Abstract
Background
At present, microRNAs and its downstream genes have been regarded as influential indicators in various malignancies. Therefore, the aim of this study was to explore the relationship and molecular mechanism of the miR‐423‐5p and its downstream gene CADM1 in the LUAD.
Methods
The pcDNA‐CADM1 was used to construct the CADM1 overexpressed cell model. The cell proliferation was determined by CCK‐8 and EdU assays and the cell metastasis was performed by wound scratch and transwell chamber assays. The relationship between miR‐423‐5p and CADM1 were determined by bioinformatics, luciferase reporter and western blot assays.
Results
The results revealed that the CADM1 was downregulated in LUAD tissues and cell lines. CADM1 overexpression markedly repressed the cell proliferation, migration and invasion. Moreover, the results of bioinformatics, luciferase reporter and WB assays showed that CADM1 was a target gene of miR‐423‐5p and the miR‐423‐5p expression was negatively associated with CADM1 in LUAD cell lines. Finally, rescue experiments revealed that downregulation of CADM1 could antagonize the functions of miR‐423‐5p inhibitor on cell proliferation and metastasis. These results indicated that miR‐423‐5p aggravated lung adenocarcinoma via downregulation of CADM1 expression.
Conclusions
Downregulation of CADM1 could antagonize the functions of miR‐423‐5p inhibitor on cell proliferation and metastasis. miR‐423‐5p aggravated lung adenocarcinoma via downregulation of CADM1 expression.
Keywords: CADM1, lung adenocarcinoma, metastasis, miR‐423‐5p, proliferation
Downregulation of CADM1 could antagonize the functions of miR‐423‐5p inhibitor on cell proliferation and metastasis. miR‐423‐5p aggravated lung adenocarcinoma via downregulation of CADM1 expression.
Introduction
Lung cancer, including non‐small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), has become one of the leading causes of death in both men and women worldwide. In recent years, lung adenocarcinoma (LUAD) has gradually become the main common subtype of NSCLC. 1 , 2 Owing to the lack of obvious early symptoms, patients with LUAD are usually diagnosed with metastasis. 3 , 4 Despite developments in clinical oncology technology, the prognosis for patients with LUAD is very poor. 5 Meanwhile, because of lack of effective biomarkers and not immediately apparent early symptoms, the clinical therapy available for LUAD is still disappointing. 6 , 7 Hence, it is essential that the progress of LUDA related gene expression and the molecular mechanism for early diagnosis and treatment is explored.
CADM1 (cell adhesion molecule‐1), also known as TSLC1, is expressed at the lateral membrane in normal epithelial cells. 8 , 9 CADM1 gene is a tumor suppressor and exerts a protect role in preventing malignant conversion and metastasis. 10 , 11 At present, more and more studies have demonstrated that CADM1 is related to cell proliferation, migration and invasion of various cancers, such as liver, 12 gastric 13 and breast cancers. 14 , 15 Of note, studies have indicated that the overexpression of CADM1 can suppress proliferation and promote apoptosis of LUAD cell lines. 16 , 17 However, the functional role of CADM1 in the progression of LUAD remains largely unclear.
MicroRNAs (miRNAs), small non‐coding RNAs containing about 22 nucleotides, have been reported to modulate the expression of downstream target genes at a post‐transcriptional level. 18 Recently,
it has been reported that miRNAs are associated with multiple physiological processes and tumorigenesis and metastasis of cancers. 19 MiR‐423‐5p has been reported to be related to the progress of lung adenocarcinoma. 4 However, the molecular mechanism of miR‐423‐5p in LUAD remains unknown. Bioinformatic analysis has confirmed that CADM1 is a potential target of miR‐423‐5p, but whether CADM1 is negatively regulated by miR‐423‐5p remains unclear.
In this study, we detected the expression levels of miR‐423‐5p and CADM1 in LUAD tissues and cell lines, and then explored the mechanism between miR‐423‐5p and CADM1. According to the outcome of the study, we concluded that miR‐423‐5p aggravated the progression of LUAD via downregulating CADM1 expression.
Methods
Collection of tissue specimens
All the LUAD tissues and normal tissues were obtained from patients with LUAD diagnosed at the Fourth Affiliated Hospital of Nanjing Medical University. The patients in the study did not receive any chemotherapy before sampling. The specimens were stored in liquid nitrogen at −80°C for this experiment. The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee of the Fourth Affiliated Hospital of Nanjing Medical University. Written informed consent was obtained from the individual or guardian partipants.
