Abstract
Background
To undertake a bioinformatics analysis to identify abnormally expressed genes [also referred to as differentially expressed genes (DEGs)] and their functions in esophageal carcinoma (ESCA).
Methods
DEGs (i.e., GSE100942, GSE17351, GSE26886, and GSE77861) were obtained from a gene expression omnibus database. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed using online tools from the Database for Annotation, Visualization and Integrated Discovery. A protein-protein interaction network was then constructed based on the Search Tool for the Retrieval of Interacting Genes website. Cytoscape software was used to identify the top 20 DEGs located in the central region of the network. For the overall survival analysis, a Kaplan–Meier analysis was conducted of the Gene Expression Profiling Interactive Analysis website, and collagen (COL) 1A2 was selected to detect the molecular mechanism of COL1A2-small interfering ribonucleic acid (siRNA) in the following ESCA cell lines: Eca109 and TE-1. Next, the expression of COL1A2-messanger ribonucleic acid was determined using real-time quantitative polymerase chain reaction. The expression of COL1A2 was also verified by Western blot. Cell proliferation was measured by colony-forming and MTT assays, and migration and invasion by the transwell assay.
Results
Based on the GEO database and screening out the hub gene, we identified that COL1A2 was abnormally expressed in ESCA. With a series of in vitro experiments, the expression of COL1A2 was defined as higher in Eca109 and TE-1.
Conclusions
COL1A2 was highly expressed in ESCA tissue samples. Additionally, the proliferation and metastasis of Eca109 and TE-1 cell lines were significantly attenuated by siRNA-COL1A2-mediated small interference. Notably, the expression level of COL1A2 was obviously related to the Akt and epithelial-mesenchymal transition (EMT) pathways.
Keywords: Esophageal carcinoma (ESCA), COL1A2, bioinformatics, extracellular matrix
Introduction
Esophageal carcinoma (ESCA) is one of the most common cancers in the world. Among malignant tumors, the incidence and mortality rates of ESCA rank eighth and sixth (1). There are two types of esophageal cancer: esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC). ESCA mainly manifests as EAC in western countries, and ESCC in Southeast Asian countries, including China (2,3). Many factors, such as smoking, drinking, and chewing betel nut, contribute to ESCA. For example, an insufficient intake of fruits and vegetables, which leads to low antioxidant levels and vitamin deficiency, also promotes the development of ESCA (4). However, the pathogenesis of ESCA is not yet fully understood. ESCA has a poor prognosis and a high mortality rate. Currently, surgical resection is the major and only recognized method for the early treatment of ESCA. Unfortunately, up to 75% of ESCA patients cannot be treated effectively due to distant metastasis, and the 5-year overall survival rate is only 30% (5). Many patients were diagnosed when they begin to eat and find it difficult to swallow; however, by this time, the best opportunity for surgical treatment has normally passed. Thus, to improve the survival time and quality of life of patients with advanced and post-operative ESCA, tumor-related differential genes need to be identified as biomarkers to create targeted drugs of higher efficacy.
Type I collagen (COL), which harbors two CYP1 chains (COL1A1) and a single CYP2 chain (COL1A2) (6,7), has unique biochemical and biomechanical properties. Some study has proved that genetic polymorphism of COL1A2 is related to susceptibility for hepatocellular carcinoma (8,9). It is a proximal structure that must be destroyed for epithelial tumor cells to invade and migrate. Increasing evidence has shown that COL1A2 is downregulated in many malignant tumors, including head and neck cancer, bladder cancer and malignant melanoma (10), but upregulated in liver, ovarian and pancreatic cancer. Thus, it can be regarded as a potential diagnostic and prognostic marker for these tumors (9,11,12). The differential expression of COL1A2 may cause different collagen-mediated effects in human malignant tumors. To date, no study appears to have examined the relationship between COL1A2 expression and ESCA. In this paper, we connected with studies of multi-center by multi-microarray analysis, and verified these results by The Cancer Genome Atlas (TCGA) database. Additionally, we searched for the biological impact of COL1A2 at the cellular level in vitro. In short, we describe the expression of COL1A2 and its molecular mechanism in ESCA.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7867).
