrophotometer.
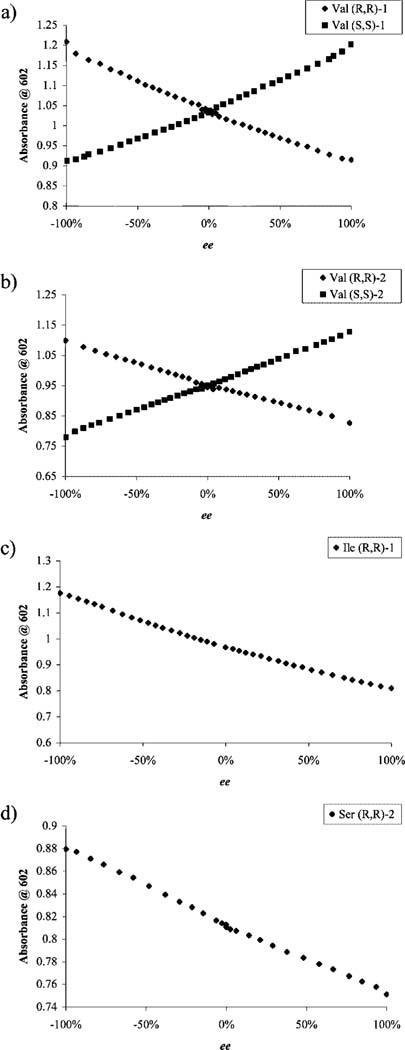
As discussed, using the two receptors presented above, we were able to enantioselectively discriminate 13 of the 17 analyzed α-amino acids. In the following phase of the project, we created ee calibration curves and tested their validity with independent test samples. As expected, [CuII((R,R)-1)]2+ and [CuII((S,S)-1)]2+ had cross-reactive ee calibration curves, since they are enantiomers, which was proven by the mirror-image plots generated (Figure 8a). A similar experiment was conducted with receptor [CuII(2)]2+ (Figure 8b). The ee calibration curves range from −100 to 100% ee, since ee was defined as ee ) ([L] − [D])/([L] + [D]).
Figure 8.
Enantiomeric excess calibration curves obtained using a UV-vis spectrophotometer. (a) Overlay of the two ee calibration curves for (R,R)-1 and (S,S)-1: absorbance at 602 nm as a function of ee for displacement experiments performed with the addition of valine (714 μM) into a solution containing CAS (10 μM), Cu(OTf)2 (200μM), and 1 (2.5 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5. (b) Overlay of two ee calibration curves for (R,R)-2 and (S,S)-2: absorbance at 602 nm as a function of ee for displacement experiments performed with the addition of valine (125 μM) into a solution containing CAS (10 μM), Cu(OTf)2 (105 μM), and 2 (8.8 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5. (c) Absorbance at 602 nm as a function of ee for displacement experiments performed with the addition of isoleucine (697 μM) into a solution containing CAS (10 μM), Cu(OTf)2 (200 μM), and (R,R)-1 (2.5mM) in 1:1 MeOH: H2O, 50 mM HEPES buffered to pH 7.5. (d) Absorbance at 602 nm as a function of ee for displacement experiments performed with the addition of serine (125 μM) into a solution containing CAS (10 μM), Cu(OTf)2 (105 μM), and (R,R)-2 (8.8 mM) in 1:1 MeOH:H2O, 50 mM HEPES buffered to pH 7.5.
Enantiomeric excess calibration curves were made for a selection of α-amino acids. The choice of amino acids used was made in an attempt to cover a range of ΔAmax values and to diversify the nature of the side chains studied. The amino acids selected for the determination of ee calibration curves with receptor [CuII((R,R)-1)]2+ were histidine, isoleucine, and valine.60 Alanine, serine, and valine were the selected amino acids for ee calibration curves to be made with receptor [CuII((R,R)-2)]2+.61 Alanine was particularly chosen because it has the smallest side chain among chiral amino acids and should represent the hardest case of enantiomer discrimination. The concentration of amino acid used in each ee calibration curve was the same as the concentration at which the respective ΔAmax value was obtained (refer to Tables S-3 and S-4 in the Supporting Information), thus maximizing the discrimination between ee values. Two representative examples of ee calibration curves (isoleucine and serine) are shown in Figure 8c,d, respectively.
