Abstract
Organisms are inherently three dimensional, thus comprehensive understanding of the complicated biological system requires analysis of organs or even whole bodies in the context of three dimensions. However, this is a tremendous task since the biological specimens are naturally opaque, a major obstacle in whole‐body and whole‐organ imaging. Tissue clearing technique provides a prospective solution and has become a powerful tool for three‐dimensional imaging and quantification of organisms. Tissue clearing technique aims to make tissue transparent by minimizing light scattering and light absorption, thus allowing deep imaging of large volume samples. When combined with diverse molecular labeling methods and high‐throughput optical sectioning microscopes, tissue clearing technique enables whole‐body and whole‐organ imaging at cellular or subcellular resolution, providing detailed and comprehensive information about the intact biological systems. Here, we give an overview of recent progress and biomedical applications of tissue clearing technique. We introduce the mechanisms and basic principles of tissue clearing, and summarize the current tissue clearing methods. Moreover, the available imaging techniques and software packages for data processing are also presented. Finally, we introduce the recent advances in applications of tissue clearing in biomedical fields. Tissue clearing contributes to the investigation of structure‐function relationships in intact mammalian organs, and opens new avenues for cellular and molecular mapping of intact human organs. We hope this review contributes to a better understanding of tissue clearing technique and can help researchers to select the best‐suited clearing protocol for their experiments.
Keywords: data processing, light scattering, optical sectioning microscope, tissue clearing technique, whole‐body imaging
Tissue clearing technique achieves tissue transparency by reducing light scattering and light absorption. Tissue clearing enables whole‐body and whole‐organ imaging and facilitates the understanding of intact organisms at system‐level. Tissue clearing contributes to the investigation of structure‐function relationships in intact mammalian organs, and opens new avenues for cellular and molecular mapping of intact human organs.
![]()
1. INTRODUCTION
Systematic investigation of the morphology and structure of the organism at single‐cell resolution is essential for elucidating the associated mechanisms in health and disease. However, extracting detailed information with a global perspective from an intact biological specimen has been a fundamental challenge in biomedicine, because biological tissues are mostly opaque, which hinders light penetration into deep tissue and restricts imaging depth and contrast (Zhu et al., 2013).
Conventional histological sectioning techniques provide two‐dimensional information about anatomical structures and cellular constituents within tissue samples (Seo et al., 2016). However, biological tissues are virtually three dimensional. For example, most individual neurons have an irregular structure with neural processes extending in multiple directions, their native structure definitely cannot be ascertained by a thin section (Furth et al., 2018). Therefore, three‐dimensional (3D) imaging is indispensable for whole‐body and whole‐organ cell profiling (Feuchtinger et al., 2016; Matsumoto et al., 2019). Noninvasive imaging techniques, such as computer tomography (CT) (Goldman, 2007), magnetic resonance imaging (MRI) (Hamaide et al., 2016), and positron emission tomography (PET) (Nader and Banks, 2014), allow detailed 3D visualization of anatomical structures in living organisms, and can be applied repeatedly on the same organism for the study of spatiotemporal changes. However, these techniques have limited resolutions, which are insufficient to extract the properties of individual cells. 3D images are traditionally acquired by serial sectioning and postimage reconstruction. Nevertheless, piecemeal reconstructions are laborious and time consuming, and may result in image distortion from folded, compressed, or stretched sections. Automated sectioning imaging techniques can avoid information loss and image alignment in manual sectioning, and have been successfully applied in both light and electron microscopes (Briggman et al., 2011; Li et al., 2018a). Yet these approaches are destructive: the sample is destroyed in a single imaging sequence. Furthermore, the complicated and expensive setup restricts wide applications of these techniques. Nonsectioning imaging techniques have also been developed. For instance, the advent of multiphoton microscope has increased the imaging depth up to hundreds of microns in opaque samples (Denk et al., 1990), but imaging resolution and contrast decline in deeper regions of the tissue due to light scattering. Therefore, multiphoton microscope is inadequate for 3D imaging of large volume tissues like brain and liver.
The recently emerging tissue clearing technique is a promising solution, which allows rapid 3D visualization of intact organs or even whole bodies. Tissue clearing technique renders tissue optically transparent and allows full penetration of light deep into the cleared tissues, hence accelerating the progress of whole‐body imaging (Figure 1). In this review, we introduce the mechanisms and basic principles of tissue clearing, describe the currently available tissue clearing methods, imaging techniques and data processing, and finally present the biomedical applications of tissue clearing.
FIGURE 1.
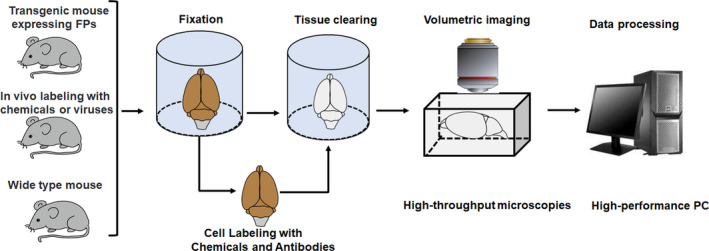
Tissue clearing technique and steps for volumetric imaging
2. MECHANISMS AND PRINCIPLES OF TISSUE CLEARING
Biological tissues are composed of heterogeneous substances with different optical properties (e.g., refractive index), such as connective tissue fibers, organelles, lipids particles, and proteins (Johnsen and Widder, 1999; Tuchin, 2005). Heterogeneity of RIs among these intrinsic components causes light scattering (Tuchin, 2005; Kim et al., 2018). The incoming light is scattered multiple times as it travels through the tissue, causing decay of light intensity in the forward direction (Tuchin, 1997). Furthermore, light absorption by endogenous light‐absorbing substances, such as heme and melanin, further attenuates the light transmission (Weissleder, 2001; Tuchin, 2015a). Light scattering and light absorption inside tissues are the main reasons of tissue opacity, which is a major obstacle for whole‐body and whole‐organ imaging (Susaki and Ueda, 2016). Controlling the optical properties of tissues may contribute to tissue transparency by physical or chemical manipulation (Tuchin et al., 1997; Vargas et al., 1999; Tuchin, 2015b). Therefore, tissue clearing aims to clarify tissue by reducing light scattering and light absorption inside tissues (Figure 2), thus facilitating volumetric imaging. The ideal tissue clearing technique should have excellent clearing capacity while preserving fluorescent proteins (FPs), native architecture, and key molecular information (Park et al., 2019).
FIGURE 2.
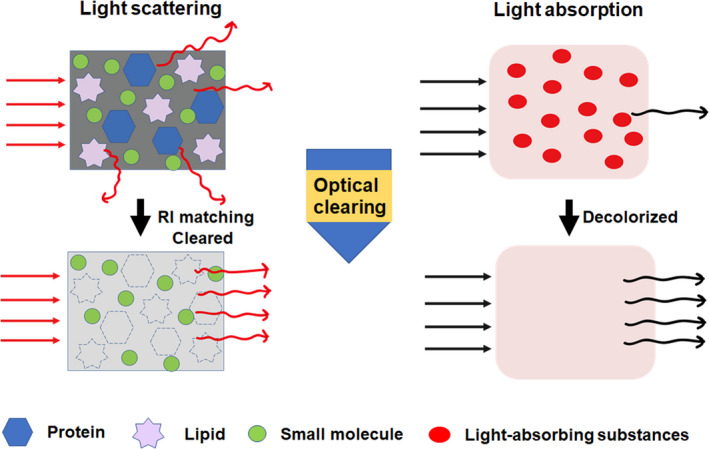
Mechanisms and principles of tissue clearing technique. Light scattering and light absorption are the leading causes of tissue opacity, which blocks light propagation. Light scattering is caused by heterogeneous components such as lipids and proteins. Light absorption is resulted from endogenous light absorbers like hemoglobin and melanin. Tissue clearing technique makes tissue optically transparent by minimizing light scattering and light absorption
2.1. Common procedures of tissue clearing
Although a large number of tissue clearing techniques now exist, they are usually conducted by two to four of the following steps: (a) tissue fixation, (b) permeabilization, (c) decolorization, and (d) RI matching (Figure 3).
FIGURE 3.
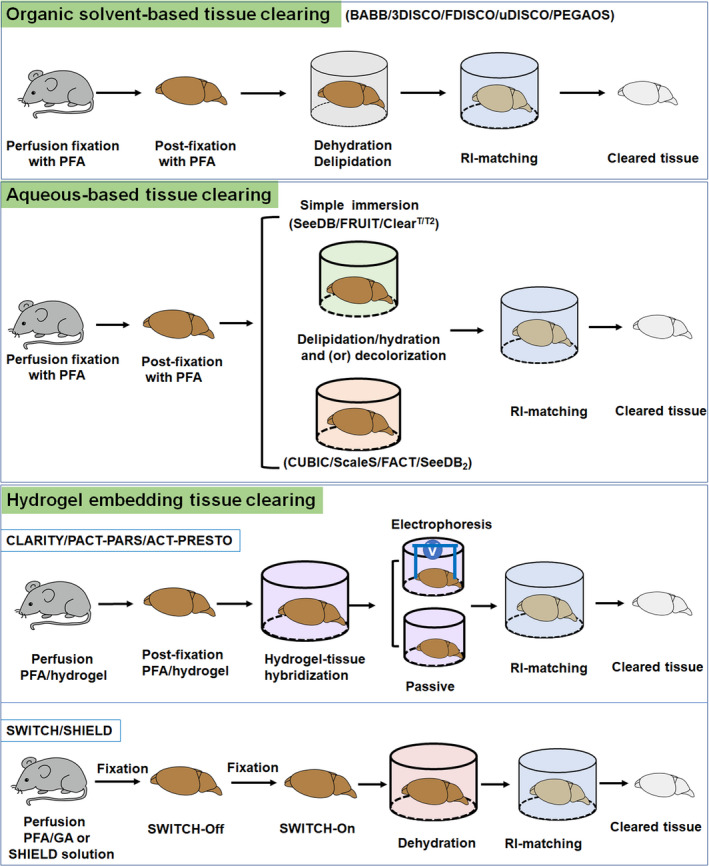
Methodology of tissue clearing technique
Tissue fixation is the primary step to preserve structural and molecular information during the clearing process. Paraformaldehyde (PFA), glutaraldehyde (GA) (Murray et al., 2015), and acrylamide hydrogel embedding (Chung et al., 2013) are usually used for tissue fixation during clearing. Permeabilization is essential for promoting the diffusion of high RI medium and antibodies into a 3D tissue. The commonly used permeabilizing reagents include solvents, hyperhydration reagents, and delipidation reagents (Tainaka et al., 2016; Richardson and Lichtman, 2017). Light absorption from endogenous pigments decays light propagation and interferes with optical observations (Weissleder, 2001). Decolorizing these light absorbers would contribute to light transmission and 3D imaging. Mismatches of RI inside tissues is the primary reason of light scattering and tissue opacity (Johnsen and Widder, 1999). Equilibrating RI of the entire tissue can reduce light scattering, thus contributes to improve clearing efficacy and imaging quality as the light penetrates deeper into the samples (Susaki and Ueda, 2016; Tainaka et al., 2016). Aromatic solvents, hydrophilic reagents, and contrast reagents have been widely used for RI matching (Table 1).
TABLE 1.
