Abstract
Background
This study aimed to investigate the SALL4 expression in lung cancer, determine if SALL4 regulates the biological functions of lung cancer cells at the cellular level, and clarify the possible mechanisms involved.
Methods
Immunohistochemistry was used to detect the SALL4 expression messenger RNA (mRNA) in 62 cases of lung cancer tissue microarray. The correlation of SALL4 with the clinical pathological parameters and overall life cycle of patients and the impact of disease-free life cycle was analyzed. Reverse transcription-polymerase chain reaction (RT-PCR) and western blotting were used to detect the SALL4 expression in lung cancer cell lines and nude mouse models. 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay, colony-forming assay, and flow cytometry were used to detect the effects of interference with SALL4 expression on lung cancer cell proliferation and transplant tumor models; the effect of interference with SALL4 expression on the growth of transplanted tumors in vivo was also examined.
Results
SALL4 was highly expressed in lung cancer tissues and cell lines and was closely related to the patient’s TNM stage and lymph node metastasis. Compared to patients with a high SALL4 expression, those with a lower SALL4 expression had a longer overall and disease-free survival. The expression of SALL4 is an independent risk factor for the prognosis of lung cancer patients. Interference with SALL4 expression can significantly inhibit cell proliferation and clonal formation. Interfering with the expression of SALL4 can arrest the cells in the G0/G1 phase by inhibiting the expression of the cell cycle–related proteins, cyclin B, cyclin E, and cyclin D1. Furthermore, wound-healing and Transwell assays showed that interference with SALL4 expression could significantly inhibit the migration and invasion of lung cancer cells, while experiments in nude mice showed that interference with SALL4 expression could significantly inhibit the size and weight of transplanted tumors.
Conclusions
SALL4 was highly expressed in lung cancer cell lines. Interference with the expression of SALL4 can effectively inhibit the proliferation, migration and invasion of lung cancer cells, promote cell cycle arrest, and play the function of tumor suppressor genes. SALL4 may be a new target for the diagnosis and treatment of lung cancer.
Keywords: Sal-like protein 4 (SALL4), lung cancer, proliferation, invasion, migration, survival
Introduction
Lung cancer is a common malignant tumor in clinical practice, with its morbidity and mortality ranking high among malignancies in China and around the world. Lung cancer can be divided into small cell lung cancer and non-small cell lung cancer, the latter of which accounts for more than 85% of total cases (1). The current traditional treatment methods for lung cancer include surgical resection, radiotherapy, and chemotherapy. However, for advanced lung cancer patients who have metastasis or low chemotherapy sensitivity, the treatment effect is not ideal with the 5-year survival rate being low (1,2). The occurrence of lung cancer is a process involving the abnormal expression of multiple oncogenes and the inactivation of tumor suppressor genes. In recent years, molecular targeted therapies based on specific gene mutation types in lung cancer patients have made great progress and improved the survival of lung cancer patients to a certain extent (3,4). Therefore, exploring the molecular mechanism of lung cancer occurrence and metastasis may provide new ideas and intervention targets for the clinical treatment of lung cancer, and may significantly enhance its efficacy.
Spalt-like transcription factor (SALL) is an important transcription factor for renewal and pluripotency. Its family contains four members: Sal-like protein 1 (SALL1), SALL2, SALL3, and SALL4 (5). SALL functions to maintain embryonic stem cells, with the SALL4 gene being considered as the main regulatory factor for members of the SALL gene family. It can participate in the self-renewal and differentiation process of embryonic stem cells by influencing epigenetic mechanisms. It plays an important role in maintaining the function of embryonic stem cells, and promoting organ formation and body development (6).In recent years, studies have found that SALL4 is abnormally expressed in many types of tumors. The high expression of SALL4 is also related to tumor recurrence, invasion and metastasis, and tumor resistance, which affects the prognosis of tumor patients (7,8). SALL4 was found to inhibit cell apoptosis and promote the abnormal proliferation of leukemia cells by downregulating the expression of apoptosis-related proteins (9). The expression of SALL4 has also been related to the prognosis of liver cancer patients, and the prognosis of patients with high SALL4 expression is generally poor (10). Finally, knockdown of SALL4 gene expression was demonstrated to inhibit the cell cycle, leading to the blockage of breast cancer cell proliferation (11).Taken together, the above research indicates that SALL4 participates in tumorigenesis and plays an important regulatory role in tumor formation. However, no systematic research on the biological function, clinical significance, and regulation mechanism of SALL4 in lung cancer has been conducted. This study thus aimed to principally explore the clinical significance of SALL4 expression in lung cancer tissues and its regulation of the biological functions of lung cancer cells. The initial results have clarified, to a degree, the mechanism by which SALL4 participates in the occurrence and development of lung cancer, and this may provide new ideas for finding new therapeutic targets in lung cancer. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7939).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all individual participants. Experiments involving patients and animals were performed under a project license (NO. 2018-54) granted by the ethics committee of Yijishan Hospital, First Affiliated Hospital of Wannan Medical College. In addition, this study complies with Chinese animal care and use guidelines.