The ee calibration curves were subjected to linear and second-degree polynomial regression. The best-fit curve (refer to Supporting Information) was used to determine the ee of test samples. Four test samples were prepared for each amino acid for which an ee calibration curve was generated. These four test samples were made independently of the ee calibration curves and analyzed after the calibration curve was generated so as to obtain real independent samples, not just a jack-knife analysis based confirmation of curve validity, as used in previous studies.54 The average absolute error was calculated for each amino acid. This error was defined as the average of the absolute difference between the actual and the experimental ee values obtained for the four test samples. The average absolute errors are shown in Table 2 for various amino acids analyzed with [CuII((R,R)-1)]2+ and [CuII((R,R)-2)]2+. The average of these values has also been calculated to be ±10.2% and ±13.6% for [CuII((R,R)-1)]2+ and [CuII((R,R)-2)]2+, respectively.
Table 2.
Average Absolute Errors for the Determination of ee of Test Samples through UV-Vis Spectrophotometric Measurements Using Two Receptors
| (a) [CuII((R,R)-1)]2+a |
(b) [CuII((R,R)-2)]2+b |
||||
|---|---|---|---|---|---|
| Amino Acid | average absolute error (%) | ΔAmax | Amino Acid | average absolute error (%) | ΔAmax |
| His | 13.0 | −1.125 | Ala | 22.6 | 0.103 |
| Ile | 5.7 | 0.392 | Ser | 12.8 | 0.120 |
| Val | 12.0 | 0.292 | Val | 5.5 | 0.331 |
Average absolute error of ee determination of four test samples for each amino acid analyzed through UV-vis measurements, and ee calibration curves made for each amino acid using receptor [CuII((R,R)-1)]2+. Test samples containing CAS (10 μM), Cu(OTf)2 (200 μM), (R,R)-1 (2.5 mM), and a mixture of l- and d-amino acid (different concentrations of amino acids were used, refer to Table S-3 in the Supporting Information) were mixed and diluted to 2 mL with 1:1 MeOH:H2O, buffered to pH 7.5 with 50 mM HEPES. For experimental values obtained, refer to Supporting Information Table S-5.
Average absolute error of ee determination of four test samples for each amino acid analyzed through UV-vis measurements, and ee calibration curves made for each amino acid using receptor [CuII((R,R)-2)]2+. Test samples containing CAS (10 μM), Cu(OTf)2 (105 μM), (R,R)-2 (8.8 mM), and a mixture of l- and d-amino acid (different concentrations of amino acids were used, refer to Table S-4 in the Supporting Information) were mixed and diluted to 2 mL with 1:1 MeOH:H2O, buffered to pH 7.5 with 50 mM HEPES. For experimental values obtained, refer to Supporting Information Table S-6.
At first glance, the overall average absolute error for [CuII((R,R)-1)]2+ may seem high, but looking at Table 2a, isoleucine has an excellent average absolute error of 5.7%. The same can be observed with analysis conducted with [CuII((R,R)-2)]2+, where the average absolute errors of serine and valine are lower than the overall absolute error of ±13.6%, and valine has an even better average absolute error of 5.5%. The overall absolute error for [CuII((R,R)-2)]2+ was skewed by alanine, which is expected to have the largest error because of its small side chain, reflecting the fact that it is the most difficult amino acid to discriminate enantioselectively.
The observed average absolute errors can be seen to roughly correlate with the ΔAmax values for each amino acid. As shown by Table 2, the amino acids that produced the largest errors were those that had smaller ΔAmax values. Histidine was an exception, but this could be due to its different binding mode (Figure 6). The correlation between the average absolute error and ΔAmax values is probably due to the fact that a large ΔAmax value makes it easier to differentiate close ee values, since their absorbance would be significantly different. This results in a lower error in determining the test samples’ ee. This would demonstrate why certain amino acids were not considered in this study even though they show a ΔAmax value close to the arbitrarily set criterion. Their ee calibration curves would be similar in shape to those shown, but there would be a smaller range of absorbance between −100% and 100% ee, leading to an inability to determine ee of unknown samples accurately. Undoubtedly, at this point the system could be fine-tuned to improve enantioselectivity in order to obtain a larger ΔAmax, which could lower the errors.