Common procedures of tissue clearing
| Procedure | Reagent | Method | References |
|---|---|---|---|
| Tissue fixation | PFA | 3DISCO | Erturk et al., 2012 |
| GA | SWITCH | Murray et al., 2015 | |
| Acrylamide hydrogel | CLARITY | Chung et al., 2013 | |
| Permeabilization | |||
| Solvents | Ethanol | BABB | Dodt et al., 2007 |
| THF, DCM | FDISCO | Qi et al., 2019 | |
| Tert‐butanol | uDISCO | Pan et al., 2016 | |
| Hydration | Urea | Scale | Hama et al., 2011 |
| Delipidation | Aminoalcohols | CUBIC | Susaki et al., 2014 |
| SDS | FACT | Xu et al., 2017 | |
| Saponin | SeeDB2 | Ke et al., 2016 | |
| Triton X−100 | Ce3D | Li et al., 2017 | |
| Decolorization | H2O2 | iDISCO | Renier et al., 2014 |
| Quadrol | PEGASOS | Jing et al., 2018 | |
| Ammonium | CUBIC | Tainaka et al., 2014 | |
| N‐alkylimidazole | CUBIC‐L/R+ | Tainaka et al., 2018 | |
| Thioglycerol | FRUIT | Hou et al., 2015 | |
| RI matching | DBE | 3DISCO | Erturk et al., 2012 |
| DPE | uDISCO | Pan et al., 2016 | |
| PEG | PEGASOS | Jing et al., 2018 | |
| Sucrose | SeeDB | Ke et al., 2013 | |
| Aromatic amide | CUBIC‐L/R+ | Tainaka et al., 2018 | |
| FocusClear | CLARITY | Chung et al., 2013 | |
| RIMS | PACT‐PARS | Yang et al., 2014 | |
| Iohexol | SeeDB2 | Ke et al., 2016 |
Abbreviations: BABB, benzyl alcohol and benzyl benzoate; DBE, dibenzyl ether; DCM, dichloromethane; DPE, diphenyl ether; GA, glutaraldehyde; PEG, polyethylene glycol; PFA, paraformaldehyde; SDS, sodium dodecyl sulfate; THF, tetrahydrofuran.
3. AVAILABLE PROTOCOLS FOR TISSUE OPTICAL CLEARING
In recent years, many attempts have been made to develop tissue clearing techniques for optical imaging, and a number of tissue clearing methods are emerging (Figure 4). Generally speaking, the current clearing methods can be classified into three groups: organic solvent‐based tissue clearing, aqueous‐based tissue clearing, and hydrogel embedding tissue clearing (Richardson and Lichtman, 2015).
FIGURE 4.
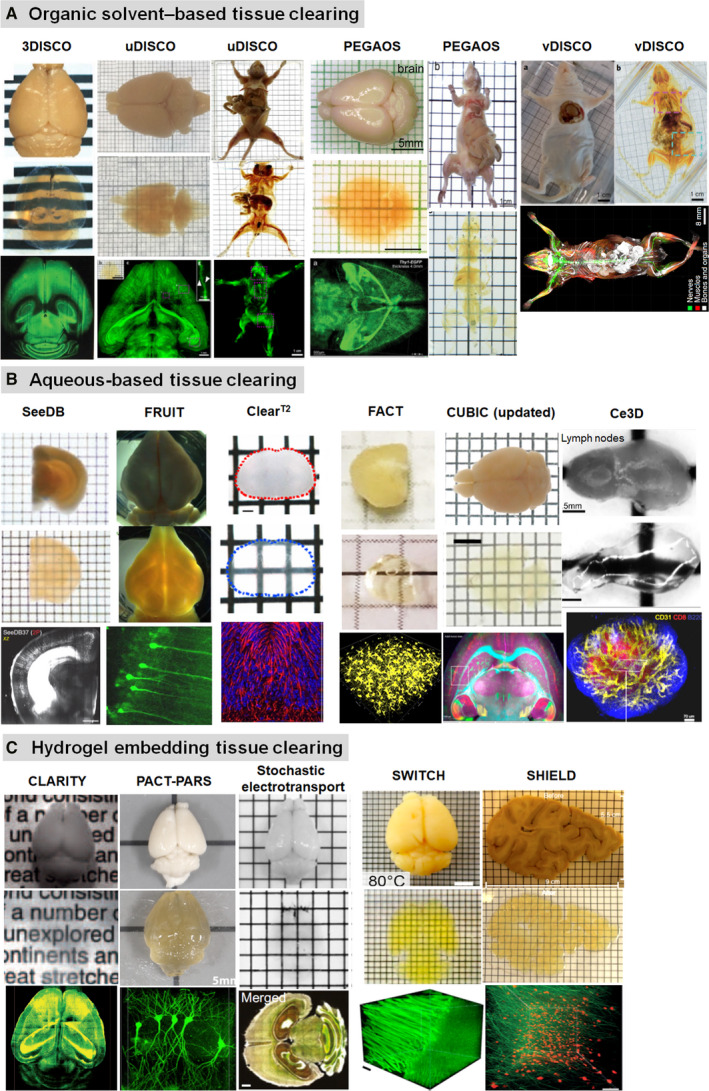
Optical clearing and imaging of diverse tissue clearing techniques. (a) Organic solvent‐based tissue clearing and volumetric imaging. Images adapted from Refs (Erturk et al., 2012; Pan et al., 2016; Jing et al., 2018; Cai et al., 2019). (b) Aqueous‐based tissue clearing and optical imaging. Images adapted from Refs (Ke et al., 2013; Kuwajima et al., 2013; Hou et al., 2015; Ke et al., 2016; Li et al., 2017; Xu et al., 2017; Tainaka et al., 2018). (c) Hydrogel embedding tissue clearing and 3D imaging. Images adapted from Refs (Chung et al., 2013; Yang et al., 2014; Kim et al., 2015; Murray et al., 2015; Park et al., 2019)
3.1. Organic solvent‐based tissue clearing
Organic solvent‐based tissue clearing methods achieve extremely excellent tissue transparency by homogenizing RI of different components with high RI organic solvents (Table 2). BABB clearing could fully clarify embryos and young mouse brain (Dodt et al., 2007), but this protocol degraded GFP signals quickly and failed to clear myelinated tissues. 3DISCO was able to achieve robust transparency of most adult rodent organs by introducing THF and DBE, and preserved FPs for a few days (Erturk et al., 2012). In contrast to 3DISCO, uDISCO enabled whole‐body clearing and imaging and maintained endogenous FPs over months (Pan et al., 2016). a‐uDISCO is a modified version of uDISCO, which improved fluorescence intensity and stability by adjusting pH condition (Li et al., 2018b). However, both uDISCO and a‐uDISCO could not efficiently clear highly colorized organs and hard tissues. To address these limitations, Zhao's team developed a polyethylene glycol (PEG)‐associated solvent system (PEGASOS), which could clear nearly all types of tissues while preserving endogenous fluorescence (Jing et al., 2018). Zhu's group developed a 3DISCO‐based clearing protocol, termed FDISCO, by temperature and pH adjustments. FDISCO effectively conserved FPs and chemical fluorescent tracers, and allowed repeated revisiting of the cleared samples over months (Qi et al., 2019). The recently developed sDISCO further retained the fluorescence signals by stabilizing DBE with antioxidant addition. The fluorescence of FPs could be maintained for more than 1 year (Alnuami et al., 2008; Hahn et al., 2019). The preservation of endogenous FPs is certainly valuable, but these proteins can also be visualized by immunolabeling. iDISCO developed by Renier et al. allowed whole‐mount immunolabeling and volumetric imaging of mouse embryos and adult organs (Renier et al., 2014). vDISCO was a whole‐body immunolabeling technology based on nanobodies. This technology enhanced the signal intensity of FPs more than 100 times, and revealed whole‐body neuronal projections in Thy1‐GFP‐M mice (Cai et al., 2019).
TABLE 2.
Comparison of organic solvent‐based tissue clearing
| Method | Tissue scale | Clearing time | FP preservation | DiI Compatible | Tissue size | Toxic | References |
|---|---|---|---|---|---|---|---|
| BABB | Whole young mouse brain | 10 days | Half day | No | Shrinkage | Yes | Dodt et al., 2007 |
| 3DISCO | Whole adult mouse brain | 3 days | A few days | No | Shrinkage | Yes | Erturk et al., 2012 |
| FDISCO | Whole adult mouse brain | 3‐4 days | Over months | No | Shrinkage | Yes | Qi et al., 2019 |
| sDISCO | Whole adult mouse brain | 4 days | More than one year | No | Shrinkage | Yes | Hahn et al., 2019 |
| uDISCO | Whole mouse body without skin | 3‐4 days | Over months | No | Shrinkage | Yes | Pan et al., 2016 |
| PEGASOS | Whole mouse body with skin | 2 weeks | Over months | No | Shrinkage | Yes | Jing et al., 2018 |
| iDISCO | Whole‐brain immunolabeling | 4 days | Many months | No | Shrinkage | Yes | Renier et al., 2014 |
| vDISCO | Whole‐body immunolabeling | 4 days | 18 months | No | Shrinkage | Yes | Cai et al., 2019 |
Although solvent‐based tissue clearing methods showed brilliant clearing performance and achieved whole‐body imaging with subcellular resolution, some drawbacks should not be neglected. Their versatility is constrained by certain deficiencies, such as substantial shrinkage of samples, toxicity of most organic solvents, and quenching of fluorescent proteins.
3.2. Aqueous‐based tissue clearing
The disadvantages of solvent‐based tissue clearing propelled researchers to develop aqueous‐based techniques that are relatively simple, safe, and friendly to FPs. The aqueous‐based tissue clearing methods can be categorized into two groups: (a) simple immersion and (b) delipidation and (or) hydration (Table 3). In simple immersion, samples are impregnated in high RI aqueous solutions and become transparent gradually. Fructose, 2,20‐thiodiethanol (TDE) and formamide are commonly used for this purpose. The fructose‐based SeeDB could clear mouse brain samples with minimal tissue deformation, but it was impractical for whole‐organ clearing (Ke et al., 2013). The SeeDB‐derived FRUIT addressed this limitation by urea addition and achieved whole‐brain clearing of adult rabbit (Hou et al., 2015). Formamide‐based ClearT/T2 could rapidly clear mouse embryos, but was insufficient to clear adult brain (Kuwajima et al., 2013). RTF was a ClearT2 variation, which achieved better clearing performance on adult brain tissues (Yu et al., 2018).
TABLE 3.
Comparison of aqueous‐based tissue clearing
| Method | Clearing capability | Tissue size | DiI Compatible | TEM Compatible | Tissue validation | References |
|---|---|---|---|---|---|---|
| Simple immersion | ||||||
| SeeDB | Weak | Constant | Yes | yes | Mouse hemi‐brain clearing | Ke et al., 2013 |
| FRUIT | Medium | Constant | Yes | N.T. | Whole adult rabbit Brain clearing | Hou et al., 2015 |
| ClearT/T2 | Weak | Constant | Yes | N.T. | Young mouse brain clearing | Kuwajima et al., 2013 |
| RTF | Medium | Constant | Yes | N.T. | Adult mouse brain blocks clearing | Yu et al., 2018 |
| Delipidation | ||||||
| FACT | Strong | Minor shrinkage | No | No | Whole adult mouse brain clearing | Xu et al., 2017 |
| SeeDB2 | Medium | Constant | Yes | No | Mouse hemi‐brain clearing | Ke et al., 2016 |
| FOCM | Strong | Constant | yes | N.T. | Mouse hemi‐brain clearing | Zhu et al., 2019 |
| Ce3D | Strong |
Minor shrinkage |
N.T. | N.T. | Whole lymph node clearing | Li et al., 2017 |
| Hydration/delipidation | ||||||
| Scale | weak | expansion | N.T. | yes | Adult mouse brain section clearing | Hama et al., 2011 |
| ScaleS | Strong |
Transient expansion |
yes | yes | Mouse hemi‐brain clearing | Hama et al., 2015 |
| CUBIC | Strong | Expansion | no | no |
Whole adult mouse Brain clearing |
Susaki et al., 2014 |
| Updated CUBIC | Very strong | Expansion | no | no |
Whole adult mouse Body clearing |
Tainaka et al., 2018 |
| UbasM | Strong | Expansion | yes | N.T. | Mouse hemi‐brain clearing | Chen et al., 2017 |
Abbreviations: N.T., not tested; TEM, transmission electron microscope.