Clinical patients
Data and samples from 30 patients (16 females and 14 males) with lung adenocarcinoma who were treated and surgically resected in the thoracic surgery department of our hospital from January 2017 to September 2019 were collected. The age ranged from 31 to 74 years, with an average of 59.2±9.8 years. All patients underwent lung cancer resection with tumor tissue specimens and paired normal tissues adjacent to the cancer (>5 cm from the edge of the cancerous tissue) being collected. None of the patients had undergone any treatment such as radiotherapy or chemotherapy before surgery. The samples were fresh samples. After being rinsed with phosphate-buffered saline (PBS), the samples were immediately put into liquid nitrogen and stored for messenger RNA (mRNA) and protein detection. All enrolled patients were informed of the details of the study, and signed an informed consent form. This study was approved by the ethics committee of our unit. Immunohistochemical staining and survival analysis included lung cancer tissue chip purchased from Shanghai Zhuoli Biotechnology Co., Ltd. (Shanghai, China). This consisted of 62 cases of lung adenocarcinoma in the survival period: 62 cases of cancer/1 point adjacent to cancer, with an operation time from July 2010 to June 2015. The follow-up time was until August 2018, for a follow-up time of3–5 years.
Cell culture and processing
Five lung cancer cell lines, A549, H157, H1650, H358, and H1703, and normal bronchial epithelial cells, 16-HBE, were purchased from the Institute of Basic Medicine, at the Chinese Academy of Medical Sciences. The cells were cultured with Roswell Park Memorial Institute 1640 (RPMI 1640) medium containing 10% fetal bovine serum (FBS) and 100 µM penicillin and streptomycin in a CO2 incubator at 37 °C and 5% CO2, and the cell suspension was distributed at different new medium. The cells in the exponential growth phase were collected in a cell culture dish, and the total mRNA and total protein in the cells were each extracted for subsequent detection.
The lung cancer A549 and H358 cells were collected in the logarithmic growth phase and inoculated onto a 6-well plate. Their cell density was adjusted to about 80% before transfection, and they were cultured in a CO2 incubator at 37 °C and 5% CO2. Transfection was carried out by Lipofectamine 2000 (Thermo Fisher Scientific, MA, USA). After 24 h of culture, G418 was used to screen positive cells. The medium was changed every 2 days. After 7 days, the culture was continued with complete medium. After cell passage, a stably transfected lung cancer cell line, SALL4, was obtained. SiRNA-SALL4 and its negative control siRNA-con were transfected into lung cancer A549 and H358 cells, and a normal control group (NC group) was set. The interference sequence and the blank control sequence were synthesized by Seymour Fisher Technology (China) Co., Ltd. (7). Specific interference sequence of 951 interference sites (5'-GACCGUUCCAGUGU AAGAUTT-3') and negative interference sequence (5'- GTTCTCCGAACGT GTCACGT-3').
Reverse transcription-polymerase chain reaction (RT-PCR) to detect the expression level of SALL4 mRNA in lung cancer tissue
(I) Approximately 0.5 g of tumor tissue was placed a mortar for grinding. During the grinding process, a small amount of liquid nitrogen was added to make the tumor tissue fully homogenized. Then, 1 mL of Trizol was added to provide the total RNA in the tissue according to the total RNA extraction kit instructions. The concentration and purity of the extracted RNA were measured with a NanoDrop 2000c ultraviolet spectrophotometer (Thermo Fisher Scientific), and the total RNA extracted was reverse transcribed to synthesize complement DNA (cDNA) according to the following reverse transcription operation kit steps. The specific reaction system for quantitative detection using the Applied Biosystem ABI 7500 RT-PCR System and SYBR Green dye method (Thermo Fisher Scientific) was as follows: 95.0 °C for 5 min, 95.0 °C for 5 s, 60.0 °C for 30 s, at 40 cycles of amplification. After the reaction was complete, the PCR product was analyzed by melting curve, the amplification curve and Ct value of each index were recorded, and the target mRNA was measured by △△Ct method.