Methods
Collection of microarray data
The expression profiles of GSE100942, GSE17351, GSE26886, and GSE77861 in 35 normal esophageal tissue samples and 25 ESCA tissue samples were extracted from the Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/geo/) based on the GPL570 platform. We then integrated four sets of raw data, and used a robust multi-array average and SVA software package to preprocess and remove the batch effect (13,14). |logFC| ≥1 and P<0.05 were used as the as the cut-off values. A total of 414 differentially expressed genes (DEGs) were identified in the ESCA tissue samples, and compared with normal esophageal tissue samples using the limma package from the R programming language (15).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses
An online annotation visualization tool and an integrated discovery database (i.e., Database for Annotation, Visualization and Integrated Discovery (DAVID), https://david.ncifcrf.gov/) were used to demonstrate the functions of the DEGs. These DEGs were further subjected to GO and KEGG analyses. A P<0.05 indicated enrichment.
Protein-protein interaction (PPI) network construction
The Search Tool for the Retrieval of Interacting Genes (STRING) (http://string-db.org) database was used to evaluate gene interactions. These interactions were visualized using Cytoscape application software (16,17). The top 20 hub genes located in the central region of the network were identified.
The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) databases were used to identify the hub genes for the survival analysis. The Gene Expression Profile Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/index.html), a mature network that uses standard processing pipelines, was employed to analyze the expression data of DEGs. Ultimately, 9,736 tumor samples and 8,587 normal samples were identified in TCGA and the GTEx databases that were then examined for DEGs in the survival analysis (18).
Tissue samples
In this study, we recruited 33 ESCA patients who had undergone esophageal cancer surgery at The First Affiliated Hospital of Soochow University between January 2019 and January 2020. Samples of cancerous tissues and adjacent tissues from these patients were collected and immediately stored in liquid nitrogen (all were single disease cases; no patients received neoadjuvant chemoradiotherapy before surgery). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). All participants signed written informed consent forms, and the study was approved by the Ethics Committee of The First Affiliated Hospital of Soochow University (2018011).
Cell lines
Human ESCA cell lines Eca109, TE-1, TE-14, kyse30 and kyse450 were purchased from Shanghai Institute of Cell Biology (Chinese Academy of Sciences, Shanghai, China). All the cells were cultured at 37 °C in an environment containing 5% carbon dioxide (CO2). Eca109, TE-1, and TE-14 cells were maintained to grow in a Dulbecco modified Eagle medium (DMEM), and kyse30 and kyse450 cells were then grown in a RPMI-1640 medium (Gibco, Carlsbad, CA, USA), both of which were supplemented with 10% fetal bovine serum (FBS).
Cell transfection
Eca109 and TE-1 cells were seeded in 96 well plates, and then divided into the following two groups: the COL1A2-small interfering ribonucleic acid (siRNA) group and the negative control (NC) group (GeneChem, Shanghai, China). In accordance with the manufacturer’s instructions, 500 ng plasmids were transfected to cultured Eca109 and TE-1 cells with 2.5 µL Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). The experiment was performed in triplicate. After a 6-hour incubation, the medium was replaced by 10% FBS (fresh medium).
RNA extraction and RT-qPCR
Total cellular ribonucleic acid (RNA) was extracted from tissues and cells using TRIzol reagent (Invitrogen, California, USA). SYBR Green II (takara, Tokyo, Japan) was used to detect RNA expression. COL1A2 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primers were provided by GenePharma (Shanghai, China). The 2−ΔΔct method was used to analyze the data. The primer sequences of COL1A2 were as follows: F: 5'-GAGACCCTTCTTACTCCTGAA-3'; and R: 5'-GTGATGTTCTGAGAGGCATAG-3'. The GAPDH primer sequences were as follows: F: 5'-GACTCATGACCACAGTCCATGC-3'; and R: 5'-AGAGGCAGGGATGATGTTCTG-3'.
Cell proliferation
In accordance with the manufacturer’s instructions, cell viability was measured using a MTT Kit (IM0280, Solarbio) for analyzing cell proliferation.
Cloning steps
Transfected Eca109 and TE-1 cells were seeded in 6-well plates at a density of 1×103 cells/well, NC-siRNA and COL1A2-siRNA groups were incubated in DMEM containing 10% FBS for 14 days. The samples were then fixed with ice-precooled formaldehyde for 30 minutes, stained with crystal violet for 1 minute, and counted.
Wound healing assay
Cells were seeded into six-well plates and cultured to a density of 90%. Then, a line wound was made by scraping a 200 µL tip across the confluent cell layer. After being washed with PBS, the cells were cultured in serum-free RPMI-1640 medium. Wound closure was visualized in five random fields using a high-power microscope (×100) after 48 h of culture.