Simple immersion methods cannot provide highly transparent samples because they do not remove lipids, which are the main source of light scattering. Tissue delipidation could improve clearing efficiency and quality (Tainaka et al., 2016). This can be accomplished with aminoalcohols or detergents like sodium dodecyl sulfate (SDS), saponin, and Triton X‐100. Fast Free‐of‐Acrylamide Clearing Tissue (FACT) was a new SDS‐based clearing method for whole‐tissue imaging (Xu et al., 2017; Wang et al., 2019a). With SDS delipidation, FACT enabled fast tissue clearing and high quality 3D imaging, and has been successfully applied for murine whole‐tissue clearing (Rezazadeh et al., 2018; Khoradmehr et al., 2019). OPTIClear was a novel RI matching solution developed for human brain tissues. With SDS treatment, OPTIClear rendered human brain blocks highly transparent without noticeable structural distortion (Lai et al., 2018). SeeDB2 is a refined version of SeeDB, which utilized iohexol and saponin to improve clearing efficiency (Ke et al., 2016). FOCM was an ultrafast optical clearing method, which could rapidly clear brain slices within 2 min without noticeable tissue deformation and fluorescent toxicity (Zhu et al., 2019). Similarly, clearing‐enhanced 3D (Ce3D) could achieved excellent transparency of most murine organs within 24 h (Li et al., 2017, 2019b). Urea has strong hyperhydration abilities, Miyawaki's group designed the Scale clearing reagent based on urea‐mediated hydration (Hama et al., 2011). Later, they upgraded Scale to improved clearing performance and developed ScaleS, which rendered mouse brain transparent without damaging FPs and subcellular structures (Hama et al., 2015). The urea‐based CUBIC enabled whole‐body clearing and imaging of adult mice by introducing aminoalcohols (Susaki et al., 2014; Tainaka et al., 2014). The CUBIC recipe has been continuously updated to improve clearing performance (e.g., CUBIC‐L/R+ and CUBIC‐X protocol) (Murakami et al., 2018; Tainaka et al., 2018). Urea‐Based Amino‐Sugar Mixture (UbasM) was another effective clearing agent, which rapidly cleared centimeter‐scale samples while preserving FPs and membrane integrity (Chen et al., 2017).
3.3. Hydrogel embedding tissue clearing
Detergents can extract lipids efficiently, but high concentrations of detergents and harsh treatments may result in loss of native biomolecules and damage of tissue architecture. Methods of coupling these biomolecules to tissue‐gel hybrids, such as CLARITY and SWITCH, can avoid these risks (Table 4).
TABLE 4.
Comparison of hydrogel embedding tissue clearing
| Method | Electrophoretic driven | Tissue validation | Clearing rate | Immunostaining | Antigenicity preservation | TEM | References |
|---|---|---|---|---|---|---|---|
| CLARITY | Yes | Whole mouse brain | Fast | Yes | Good | No | Chung et al., 2013 |
| ACT‐PRESTO | Yes | Mouse and rabbit brain | Fast | Yes | Good | No | Lee et al., 2016 |
| PRE‐CLARITY | Yes | Whole mouse brain | Fast | Yes | Good | No | Du et al., 2019 |
| Stochastic electrotransport | Yes | Whole mouse brain | Very fast | Yes | Good | No | Kim et al., 2015 |
| passive CLARITY | No | Whole mouse brain | Slow | Yes | Good | No | Tomer et al., 2014 |
| PACT‐PARS | No | Whole mouse body | Fast | Yes | Good | No | Yang et al., 2014 |
| bone CLARITY | No | Bones of trunk and limbs | Fast | Yes | Good | No | Greenbaum et al., 2017 |
| SWITCH | No | Rat brain human sample | Very fast | Yes | Excellent | No | Murray et al., 2015 |
| SHIELD | No | Mouse brain human block | Very fast | Yes | Excellent | No | Park et al., 2019 |
In CLARITY, the utilization of hydrogel embedding and electrophoretic tissue clearing (ETC) contribute to rapidly achieve uniformly transparent tissues with minimal structural disturbance and biomolecule loss (Chung et al., 2013). Since then many variants of CLARITY have been presented, such as ACT‐PRESTO and PRE‐CLARITY, which modified the ETC process and device to accelerate clearing process and increase the depth and speed of antibody penetration (Lee et al., 2016; Du et al., 2019). Although ETC system facilitated lipid removal, high electric fields needed for fast clearing might cause structural destruction and tissue degradation. This limitation could be addressed by stochastic electrotransport, which accelerated both clearing and staining process of intact tissues while preserving structural integrity (Kim et al., 2015). ETC‐independent approaches like passive CLARITY could also bypass these risks (Tomer et al., 2014). However, the slow clearing rate of passive CLARITY limited its application in whole‐body clearing. Another passive clarity technique (PACT) was then presented, which replaced ETC with perfusion‐assisted agent release in situ (PARS). PACT‐PARS allowed rapid whole‐body clearing and immunolabeling in adult rodents (Yang et al., 2014). Later, modified CLARITY protocols for specific experimental objectives were also proposed, such as bone CLARITY (Greenbaum et al., 2017) for bone tissue clearing, and EDC‐CLARITY for RNA preservation and detection in clarified tissues (Sylwestrak et al., 2016). In addition to acrylamide‐based hydrogel‐tissue hybridization, other gelation mechanisms have also been reported. Chung's group introduced SWITCH technique, which maintained tissue architecture and antigenicity under harsh clearing condition by synchronizing GA‐tissue‐gel formation (Murray et al., 2015). The SWITCH‐mediated SHIELD further preserved key molecular information in intact tissues by using polyfunctional crosslinkers (Park et al., 2019).
4. AVAILABLE MICROSCOPY FOR IMAGING OPTICALLY TRANSPARENT TISSUES
With advances of fluorescent light microscopy, many optical sectioning techniques for 3D imaging have been developed. Of these techniques, two‐photon microscopy is less sensitive to light scattering, thus is best suited for deep imaging in noncleared tissues. However, light scattering is minimized in cleared samples, implying that imaging depth is no longer restricted by light penetration but by microscopy (Richardson and Lichtman, 2015). Therefore, researchers need to reselect the most suited microscopy for their cleared specimens (Table 5). Equipped with optimized objectives (working distance >5 mm, NA >0.9), both confocal and two‐photon microscopy permit 3D cellular imaging of cleared samples at high resolution and contrast (Conchello and Lichtman, 2005; Helmchen and Denk, 2005; Lidke and Lidke, 2012; Richardson and Lichtman, 2015). The main disadvantage of confocal and two‐photon microscopy is the slow scanning speed. Both of them utilize point detector to scan specimens point by point, resulting in limited image acquisition rate (Mertz, 2011). Therefore, they are only suited for high‐resolution imaging of localized region within cleared tissues (Richardson and Lichtman, 2015). Unlike point‐scanning microscopy, light sheet fluorescence microscopy (LSFM) selectively illuminates the entire x‐y plane of interest from the side of samples with a thin light sheet, while region in out‐of‐focal plane is free of excitation, thus significantly reducing photobleaching and background signal (Susaki and Ueda, 2016; Hillman et al., 2019). LSFM enables volumetric imaging at much higher speeds, higher signal‐to‐noise ratios, and lower phototoxicity compared to point‐scanning Methods (Keller and Ahrens, 2015; Hillman et al., 2019). Therefore, LSFM is more practical for whole‐body and whole‐organ imaging. Currently, several variants of LSFM have been developed to facilitate rapid high‐resolution 3D imaging of larger intact tissues, including OCPIM, iSPIM, SCAPE/OPM, and LSTM (Migliori et al., 2018). Stimulated Raman scattering (SRS) microscopy allows fast super‐multiplex optical imaging without histological staining (Wei et al., 2017; Wei et al., 2019). Recently, SRS microscopy was also combined with tissue clearing to facilitate deep volumetric imaging (Wei et al., 2019; Li et al., 2019a).
TABLE 5.
Comparison of microscopy techniques for cleared samples
| Microscopy | Advantage | Disadvantage | Reference |
|---|---|---|---|
| Confocal microscopy | 1. Achieving 3D imaging at high image contrast and resolution; 2. Enabling multispectral imaging without cross‐excitation; 3. Obtaining multicolor confocal images quickly. | 1. Slow scanning speed; 2. The out‐of‐focus planes can also be illuminated during imaging, resulting in phototoxicity and sample photobleaching. | Conchello and Lichtman, 2005; Richardson and Lichtman, 2015 |
| Two‐photon microscopy | 1. Utilizing nonlinear techniques to generate signal contrast, reducing sample photobleaching; 2. Less sensitive to light scattering; 3. Adoption of near‐infrared light enhances tissue penetration and generates less phototoxicity and autofluorescence. | Utilization of laser scanning results in limited image acquisition rate. | Helmchen and Denk, 2005; Mertz, 2011; Lidke and Lidke, 2012 |
| Light sheet microscopy | 1. Allowing volumetric imaging at much higher speeds and signal‐to‐noise ratios; 2. Selective lower power illumination leads to less photobleaching and low phototoxicity. | Inability to image samples with sufficient pixel density at low magnification. | Keller and Ahrens, 2015; Hillman et al., 2019 |
| SRS microscopy | 1. Enabling fast super‐multiplex optical imaging without histological staining; 2. Utilization of nonlinear Raman effects improves the sensitivity and biocompatibility of spontaneous Raman imaging. | The penetration depth of SRS microscopy is limited in highly scattering tissues. | Wei et al., 2017; Wei et al., 2019 |
5. DATA STORAGE AND PROCESSING
Gigabytes or even terabytes of images are produced during large‐scale volumetric imaging. Handling these massive datasets poses a great challenge for both software and hardware. Therefore, a high‐performance workstation is truly necessary for data storage and analysis. The computer used for data processing is advised to maximize the RAM (128 GB or more) and afford multicore CPU as well as professional grade hardware with multiple terabytes. Solid‐state drive (SSD) and graphics processing units (GPU) are also essential (Watson and Watkins, 2019). At present, a number of commercially available software have been exploited to assist researchers in dealing with these large volumetric data, including Imaris, Vision4D, Image‐Pro Plus, and Amira. Some open‐source software packages are also developed, such as Fiji/ImageJ, BigDataViewer, and NeuroTrace (Susaki and Ueda, 2016; Watson and Watkins, 2019).
6. APPLICATIONS OF TISSUE CLEARING IN BIOMEDICAL FIELDS
Tissue clearing technique enables large‐scale volumetric imaging of intact tissues and accelerates our understanding of the complex organisms at the system level. In recent years, tissue clearing technique has been widely applied in biomedical fields.
6.1. Applications in neuroscience
Neuroscience has always been the focus of life science. High‐resolution volumetric imaging is indispensable for mapping the structural and functional connectivity of neurons. Coupled with advanced microscopies, tissue clearing techniques facilitate the elucidation of mechanisms underlying the neuromodulation of proper behavior and contribute to reveal the pathogenesis of neurological diseases.