The primer sequence for the internal reference GAPDH was as follows:
Forward: 5'-CGGAACCCGTCAGATCGTAT-3';
Reverse: 5'-AGCTTTCGATGCAGCTAGAC-3';
SALL4 primer sequence:
Forward: 5'-CCGCGCTAGAGCATGGAT-3';
Reverse: 5'-GTGAAAGTCTGCGGAGG-3'.
MTT assay to detect cell proliferation
The lung cancer cells that were growing well after transfection were collected and inoculated in a 96-well plate. The inoculation volume per well was 200 µL, with each well containing about 2,000 cells. The wells were placed in a CO2 constant-temperature incubator at 37 °C and 5% CO2, and cultured overnight. Next, 200 µL of MTT solution (5 mg/mL) was added to each well at 12, 24, 36, 48, and 72 h; wells were incubated at 37 °C for 4 h, after which time 50 µL of dimethyl sulfoxide (DMSO) was added to each well and mixed gently. A microplate reader was used to detect the A490 nm. After the absorbance values at different time points were continuously measured, the maximum and minimum values in each group of test results were removed. A growth curve was drawn according to the data obtained.
Clone formation experiment
Cells in good growth condition were trypsinized to make a single cell suspension, and the cells were resuspended in complete medium and inoculated into a 6-well plate at a cell density of 3×103 cells/well. The culture was changed every 2–3 days with the cell growth being regularly observed. After about 2 weeks, when a clonal cell cluster that was visible to the naked eye had formed, the culture was stopped and the medium discarded. After a wash with PBS buffer, 4% paraformaldehyde added Cells were fixed for 20 min, and then washed with PBS buffer twice. Crystal violet solution was added for staining, and the solution was rinsed with water three times after staining. A cell map consisting of a cell cluster of ≥50 cells as the number of clones was collected. The clone formation rate was then analyzed, and the experiment was repeated three times.
Wound-healing assay to detect cell migration ability
Cells in good growth condition were collected from each transfection group, and inoculated in a 6-well plate after trypsin digestion. Plates contained about 5× cells or 2 mLof cells per well. Cells were cultured in a constant-temperature incubator at 37 °C and 5% CO2 overnight. The next day, the old cell culture medium was discarded, and the cells were placed under an inverted microscope. The small tip of a micropipette was used to make a scratch on the back, perpendicular to the horizontal line. The cells were then washed with sterile PBS buffer three times, and the cells scraped during the scratching process were also washed. An appropriate amount of serum-free medium was added to the mixture, which was observed under a microscope (Shanghai Yuguang Instrument Co., Ltd., China) with a photo taken of the cell position at 0 h. The cells were cultured in a constant temperature incubator at 37 °C and 5% CO2. The cells were photographed at 48 h, and the cell position was recorded. Image J 1.46 software was used to process the photographed pictures and calculate the migration distance and cell migration rate of each group of cells.
Transwell assay to detect cell invasion ability
Before the start of the experiment, a pipette was used to remove the remaining liquid in the culture plate, and then new serum-free medium was added (50 µL per well) to each well, which were left at room temperature for 30 min. The 50 mg/L of Matrigel was mixed with the diluent in a 1:1 ratio, and the upper chamber surface of the bottom membrane of the Transwell chamber was coated by mixture, and left at 4 °C to air dry. The cells were cultivated with serum-free cell culture medium 12–24 h in advance, so that the cells were in a state of starvation, and then cells in the logarithmic growth phase were collected, washed two to three times with PBS buffer, digested with trypsin, centrifuged, and resuspended. For cell counting, the cell density was adjusted to 1×106 cells/mL. Next, 600 µL of culture medium containing 20% FBS was added to the 24-well plate prepared in advance, which was placed in the lower chamber of the Transwell. After this, 100 µL of cell suspension were carefully added to the cell compartment of the Transwell, with care taken to avoid air bubbles during cell inoculation. Repeat observation after incubation for 12–48 h. Next, the 24-well plate was taken out of the Transwell chamber, and 600 µL of methanol was added for fixation. After 30 min, the culture medium was removed by a pipette, and washed with PBS buffer two to three times, to remove methanol and any cells that had not been invaded. The remaining cells were stained with 0.1% crystal violet for10 min. After three washes with PBS, five fields of view were randomly selected under the microscope to count the cells, and the average value was calculated based on the three multiple wells.