Transwell assay
A transwell chamber (diameter 8 µm, Corning, NY, USA) was used to evaluate the migration of Eca109 cells and TE-1 cells. 5×104 transfected cells were inoculated into the upper chamber, and a medium supplemented with 10% FBS was added to the lower chamber. After 24 hours of culturing at 37 °C and 5% CO2, the samples were fixed for 30 minutes with ice-precooled formaldehyde and stained with crystal violet for 1 minute at room temperature. The cells on the upper surface of the polycarbonate membrane were removed with a cotton swab. To evaluate the migrative ability, the cell count was performed under a high-power microscope (×100), and at least three randomly selected microscopic fields were counted.
The Matrigel invasion chamber (BD Biosciences, San Jose, CA, USA) was used to determine the invasive ability of the cells. 1×105 cells were inoculated into the upper chamber that contained the Matrigel and DMEM without 10% FBS. 500 µL of DMEM supplemented with 10% FBS was added to the lower chamber. After an incubation period of 72 hours at 37 °C and 5% CO2, the cells were fixed for 30 minutes with ice-precooled formaldehyde at room temperature and stained with crystal violet for 1 minute. Next, a cotton swab was used to wipe off the non-invading cells. The number of cells invading the inferior chamber was recorded under a high-power microscope (×100). The experiment was repeated three times.
Western blot
The cells were lysed by a radioimmunoprecipitation assay buffer (Epizyme Biotech, Shanghai, China), and the protein concentration was determined by a bicinchoninic acid assay test (Epizyme Biotech, Shanghai, China). Western blotting and the bio rad ChemiDoc MP imaging system (Hercules, CA, USA) were used in accordance with the manufacturer’s instructions. COL1A2 (1:1,000, yb-16835r, YBscience, Shanghai, China) was the first antibody used. GAPDH was used as the control.
Data statistics
In this study, SPSS 19.0 software (IBM, USA) was used for the statistical analysis, and each experiment was repeated at least three times. The data were expressed as mean ± standard deviation. A student’s t-test was used to estimate the differences between the groups. The listed data were expressed in percentages or ratios and checked by a chi-squared test. A value of P<0.05 was regarded as statistically significant.
Results
Identification and enrichment analysis of DEGs
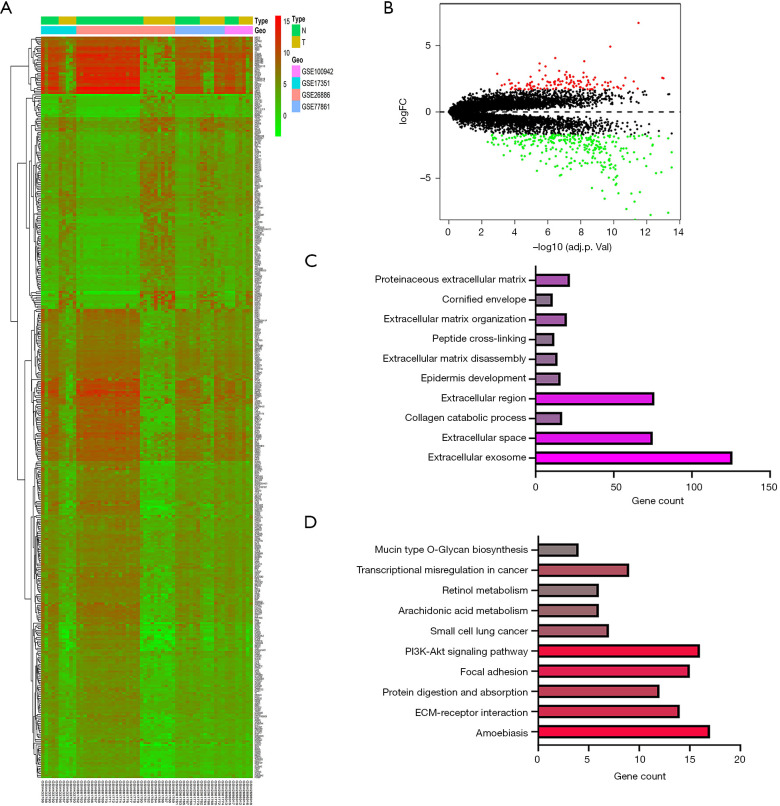
After the pre-treatment and the removal of batch effects, the DEGs (i.e., GSE100942, GSE17351, GSE26886, and GSE77861) were analyzed using a limma software package (see Figure 1A). Using a P<0.05 and |logFC| ≥1 as the cut-off values, 414 DEGs were identified in the ESCA tissues, including 120 upregulated and 294 downregulated DEGs. According to the value of |logFC|, the volcano map showed that the red-labeled genes (upregulated) and the green-labeled genes (downregulated) (see Figure 1B). The DAVID tool was used for the GO and KEGG analyses (P<0.05). The results of the GO analysis showed that the DEGs were significantly enriched in exosomes, extracellular spaces, and extracellular regions (see Figure 1C). The results of the KEGG analysis showed that the DEGs were mainly enriched in the PI3K-Akt signaling pathway, the extracellular matrix (ECM)-receptor interaction, and protein digestion and absorption (see Figure 1D).