Ueda's group achieved virtual‐multiplex whole‐brain imaging with single‐cell resolution by combining CUBIC‐L/R+ protocol and fluorescently labeled mouse samples (Tainaka et al., 2018). They also constructed a point‐based mouse brain atlas with single‐cell annotation (CUBIC‐Atlas) using CUBIC‐X expansion protocol and customized LSFM (Figure 5A). CUBIC‐Atlas enabled whole‐brain cell profiling and revealed inhomogeneous whole‐brain development. Probabilistically mapping of pharmacologically activated cells onto CUBIC‐Atlas uncovered a novel functional subarea in the hippocampal dentate gyrus. CUBIC‐Atlas provided a new platform for 3D brain mapping and for unbiased system‐level cellular analysis (Murakami et al., 2018). They then presented a whole‐organ cell profiling pipeline by combining advanced CUBIC protocol, customized LSFM, and cell‐nucleus‐detection algorithm. With this protocol, they recounted the cell number of the adult whole mouse brain, and quantified the whole‐brain infection efficiency of AAV‐PHP.eB (NSE‐H2B‐mCherry) (Matsumoto et al., 2019). CUBIC was also applied to trace spatial projections of sweet and bitter cortical neurons in mouse brains (Wang et al., 2018). Liebmann et al. studied plaque formation in Alzheimer's disease (AD) mouse model using iDISCO. They demonstrated 3D spatial distribution of beta‐amyloid plaques together with microglia and vasculature (Figure 5B). The combination of volume imaging and ClearMap analysis allowed them to accurately quantify the amyloid plaques within the intact AD mouse brain. They also evaluated the 3D amyloid patterns (TAPs) in archived human brain tissues, which exhibited large variation and complexity compared to those in mouse models (Liebmann et al., 2016). Hildebrand's group introduced a novel cytoarchitectonic labeling approach for cleared human samples, termed MASH (Multiscale Architectonic Staining of Human cortex). Combining MASH and modified DISCO protocol, they achieved optical clearing and labeling of 5‐mm‐thick archival adult human brain samples (formalin fixed for 14 to 30 months). Furthermore, they characterized the 3D cortical architecture, and classified cytoarchitectonic layers over the entire cortical depth at cellular resolution (Hildebrand et al., 2019).
FIGURE 5.
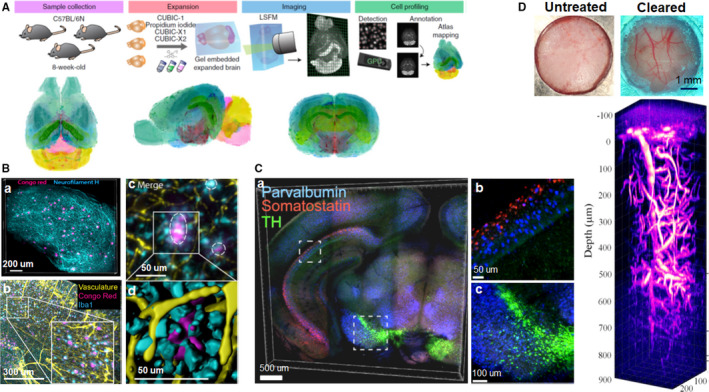
Applications of tissue clearing in neuroscience. (A) Overview of construction of mouse brain atlas with single‐cell annotation (CUBIC‐Atlas), and the volume‐rendered images of the CUBIC‐Atlas from the horizontal, sagittal, and coronal view. (B) 3D visualization of amyloid plaques in AD using iDISCO clearing. a, Amyloid plaques and neurofilament H imaging in a cleared cortex slice of 10‐month‐old 2xTg AD mouse (500 μm thick). b, Maximum projection of vasculature, amyloid plaques, and microglia staining from an 11‐month‐old 2xTg AD mouse brain (1 mm thick). The insert is the magnification of the indicated region (white rectangle). c, The magnified optical section from the AD brain in b, the circles indicate the plaques surrounded by reactive microglia and vessels. d, The magnified surface render in c. (C) Multiplexed detection of mRNAs in EDC‐CLARITY. a, Multiplexed in situ hybridization of coronal section (0.5 mm thick) using somatostatin (red), parvalbumin (blue), and tyrosine hydroxylase (green) probe sets. b, c, Magnified view of indicated boxes. (D) 3D imaging of mouse brain vasculature labeled with Texas Red through the transparent skull window. Images adapted from Refs (Liebmann et al., 2016; Sylwestrak et al., 2016; Murakami et al., 2018; Chen et al., 2019)
To achieve volumetric nucleic acid labeling of large tissue volumes, Sylwestrak et al. developed a technique using carbodiimide compound (EDC)‐based chemistry for enhanced and stable RNA preservation in CLARITY‐cleared tissue. Coupled with DNA‐based amplification system, this technique enabled robust detection of diverse coding RNA variants and noncoding RNAs in clarified mouse and human tissues, and also allowed multiplexed RNA labeling in EDC‐CLARITY tissue (Figure 5C) (Sylwestrak et al., 2016). Morawski's group presented a tailored histological method for hMRI validation based on adapted CLARITY and optical 3D imaging. They achieved overall transparency of postmortem human brain tissue with the thickness of up to 8 mm, and achieved efficient immunostaining of 5‐mm‐thick human postmortem brain tissue. With customized image processing tools, they demonstrated cell type classification, fiber orientation, and spatial distribution of pyramidal and nonpyramidal cells within the cortex. Combined with MRI, this tailored histological approach provided a promising tool for validating MRI‐based histology in humans (Morawski et al., 2018).
Noninvasive imaging of cerebral vasculature and neurocytes in the living brain is an indispensable method to investigate cerebrocortical structure and function. Using skull optical clearing agents (OCAs), Zhu's group developed a transparent skull window to study cortical vessels without craniotomy. Combined with several optical imaging techniques, the skull optical clearing window (SOCW) not only enhanced imaging contrast and depth but also allowed monitoring the cortical hemodynamic changes after middle cerebral artery occlusion surgery (Zhang et al., 2018). In addition, they utilized the SOCW technique to repeatedly image dendritic spines, microglia processes, and microvasculature in the superficial cortical layers. They also applied this method to monitor the plastic changes of dendrites and microglia after laser ablation (Zhao et al., 2018). With hyperspectral SRS imaging of a mouse skull, Chen et al. developed an optimized clearing strategy, which converted the turbid skull into an optically transparent skull window. Coupled with three‐photon microscope, the optimal clearing method enabled 3D imaging of mouse vascular structures at a depth of 850 μm beneath the cleared skull window (Figure 5D) (Chen et al., 2019). Overall, skull optical clearing technique provided an alternative way for imaging cortical structures in vivo without craniotomy, rendering a better understanding of pathophysiologic processes of neurological disorders.
6.2. Applications in soft tissue organs
Tissue clearing has also been widely applied in soft tissues, such as vasculature, kidney, pancreas, and organoids. To characterize the neuro‐insular network of pancreas, Tang's group prepared transparent pancreas of young mice with optical clearing agent (RapiClear). They achieved 3D panoramic visualization of pancreatic microstructure, islet distribution, neurovascular network, and neural network remodeling in disease (Tang et al., 2018). Fowler et.al utilized the modified transparent tissue tomography (T3) to generate 3D reconstructions of entire human pancreas. They visualized islet cellular composition and architecture as well as native pancreatic microenvironment, including ducts, capillaries, and the extracellular matrix. They also exhibited that intraislet capillary density was positively related to the islet size while islet sphericity was negatively related to the islet size (Fowler et al., 2018). Cao et al. presented a 3D volume fluorescence‐imaging protocol to visualize the entire vascular network in mouse adipose tissues by combining 3DISCO/iDISCO clearing, immunolabeling, and LSFM imaging. They evaluated the pathological remodeling of intra‐adipose vasculatures under obese conditions. In addition, they assessed vascular plasticity in white adipose tissues (WATs) in response to a cold challenge (Figure 6A), and found that the cold‐induced vascular plasticity was regulated by the sympathetic‐derived catecholamine signal and was associated with beiging process of WAT (Cao et al., 2018). Using CUBIC clearing protocol, Hasegawa et.al achieved comprehensive 3D imaging of adult mouse kidney. They visualized the spatial distribution of sympathetic nerves (Figure 6B), and revealed 3D structure of collecting duct, early segments of proximal tubules, and glomeruli of mouse kidney. Moreover, they confirmed sympathetic denervation and norepinephrine decrease after ischemia‐reperfusion injury (Hasegawa et al., 2019). CUBIC‐kidney provided a macroscopic perspective for kidney research. Mzinza et al. introduced 3DISCO/iDISCO and LSFM to assess bronchus‐associated lymphoid tissue (BALT) in mice. They demonstrated qualitative and quantitative analysis of BALT induced by modified vaccinia virus Ankara (MVA) in whole mouse lung lobes, and characterized the development and organization of heat‐inactivated E. coli‐induced BALT (Mzinza et al., 2018). Vargas's team combined optimized BABB clearing and large‐scale 3D microscopy to study murine lung fibrosis. They revealed 3D fibrillar collagen network in whole murine lungs at high resolution, and quantified collagen deposition at the airway level throughout the entire lungs (Ochoa et al., 2018).
FIGURE 6.
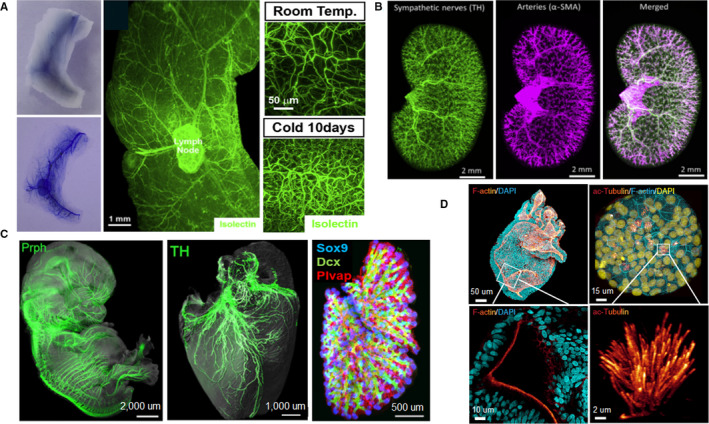
Applications of tissue clearing in soft tissue organs. (a)Tridimensional imaging of the vasculatures in white adipose tissues (WAT) after optical clearing, and the vascular plasticity in WAT (right) in response to cold challenge. (b) Spatial distribution of sympathetic nerves (TH, green) and arteries (a‐SMA, magenta) in the kidney. (c) 3D Analysis of the developing peripheral nerves, heart, and lung in solvent‐cleared human embryos. (d) 3D whole‐mount images (top) and magnified view (bottom) of human colonic and airway organoids. Images adapted from Refs (Belle et al., 2017; Cao et al., 2018; Hasegawa et al., 2019; Dekkers et al., 2019)
An accurate atlas of human embryo is indispensable for better understanding of embryogenesis and developmental biology under healthy and pathological conditions. Belle et al. combined 3DISCO/iDISCO+clearing, immunostaining, and LSFM to build a 3D cell atlas of human development during early gestation (Belle et al., 2017). They generated high‐resolution 3D images of human embryos by performing whole‐mount immunostaining on 36 human embryos and fetuses with over 70 antibodies. Furthermore, they demonstrated the dynamic development of peripheral nervous, vascular, muscular, cardiopulmonary, and urogenital systems during the first trimester of gestation (Figure 6C). Organoid systems reproduce cellular organization and architectural microenvironment of biological tissues, and have proved to be a powerful tool for modeling diseases and developing personalized or regenerative medicine (Rios and Clevers, 2018; Costa et al., 2019). Rios's group presented a simple clearing protocol based on fructose–glycerol for 3D visualization of entire organoids (Figure 6D). They achieved high‐resolution volumetric imaging of various intact organoids, including human colon, airway, fetal liver, breast tumor, as well as mouse small intestine, and mammary gland organoids (Dekkers et al., 2019). Cora et al. optimized the passive clearing technique (PACT) for 3D imaging of intact retinal organoids. Using cleared retinal organoids, they demonstrated the fine morphology and structural organization of photoreceptor cells and bipolar cell layers. They also visualized neural processes and synaptic interconnections of photoreceptor cells in three dimensions (Cora et al., 2019).
6.3. Applications in hard tissue organs
Hard tissues like bones and teeth contain rich minerals and low lipids. Thus, tissue clearing techniques specially developed for soft tissues are not applicable to hard tissues. Recently, several optimized tissue clearing protocols have been presented to clarify hard tissues.