Western blotting
An appropriate amount of pre-cooled RIPA cell lysate and 10 µL of phenylmethylsulfonyl fluoride (PMSF) protease inhibitor was added to the tissue or cell fluid. The total protein was extracted according to the total protein extraction kit P1250 (Priele Genetic Technology Co., LTD., Beijing, China) instructions, and the operating steps of the Bicinchoninic acid disodium (BCA) detection kit (Beijing Leigen Biotechnology Co. LTD., China) were followed to determine the protein concentration in the sample. According to the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) preparation ratio, 10% separating gel and 5% concentrated gel were prepared and left to stand at room temperature for 30 min. Then, 10 µL of protein sample (50 µg) was added to each sample well, and protein electrophoresis was performed under the preset conditions of 30 min at 90 V. After the electrophoresis was completed, it was transferred to a polyvinylidene fluoride (PVDF) membrane, and 5% skimmed milk powder was placed on a shaker for blocking for 1 h. The mouse anti-human SALL4 primary antibody (diluted with 1:500) was incubated in a shaking table at 4 °C overnight. After washing with TBST for 3 times, the goat anti-rat secondary antibody (diluted with 1:5,000) with horseradish peroxidase-labeled was added and incubated at room temperature for 2 h and then washed with TBST. The images were exposed with enhanced chemiluminescence (ECL) developer and analyzed with microscope.
Subcutaneous transplantation tumor experiment in nude mice
BALB/c male nude mice weighing 18–20 g and 4–6 weeks old were purchased from the Shanghai Experimental Animal Center of the Chinese Academy of Sciences and raised in the specific pathogen-free (SPF) Animal Experimental Center. Here, the mice were kept in a 12-h alternating light-dark cycle at a relative humidity of 50%±10%, and a temperature of 22±2 °C. The enclosure was disinfected with ultraviolet radiation regularly. Nude mice were allowed to drink freely, and were given special mice pellets for sterilization.
Fast-growing A549 cells in a short interfering RNA (siRNA)-con group and in the and siRNA-SALL4 group were trypsinized and washed twice with PBS buffer for preparation of a single-cell suspension, with the cell density being adjusted to 1×107 cells/mL. The armpits of nude mice were sterilized with iodophor, 200 µL of cell suspension was drawn with a disposable sterile syringe (with the syringe being gently shook to ensure the cells were evenly distributed), and the cell suspension was injected into the armpits of nude mice. When a naked mouse had a transplanted tumor that was visible to the naked eye, the long and short diameters of the transplanted tumor were measured every 3 days with a Vernier caliper to calculate the volume of the transplanted tumor. The volume calculation was as follows: V (mm3) = long diameter × short diameter × short diameter × 0.5. A tumor growth curve was drawn according to the tumor volume. On the 28th day of subcutaneous injection, the nude mice were anesthetized and sacrificed. The skin around the transplanted tumor was disinfected with alcohol. The transplanted tumors were completely excised with medical scissors and tweezers. After the mucous tissues around the tumor were cleaned up, the tumor was weighed and recorded. The transplanted tumor of each nude mouse was placed on a white paper from large to small, and photographed. The separated transplanted tumor tissue was frozen in liquid nitrogen and stored in a refrigerator at −80 °C until use. RT-PCR and western blot were used to detect the expression levels of SALL4, and related mRNA and protein in transplanted tumor tissues.
Immunohistochemical staining
After the tumor tissue was removed from the nude mice, it was fixed with 10% formalin, embedded in paraffin, sliced, and blocked by peroxidase; then, the antibody was fixed at a concentration recommended by the antibody specification. The antibody was diluted by dilution solution, 50–100 µL of antibody working solution was added according to the area of the tissue section, and the solution was incubated overnight in a refrigerator at 4 °C. After this, the protein expression was observed, with the distribution and cell location in the tissue in each slice being viewed by a microscope (Shanghai Yuguang Instrument Co., Ltd., China). Pictures were taken of three randomly selected fields of view for the record.