Cell culture and transfection
Human bronchial epithelial cell (BEAS‐2B) and human LUAD cell lines (A549, H1299, NCI‐H1975 and SPC‐A1) were obtained from the Chinese Academy of Science (Shanghai, China). Cells were cultured with DMEM (Keygen, Nanjing, China) in a humidified atmosphere at 37°C with 5% CO2 and saturated humidity. Small hairpin RNAs (shRNAs) targeting CADM1 (sh‐CADM1), shRNA negative control (sh‐NC), miRNA mimics (miR‐423‐5p), miRNA inhibitors (miR‐423‐5p), negative control mimics (NC mimics) and miRNA negative control inhibitors (NC inhibitors) were purchased from GenePharma (Shanghai, China). These oligonucleotides or plasmids were transfected into A549 and H1299 cells using Lipofectamine 2000 (Thermofisher, USA).
Overexpression of CADM1 in A549 and H1299 cells
Plasmid pcDNA‐CADM1 was constructed by introducing a BamHI‐EcoRI fragment containing CADM1 cDNA into the same sites in pcDNA 3.1. The pcDNA‐CADM1 was transfected into A549 and H1299 cells by using Lipofectamine 2000 (Thermofisher, USA). pcDNA‐control was used as a negative control.
RNA extraction and RT‐qPCR
The total RNAs of cells and tissues were extracted using TRIZOL reagent (Keygen, Nanjing, China), and converted into cDNA employing the PrimeScript RT reagent kit (Takara, Tokyo, Japan). RT‐qPCR assays were performed with SYBR Green PCR Master Mix (Takara), followed by 2−ΔΔCT method analysis. The internal control was GADPH and U6. The primer sets for each gene are listed below.
CADM1, forward 5'‐CCACAGGTGATGGGCAGAAT‐3′, reverse 5'‐TTCCTGTGGGGGATCGGTAT‐3′; miR‐423‐5p, forward 5'‐CGAAGTTCCCTTTGTCATCCT‐3′, reverse 5'‐GTGCAGGGTCCGAGGTATTC‐3′; GADPH, forward 5'‐GAGAAGGCTGGGGCTCATTT‐3′, reverse 5'‐AGTGATGGCATGGACTGTGG‐3′; U6, forward 5'‐CTCGCTTCGGCAGCACA‐3′, reverse 5'‐AACGCTTCACGAATTTGCGT‐3′.
Western blot assay
Total proteins were extracted using a RIPA kit (Beyotime, Shanghai, China), and BCA assay kit (Beyotime, Shanghai, China) to detect total protein concentration. Prepared protein samples were separated in SDS‐PAGE (Keygen, Nanjing, China), and transferred into 0.22 μm PVDF (Beyotime, Shanghai, China). The membranes were incubated with primary antibodies (Abcam, Cambridge, MA, UK) listed as follows: rabbit antibodies against CADM1 (ab3910), Cox‐2 (ab15191), MMP‐2 (ab97779), MMP‐9 (ab38898) and β‐actin (ab8227). After 4°C incubation overnight, the membranes were incubated with secondary horseradish peroxidase (HRP) antibodies (Sigma, Aldrich). The protein bands were observed using the Image Lab (Bio‐Rad Laboratories, CA, USA).
Cell counting kit‐8 (CCK‐8) assay
For cell viability, 1 × 105 cells were seeded for 0, 24, 48 and 72 hours. Then, 10 μL CCK‐8 reagent (Beyotime, Shanghai, China) was added and the cell proliferation was evaluated. The optical density was detected at 490 nm by a microplate reader (BioTek Instruments Inc., Winooski, VT, USA).
EdU assay
2 × 105 cells per well were plated for 48 hours, incubated with EdU (50 μM) for another two hours, and then fixed with 4% paraformaldehyde for 30 minutes. The nuclei were then counterstained with DAPI. Cell proliferation was monitored under fluorescence microscopy (Leica, Wetzlar, Germany) at a magnification of 200×.
Wound scratch assay
The migration abilities of the LUAD cells were determined by wound scratch assay. First, the transfected cells were cultured for 24 hours. When the treated cells reached confluence, a straight line scratched on the cell monolayer and width of the scratch was recorded under a microscope. The migrated cells and wound healing images were visualized at 24 and 48 hours using the image analysis and detection system.
Transwell migration assay
The migration and invasion abilities of the LUAD cells were determined through ranswell migration assays. For the migration assay, 1 × 105 cells per well were added into the transwell chambers. For the invasion assay, matrigel was diluted with medium and 1 × 105 cells per well were cultured into transwell chambers. The invasive or migratory cells were fixed and stained with crystal violet for 15 minutes. Finally, four fields of view were photographed under a microscope (BX53, Olympus, Tokyo, Japan) and images were recorded.