Figure 1.
Differentially expressed genes (DEGs) identified in GSE100942, GSE17351, GSE26886, and GSE77861. (A) Heatmap of the 414 DEGs according to the value of |logFC|; (B) volcano map of differently expressed genes between esophageal carcinoma (ESCA) tissues and normal esophageal tissues; (C) the top 10 Gene Ontology (GO) terms in the enrichments analysis of the DEGs; (D) the top 10 GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in the enrichment analysis of the DEGs.
PPI network analysis, validation of differences and survival analysis in GEPIA
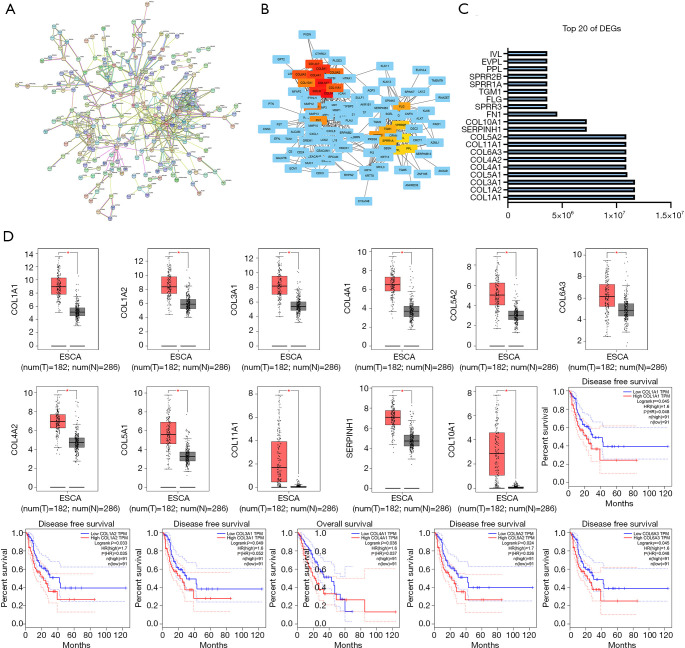
Using the STRING website, a PPI network of 414 DEGs, including 240 nodes and 532 edges, was constructed (see Figure 2A). This network was further visualized using Cytoscape software, and 20 hub genes located in the network center were identified (see Figure 2B,C). The top 11 DEGs underwent a differential and survival analysis on the GEPIA website. The results showed that the expression of COL1A1, COL1A2, COL3A1, COL4A1, COL4A2, COL5A1, COL5A2, COL6A3, COL10A1, COL11A1, and SERPINH1 was significantly higher in cancer tissues than normal tissues (P<0.05). The disease-free survival (DFS) rate of low-risk groups of COL1A1, COL1A2, COL3A1, COL5A2, COL6A3 was significantly higher than the DFS rate of high-risk groups (P<0.05), and the overall survival (OS) rate of low-risk groups COL4A1 group was also higher than the OS rate of high-risk group (P<0.05) (see Figure 2D).
Figure 2.
Differential expression and survival curves of the top 20 hub genes. (A) PPI network of differentially expressed genes (DEGs) based on the Search Tool for the Retrieval of Interacting Genes (STRING) website; (B) the top 20 hub genes were visualized on the Cytoscape application, and the scores (C) were performed; (D) differential expression and survival curves.