Morrison's team modified Murray's clearing to clear bisected long bones. They obtained deep imaging of bone marrow to a depth of up to 600 um using confocal microscopy, and identified the location of dividing and nondividing haematopoietic stem cells throughout the bone marrow (Acar et al., 2015). Greenbaum et al. introduced Bone CLARITY to detect Sox9‐tdTomato+osteoprogenitor cells in the bone tissues of adult transgenic mice (Figure 7A). Coupled with a customized LSFM and the data processing pipeline, Bone CLARITY mapped 3D spatial distribution of Sox9‐tdTomato+osteoprogenitor cells in their native microenvironment, and detected their responses to sclerostin antibody treatment. Bone CLARITY provided a platform for studying the physiological processes of intact bone tissues in healthy and disease (Greenbaum et al., 2017). Wang et al. developed BoneClear method for robust clearing and immunolabeling of intact mouse bone tissues. With BoneClear, they fully cleared mouse intact femurs and unsectioned vertebral column and hindpaws. They also achieved 3D visualization of neural anatomy and pathology with whole‐tissue immunolabeling of mouse femurs and hindpaws (Wang et al., 2019b).
FIGURE 7.
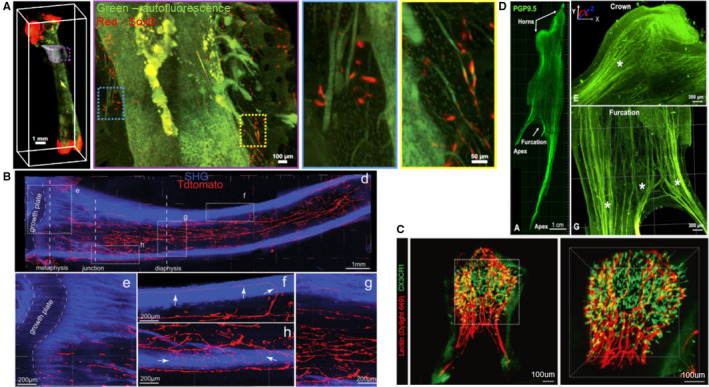
Applications of tissue clearing in hard tissue organs. (a) Maximum intensity projection (MIP) fluorescence image of the femur revealing Sox9+ cells distribution in the vicinity of the third trochanter. Dotted boxed regions in the MIP represent progressive magnification. (b) Spatial distribution of nerves and arteries within the marrow space of the tibia. Dotted areas are magnified in e‐g. Arrows in f and h show nerve fibers penetrating the cortical bone. SHG, second harmonic generation signal. (c) Volumetric imaging of immune cell distributions in an upper third molar from a CX3CR1‐GFP transgenic mouse, with lectin labeled (red) endothelial cells in blood vessels. Maximal intensity Z‐projection image (left) and 3D reconstruction image (right) in the dotted area. (d) Tridimensional innervation of a human premolar (left) and high‐magnification images (right) of distinctive nerve splitting patterns in coronal and furcation direction. Images adapted from Refs (Greenbaum et al., 2017; Jing et al., 2018; Franca et al., 2019; Hong et al., 2019)
Zhao's group designed a PEGASOS clearing protocol, which could cleared almost all types of tissues including brain, liver, bones, and teeth (Figure 7B) (Jing et al., 2018). They applied the PEGASOS method to investigate the dental implant‐bone interface and achieved multichannel 3D imaging of implant‐tissue interface at high resolution. They revealed that angiogenesis process occurred at both titanium and stainless steel implant‐bone interfaces, while osseointegration only progressed surrounding titanium implants. The PEGASOS method provided a useful tool for studying implant‐tissue interface (Yi et al., 2019). Later, they combined the PEGASOS‐based 3D imaging and various labeling strategies to study mouse postnatal craniofacial bone development. They demonstrated a strong spatial association between craniofacial osteogenesis and angiogenesis during the entire postnatal development. Additionally, Gli1+ mesenchymal stem cells within sutures were also closely coupled with osteogenic activity and vasculature in adult craniofacial bones (Luo et al., 2019).
Dental pulp plays a significant role in maintaining tooth integrity and function. Extensive efforts have been made to investigate the structure and function relationships of the dental pulp tissues. França et al. applied CLARITY to study the neurovascular components of human dental pulp. They achieved 3D visualization of entire neuronal and vascular networks in human dental pulp (Figure 7C). Furthermore, they characterized the complex morphological interconnection of the dental innervation and vasculature, which improved the understanding of physiologic and pathologic conditions affecting the teeth (Franca et al., 2019). Hong et.al modified the Murray's clearing approach for rapid clearing of the intact mouse teeth. They achieved 3D cellular visualization of the entire microvasculature as well as 3D distribution of immune cells inside the mouse dental pulp (Figure 7D). In addition, they visualized the recruitment of granulocytes and microvasculature changes in a carious tooth at cellular level (Hong et al., 2019). To better understand the auditory perception and transduction, Okabe's group constructed 3D cellular mapping of the organ of Corti by combining modified ScaleS clearing protocol and machine learning‐based automatic cell detection. They achieved high‐fidelity recording and analysis of hair cells throughout the organ of Corti, including density, position, and morphology. They also evaluated subcellular components and structural integrity under normal and pathological conditions, uncovering different effects of aging and noise on spatial characteristics of hair cell loss (Urata et al., 2019).
6.4. Applications in tumor research
Solid tumor metastasis determines the survival of cancer patients. To overcome tumor metastasis, it is imperative to detect the metastatic progression and microenvironments of tumor cells at early stages. Ueda's group applied CUBIC‐based cancer (CUBIC‐cancer) analysis to investigate cancer metastasis within 13 mouse models using nine cancer cell lines (Figure 8A). CUBIC‐cancer analysis enabled comprehensive spatiotemporal profiling of cancer metastasis at single‐cell resolution throughout the mouse body, and could distinguish different patterns of metastasis formation. In addition, CUBIC‐cancer analysis enabled therapeutic evaluation of antitumor drugs on cancer metastasis and bridged the spatial resolution gap between in vivo imaging and 2D histology. CUBIC‐cancer analysis provided a novel platform for analyzing cancer metastasis throughout the body (Kubota et al., 2017). Targeted delivery of nanoparticles to micrometastases could offer a way to stop tumor growth at early phases. Kingston et al. combined tissue clearing, 3D microscopy and machine learning‐based automated image analysis to analyze nanoparticle interaction with micrometastases. They assessed the physiological features of micrometastases and track the delivery, penetration, and distribution of nanoparticles within individual micrometastases. Using a machine learning algorithm, they could predict nanoparticle delivery to metastatic tumors based on their physiology. This technique enabled personalized treatment for patients based on the specific physiology of micrometastases (Kingston et al., 2019).
FIGURE 8.
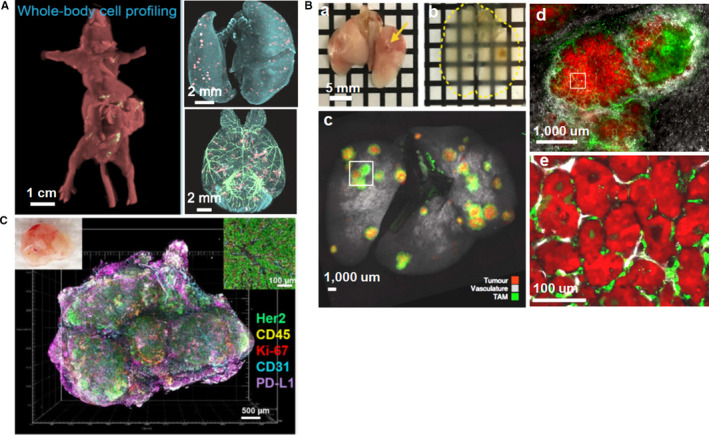
Applications of tissue clearing in tumor research. (A) Whole‐body and whole‐organ (lung and brain) imaging of cancer metastasis at single‐cell resolution. (B) Whole lung clearing allows identification of tumors and visualization of tumor burden. a‐b, Whole lung clearing and identification of small tumors (small arrows); c, Whole lung imaging of KP tumour‐bearing mouse; d, Magnification of a tumor from box in c; e, Magnification imaging from box in d. (C) 3D reconstruction image of multiple tumor immune microenvironment components and biomarkers in an intact mammary tumor from a BALBNeuT mouse. Images adapted from Refs (Cuccarese et al., 2017; Kubota et al., 2017; Lee et al., 2017)
Investigation of heterogeneity is crucial for tumor diagnosis and treatment, because it is associated with carcinogenesis, metastasis, and drug tolerance (Waclaw et al., 2015). Integrating the iDISCO protocol and LSFM, Tanaka et al. developed a tumor‐phenotyping pipeline to study and diagnose human formalin‐fixed paraffin‐embedded (FFPE) tumors in three dimensions. With this pipeline, they achieved whole‐tissue phenotyping of both mouse and human FFPE tumors. They demonstrated the inner landscape and spatial features of the solid tumors, and revealed distinct patterns of cancer heterogeneity (Tanaka et al., 2017). Investigation of tumour immune microenvironment provides much valuable information on tumor biology. Cuccarese et al. used tissue clearing technique and whole lung imaging protocol to study the spatial distribution of tumor‐associated macrophages (TAM) and their responses to drug delivery in mouse pulmonary carcinoma. They analyzed lung tumor burden and visualized TAM characteristics at cellular resolution (Figure 8B). Using different pulmonary tumor models, they revealed notable heterogeneity of TAM density and infiltration, and demonstrated therapeutic response to different drug delivery. These findings facilitate the study of tumor and host‐cell heterogeneity in the entire organs (Cuccarese et al., 2017).
Lee et al. presented transparent tissue tomography (T3) for 3D mapping and analysis of the breast tumor microenvironment within Her2 transgenic mouse models (Figure 8C). Using T3 protocol, they revealed heterogeneous expression of programmed death‐ligand 1 (PD‐L1) throughout the tumors, and demonstrated the heterogeneity of tumor immune microenvironment by simultaneously evaluating multiple tumor parameters (Lee et al., 2017). Afterwards, they applied T3 to track macromolecular drug delivery in mouse mammary tumors. They determined the unique distributions and spatial pharmacokinetics of anti‐PD‐L1 antibody in different tumor microenvironments (Lee et al., 2019). To predict drug uptake and therapeutic responses in tumors, Esposito et al. present a framework by combining mathematical modelling, tissue clearing and in vivo imaging techniques. They applied this framework to simulate steady‐state fluid dynamics and the delivery of MRI contrast agent Gd‐DTPA, and model drug uptake and response to OXi4503 treatment. Their results validated the feasibility and accuracy of this framework and demonstrated the interactions between individual tumor and therapeutic agents (d'Esposito et al., 2018).
7. DISCUSSION
Tissue clearing technique enables high‐resolution volumetric imaging and contribute to reveal anatomical and functional connectivity within intact organisms, thus providing a powerful tool for systematical understanding of the complicated biological systems at multiple levels. In recent years, a variety of tissue clearing techniques have sprung up and each has distinct applications.
The major advantage of organic solvent‐based clearing is the high clearing efficiency and quality, making it more suited for clearing larger tissue volumes. Both uDISCO and PEGASOS protocols render the whole adult rodent body highly transparent and preserve endogenous fluorescent proteins, thus they are the first choice for whole‐body and whole‐organ clearing (Pan et al., 2016; Jing et al., 2018). Aqueous reagent‐based clearing methods like updated CUBIC protocol also efficiently clear the entire mouse body as well as large primate and human tissues through decalcification and decolorization (Tainaka et al., 2018). Moreover, the hydrogel embedding‐based PACT‐PARS and ACT‐PRESTO can render whole mouse body transparent (Yang et al., 2014; Lee et al., 2016). However, the limited clearing quality and the need for special equipment restrict their applications in whole‐body clearing. Lipophilic dyes (e.g., DiI and CM‐Dil) are commonly used neural tracers and play an important role in the study of neural circuits. Solvent‐based clearing methods are not compatible with lipophilic tracers because the membrane integrity is destroyed during delipidation step. Passive immersion clearing methods, such as SeeDB, FRUIT, and Scale (Hama et al., 2011; Ke et al., 2013; Hou et al., 2015), are simple, economical, and have a good compatibility with lipophilic tracers, yet their clearing efficiency is not as high as solvent‐based or other aqueous‐based clearing methods. Therefore, they are only applicable to clear lipophilic tracer‐labeled tissue slices. By contrast, SWITCH and ScaleS, which clear tissues with high transparency while preserving membrane integrity (Hama et al., 2015; Murray et al., 2015), are more suited for clearing larger samples labeled with lipophilic dyes. It is critical to preserve the structural and molecular integrity of samples during the clearing process. Gel‐embedding clearing procedures, such as CLARITY, PACT‐PARS, SWITCH, and SHIELD (Chung et al., 2013; Yang et al., 2014; Murray et al., 2015; Park et al., 2019), preserve fluorescent proteins, tissue architectures, antigenicity, and transcripts through covalent cross‐linking, thus can be applied to RNA transcripts detection and ultrastructure analysis. However, they are unable to fully clear hard tissues like bones and teeth. Accordingly, no individual clearing technique is ideally suited for all applications, researchers need to select the best‐suited clearing scheme for their research objectives, considering clearing efficiency, key information preservation, as well as type and volume of tissues.