Statistical methods
All data were analyzed using SPSS19.0 and Graphpad prism 6.0 software. Quantitative data are expressed as mean ± standard error (). The comparison between the two groups was performed by t test. The count data were tested by a two-tailed test. Kaplan-Meier method was used to plot the patient survival curve. With a P value <0.05 indicating statistical significance.
Results
SALL4 was highly expressed in lung cancer tissues and was related to the clinicopathological characteristics of patients
In order to detect the expression of SALL4 in lung cancer tissues, RT-PCR was first used to detect the expression level of SALL4 mRNA in 30 lung cancer tissues and their adjacent tissues. As shown in Figure 1A, the expression level of SALL4 mRNA in tumor tissues was significantly higher than that in the normal adjacent tissues (P<0.05). The expression of SALL4 protein in eight pairs of samples was further examined by western blotting, as shown in Figure 1B: the expression level of SALL4 protein in eight tumor tissues was significantly higher than that in normal adjacent tissues (P<0.05). This shows that the expression of SALL4 was significantly upregulated in lung cancer tissues.
Figure 1.
SALL4 was highly expressed in lung cancer tissues. (A) RT-PCR was used to detect the expression level of SALL4 mRNA in lung cancer tissues and adjacent normal tissues, (B) Western blot was used to detect the expression level of SALL4 in eight lung cancer tissues and corresponding adjacent normal tissues. (C) Immunohistochemical detection of SALL4 expression in lung cancer tissues and corresponding adjacent normal tissues. The scale represents 100 µm. (D,E) Kaplan-Meier analysis of the relationship between the expression of SALL4 and the overall life cycle and disease-free life cycle of lung cancer patients. *, P<0.05. SALL4, Sal-like protein 4; RT-PCR, reverse transcription-polymerase chain reaction; mRNA, messenger RNA.
Further immunohistochemical analysis of SALL4 expression in 62 cases of cancer tissues and the corresponding adjacent normal tissues was conducted to determine the clinical pathological correlation of the patients. As shown in Figure 1C, SALL4 protein was mainly located in the nucleus and occasionally located in the cytoplasm, appearing as brownish-yellow particles. The positive rate of SALL4 expression in normal adjacent tissues was 19.4% (12/62), while the positive rate of expression in tumor tissues was 67.7% (42/62), which was significantly higher (P<0.05). As shown in Table 1, the expression of SALL4 was closely related to the TNM stage and lymph node metastasis (P<0.05); high SALL4 expression was correlated with the patient’s gender, age, smoking status, tumor size, and degree of differentiation, but not significantly so (P>0.05). The number of lymph node metastases in the high SALL4 expression group and the number of cases in patients with stage III–IV TNM were significantly higher than those in the low SALL4 expression group, indicating that SALL4 expression affects the disease progression of lung cancer patients. Kaplan-Meier survival curve showed that the overall survival period and disease-free survival period of patients with high SALL4 expression were significantly lower than those with low SALL4 expression (P<0.05) (Figure 1D,E). Univariate and multivariate regression models were used to analyze independent risk factors for the prognosis of lung cancer patients. The results of univariate analysis showed that SALL4 expression, tumor differentiation, TNM staging, and lymph node metastasis were influencing factors for the overall survival of lung cancer patients. Further multivariate analysis showed that SALL4 expression, TNM staging, and lymph node metastasis were independent risk factors affecting the overall survival of lung cancer patients (Table 2).
Table 1. Clinicopathological correlation analysis between SALL4 expression and patients.
| Clinical features | SALL4 expression | P | |
|---|---|---|---|
| Low expression (n=20) | High expression (n=42) | ||
| Gender | 0.239 | ||
| Male | 11 | 25 | |
| Female | 9 | 17 | |
| Age | 0.946 | ||
| <50 | 7 | 15 | |
| ≥50 | 13 | 27 | |
| Differentiation | 0.137 | ||
| Poorly differentiated | 15 | 29 | |
| Well differentiated | 5 | 13 | |
| Smoking | 0.322 | ||
| Yes | 8 | 17 | |
| No | 12 | 25 | |
| Tumor size | 0.418 | ||
| ≤3 cm | 7 | 19 | |
| >3 cm | 13 | 23 | |
| TNM staging | 0.012* | ||
| I–II | 15 | 20 | |
| III–IV | 5 | 22 | |
| Lymph node | 0.002* | ||
| Metastasis | 14 | 19 | |
| No | 6 | 23 | |
*, P<0.05. SALL4, Sal-like protein 4.