Expression of COL1A2 in tissues and cells
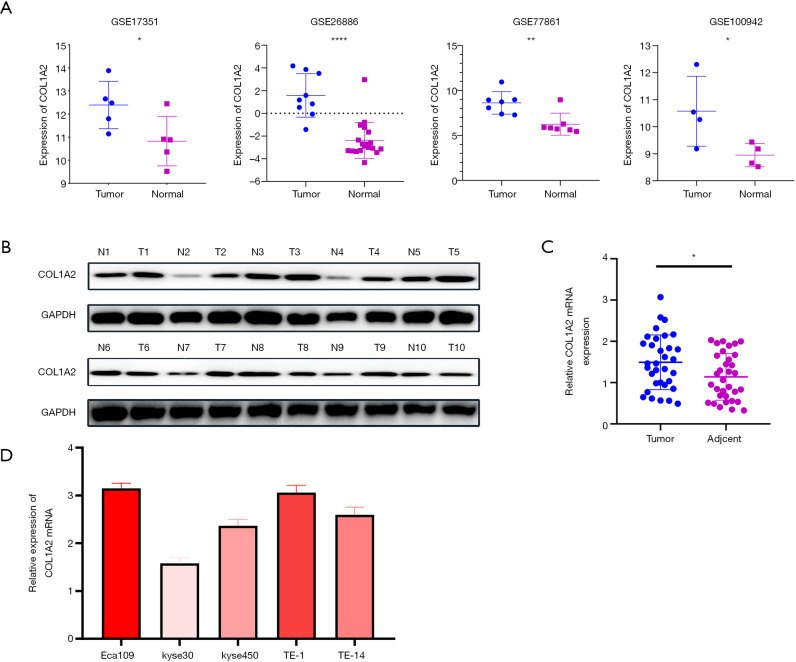
We found that the expression level of COL1A2 in cancer tissues was higher than that in normal tissues (P<0.05) in the four groups of GSE100942, GSE17351, GSE26886, and GSE77861 (see Figure 3A). In the 10 pairs of cancer tissue and adjacent tissue samples, the expression level of COL1A2 in cancer tissues was significantly higher than that in adjacent tissues (see Figure 3B,C). Clinical data suggested that the expression of COL1A2 was significantly correlated with pathologic stage and tumor type (P<0.05) (see Table 1). Among the selected ESCA cell lines Eca109, TE-1, kyse30, kyse450, and TE-14, COL1A2 showed the highest expression in Eca109 and TE-1 cells (see Figure 3D). Thus, these cell lines were selected for the next experiment.
Figure 3.
Expression of COL1A2 in tissue and cell lines. (A) In four sets of microarray data, GSE100942, GSE17351, GSE26886, and GSE77861, the expression of COL1A2 was higher in cancer tissues than in normal tissues (P<0.05); (B,C) in clinical samples, the mRNA expression of COL1A2 was significantly lower in normal tissues than in tumor tissues; (D) the expression of COL1A2 in cell lines Eca109, TE-1, Ec5, Ec6, and TE-14. *, P<0.05, **P<0.01, ****P<0.0001.
Table 1. Expression of COL1A2 according to patients’ clinical features.
| Factors | COL1A2 mRNA expression | P value | |
|---|---|---|---|
| High (n=26) | Low (n=26) | ||
| Gender | 0.095 | ||
| Male | 17 | 11 | |
| Female | 9 | 15 | |
| Age(years) | 0.271 | ||
| ≥50 | 6 | 3 | |
| <50 | 20 | 23 | |
| Tumor size (cm) | 0.336 | ||
| >5 | 8 | 5 | |
| ≤5 | 18 | 21 | |
| Lymph node metastasis | 0.011* | ||
| Positive | 7 | 16 | |
| Negative | 19 | 10 | |
| Differentiation | 0.010* | ||
| Good/moderate | 15 | 6 | |
| Poor | 11 | 20 | |
*, P<0.05.
The expression level of COL1A2 affected the proliferation, migration and invasion of cells
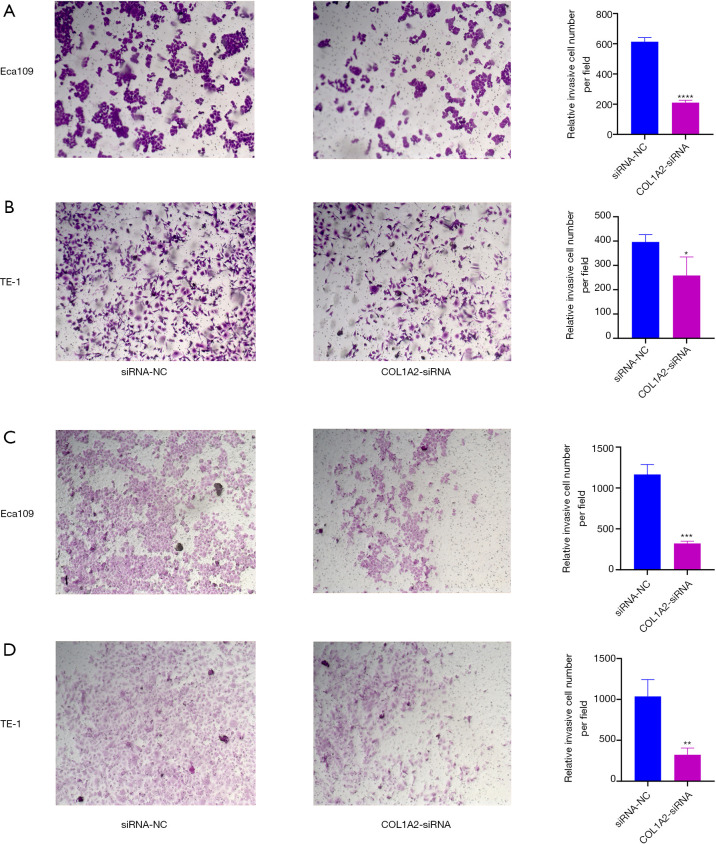
To determine the effect of COL1A2 knockout on cellular processes, real-time quantitative polymerase chain reaction (RT-qPCR). and Western blot were performed. The results showed that the messenger ribonucleic (mRNA) and protein levels of COL1A2 in Eca109 and TE-1 cell lines were significantly inhibited (P<0.05) (see Figure 4A, Figure S1). The MTT assay showed that the proliferation of ECA109 and TE-1 cells was inhibited by COL1A2-siRNA (P<0.05) (see Figure 4B,C), and the colony count of the COL1A2-siRNA group was significantly lower than that of the siRNA-NC group (P<0.01) (see Figure 4D). To examine the effect of COL1A2 on invasion and migration, we performed wound healing assay, transwell and Matrigel assay. The wound-healing ability of the COL1A2-siRNA group was obviously less than that of the siRNA-NC group (P<0.01) (see Figure S2). The results of migration test showed that the migratory cells in the COL1A2-siRNA group were less than those in siRNA-NC group (P<0.01) (see Figure 5A,B). In the invasion test, the invasive cells in COL1A2-siRNA group were also less than those in the siRNA-NC group (P<0.01) (see Figure 5C,D). Additionally, COL1A2-siRNA was found to change the protein content of p-Akt and vimentin (S1). In conclusion, the low expression of COL1A2 significantly reduced the migration and invasion of Eca109 and TE-1 cells.