Despite its rapid development, tissue clearing technique is still facing some challenges that limit its large‐scale application in large‐scale volumetric imaging. First, whole human organ clearing has always been challenging, especially for the adult brain tissue. Tissue clearing techniques dedicatedly developed for rodent samples exhibits limited capacity in clearing human organs, because of the difference of tissue size and physiochemical properties between rodents and humans. Although with much efforts, tissue transparency was limited to small pieces of human organs (Lai et al., 2018). Fortunately, the newly developed clearing method SHANEL achieved clearing and labeling of whole adult human brain, paving the way for charting the cellular and molecular architecture of intact adult human organs (Zhao et al., 2020). However, the clearing speed of SHANEL is slightly slow, the total processing time for one adult human brain is up to 4 months, and the cost is relatively high. In the future, nontoxic, inexpensive and more effective clearing reagents should be developed to improve clearing efficiency in human fresh biopsy specimens and post mortem tissues. Second, low antibody penetration hinders the wide adoption of volumetric immunolabeling and imaging. The large molecular weight of antibody restricts its rapid penetration throughout the tissues, resulting in extremely long incubation times and nonuniform tissue staining. Although clearing methods such as AbScale, PACT‐PARS, ACT‐PRESTO, Ce3D (Yang et al., 2014; Hama et al., 2015; Lee et al., 2016; Li et al., 2017) have improved antibody penetration deep inside the tissue, the immunostaining depth is confined to several millimeters. Therefore, it is urgently needed to enhance antibody tissue penetration to achieve whole‐body and whole‐organ immunolabelling. Use of Fab antibody fragments, nucleic acid aptamers, or nanobodies should facilitate antibody diffusion and staining homogeneity (Richardson and Lichtman, 2015). vDISCO adopted nanobodies for whole‐body immunolabeling and achieved panoptic imaging of neuronal projections using transparent Thy1‐GFP mice (Cai et al., 2019). However, the current nanobodies are designed mainly for in vivo applications but not for fixed samples, and the types of available nanobodies are limited at present. Therefore, it is imperative to produce diverse nanobodies for tissue volume immunolabeling in the future. Furthermore, microwave or electrophoresis technology may also promote 3D immunolabeling of intact organisms (Ferris et al., 2009; Cheng and Xie, 2015). Third, high‐resolution deep imaging is challenging. LSFM is an attractive choice for volumetric 3D imaging due to high‐speed image acquisition, good signal‐to‐noise ratios, and low phototoxicity. However, LSFM employs low NA objectives to provide a wide field of view, thus the spatial resolution is poor, especially the axial resolution. Reflective imaging technique has been developed to improve spatiotemporal resolution of LSFM (Wu et al., 2017). Moreover, specialized objective lenses with high NA and longer working distance are also crucial for LSFM to image intact organisms at subcellular resolution. Apart from imaging resolution, the subsequent data processing is also troublesome. Dedicated workstations for quantitative analysis of extremely large datasets are urgently needed. Currently, a variety of commercial and open‐source software packages have been exploited to handle these datasets. However, each of them concentrates on particular challenges. A computational framework possessing comprehensive imaging analysis tools should be developed, for better processing and understanding these massive datasets. Finally, tissue clearing techniques have been widely used for ex vivo tissues, but they are still in their infancy about in vivo applications. To date, in vivo optical transparency is only achieved in the skin and skull bone (Inyushin et al., 2019). The optical clearing agents (OCAs) developed for ex vivo tissues are usually toxic, chemical aggressive, and irreversible. The safety and biocompatibility issues limit applications of OCAs in living organisms. At present, the OCAs used for in vivo biological tissues are confined to aqueous solutions, such as glycerol, sugars, propylene glycol, or polyethylene glycol (Costantini et al., 2019). In vivo tissue optical clearing shows enormous potential for in vivo biomedical imaging, clinical diagnosis, and therapy. Much efforts need to be made to eliminate the potential detrimental effects of OCAs and develop safe optical clearing methods for in vivo applications on humans.
Undoubtedly, tissue clearing technique will continue to evolve in the near future. With advances in novel clearing reagents, 3D immunolabeling, volumetric imaging, and massive dataset analysis, tissue clearing will have a broader range of applications and pave the way for biomedical scientists to achieve whole‐body cell profiling.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Ting Tian designed the content of this review and drafted the manuscript. Zhaoyang Yang and Xiaoguang Li revised the manuscript. Xiaoguang Li approved the manuscript.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (Grant 31730030), and Ministry of Science and Technology of China (Grants 2017YFC1104001, 2017YFC1104002), and Beijing Science and Technology Program (Grant Z181100001818007), and National Natural Science Foundation of China (Grants 31670988, 31971279), and Beijing Natural Science Foundation Program (Grant KZ201810025030), and National Natural Science Foundation of China (Grant 31771053, 31900749, 31970970, 31900980, 31320103903).
Tian T, Yang Z, Li X, et al. Tissue clearing technique: Recent progress and biomedical applications. J Anat.2021;238:489–507. 10.1111/joa.13309
REFERENCES
- Acar, M. , Kocherlakota, K.S. , Murphy, M.M. , Peyer, J.G. , Oguro, H. , Inra, C.N. et al (2015) Deep imaging of bone marrow shows non‐dividing stem cells are mainly perisinusoidal. Nature, 526, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alnuami, A.A. , Zeedi, B. , Qadri, S.M. & Ashraf, S.S. (2008) Oxyradical‐induced GFP damage and loss of fluorescence. International Journal of Biological Macromolecules, 43, 182–186. 10.1016/j.ijbiomac.2008.05.002 [DOI] [PubMed] [Google Scholar]
- Belle, M. , Godefroy, D. , Couly, G. , Malone, S.A. , Collier, F. , Giacobini, P. et al (2017) Tridimensional visualization and analysis of early human development. Cell, 169, 161–173. 10.1016/j.cell.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Briggman, K.L. , Helmstaedter, M. & Denk, W. (2011) Wiring specificity in the direction‐selectivity circuit of the retina. Nature, 471, 183–188. 10.1038/nature09818 [DOI] [PubMed] [Google Scholar]
- Cai, R. , Pan, C. , Ghasemigharagoz, A. , Todorov, M.I. , Forstera, B. , Zhao, S. et al (2019) Panoptic imaging of transparent mice reveals whole‐body neuronal projections and skull‐meninges connections. Nature Neuroscience, 22, 317–327. 10.1038/s41593-018-0301-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y. , Wang, H. , Wang, Q. , Han, X. & Zeng, W. (2018) Three‐dimensional volume fluorescence‐imaging of vascular plasticity in adipose tissues. Mol Metab, 14, 71–81. 10.1016/j.molmet.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Li, G. , Li, Y. , Li, Y. , Zhu, H. , Tang, L. et al (2017) UbasM: An effective balanced optical clearing method for intact biomedical imaging. Scientific Reports, 7, 12218 10.1038/s41598-017-12484-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Liu, S. , Liu, H. , Tong, S. , Tang, H. , Zhang, C. et al (2019) Coherent Raman scattering unravelling mechanisms underlying skull optical clearing for through‐skull brain imaging. Analytical Chemistry, 91, 9371–9375. 10.1021/acs.analchem.9b02624 [DOI] [PubMed] [Google Scholar]
- Cheng, J.X. & Xie, X.S. (2015) Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science, 350(6264), aaa8870 10.1126/science.aaa8870 [DOI] [PubMed] [Google Scholar]
- Chung, K. , Wallace, J. , Kim, S.Y. , Kalyanasundaram, S. , Andalman, A.S. , Davidson, T.J. et al (2013) Structural and molecular interrogation of intact biological systems. Nature, 497, 332–337. 10.1038/nature12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conchello, J.A. & Lichtman, J.W. (2005) Optical sectioning microscopy. Nature Methods, 2, 920–931. 10.1038/nmeth815 [DOI] [PubMed] [Google Scholar]
- Cora, V. , Haderspeck, J. , Antkowiak, L. , Mattheus, U. , Neckel, P.H. , Mack, A.F. et al (2019) A cleared view on retinal organoids. Cells, 8, 391 10.3390/cells8050391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, E.C. , Silva, D.N. , Moreira, A.F. & Correia, I.J. (2019) Optical clearing methods: An overview of the techniques used for the imaging of 3D spheroids. Biotechnology and Bioengineering, 116, 2742–2763. 10.1002/bit.27105 [DOI] [PubMed] [Google Scholar]
- Costantini, I. , Cicchi, R. , Silvestri, L. , Vanzi, F. & Pavone, F.S. (2019) In‐vivo and ex‐vivo optical clearing methods for biological tissues: review. Biomed Opt Express, 10, 5251–5267. 10.1364/BOE.10.005251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuccarese, M.F. , Dubach, J.M. , Pfirschke, C. , Engblom, C. , Garris, C. , Miller, M.A. et al (2017) Heterogeneity of macrophage infiltration and therapeutic response in lung carcinoma revealed by 3D organ imaging. Nature Communications, 8, 14293 10.1038/ncomms14293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Esposito, A. , Sweeney, P.W. , Ali, M. , Saleh, M. , Ramasawmy, R. , Roberts, T.A. et al (2018) Computational fluid dynamics with imaging of cleared tissue and of in vivo perfusion predicts drug uptake and treatment responses in tumours. Nat Biomed Eng, 2, 773–787. 10.1038/s41551-018-0306-y [DOI] [PubMed] [Google Scholar]
- Dekkers, J.F. , Alieva, M. , Wellens, L.M. , Ariese, H.C.R. , Jamieson, P.R. , Vonk, A.M. et al (2019) High‐resolution 3D imaging of fixed and cleared organoids. Nature Protocols, 14, 1756–1771. 10.1038/s41596-019-0160-8. [DOI] [PubMed] [Google Scholar]
- Denk, W. , Strickler, J.H. & Webb, W.W. (1990) Two‐photon laser scanning fluorescence microscopy. Science, 248, 73–76. 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Dodt, H.U. , Leischner, U. , Schierloh, A. , Jahrling, N. , Mauch, C.P. , Deininger, K. et al (2007) Ultramicroscopy: Three‐dimensional visualization of neuronal networks in the whole mouse brain. Nature Methods, 4, 331–336. 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- Du, H. , Hou, P. , Wang, L. , Wang, Z. & Li, Q. (2019) Modified CLARITY achieving faster and better intact mouse brain clearing and immunostaining. Scientific Reports, 9, 10571 10.1038/s41598-019-46814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erturk, A. , Becker, K. , Jahrling, N. , Mauch, C.P. , Hojer, C.D. , Egen, J.G. et al (2012) Three‐dimensional imaging of solvent‐cleared organs using 3DISCO. Nature Protocols, 7, 1983–1995. 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- Ferris, A.M. , Giberson, R.T. , Sanders, M.A. & Day, J.R. (2009) Advanced laboratory techniques for sample processing and immunolabeling using microwave radiation. Journal of Neuroscience Methods, 182, 157–164. 10.1016/j.jneumeth.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Feuchtinger, A. , Walch, A. & Dobosz, M. (2016) Deep tissue imaging: a review from a preclinical cancer research perspective. Histochemistry and Cell Biology, 146, 781–806. 10.1007/s00418-016-1495-7. [DOI] [PubMed] [Google Scholar]