Dual‐luciferase reporter gene assay
The sequences of wild‐type CADM1 (CADM1‐WT) and mutant type (CADM1‐Mut) were inserted into pmirGLO reporter vector (Beyotime, Shanghai, China). Then, A549 and H1299 cells were cotransfected with mimics or mimic‐NC together with CADM1‐WT or CADM1‐Mut. After transfection for 48 hours, the activity of luciferase was measured by dual‐luciferase reporter assay system (Promega, Shanghai, China) and exhibited as firefly luciferase intensity calibrated to Renilla luciferase activity.
Statistical analysis
Data analysis was implemented by SPSS 19.0 (IBM Crop., Armonk, NY, USA) and is shown as mean ± the standard deviation. Specifically, t‐test and the one‐way ANOVA test were performed to detect differences between two or more groups. Kaplan‐Meier survival analysis was used to evaluate the overall survival (OS) of LUAD patients with expression of CADM1. P < 0.05 was considered statistically significant.
Results
CADM1 was downregulated in LUAD tissues and cell lines
To evaluate the expression of CADM1 in LUAD tissues and normal tissues, RT‐qPCR was employed. As shown in Fig 1a, the level of CADM1 was obviously downregulated in LUAD tissues compared with normal tissues. Next, we measured the expression level of CADM1 in human bronchial epithelial cell (BEAS‐2B) and human LUAD cell lines (A549, H1299, NCI‐H1975 and SPC‐A1) by RT‐qPCR. Compared with the BEAS‐2B cell line, CADM1 expression was markedly decreased (Fig 1b). Meanwhile, the protein expression level of CADM1 in human alveolar epithelial cells (HPAEpiC), human bronchial epithelial cell (BEAS‐2B) and human LUAD cell lines (A549, H1299, NCI‐H1975 and SPC‐A1) by RT‐qPCR were detected by western blot. The protein expression level of CADM1 was obviously reduced when compared with HPAEpiC and BEAS‐2B (Fig 1c). Among them, the A549 cell line and H1299 cell line exhibited the lowest CADM1expression. Hence, we chose these two cell lines (A549 and H1299) for further study. Finally, Kaplan‐Meier analysis of the survival of patients was performed to evaluate CADM1 expression in LUAD. As expected, compared to the patients with high expression of CADM1, the patients who had low expression of CADM1 had a shorter overall survival (Fig 1d).
Figure 1.
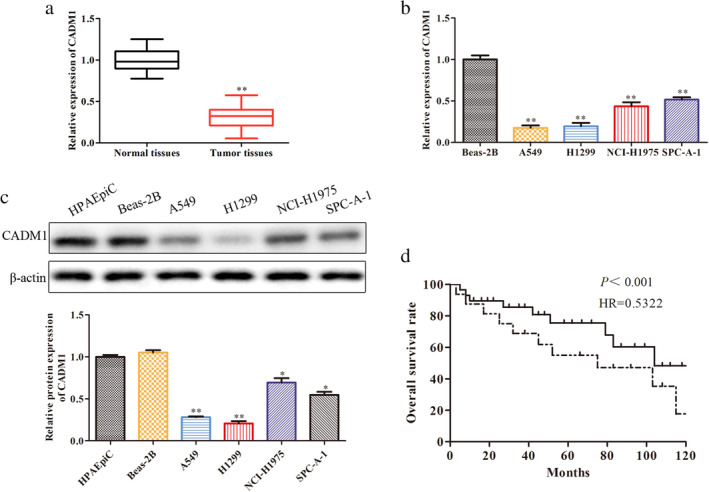
CADM1 is downregulated in LUAD tissues and cell lines. (a) Relative expression levels of CADM1 in LUAD tissues and normal tissues were detected by RT‐qPCR; **P < 0.01 versus normal tissues group. (b) Relative expression levels of CADM1 in different LUAD cell lines were detected by RT‐qPCR. (c) Relative proteins levels of CADM1 in different LUAD cell lines were detected by western blot; **P < 0.01 versus BEAS‐2B group. (d) Overall survival (OS) was used to evaluate the association between CADM1 expression levels with LUAD patients’ prognosis  , High expression;
, High expression;  , Low expression.
, Low expression.