Figure 4.
COL1A2 promotes cell colony and growth of Eca109 and TE-1. COL1A2 mRNA (A) was assessed using RT-qPCR after siCOL1A2 small interference (P<0.001). (B,C) Cell colony of Eca109 and TE-1 cells was stained by Crystal violet. (D) Cell growth of Eca109 and TE-1 after COL1A2-siRNA and siRNA-NC infection from day 1 to 5. *P<0.05, ***P<0.001.
Figure 5.
Cell invasion of Eca109 and TE-1 cell lines after COL1A2-siRNA and NC-siRNA infection were examined through the transwell assay. Cell migration analysis in Eca109 (A) and TE-1 (B) cells with COL1A2-siRNA and NC-siRNA infection. Cell invasion analysis in Eca109 (C) and TE-1 (D) cells with COL1A2-siRNA and NC-siRNA infection. The samples were stained with crystal violet, and the number of cells was recorded under a high-power microscope (×100). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Discussion
The incidence of ESCA in China remains significantly higher than that in western countries; however, great progress has been made in its treatment (4). Surgery, medical radiotherapy, chemotherapy, and emerging gene detection targeted therapy are used to treat ESCA (19). However, these treatments may result in some side effects, such as esophageal anastomotic leakage, anastomotic stenosis, infection, and other post-operative complications (20). In relation to targeted therapy, drugs are limited and some are ineffective, especially for ESCC (21). This study explored the expression of COL1A2 and its cellular pathways in ESCA with the aim of finding a new therapeutic target.
Using the GEO, TCGA, and GTEx databases, and undertaking GO and KEGG analyses of the PPI network, the DEGs in ESCA were identified. COL1A2 located in the center of the network was identified as a hub gene for further study. COL1A1 and COL1A2 are two types of type I COL (22). Some research has shown that the COL1A1 promoter region was methylated in renal cell carcinoma and hepatocellular carcinoma (23,24). However, the mRNA expression of COL1A1 and COL1A2 increased in colorectal cancer and concomitant tumors (25,26). In pancreatic cancer, COL1A2 was also highly expressed (27). COL1A2 mRNA expression was also found to be downregulated in melanoma (28). In esophageal cancer, miR-133a-3p was negatively correlated with COL1A1. A large number of cell functional experiments revealed that miR-133a-3p could directly target COL1A1 and suppress its expression; thus, inhibiting proliferation and metastasis, and promoting the apoptosis of ESCC cells (29).