Table 2. Univariate and multivariate regression models were used to analyze independent risk factors for the prognosis of lung cancer patients.
| Parameter | Single factor analysis | Multiple factor analysis | |||||
|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P | Hazard ratio | 95% CI | P | ||
| Age | 0.136 | 0.788–1.825 | 0.873 | – | – | – | |
| Gender | 0.935 | 0.466–1.732 | 0.732 | – | – | – | |
| Differentiation | 1.182 | 0.812–2.537 | 0.025* | 0.937 | 0.524–1.502 | 0.233 | |
| TNM staging | 1.922 | 0.878-3.212 | 0.022* | 2.231 | 0.948–4.056 | 0.038* | |
| Smoking or with lymph node | 1.485 | 1.016-2.218 | 0.502 | 1.595 | 1.102–2.127 | 0.721 | |
| Metastasis | 1.588 | 1.094–2.366 | 0.008* | 1.219 | 0.866–3.815 | 0.016* | |
| SALL4 expression | 2.836 | 1.307–3.845 | 0.016* | 2. Ooo334 | 1.233–3.729 | 0.022* | |
*, P<0.05. SALL4, Sal-like protein 4.
Expression of SALL4 in lung cancer cell lines
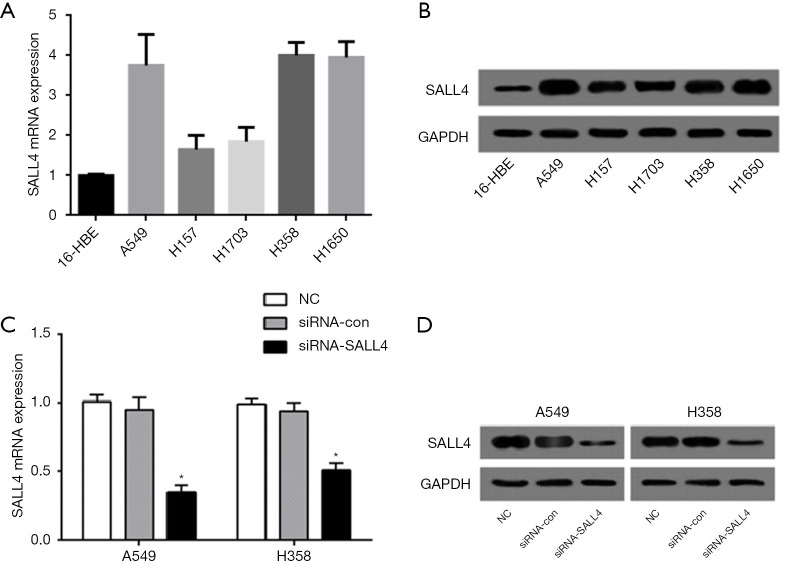
In order to further study the specific functions of SALL4 in lung cancer, RT-PCR was first used to detect the expression of SALL4 in five lung cancer cell lines, A549, H157, H1650, H358, and H1703, and normal 16-HBE bronchial epithelial cells. Compared with 16-HBE cells, the expression level of SALL4 in various lung cancer cell lines was significantly increased (P<0.05) (Figure 2A). Western blotting detected the expression of SALL4 in each cell line, and the results were consistent with those of RT-PCR, in that the expression level of SALL4 protein in the five lung cancer cell lines was significantly higher than that in the 16-HBE cells (P<0.05) (Figure 2B). Those A549 and H358 cells with a higher degree of upregulation were selected as the follow-up objects for functional research.
Figure 2.
Establishment of a lung cancer cell line that interferes with SALL4. (A) RT-PCR detection of SALL4 expression in five lung cancer cell lines, A549, H157, H1650, H358, and H1703, and normal 16-HBE bronchial epithelial cells. (B) Western blotting detection of SALL4 expression in lung cancer cell lines and normal bronchial epithelial cells. (C) RT-PCR detection of the effect of siRNA-SALL4 transfection on SALL4 mRNA expression in A549 and H358 cells. (D) Western blotting detection of transfection siRNA and the effect of SALL4 on the expression of SALL4 in A549 and H358 cells. *, P<0.05. SALL4, Sal-like protein 4; RT-PCR, reverse transcription-polymerase chain reaction; mRNA, messenger RNA.