Overexpression of CADM1 inhibits progression of LUAD
To investigate the function of CADM1 in LUAD progression, pcDNA‐control and pcDNA‐CADM1 were transfected into A549 and H1299 cells, respectively. The results showed that the expression of CADM1 was raised after transfection with pcDNA‐CADM1 when compared with pcDNA‐control in A549 and H1299 cells (Fig 2a). Meanwhile, CCK‐8 and EdU assays revealed that pc‐CADM1 markedly suppressed proliferation of A549 and H1299 cells (Fig 2b,c). Coincident with the above results, pc‐CADM1 also suppressed cell migration and invasion both in A549 and H1299 cell lines (Fig 2d and e). In addition, the Cox‐2, MMP‐2 and MMP‐9 proteins expression were markedly downregulated when CADM1 was overexpressed (Fig 2f). Therefore, overexpression of CADM1 inhibited the progression of LUAD.
Figure 2.
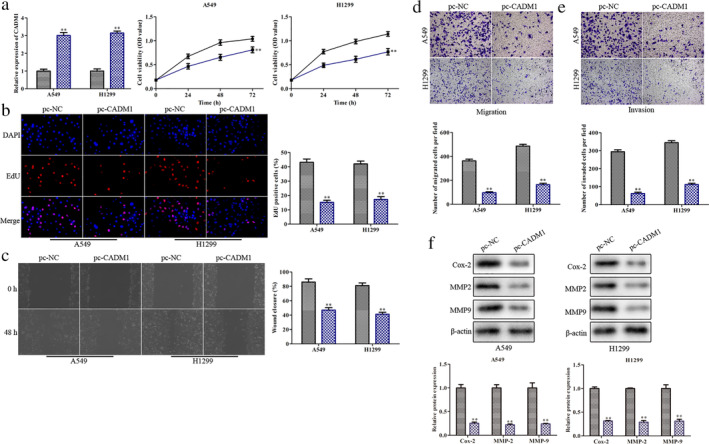
Overexpression of CADM1 inhibits the progression of LUAD. A549 and H1299 cells were transfected with pcDNA‐CADM1 or pcDNA negative control. (a) RT‐qPCR analysis of the transfection efficiency and CCK‐8 assay for cell viability  , pc‐NC;
, pc‐NC;  , pc‐CADM1;
, pc‐CADM1;  , pc‐NC;
, pc‐NC;  , pc‐CADM1;
, pc‐CADM1;  , pc‐NC;
, pc‐NC;  , pc‐CADM1. (b) EdU assay for cell proliferation
, pc‐CADM1. (b) EdU assay for cell proliferation  , pc‐NC;
, pc‐NC;  , pc‐CADM1; (c) Cell capacity of migration (scratch test)
, pc‐CADM1; (c) Cell capacity of migration (scratch test)  , pc‐NC;
, pc‐NC;  , pc‐CADM1. (d) Transwell assay for cell migration
, pc‐CADM1. (d) Transwell assay for cell migration  , pc‐NC;
, pc‐NC;  , pc‐CADM1; (e) Transwell assay for cell invasion
, pc‐CADM1; (e) Transwell assay for cell invasion  , pc‐NC;
, pc‐NC;  , pc‐CADM1; (f) Western blot assay of the expression level of migration related proteins (Cox‐2, MMP‐2 and MMP‐9)
, pc‐CADM1; (f) Western blot assay of the expression level of migration related proteins (Cox‐2, MMP‐2 and MMP‐9)  , pc‐NC;
, pc‐NC;  , pc‐CADM1;
, pc‐CADM1;  , pc‐NC;
, pc‐NC;  , pc‐CADM1; **P < 0.01 versus pc‐NC group.
, pc‐CADM1; **P < 0.01 versus pc‐NC group.
MiR‐423‐5p targets CADM1 and causes post‐transcriptional repression
To study CADM1‐related miRNAs, we predicted the differentially expressed miRNAs which was related with the progression of LUAD by bioinformatic analysis (ENCORI, miRwalk and Pubmed). The results indicated that 10 miRNAs were expressed differently between the normal and tumor tissues (Fig 3a). Among them, miR‐423‐5p was highly expressed which indicated that CADM1 was regarded as a potential target of miR‐423‐5p in LUAD. As shown in Fig 3b, the 3'‐UTR of the CADM1 contained a putative binding site of miR‐423‐5p. Subsequently, luciferase reporter analysis was employed to validate the coactions between miR‐423‐5p and CADM1. The results showed that miR‐423‐5p mimic could inhibit the luciferase activity of CADM1‐WT compared with mimic‐NC. However, no obvious changes were observed in the luciferase activity of CADM1‐Mut (Fig 3c). Coincident with the results, the expression levels of CADM1 were increased apparently at transcriptional and translational levels in A549 and H1299 cell lines transfected with miR‐423‐5p inhibitor (Fig 3d and e). In addition, we also verified miR‐423‐5p expression in LUAD cell lines and LUAD tissues by RT‐qPCR. The results indicated that miR‐423‐5p expression was increased in LUAD cell lines and LUAD tissues when compared with normal cell line and normal tissues (Fig 3f). Finally, Kaplan‐Meier analysis of the survival of patients was performed to evaluate miR‐423‐5p expression in LUAD. As expected, compared to the patients with low expression of miR‐423‐5p, the patients with high expression of miR‐423‐5p had a shorter overall survival (Fig 3g). Hence, we confirmed that CADM1 was a target gene of miR‐423‐5p that was negatively regulated by miR‐423‐5p.