In the tumor microenvironment, ECM plays an important role in the occurrence and development of tumors. The activation of ECM is a marker for cancer cells to form tumors. The major component of ECM is COL (mainly type I COL. COL1A1 and COL1A2, two subtypes of type I COL, participate in the adhesion, proliferation and differentiation of cancer cells (30,31). To date, most research has focused on bone-related COL1A2, as the bone is a fertile soil for metastatic cancer cells. In pathological conditions, once cancer cells enter the bone matrix, bone matrix confusion and abnormal cell behavior occur. Tumor invasion is greatly promoted because of interacting with tumor microenvironment, while the integrity and kinetic stability of ECM are destroyed (32). This is also consistent with clinical findings that COL1A2 expression is related to tumor staging.
Thrombospondin-1 (TSP1) can bind to cluster of differentiation 36 receptors to inhibit angiogenesis, and induce apoptosis (33). The Atlas of Genetics and Cytogenetics in Oncology and Hematology specifically states that endogenous TSP1 reduces the mRNA level of COL1A2. Thus, the increased angiogenesis in tissues TSP1 may be caused by the overexpression of COL1A2 (34). Thus, TSP1 may contribute to tumor growth.
This study had a number of limitations. The upstream and downstream regulatory pathways of COL1A2 have not yet been uncovered. Further, the specific component of ECM that plays an important role in cancer progression and metastasis needs to be identified. Other pathways involving COL1A2 in cancer cells should also be subject to further study to explore its extra functions.
In conclusion, COL1A2 was highly expressed in ESCA tissue samples. Additionally, the proliferation and metastasis of the Eca109 and TE-1 cell lines were significantly attenuated by siRNA-COL1A2-mediated small interference. Further, the expression level of COL1A2 was obviously related to Akt and EMT pathways.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors would like to thank the Department of Thoracic Surgery, The First Affiliated Hospital of Soochow University.
Funding: This work was supported by The National Natural Science Foundation of China (grant no. 81800279), Natural Science Foundation of Jiangsu Province (grant no. BK20180197) and Jiangsu Provincial Research Foundation for Basic Research (grant no. BK20191174).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). All participants signed written informed consent forms, and the study was approved by the Ethics Committee of The First Affiliated Hospital of Soochow University (2018011).
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7867
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7867). The authors have no conflicts of interest to declare.
(English Language Editor: D. Huleatt)
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Lin Y, Totsuka Y, He Y, et al. Epidemiology of esophageal cancer in Japan and China. J Epidemiol 2013;23:233-42. 10.2188/jea.JE20120162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Horst S, de Maat MFG, van der Sluis PC, et al. Extended thoracic lymph node dissection in robotic-assisted minimal invasive esophagectomy (RAMIE) for patients with superior mediastinal lymph node metastasis. Ann Cardiothorac Surg 2019;8:218-25. 10.21037/acs.2019.01.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Short MW, Burgers KG, Fry VT. Esophageal Cancer. Am Fam Physician 2017;95:22-8. [PubMed] [Google Scholar]
- 5.Lagergren J, Lagergren P. Recent developments in esophageal adenocarcinoma. CA Cancer J Clin 2013;63:232-48. 10.3322/caac.21185 [DOI] [PubMed] [Google Scholar]
- 6.Bella J, Hulmes DJ. Fibrillar Collagens. Subcell Biochem 2017;82:457-90. 10.1007/978-3-319-49674-0_14 [DOI] [PubMed] [Google Scholar]
- 7.Nissen NI, Karsdal M, Willumsen N. Collagens and Cancer associated fibroblasts in the reactive stroma and its relation to Cancer biology. J Exp Clin Cancer Res 2019;38:115. 10.1186/s13046-019-1110-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Z, Jiang Y, Chen S, et al. An insertion/deletion polymorphism in the 3' untranslated region of type I collagen a2 (COL1A2) is associated with susceptibility for hepatocellular carcinoma in a Chinese population. Cancer Genet 2011;204:265-9. 10.1016/j.cancergen.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 9.Ji J, Zhao L, Budhu A, et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol 2010;52:690-7. 10.1016/j.jhep.2009.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misawa K, Kanazawa T, Misawa Y, et al. Hypermethylation of collagen α2 (I) gene (COL1A2) is an independent predictor of survival in head and neck cancer. Cancer Biomark 2011-2012;10:135-44. 10.3233/CBM-2012-0242 [DOI] [PubMed] [Google Scholar]