Western blot and RT-PCR verified the transfection efficiency. The results showed that the expression level of SALL4 in lung cancer A549 and H358 cells was significantly reduced after the transfection of siRNA-SALL4, and the difference was statistically significant compared with the siRNA-con group and NC group (P<0.05) (Figure 2C,D). This indicated that the transfection was successful and the expression factor does not cause cytotoxicity to lung cancer cells.
The effect of interference with SALL4 expression on the proliferation of lung cancer cells
MTT assay was used to determine the effect of interference with SALL4 on cell proliferation activity at different time points. As shown in Figure 3A, in the two cell lines, the OD values of the A549 and H358 cells in the si-SALL4 group at 490 nm were lower than those of the siRNA-con group at 48 h, and were significantly lower than those of the siRNA-con group at 72 h (P<0.05). The cell clone formation experiment was used to analyze the clone formation of lung cancer cells by knocking down SALL4. As shown in Figure 3B, after the expression of SALL4 was knocked down, the number of cell clones of the A549 and H358 lung cancer cells was significantly reduced, as compared to that of the siRNA-con group and NC group (P<0.05). The above results suggested that inhibiting the expression of SALL4 can reduce the proliferation activity of lung cancer cells.
Figure 3.
MMT and clone formation experiments to detect the effect of knockdown of SALL4 expression on cell proliferation activity. (A) MTT assay was used to detect cell proliferation ability. (B) Plate cloning experiment was used to detect cell clone formation ability (Crystal violet 100×). *, P<0.05. SALL4, Sal-like protein 4.
The effect of interference with SALL4 expression on cell cycle and related protein expression
Flow cytometry was used to detect the cell cycle distribution of cells in each group. After the expression of SALL4 was suppressed (Figure 4A,B), the proportion of S-phase cells in A549 and H358 cells was significantly lower than that of the siRNA-con and NC groups (P<0.05). The proportion of cells in the G0/G1 phase was significantly higher than that in the siRNA-con and NC groups (P<0.05), indicating that knocking down the expression of SALL4 causes the A549 and H358 cells to arrest in the G0/G1 phase. Western blot was used to detect the expression of cell cycle–related proteins. As shown in Figure 4C,D, knocking down the expression of SALL4 in A549 and H358 lung cancer cells could significantly inhibit the expression levels of cyclin B, cyclin E, and cyclin D1 proteins. The difference between the two groups was statistically significant (P<0.05). This indicated that knocking down the expression of SALL4 could reduce the expression of the cell cycle–related proteins cyclin B, cyclin E, and cyclin D1, and block the cell cycle at G0/G1.
Figure 4.
The effect of interference with SALL4 expression on the cell cycle. (A) The effect of interference with SALL4 expression on the cell cycle was detected by flow cytometry. (B) The statistical analysis of the results of flow cytometry. (C) The effect of interference with SALL4 expression on cycle-related protein expression in A549 cells. (D) The influence of interference with SALL4 expression on the expression of cycle-related proteins in H358 cells; *, P<0.05. SALL4, Sal-like protein 4.
The effect of interference with SALL4 expression on cell migration and invasion
Wound-healing assay was used to detect the effect of knocking down the expression of SALL4 on cell migration. The width of the scratch was measured at 0 and 48 h. As shown in Figure 5A,B, in the A549 and H358 cells, the migration distance of the cells in the siRNA-SALL4 group was significantly shorter than that in the siRNA-con group and the NC group (P<0.01); that is, the width of the scratches of the siRNA-SALL4 group healed at the slowest speed, indicating that inhibition of SALL4 expression could reduce the migration ability of A549 and H358 cells.
Figure 5.
The effect of interference with SALL4 expression on the scratching and invasion ability of A549 and H358 cells. (A,B) The effect of interference with SALL4 expression on the migration of A549 and H358 cells. (C,D) The effect of interference with SALL4 expression on the invasion of A549 and H358 cells (100×, Crystal violet). **, P<0.01. SALL4, Sal-like protein 4.