Figure 3.
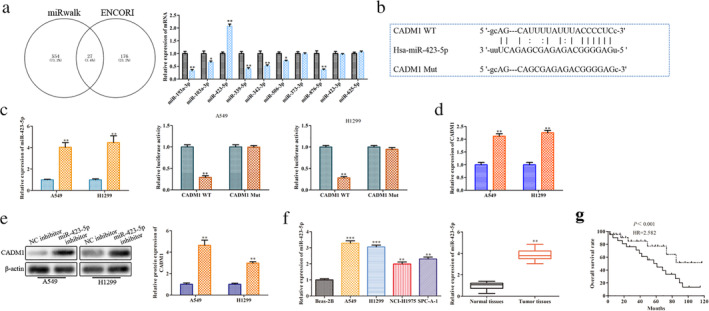
miR‐423‐5p targets CADM1 and causes post‐transcriptional suppression. A549 and H1299 cells were transfected with miR‐423‐5p mimic, miR‐423‐5p inhibitor, NC‐mimic and NC‐inhibitor. (a) Relative expression levels of 10 differentially expressed miRNAs  , Normal tissue;
, Normal tissue;  , Tumor tissue; *P < 0.05, **P < 0.01 versus normal tissues group. (b) The predicted miR‐423‐5p binding sites in the 3'‐UTR of CADM1 and the corresponding sequence were shown. (c) RT‐qPCR analysis of the transfection efficiency and relative values of luciferase signal
, Tumor tissue; *P < 0.05, **P < 0.01 versus normal tissues group. (b) The predicted miR‐423‐5p binding sites in the 3'‐UTR of CADM1 and the corresponding sequence were shown. (c) RT‐qPCR analysis of the transfection efficiency and relative values of luciferase signal  , NC;
, NC;  , miR‐423‐5p mimic;
, miR‐423‐5p mimic;  , NC mimic;
, NC mimic;  , miR‐423‐5p mimic;
, miR‐423‐5p mimic;  , NC mimic;
, NC mimic;  , miR‐423‐5p mimic; **P < 0.01 versus NC mimic group. (d) Relative mRNA expression level of CADM1 in different transcription groups was detected by RT‐qPCR
, miR‐423‐5p mimic; **P < 0.01 versus NC mimic group. (d) Relative mRNA expression level of CADM1 in different transcription groups was detected by RT‐qPCR  , NC inhibitor;
, NC inhibitor;  , miR‐423‐5p inhibitor. (e) Relative protein expression level of CADM1 in different transcription groups was detected by western blot analysis
, miR‐423‐5p inhibitor. (e) Relative protein expression level of CADM1 in different transcription groups was detected by western blot analysis  , NC inhibitor;
, NC inhibitor;  , miR‐423‐5p inhibitor; **P < 0.01 versus NC inhibitor group. (f) Relative expression levels of miR‐423‐5p in LUAD cells and LUAD tissues were detected by RT‐qPCR; **P < 0.01, ***P < 0.001 versus BEAS‐2B group. (g) Overall survival (OS) was used to evaluate the association between miR‐423‐5p expression levels with LUAD patients’ prognosis
, miR‐423‐5p inhibitor; **P < 0.01 versus NC inhibitor group. (f) Relative expression levels of miR‐423‐5p in LUAD cells and LUAD tissues were detected by RT‐qPCR; **P < 0.01, ***P < 0.001 versus BEAS‐2B group. (g) Overall survival (OS) was used to evaluate the association between miR‐423‐5p expression levels with LUAD patients’ prognosis  , High expression;
, High expression;  , Low expression.
, Low expression.