The average error obtained with the two proposed receptors falls within an adequate range, compatible with the primary goal of this research, which is to explore the use of eIDA as a method for preliminary determination of ee in a HT fashion. In practice, potentially only those samples showing a 90% ee or above would be examined using a more accurate method, such as chiral HPLC. Processes yielding low ee would not be analyzed further, thus saving significant time and expense. As we discuss in the following paper, we feel that absolute errors lower than approximately 15% are acceptable upon consulting with various individuals in pharmaceutical firms.56
As stated above, fine-tuning of receptors could eventually enable the determination of ee for all α-amino acids and possibly afford a lower error. However, our academic goal was not to optimize the assays but rather to demonstrate the principle. The next stage of the project, which is reported in the following paper, was to move to a HTS platform by implementing eIDA’s advantages compared to traditional methods of determining ee.56 As we show, moving to a rapid HTS technique does not lead to a deterioration of the error and significantly increases the speed of analysis.
Summary
In this study, enantioselective indicator displacement assays (eIDAs) have been shown to be able to determine enantiomeric excess values of α-amino acids with adequate accuracy on a conventional UV-vis spectrophotometer. With only two receptors ([CuII((R,R)-1)]2+, [CuII((R,R)-2)]2+) and one indicator (CAS), we were able to enantioselectively discriminate 13 of the 17 analyzed α-amino acids. Enantiomeric excess was determined for true test samples using ee calibration curves to demonstrate the capabilities of eIDAs. Analysis using receptor [CuII((R,R)-1)]2+ afforded an overall average error of ±10.2%, whereas analysis by receptor [CuII((R,R)-2)]2+ showed an overall average error of ±13.6%. Also, eIDAs were able to enantioselectively discriminate the enantiomers of histidine and valine using “naked eye” detection.
Therefore, enantiomeric indicator displacement assays are excellent for HT determination of ee due to the use of optical methods and associated inexpensive instrumentation. Transitioning to a microplate reader is possible and easy, allowing for a more rapid analysis time, which could overcome the limitation of current methods for determining ee in a HT manner (see the following paper).56
Supplementary Material
Acknowledgment.
We gratefully acknowledge financial support from the National Institutes of Health (GM77437) and the Welch Foundation.
Footnotes
Supporting Information Available: Experimental details, crystallographic data, spectral data, displacement curves, calibration curves, and ee determinations. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).FDA, Chirality 1992, 4, 338–340. [DOI] [PubMed] [Google Scholar]
- (2).Marchelli R; Dossena A; Palla G. Trends Food Sci. Technol 1996, 7, 113–119. [Google Scholar]
- (3).Kim CH; Song YM; Baick SC J. Anim. Sci. Technol 2004, 46, 91–96. [Google Scholar]
- (4).Reetz MT Angew. Chem., Int. Ed 2001, 40, 284–310. [PubMed] [Google Scholar]