- 11.Wu YH, Chang TH, Huang YF, et al. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene 2014;33:3432-40. 10.1038/onc.2013.307 [DOI] [PubMed] [Google Scholar]
- 12.Shintani Y, Hollingsworth MA, Wheelock MJ, et al. Collagen I promotes metastasis in pancreatic cancer by activating c-Jun NH(2)-terminal kinase 1 and up-regulating N-cadherin expression. Cancer Res 2006;66:11745-53. 10.1158/0008-5472.CAN-06-2322 [DOI] [PubMed] [Google Scholar]
- 13.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249-64. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- 14.Varma S. Blind estimation and correction of microarray batch effect. PLoS One 2020;15:e0231446. 10.1371/journal.pone.0231446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-d8. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su G, Morris JH, Demchak B, et al. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics 2014;47:8.13.1-24. [DOI] [PMC free article] [PubMed]
- 18.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-w102. 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo H, Song H, Mao R, et al. Targeting valosin-containing protein enhances the efficacy of radiation therapy in esophageal squamous cell carcinoma. Cancer Sci 2019;110:3464-75. 10.1111/cas.14184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu ML, Lin JB, Li X, et al. Current state of esophageal cancer surgery in China: a national database analysis. BMC Cancer 2019;19:1064. 10.1186/s12885-019-6191-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong Y, Ding ZY. PD-1 Inhibitors in the Advanced Esophageal Cancer. Front Pharmacol 2019;10:1418. 10.3389/fphar.2019.01418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Exposito JY, Valcourt U, Cluzel C, et al. The fibrillar collagen family. Int J Mol Sci 2010;11:407-26. 10.3390/ijms11020407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibanez de Caceres I, Dulaimi E, Hoffman AM, et al. Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer Res 2006;66:5021-8. 10.1158/0008-5472.CAN-05-3365 [DOI] [PubMed] [Google Scholar]
- 24.Hayashi M, Nomoto S, Hishida M, et al. Identification of the collagen type 1 α 1 gene (COL1A1) as a candidate survival-related factor associated with hepatocellular carcinoma. BMC Cancer 2014;14:108. 10.1186/1471-2407-14-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou X, Feng B, Dong T, et al. Up-regulation of type I collagen during tumorigenesis of colorectal cancer revealed by quantitative proteomic analysis. J Proteomics 2013;94:473-85. 10.1016/j.jprot.2013.10.020 [DOI] [PubMed] [Google Scholar]
- 26.Liang Y, Diehn M, Bollen AW, et al. Type I collagen is overexpressed in medulloblastoma as a component of tumor microenvironment. J Neurooncol 2008;86:133-41. 10.1007/s11060-007-9457-5 [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Liu J, Wei X, et al. A feature-based analysis identifies COL1A2 as a regulator in pancreatic cancer. J Enzyme Inhib Med Chem 2019;34:420-8. 10.1080/14756366.2018.1484734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonazzi VF, Nancarrow DJ, Stark MS, et al. Cross-platform array screening identifies COL1A2, THBS1, TNFRSF10D and UCHL1 as genes frequently silenced by methylation in melanoma. PLoS One 2011;6:e26121. 10.1371/journal.pone.0026121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin Y, Du L, Li X, et al. miR-133a-3p suppresses cell proliferation, migration, and invasion and promotes apoptosis in esophageal squamous cell carcinoma. J Cell Physiol 2019;234:12757-70. 10.1002/jcp.27896 [DOI] [PubMed] [Google Scholar]
- 30.Kokenyesi R, Murray KP, Benshushan A, et al. Invasion of interstitial matrix by a novel cell line from primary peritoneal carcinosarcoma, and by established ovarian carcinoma cell lines: role of cell-matrix adhesion molecules, proteinases, and E-cadherin expression. Gynecol Oncol 2003;89:60-72. 10.1016/S0090-8258(02)00152-X [DOI] [PubMed] [Google Scholar]
- 31.Ritty TM, Herzog J. Tendon cells produce gelatinases in response to type I collagen attachment. J Orthop Res 2003;21:442-50. 10.1016/S0736-0266(02)00200-0 [DOI] [PubMed] [Google Scholar]
- 32.Kolb AD, Bussard KM. The Bone Extracellular Matrix as an Ideal Milieu for Cancer Cell Metastases. Cancers (Basel) 2019;11:1020. 10.3390/cancers11071020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Q, Finley SD. Predictive model identifies strategies to enhance TSP1-mediated apoptosis signaling. Cell Commun Signal 2017;15:53. 10.1186/s12964-017-0207-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huret JL, Dessen P, Bernheim A. Atlas of Genetics and Cytogenetics in Oncology and Haematology, updated. Nucleic Acids Res 2001;29:303-4. 10.1093/nar/29.1.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as