Downregulation of CADM1 rescues miR‐423‐5p inhibitor‐induced suppression of LUAD cell activation
To further verify if CADM1 mediating miR‐423‐5p induced cell proliferation, migration and invasion, we transfected sh‐CADM1 into A549 and H1299 cells treated with miR‐423‐5p inhibitor. First, the relative expression levels of CADM1 and miR‐423‐5p were examined to detect if CADM1 and miR‐423‐5p interference were successful. As a result, the CADM1 and miR‐423‐5p expression was significantly downregulated in sh‐CADM1 group and miR‐423‐5p inhibitor group, respectively (Fig 4a), indicating a successful interference was established. From the results of the CCK‐8 and EdU assays, we found that the sh‐CADM1 inhibited the decrease of cell viability and cell proliferation caused by miR‐423‐5p inhibitor (Fig 4a and b). Wound scratch and transwell chamber assays showed that although the migration and invasion of the A549 and H1299 cells were inhibited by miR‐423‐5p inhibitor, this alteration was reversed by knockdown of CADM1 (Fig 4c–e). These results indicated that downregulation of CADM1 inhibited cell migration and invasion caused by miR‐423‐5p inhibitor.
Figure 4.
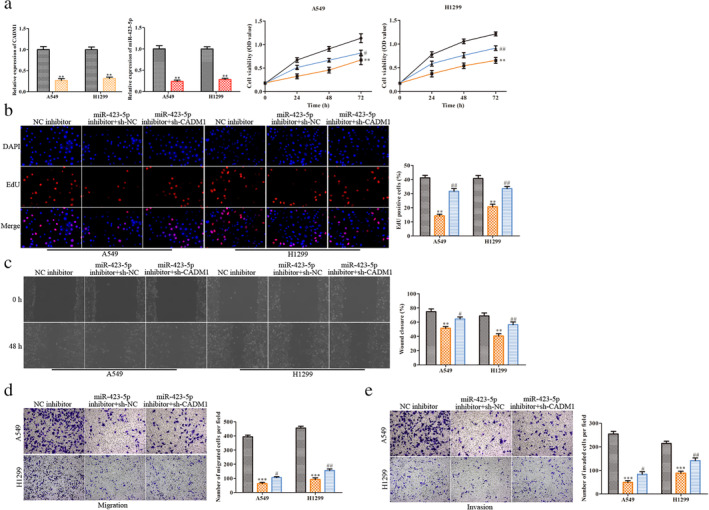
Downregulation of CADM1 rescues miR‐423‐5p inhibitor‐induced suppression of LUAD cell activation. Either miR‐423‐5p inhibitor or CADM1 shRNA were transfected into the A549 and H1299 cells. (a) RT‐qPCR was performed to evaluate the efficiency of miR‐423‐5p inhibitor and CADM1 shRNA and CCK‐8 assay for cell viability  , sh‐NC;
, sh‐NC;  , sh‐CADM1;
, sh‐CADM1;  , NC inhibitor;
, NC inhibitor;  , miR‐423‐5p inhibitor;
, miR‐423‐5p inhibitor;  , NC inhibitor;
, NC inhibitor;  , miR‐423‐5p inhibitor + sh‐NC;
, miR‐423‐5p inhibitor + sh‐NC;  , miR‐423‐5p inhibitor + sh‐CADM1;
, miR‐423‐5p inhibitor + sh‐CADM1;  , NC inhibitor;
, NC inhibitor;  , miR‐423‐5p inhibitor + sh‐NC;
, miR‐423‐5p inhibitor + sh‐NC;  , miR‐423‐5p inhibitor + sh‐CADM1. (b) EdU assay for cell proliferation
, miR‐423‐5p inhibitor + sh‐CADM1. (b) EdU assay for cell proliferation  , NC inhibitor;
, NC inhibitor;  , miR‐423‐5p inhibitor + sh‐NC;
, miR‐423‐5p inhibitor + sh‐NC;  , miR‐423‐5p inhibitor + sh‐CADM1. (c) Cell capacity of migration (scratch test)
, miR‐423‐5p inhibitor + sh‐CADM1. (c) Cell capacity of migration (scratch test)  , NC inhibitor;
, NC inhibitor;  , miR‐423‐5p inhibitor + sh‐NC;
, miR‐423‐5p inhibitor + sh‐NC;  , miR‐423‐5p inhibitor + sh‐CADM1. (d) Transwell assay for cell migration
, miR‐423‐5p inhibitor + sh‐CADM1. (d) Transwell assay for cell migration  , NC inhibitor;
, NC inhibitor;  , miR‐423‐5p inhibitor + sh‐NC;
, miR‐423‐5p inhibitor + sh‐NC;  , miR‐423‐5p inhibitor + sh‐CADM1. (e) Transwell assay for cell invasion
, miR‐423‐5p inhibitor + sh‐CADM1. (e) Transwell assay for cell invasion  , NC inhibitor;
, NC inhibitor;  , miR‐423‐5p inhibitor + sh‐NC;
, miR‐423‐5p inhibitor + sh‐NC;  , miR‐423‐5p inhibitor + sh‐CADM1. **P < 0.01, ***P < 0.001 versus NC inhibitor group; #
P < 0.05, ##
P < 0.01 versus miR‐423‐5p inhibitor + SH‐NC group.