- (5).Traverse JF; Snapper ML Drug DiscoVery Today 2002, 7, 1002–1012. [DOI] [PubMed] [Google Scholar]
- (6).Kuntz KW; Snapper ML; Hoveyda AH Curr. Opin. Chem. Biol 1999, 3, 313–319. [DOI] [PubMed] [Google Scholar]
- (7).Charbonneau V; Ogilvie WW Mini-Rev. Org. Chem 2005, 2, 313–332. [Google Scholar]
- (8).Finn MG Chirality 2002, 14, 534–540. [DOI] [PubMed] [Google Scholar]
- (9).Reetz MT; Eipper A; Tielmann P; Mynott R. Adv. Synth. Catal 2002, 344, 1008–1016. [Google Scholar]
- (10).Reetz MT; Tielmann P; Eipper A; Ross A; Schlotterbeck G. Chem. Commun 2004, 1366–1367. [DOI] [PubMed] [Google Scholar]
- (11).Reetz MT; Kuhling KM; Deege A; Hinrichs H; Belder D. Angew. Chem., Int. Ed 2000, 39, 3891–3893. [DOI] [PubMed] [Google Scholar]
- (12).Wang J; Liu KY; Wang L; Bai JL Chin. Chem. Lett 2006, 17, 49–52. [Google Scholar]
- (13).Blomberg LG; Wan H. Electrophoresis 2000, 21, 1940–1952. [DOI] [PubMed] [Google Scholar]
- (14).Van Delden RA; Feringa BL Angew. Chem., Int. Ed 2001, 40, 3198–3200. [DOI] [PubMed] [Google Scholar]
- (15).Walba DM; Eshdat L; Korblova E; Shao R; Clark NA Angew. Chem., Int. Ed 2007, 46, 1473–1475. [DOI] [PubMed] [Google Scholar]
- (16).Schrader W; Eipper A; Pugh DJ; Reetz MT Can. J. Chem 2002, 80, 626–632. [Google Scholar]
- (17).Sawada M; Takai Y; Yamada H; Hirayama S; Kaneda T; Tanaka T; Kamada K; Mizooku T; Takeuchi S; Ueno K; Hirose K; Tobe Y; Naemura KJ Am. Chem. Soc 1995, 117, 7726–7736. [Google Scholar]
- (18).Felten C; Foret F; Minarik M; Goetzinger W; Karger BL Anal. Chem 2001, 73, 1449–1454. [DOI] [PubMed] [Google Scholar]
- (19).Reetz MT; Hermes M; Becker MH Appl. Microbiol. Biotechnol 2001, 55, 531–536. [DOI] [PubMed] [Google Scholar]
- (20).Millot N; Borman P; Anson MS; Campbell IB; Macdonald SJF; Mahmoudian M. Org. Process Res. Dev 2002, 6, 463–470. [Google Scholar]
- (21).Abato P; Seto CT J. Am. Chem. Soc 2001, 123, 9206–9207. [DOI] [PubMed] [Google Scholar]
- (22).Korbel GA; Lalic G; Shair MD J. Am. Chem. Soc 2001, 123, 361–362. [DOI] [PubMed] [Google Scholar]
- (23).Onaran MB; Seto CT J. Org. Chem 2003, 68, 8136–8141. [DOI] [PubMed] [Google Scholar]
- (24).Sprout CM; Seto CT Org. Lett 2005, 7, 5099–5102. [DOI] [PubMed] [Google Scholar]
- (25).Chen Y; Shimizu KD Org. Lett 2002, 4, 2937–2940. [DOI] [PubMed] [Google Scholar]
- (26).Reetz MT; Becker MH; Klein H-W; Stockigt D. Angew. Chem., Int. Ed 1999, 38, 1758–1761. [DOI] [PubMed] [Google Scholar]
- (27).Lin J; Zhang HC; Pu L. Org. Lett 2002, 4, 3297–3300. [DOI] [PubMed] [Google Scholar]
- (28).Lee SJ; Lin WJ Am. Chem. Soc 2002, 124, 4554–4555. [DOI] [PubMed] [Google Scholar]
- (29).Ahn KH; Ku H.-y.; Kim Y; Kim S-G; Kim YK; Son HS; Ku JK Org. Lett 2003, 5, 1419–1422. [DOI] [PubMed] [Google Scholar]
- (30).Mei X; Wolf CJ Am. Chem. Soc 2006, 128, 13326–13327. [DOI] [PubMed] [Google Scholar]
- (31).Wolf C; Liu S; Reinhardt BC Chem. Commun 2006, 4242–4244. [DOI] [PubMed] [Google Scholar]
- (32).Kim YK; Lee HN; Singh NJ; Choi HJ; Xue JY; Kim KS; Yoon J; Hyun MH J. Org. Chem 2008, 73, 301–304. [DOI] [PubMed] [Google Scholar]
- (33).Corradini R; Paganuzzi C; Marchelli R; Pagliari S; Sforza S; Dossena A; Galaverna G; Duchateau AJ Mater. Chem 2005, 15, 2741–2746. [DOI] [PubMed] [Google Scholar]