- Fowler, J.L. , Lee, S.S. , Wesner, Z.C. , Olehnik, S.K. , Kron, S.J. & Hara, M. (2018) Three‐dimensional analysis of the human pancreas. Endocrinology, 159, 1393–1400. 10.1210/en.2017-03076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca, C.M. , Riggers, R. , Muschler, J.L. , Widbiller, M. , Lococo, P.M. , Diogenes, A. et al (2019) 3D‐imaging of whole neuronal and vascular networks of the human dental pulp via CLARITY and light sheet microscopy. Scientific Reports, 9, 10860 10.1038/s41598-019-47221-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth, D. , Vaissiere, T. , Tzortzi, O. , Xuan, Y. , Martin, A. , Lazaridis, I. et al (2018) An interactive framework for whole‐brain maps at cellular resolution. Nature Neuroscience, 21, 139–149. 10.1038/s41593-017-0027-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, L.W. (2007) Principles of CT and CT technology. J Nucl Med Technol, 35, 115–128. 10.2967/jnmt.107.042978. [DOI] [PubMed] [Google Scholar]
- Greenbaum, A. , Chan, K.Y. , Dobreva, T. , Brown, D. , Balani, D.H. , Boyce, R. et al (2017) Bone CLARITY: Clearing, imaging, and computational analysis of osteoprogenitors within intact bone marrow. Science Translational Medicine, 9, eaah6518. [DOI] [PubMed] [Google Scholar]
- Hahn, C. , Becker, K. , Saghafi, S. , Pende, M. , Avdibašić, A. , Foroughipour, M. et al (2019) High resolution imaging of fluorescent whole mouse brains using stabilised organic media (sDISCO). Journal of Biophotonics, 12, e201800368 10.1002/jbio.201800368. [DOI] [PubMed] [Google Scholar]
- Hama, H. , Hioki, H. , Namiki, K. , Hoshida, T. , Kurokawa, H. , Ishidate, F. et al (2015) ScaleS: an optical clearing palette for biological imaging. Nature Neuroscience, 18, 1518–1529. 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- Hama, H. , Kurokawa, H. , Kawano, H. , Ando, R. , Shimogori, T. , Noda, H. et al (2011) Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nature Neuroscience, 14, 1481–1488. 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Hamaide, J. , Groof, G.D. & Linden, A.V.D. (2016) Neuroplasticity and MRI: A perfect match. NeuroImage, 131, 13–28. 10.1016/j.neuroimage.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Hasegawa, S. , Susaki, E.A. , Tanaka, T. , Komaba, H. , Wada, T. , Fukagawa, M. et al (2019) Comprehensive three‐dimensional analysis (CUBIC‐kidney) visualizes abnormal renal sympathetic nerves after ischemia/reperfusion injury. Kidney International, 96, 129–138. 10.1016/j.kint.2019.02.011. [DOI] [PubMed] [Google Scholar]
- Helmchen, F. & Denk, W. (2005) Deep tissue two‐photon microscopy. Nature Methods, 2, 932–940. 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Hildebrand, S. , Schueth, A. , Herrler, A. , Galuske, R. & Roebroeck, A. (2019) Scalable labeling for cytoarchitectonic characterization of large optically cleared human neocortex samples. Scientific Reports, 9, 10880 10.1038/s41598-019-47336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman, E.M.C. , Voleti, V. , Li, W. & Yu, H. (2019) Light‐sheet microscopy in neuroscience. Annual Review of Neuroscience, 42, 295–313. 10.1146/annurev-neuro-070918-050357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S. , Lee, J. , Kim, J.M. , Kim, S.Y. , Kim, H.R. & Kim, P. (2019) 3D cellular visualization of intact mouse tooth using optical clearing without decalcification. International Journal of Oral Science, 11, 25 10.1038/s41368-019-0056-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, B. , Zhang, D. , Zhao, S. , Wei, M. , Yang, Z. , Wang, S. et al (2015) Scalable and DiI‐compatible optical clearance of the mammalian brain. Frontiers in Neuroanatomy, 9, 19 10.3389/fnana.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inyushin, M. , Meshalkina, D. , Zueva, L. & Zayas‐Santiago, A. (2019) Tissue transparency in vivo. Molecules, 24, 2388 10.3390/molecules24132388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, D. , Zhang, S. , Luo, W. , Gao, X. , Men, Y. , Ma, C. et al (2018) Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Research, 28, 803–818. 10.1038/s41422-018-0049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen, S. & Widder, E.A. (1999) The physical basis of transparency in biological tissue: ultrastructure and the minimization of light scattering. Journal of Theoretical Biology, 199, 181–198. 10.1006/jtbi.1999.0948. [DOI] [PubMed] [Google Scholar]
- Ke, M.T. , Fujimoto, S. & Imai, T. (2013) SeeDB: A simple and morphology‐preserving optical clearing agent for neuronal circuit reconstruction. Nature Neuroscience, 16, 1154–1161. 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- Ke, M.T. , Nakai, Y. , Fujimoto, S. , Takayama, R. , Yoshida, S. , Kitajima, T.S. et al (2016) Super‐resolution mapping of neuronal circuitry with an index‐optimized clearing agent. Cell Reports, 14, 2718–2732. 10.1016/j.celrep.2016.02.057. [DOI] [PubMed] [Google Scholar]
- Keller, P.J. & Ahrens, M.B. (2015) Visualizing whole‐brain activity and development at the single‐cell level using light‐sheet microscopy. Neuron, 85, 462–483. [DOI] [PubMed] [Google Scholar]
- Khoradmehr, A. , Mazaheri, F. , Anvari, M. & Tamadon, A. (2019) A simple technique for three‐dimensional imaging and segmentation of brain vasculature using fast free‐of‐acrylamide clearing tissue in murine. Cell Journal, 21, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H. , Jang, M.J. , Choi, J. , Lee, E. , Song, K.D. , Cho, J. et al (2018) Optimizing tissue‐clearing conditions based on analysis of the critical factors affecting tissue‐clearing procedures. Scientific Reports, 8, 12815 10.1038/s41598-018-31153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.Y. , Cho, J.H. , Murray, E. , Bakh, N. , Choi, H. , Ohn, K. et al (2015) Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc Natl Acad Sci U S A, 112, E6274–6283. 10.1073/pnas.1510133112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston, B.R. , Syed, A.M. , Ngai, J. , Sindhwani, S. & Chan, W.C.W. (2019) Assessing micrometastases as a target for nanoparticles using 3D microscopy and machine learning. Proc Natl Acad Sci U S A, 116, 14937–14946. 10.1073/pnas.1907646116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota, S.I. , Takahashi, K. , Nishida, J. , Morishita, Y. , Ehata, S. , Tainaka, K. et al (2017) Whole‐body profiling of cancer metastasis with single‐cell resolution. Cell Reports, 20, 236–250. 10.1016/j.celrep.2017.06.010. [DOI] [PubMed] [Google Scholar]
- Kuwajima, T. , Sitko, A.A. , Bhansali, P. , Jurgens, C. , Guido, W. & Mason, C. (2013) ClearT: a detergent‐ and solvent‐free clearing method for neuronal and non‐neuronal tissue. Development, 140, 1364–1368. 10.1242/dev.091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, H.M. , Liu, A.K.L. , Ng, H.H.M. , Goldfinger, M.H. , Chau, T.W. , DeFelice, J. et al (2018) Next generation histology methods for three‐dimensional imaging of fresh and archival human brain tissues. Nature Communications, 9, 1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E. , Choi, J. , Jo, Y. , Kim, J.Y. , Jang, Y.J. , Lee, H.M. et al (2016) ACT‐PRESTO: Rapid and consistent tissue clearing and labeling method for 3‐dimensional (3D) imaging. Scientific Reports, 6, 18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.S. , Bindokas, V.P. & Kron, S.J. (2017) Multiplex three‐dimensional optical mapping of tumor immune microenvironment. Scientific Reports, 7, 17031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.S. , Bindokas, V.P. & Kron, S.J. (2019) Multiplex three‐dimensional mapping of macromolecular drug distribution in the tumor microenvironment. Molecular Cancer Therapeutics, 18, 213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Lin, P. , Tan, Y. & Cheng, J.X. (2019a) Volumetric stimulated Raman scattering imaging of cleared tissues towards three‐dimensional chemical histopathology. Biomed Opt Express, 10, 4329–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Germain, R.N. & Gerner, M.Y. (2017) Multiplex, quantitative cellular analysis in large tissue volumes with clearing‐enhanced 3D microscopy (Ce3D). Proc Natl Acad Sci U S A, 114, E7321–E7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Germain, R.N. & Gerner, M.Y. (2019b) High‐dimensional cell‐level analysis of tissues with Ce3D multiplex volume imaging. Nature Protocols, 14, 1708–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Yu, B. , Sun, Q. , Zhang, Y. , Ren, M. , Zhang, X. et al (2018a) Generation of a whole‐brain atlas for the cholinergic system and mesoscopic projectome analysis of basal forebrain cholinergic neurons. Proc Natl Acad Sci U S A, 115, 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Xu, J. , Wan, P. , Yu, T. & Zhu, D. (2018b) Optimization of GFP fluorescence preservation by a modified uDISCO clearing protocol. Frontiers in Neuroanatomy, 12, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke, D.S. & Lidke, K.A. (2012) Advances in high‐resolution imaging–techniques for three‐dimensional imaging of cellular structures. Journal of Cell Science, 125, 2571–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebmann, T. , Renier, N. , Bettayeb, K. , Greengard, P. , Tessier‐Lavigne, M. & Flajolet, M. (2016) Three‐dimensional study of Alzheimer's disease hallmarks using the iDISCO clearing method. Cell Reports, 16, 1138–1152. 10.1016/j.celrep.2016.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, W. , Yi, Y. , Jing, D. , Zhang, S. , Men, Y. , Ge, W.P. et al (2019) Investigation of postnatal craniofacial bone development with tissue clearing‐based three‐dimensional imaging. Stem Cells and Development, 28, 1310–1321. 10.1089/scd.2019.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, K. , Mitani, T.T. , Horiguchi, S.A. , Kaneshiro, J. , Murakami, T.C. , Mano, T. et al (2019) Advanced CUBIC tissue clearing for whole‐organ cell profiling. Nature Protocols, 14, 3506–3537. 10.1038/s41596-019-0240-9. [DOI] [PubMed] [Google Scholar]
- Mertz, J. (2011) Optical sectioning microscopy with planar or structured illumination. Nature Methods, 8, 811–819. 10.1038/nmeth.1709. [DOI] [PubMed] [Google Scholar]
- Migliori, B. , Datta, M.S. , Dupre, C. , Apak, M.C. , Asano, S. , Gao, R. et al (2018) Light sheet theta microscopy for rapid high‐resolution imaging of large biological samples. BMC Biology, 16, 57 10.1186/s12915-018-0521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawski, M. , Kirilina, E. , Scherf, N. , Jager, C. , Reimann, K. , Trampel, R. et al (2018) Developing 3D microscopy with CLARITY on human brain tissue: Towards a tool for informing and validating MRI‐based histology. NeuroImage, 182, 417–428. 10.1016/j.neuroimage.2017.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, T.C. , Mano, T. , Saikawa, S. , Horiguchi, S.A. , Shigeta, D. , Baba, K. et al (2018) A three‐dimensional single‐cell‐resolution whole‐brain atlas using CUBIC‐X expansion microscopy and tissue clearing. Nature Neuroscience, 21, 625–637. 10.1038/s41593-018-0109-1. [DOI] [PubMed] [Google Scholar]