, miR‐423‐5p inhibitor + sh‐CADM1. **P < 0.01, ***P < 0.001 versus NC inhibitor group; #
P < 0.05, ##
P < 0.01 versus miR‐423‐5p inhibitor + SH‐NC group.
Discussion
In recent years, CADM1 has been considered as a tumor suppressor in lung adenocarcinoma (LUAD) and its expression has been previously reported to be closely associated with the progression of lung adenocarcinoma, which suggests that CADM1 is an effective target for LUAD. 8 , 9 , 10 , 11 , 12 , 13 , 14 In the current study, we evaluated the expression levels of CADM1 in LUAD tissues and cell lines. As expected, CADM1 was aberrantly downregulated in LUAD tissues and cell lines. This outcome is in accordance with previous data that CADM1 was found to be downregulated in human cancers. 12 , 14 We also found that compared to the patients with high expression of CADM1, the patients who had low expression of CADM1 had a shorter overall survival. In addition, we discovered that overexpression of CADM1 inhibited proliferation, migration and invasion of A549 and H1299 cells. Therefore, we concluded from our results that the CADM1 could become a potential biomarker and therapeutic target in the treatment of LUAD.
CADM1 is a target gene of many miRNAs, such as miR‐155, 14 miR‐1246, 20 miR‐21, 21 miR‐214 22 and miR‐196b. 23 For example, it has been previously reported that miR‐155‐3p promoted breast cancer progression by modulating CADM1. 14 Meanwhile, multiple studies have determined that CADM1 regulates tumor development of various malignancies. For example, CADM1 has been reported to inhibit ovarian cancer cell proliferation and migration by regulating the PI3K/Akt/mTOR pathway. 24 In hepatocellular carcinoma (HCC), CADM1 has been shown to regulate the G1/S transition by the Rb‐E2F pathway to suppress the development of HCC. 25 Therefore, CADM1 plays a vital role in the development of cancers.
In this study, we aimed to explore the molecular mechanism between CADM1 and miR‐423‐5p. The expression levels of miR‐423‐5p and CADM1 were investigated in LUAD cell lines. We found that MiR‐423‐5p expression increased while CADM1 expression decreased, which suggested that there was a potential interaction between miR‐423‐5p and CADM1 in LUAD. This was further confirmed by bioinformatic analysis, luciferase report assay and WB analysis. These results indicate that miR‐423‐5p is negatively correlated with CADM1 expression in LUAD. In addition, downregulation of CADM1 rescues cell proliferation and inhibits cell migration and invasion caused by miR‐423‐5p inhibitor.
In conclusion, this study revealed that the CADM1 was markedly downregulated in LUAD tissues and cells, and overexpression of CADM1 suppressed the progression of LUAD. We first elucidated that miR‐423‐5p negatively regulated CADM1 expression and then aggravated the progression of LUAD. MiR‐423‐5p/CADM1 axis might therefore serve as biomarkers or potential therapeutic targets for the treatment of LUAD.
Disclosure
All authors declare that there are no competing financial interests.