- (34).Pu L. Chem. ReV 2004, 104, 1687–1716. [DOI] [PubMed] [Google Scholar]
- (35).Anslyn EV J. Org. Chem 2007, 72, 687–699. [DOI] [PubMed] [Google Scholar]
- (36).Tsubaki K; Tanima D; Nuruzzaman M; Kusumoto T; Fuji K; Kawabata TJ Org. Chem 2005, 70, 4609–4616. [DOI] [PubMed] [Google Scholar]
- (37).Qing G-Y; He Y-B; Zhao Y; Hu C-G; Liu S-Y; Yang X. Eur. J. Org. Chem 2006, 1574–1580. [Google Scholar]
- (38).Lee C-S; Teng P-F; Wong W-L; Kwong H-L; Chan AS C. Tetrahedron 2005, 61, 7924–7930. [Google Scholar]
- (39).Kubo Y; Maeda S.y.; Tokita S; Kubo M. Nature 1996, 382, 522–524. [Google Scholar]
- (40).Zhao J; James TD J. Mater. Chem 2005, 15, 2896–2901. [Google Scholar]
- (41).Zhu L; Zhong Z; Anslyn EV J. Am. Chem. Soc 2005, 127, 4260–4269. [DOI] [PubMed] [Google Scholar]
- (42).Piatek AM; Bomble YJ; Wiskur SL; Anslyn EV J. Am. Chem. Soc 2004, 126, 6072–6077. [DOI] [PubMed] [Google Scholar]
- (43).Lavigne JJ; Anslyn EV Angew. Chem., Int. Ed 1999, 38, 3666–3669. [PubMed] [Google Scholar]
- (44).Nguyen BT; Anslyn EV Coord. Chem. ReV 2006, 250, 3118–3127. [Google Scholar]
- (45).Zhang T; Anslyn EV Org. Lett 2007, 9, 1627–1629. [DOI] [PubMed] [Google Scholar]
- (46).McCleskey SC; Floriano PN; Wiskur SL; Anslyn EV; McDevitt JT Tetrahedron 2003, 59, 10089–10092. [Google Scholar]
- (47).Zhang T; Anslyn EV Tetrahedron 2004, 60, 11117–11124. [Google Scholar]
- (48).Nguyen BT; Wiskur SL; Anslyn EV Org. Lett 2004, 6, 2499–2501. [DOI] [PubMed] [Google Scholar]
- (49).Lavigne JJ; Anslyn EV Angew. Chem., Int. Ed 2001, 40, 3118–3130. [DOI] [PubMed] [Google Scholar]
- (50).Brueckner H; Jaek P; Langer M; Godel H. Amino Acids 1992, 2, 271–284. [DOI] [PubMed] [Google Scholar]
- (51).Ma J-A Angew. Chem., Int. Ed 2003, 42, 4290–4299. [DOI] [PubMed] [Google Scholar]
- (52).Seyden-Penne J. Chiral auxiliaries and ligands in asymmetric synthesis; Wiley: New York, 1995. (English translation of Synthesis et catalyse asymmetriques). [Google Scholar]
- (53).Nájera C; Sansano JM Chem. ReV 2007, 107, 4584–4671. [DOI] [PubMed] [Google Scholar]
- (54).Folmer-Andersen JF; Lynch VM; Anslyn EV J. Am. Chem. Soc 2005, 127, 7986–7987. [DOI] [PubMed] [Google Scholar]
- (55).Folmer-Andersen JF; Kitamura M; Anslyn EV J. Am. Chem. Soc 2006, 128, 5652–5653. [DOI] [PubMed] [Google Scholar]
- (56).Leung D; Anslyn EV J. Am. Chem. Soc 2008, 130, 12328–12333 (following paper in this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Mimoun H; de Laumer JY; Giannini L; Scopelliti R; Floriani CJ Am. Chem. Soc 1999, 121, 6158–6166. [Google Scholar]
- (58). [Refer to Supporting Information, Figure S-1, for displacement isotherms of amino acids analyzed with [CuII((R,R)-1)]2+.]
- (59). [Refer to Supporting Information, Figure S-2, for displacement isotherms of amino acids analyzed with [CuII((R,R)-2)]2+.]
- (60). [Refer to Supporting Information, Figure S-3, for ee calibration curves of a selection of amino acids analyzed with [CuII((R,R)-1)]2+ on a UV-vis spectrophotometer.]
- (61). [Refer to Supporting Information, Figure S-4, for ee calibration curves of a selection of amino acids analyzed with [CuII((R,R)-2)]2+ on a UV-vis spectrophotometer.]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.