- Murray, E. , Cho, J.H. , Goodwin, D. , Ku, T. , Swaney, J. , Kim, S.Y. et al (2015) Simple, scalable proteomic imaging for high‐dimensional profiling of intact systems. Cell, 163, 1500–1514. 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mzinza, D.T. , Fleige, H. , Laarmann, K. , Willenzon, S. , Ristenpart, J. , Spanier, J. et al (2018) Application of light sheet microscopy for qualitative and quantitative analysis of bronchus‐associated lymphoid tissue in mice. Cellular & Molecular Immunology, 15, 875–887. 10.1038/cmi.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader, M.A. & Banks, M.L. (2014) Environmental modulation of drug taking: Nonhuman primate models of cocaine abuse and PET neuroimaging. Neuropharmacology, 76, 510–517. 10.1016/j.neuropharm.2013.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa, L.F. , Kholodnykh, A. , Villarreal, P. , Tian, B. , Pal, R. , Freiberg, A.N. et al (2018) Imaging of murine whole lung fibrosis by large scale 3D microscopy aided by tissue optical clearing. Scientific Reports, 8, 13348 10.1038/s41598-018-31182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, C. , Cai, R. , Quacquarelli, F.P. , Ghasemigharagoz, A. , Lourbopoulos, A. , Matryba, P. et al (2016) Shrinkage‐mediated imaging of entire organs and organisms using uDISCO. Nature Methods, 13, 859–867. 10.1038/nmeth.3964. [DOI] [PubMed] [Google Scholar]
- Park, Y.G. , Sohn, C.H. , Chen, R. , McCue, M. , Yun, D.H. , Drummond, G.T. et al (2019) Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nature Biotechnology, 37, 73–83. 10.1038/nbt.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y. , Yu, T. , Xu, J. , Wan, P. , Ma, Y. , Zhu, J. et al (2019) FDISCO: Advanced solvent‐based clearing method for imaging whole organs. Science Advances, 5(1), eaau8355 10.1126/sciadv.aau8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier, N. , Wu, Z. , Simon, D.J. , Yang, J. , Ariel, P. & Tessier‐Lavigne, M. (2014) iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell, 159, 896–910. 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Rezazadeh, F.M. , Saedi, S. , Rahmanifar, F. , Namavar, M.R. , Dianatpour, M. , Tanideh, N. et al (2018) Fast free of acrylamide clearing tissue (FACT) for clearing, immunolabelling and three‐dimensional imaging of partridge tissues. Microscopy Research and Technique, 81, 1374–1382. 10.1002/jemt.23078. [DOI] [PubMed] [Google Scholar]
- Richardson, D.S. & Lichtman, J.W. (2015) Clarifying tissue clearing. Cell, 162, 246–257. 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, D.S. & Lichtman, J.W. (2017) SnapShot: Tissue clearing. Cell, 171, 496–496.e1. 10.1016/j.cell.2017.09.025. [DOI] [PubMed] [Google Scholar]
- Rios, A.C. & Clevers, H. (2018) Imaging organoids: A bright future ahead. Nature Methods, 15, 24–26. 10.1038/nmeth.4537. [DOI] [PubMed] [Google Scholar]
- Seo, J. , Choe, M. & Kim, S.Y. (2016) Clearing and labeling techniques for large‐scale biological tissues. Molecules and Cells, 39, 439–446. 10.14348/molcells.2016.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki, E.A. , Tainaka, K. , Perrin, D. , Kishino, F. , Tawara, T. , Watanabe, T.M. et al (2014) Whole‐brain imaging with single‐cell resolution using chemical cocktails and computational analysis. Cell, 157, 726–739. 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- Susaki, E.A. & Ueda, H.R. (2016) Whole‐body and whole‐organ clearing and imaging techniques with single‐cell resolution: toward organism‐level systems biology in mammals. Cell Chem Biol, 23, 137–157. 10.1016/j.chembiol.2015.11.009. [DOI] [PubMed] [Google Scholar]
- Sylwestrak, E.L. , Rajasethupathy, P. , Wright, M.A. , Jaffe, A. & Deisseroth, K. (2016) Multiplexed intact‐tissue transcriptional analysis at cellular resolution. Cell, 164, 792–804. 10.1016/j.cell.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tainaka, K. , Kubota, S.I. , Suyama, T.Q. , Susaki, E.A. , Perrin, D. , Ukai‐Tadenuma, M. et al (2014) Whole‐body imaging with single‐cell resolution by tissue decolorization. Cell, 159, 911–924. 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- Tainaka, K. , Kuno, A. , Kubota, S.I. , Murakami, T. & Ueda, H.R. (2016) Chemical principles in tissue clearing and staining protocols for whole‐body cell profiling. Annual Review of Cell and Developmental Biology, 32, 713–741. 10.1146/annurev-cellbio-111315-125001. [DOI] [PubMed] [Google Scholar]
- Tainaka, K. , Murakami, T.C. , Susaki, E.A. , Shimizu, C. , Saito, R. , Takahashi, K. et al (2018) Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Reports, 24, 2196–2210.e9. 10.1016/j.celrep.2018.07.056. [DOI] [PubMed] [Google Scholar]
- Tanaka, N. , Kanatani, S. , Tomer, R. , Sahlgren, C. , Kronqvist, P. , Kaczynska, D. et al (2017) Whole‐tissue biopsy phenotyping of three‐dimensional tumours reveals patterns of cancer heterogeneity. Nature Biomedical Engineering, 1, 796–806. 10.1038/s41551-017-0139-0. [DOI] [PubMed] [Google Scholar]
- Tang, S.C. , Shen, C.N. , Lin, P.Y. , Peng, S.J. , Chien, H.J. , Chou, Y.H. et al (2018) Pancreatic neuro‐insular network in young mice revealed by 3D panoramic histology. Diabetologia, 61, 158–167. 10.1007/s00125-017-4408-y. [DOI] [PubMed] [Google Scholar]
- Tomer, R. , Ye, L. , Hsueh, B. & Deisseroth, K. (2014) Advanced CLARITY for rapid and high‐resolution imaging of intact tissues. Nature Protocols, 9, 1682–1697. 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuchin, V.V. , Maksimova, I.L. , Zimnyakov, D.A. , Kon, I.L. , Mavlyutov, A.H. & Mishin, A.A. (1997) Light propagation in tissues with controlled optical properties. Journal of Biomedial Optics, 2, 401–417. 10.1117/12.281502. [DOI] [PubMed] [Google Scholar]
- Tuchin, V.V. (1997) Light scattering study of tissues. Physics‐Uspekhi, 40, 495–515. [Google Scholar]
- Tuchin, V.V. (2005) Optical clearing of tissues and blood. Bellingham, WA: SPIE Press. [Google Scholar]
- Tuchin, V.V. (2015a) Tissue optics and photonics: Light‐tissue interaction. Journal of Biomedical Photonics & Engineering, 1, 98–134. [Google Scholar]
- Tuchin, V.V. (2015b) Tissue optics: Light scattering methods and instruments for medical diagnostics (3rd edn). Bellingham, WA: SPIE Press. [Google Scholar]
- Urata, S. , Iida, T. , Yamamoto, M. , Mizushima, Y. , Fujimoto, C. , Matsumoto, Y. et al (2019) Cellular cartography of the organ of Corti based on optical tissue clearing and machine learning. Elife, 8, e40946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, G. , Chan, E.K. , Barton, J.K. , Rylander, H.G. & Welch, A.J. (1999) Use of an agent to reduce scattering in skin. Lasers in Surgery and Medicine, 24, 133–141. [DOI] [PubMed] [Google Scholar]
- Waclaw, B. , Bozic, I. , Pittman, M.E. , Hruban, R.H. , Vogelstein, B. & Nowak, M.A. (2015) A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature, 525, 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Khoradmehr, A. & Tamadon, A. (2019a) FACT or PACT: A comparison between free‐acrylamide and acrylamide‐based passive sodium dodecyl sulfate tissue clearing for whole tissue imaging. Cell Journal, 21, 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Gillis‐Smith, S. , Peng, Y. , Zhang, J. , Chen, X. , Salzman, C.D. et al (2018) The coding of valence and identity in the mammalian taste system. Nature, 558, 127–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Liu, K. , Yang, L. , Wang, H. & Yang, J. (2019b) BoneClear: Whole‐tissue immunolabeling of the intact mouse bones for 3D imaging of neural anatomy and pathology. Cell Research, 29, 870–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, A.M. & Watkins, S.C. (2019) Massive volumetric imaging of cleared tissue: The necessary tools to be successful. International Journal of Biochemistry & Cell Biology, 112, 76–78. 10.1016/j.biocel.2019.05.007. [DOI] [PubMed] [Google Scholar]
- Wei, L. , Chen, Z. , Shi, L. , Long, R. , Anzalone, A.V. , Zhang, L. et al (2017) Super‐multiplex vibrational imaging. Nature, 544, 465–470. 10.1038/nature22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, M. , Shi, L. , Shen, Y. , Zhao, Z. , Guzman, A. , Kaufman, L.J. et al (2019) Volumetric chemical imaging by clearing‐enhanced stimulated Raman scattering microscopy. Proc Natl Acad Sci U S A, 116, 6608–6617. 10.1073/pnas.1813044116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder, R. (2001) A clearer vision for in vivo imaging. Nature Biotechnology, 19, 316–317. 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- Wu, Y. , Kumar, A. , Smith, C. , Ardiel, E. , Chandris, P. , Christensen, R. et al (2017) Reflective imaging improves spatiotemporal resolution and collection efficiency in light sheet microscopy. Nature Communications, 8, 1452 10.1038/s41467-017-01250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, N. , Tamadon, A. , Liu, Y. , Ma, T. , Leak, R.K. , Chen, J. et al (2017) Fast free‐of‐acrylamide clearing tissue (FACT)‐an optimized new protocol for rapid, high‐resolution imaging of three‐dimensional brain tissue. Scientific Reports, 7, 9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. , Treweek, J.B. , Kulkarni, R.P. , Deverman, B.E. , Chen, C.K. , Lubeck, E. et al (2014) Single‐cell phenotyping within transparent intact tissue through whole‐body clearing. Cell, 158, 945–958. 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, Y. , Men, Y. , Jing, D. , Luo, W. , Zhang, S. , Feng, J.Q. et al (2019) 3‐dimensional visualization of implant‐tissue interface with the polyethylene glycol associated solvent system tissue clearing method. Cell Proliferation, 52, e12578 10.1111/cpr.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T. , Zhu, J. , Li, Y. , Ma, Y. , Wang, J. , Cheng, X. et al (2018) RTF: A rapid and versatile tissue optical clearing method. Scientific Reports, 8, 1964 10.1038/s41598-018-20306-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Feng, W. , Zhao, Y. , Yu, T. , Li, P. , Xu, T. et al (2018) A large, switchable optical clearing skull window for cerebrovascular imaging. Theranostics, 8, 2696–2708. 10.7150/thno.23686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S. , Todorov, M.I. , Cai, R. , Maskari, R.A. , Steinke, H. , Kemter, E. et al (2020) Cellular and molecular probing of intact human organs. Cell, 180, 796–812.e19. 10.1016/j.cell.2020.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.J. , Yu, T.T. , Zhang, C. , Li, Z. , Luo, Q.M. , Xu, T.H. et al (2018) Skull optical clearing window for in vivo imaging of the mouse cortex at synaptic resolution. Light: Science & Applications, 7(2), 17153 10.1038/lsa.2017.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, D. , Larin, K.V. , Luo, Q. & Tuchin, V.V. (2013) Recent progress in tissue optical clearing. Laser & Photonics Reviews, 7(5), 732–757. 10.1002/lpor.201200056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. , Huang, L. , Zheng, Y. , Song, Y. , Xu, Q. , Wang, J. et al (2019) Ultrafast optical clearing method for three‐dimensional imaging with cellular resolution. Proceedings of the National Academy of Sciences, 116(23), 11480–11489. 10.1073/pnas.1819583116. [DOI] [PMC free article] [PubMed] [Google Scholar]