Acknowledgments
This research was funded by National Natural Science Foundation of China (NO. 81670013). We deeply appreciate the support of all participants in the study.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. Cancer J Clin 2019; 69 (1): 7–34. [DOI] [PubMed] [Google Scholar]
- 2. Hasan N, Kumar R, Kavuru MS. Lung cancer screening beyond low‐dose computed tomography: The role of novel biomarkers. Lung 2014; 192 (5): 639–48. [DOI] [PubMed] [Google Scholar]
- 3. Gridelli C, Rossi A, Carbone DP et al Non‐small‐cell lung cancer. Nat Rev Dis Primers 2015; 1: 15009. [DOI] [PubMed] [Google Scholar]
- 4. Li W, Zhang B, Jia YC et al LncRNA LOXL1‐AS1 regulates the tumorigenesis and development of lung adenocarcinoma through sponging miR‐423‐5p and targeting MYBL2. Cancer Med 2020; 9 (2): 689–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li P, Liu HQ, Li YM, Wang Y, Zhao L, Wang H. MiR‐339‐5p inhibits lung adenocarcinoma invasion and migration by directly targeting BCL6. Oncol Lett 2018; 16 (5): 5785–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ni M, Shi XL, Qu ZG, Jiang H, Chen ZQ, Hu J. Epithelial mesenchymal transition of non‐small‐cell lung cancer cells A549 induced by SPHK1. Asian Pac J Trop Med 2015; 8 (2): 142–6. [DOI] [PubMed] [Google Scholar]
- 7. Hu HH, Xu XD, Lu F et al Exosome‐derived miR‐486‐5p regulates cell cycle, proliferation and metastasis in lung adenocarcinoma via targeting NEK2. Front Bioeng Biotechnol 2020; 8: 259. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8. Masuda M, Yageta M, Fukuhara H et al The tumor suppressor protein TSLC1 involved in cell‐cell adhesion. J Biol Chem 2002; 277 (34): 31014–9. [DOI] [PubMed] [Google Scholar]
- 9. Ito T, Nakamura A, Tanaka I et al CADM1 associates with hippo pathway core kinases; membranous co‐expression of CADM1 and LATS2 in lung tumors predicts goof prognosis. Cancer Sci 2019; 110 (7): 2284–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartsough EJ, Weiss MB, Heilman SA et al CADM1 is a TWIST1‐regulated suppressor of invasion and survival. Cancer Death Dis 2019; 10 (4): 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuramochi M, Fukuhara H, Nobukuni T et al TSLC1 is a tumor‐suppressor gene in human non‐small‐cell lung cancer. Nat Genet 2001; 27 (4): 427–30. [DOI] [PubMed] [Google Scholar]
- 12. Wang F, Qi X, Li ZX, Jin SQ, Xie Y, Zhong HS. LncRNA CADM1‐AS1 inhibits cell‐cycle progression and invasion via PTEN/ATK/GSK‐3β axis in hepatocellular carcinoma. Cancer Manage Res 2019; 11: 3813–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi XY, Sun YZ, Li HY. LncRNA CADM1‐AS1 serve as new prognostic biomarker for gastric cancer. Eur Rev Med Pharm Sci 2019; 23: 232–8. [DOI] [PubMed] [Google Scholar]
- 14. Zhang GC, Zhang LL, Luo H, Wang SB. MicroRNA‐155‐3p promotes breast cancer progression through down‐regulating CADM1. OncoTargets Ther 2019; 12: 7993–8002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Saito M, Goto A, Abe N et al Decreased expression of CADM1 and CADM4 are associated with advanced stage breast cancer. Oncol Lett 2018; 15 (2): 2401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goto A, Niki T, Chi‐Pin L. Loss of TSLC1 expression in lung adenocarcinoma: Relationships with histological subtypes, sex and prognostic significance. Cancer Sci 2005; 96 (8): 480–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mao XL, Seidlitz E, Truant R, Hitt M, Ghosh HP. Re‐expression of TSLC1 in a non‐small‐cell lung cancer cell line induces apoptosis and inhibits tumor growth. Oncogene 2004; 22 (33): 5632–42. [DOI] [PubMed] [Google Scholar]
- 18. Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell 2009; 136 (2): 215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009; 10 (10): 704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun Z, Meng CT, Wang SH et al MicroRNA‐1246 enhances migration and invasion through CADM1 in hepatocellular carcinoma. BMC Cancer 2014; 14: 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cao W, Shi P, Ge JJ. MiR‐21 enhances cardiac fibrotic remodeling and fibroblast proliferation via CADM1/STAT3 pathway. BMC Cardiovasc Disord 2017; 17: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cai HQ, Miao MY, Wang ZG. MiR‐214‐3p promotes the proliferation, migration and invasion of osteosarcoma cells by targeting CADM1. Oncol Lett 2018; 16 (2): 2620–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang HL, Zhou R, Liu J et al MicroRNA‐196b inhibits late apoptosis of pancreatic cancer cells by targeting CADM1. Sci Rep 2017; 7: 11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Si X, Xu F, Xu F, Wei M, Ge Y, Chenge S. CADM1 inhibits ovarian cancer cell proliferation and migration by potentially regulating the PI3K/Akt/mTOR pathway. Biomed Pharmacother 2020; 123: 109717. [DOI] [PubMed] [Google Scholar]
- 25. Zhang W, Xie HY, Ding SM et al CADM1 regulates the G1/S transition and represses tumorigenicity through the Rb‐E2F pathway in hepatocellular carcinoma. Hepat Pancr Dis Int 2016; 15 (3): 289–96. [DOI] [PubMed] [Google Scholar]