Transwell assay was used to detect cell invasion ability. As shown in Figure 5C,D, the average cells number per field of view in the siRNA-SALL4 group was 65.44±5.06, the average number of cells per field was 145.11±8.21 in the NC group, and the average number of cells per field was 136±5.21 in the siRNA-con group. In H358 cells, the average number of cells per field of view in the siRNA-SALL4 group was 54.95±4.05, and the average number of cells per field of view in the NC and siRNA-con groups was 132.65±9.09 and 122.65±11.09, respectively. We could thus surmise that the number of lung cancer cells passing through the membrane in the siRNA-SALL4 group was significantly lower than that in the siRNA-con group and NC group (P<0.05). This shows that inhibiting the expression of SALL4 could reduce the invasion ability of lung cancer cells.
The effect of interference with SALL4 expression on cell proliferation in transplanted tumors in nude mice
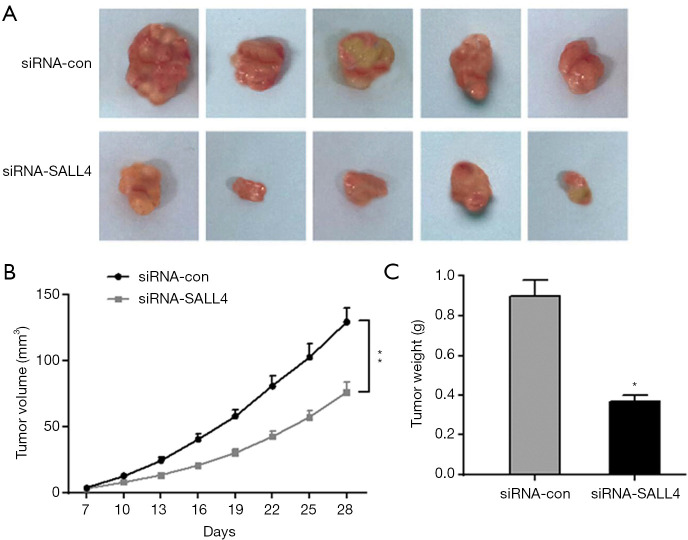
During the establishment of the nude mouse subcutaneous tumor model, the nude mice in each group were in good mental state, no deaths occurred, and the nude mice had shiny skin, sensitive responses, and normal daily intake of food and drinking water. The tumor body could be observed from the seventh day after the nude mouse xenograft model was established, and the volume of the xenograft tumor at different time points was recorded, with a corresponding the xenograft growth curve being drawn. From the 16th day, the growth rate of the xenograft tumor in the siRNA-SALL4 group was significantly lower than that in the siRNA-con group (P<0.05). On the fourth week, the nude mice were sacrificed, and the entire transplanted tumor tissue was taken. The weight of each nude mouse transplanted tumor was weighed, and the average value was calculated. The results showed that the tumor weight of the transplanted tumor in the siRNA-SALL4 group was significantly lower than that in the siRNA-con group. As shown in Figure 6, inhibiting SALL4 expression could reduce transplantation in nude mice.
Figure 6.
Subcutaneous tumor formation in nude mice was used to detect the effect of SALL4 knockdown on the tumorigenicity of cells. (A) Comparison of tumor morphology 28 days after subcutaneous inoculation. (B) Changes in tumor volume 7 days after subcutaneous inoculation of cells. (C) Tumor detection on the formed tumor tissue after killing the nude mice. *, P<0.05, **, P<0.01. SALL4, Sal-like protein 4.
Discussion
In non-small cell lung cancer, the lung adenocarcinoma generally manifests as a mucous gland that grows in the small bronchial tubes at the edge of the lung and is more common in women. A considerable part of lung adenocarcinoma is at an advanced stage when it is discovered, the tumor cells have metastasized, and the sensitivity to chemotherapy is moderate. This leads to a lower 5-year survival rate for patients with lung adenocarcinoma (12,13). In recent years, the research on the targeted therapy of lung cancer has intensified and developed rapidly. Targeted drugs can specifically bind to the target protein in lung cancer cells to inhibit the proliferation, migration, and invasion of lung cancer cells, and promote cell apoptosis, thus effectively achieving a lung cancer treatment (14,15). Because the targeted therapy method has the advantage of high specificity, in that the drug is less harmful to normal cells, an increasing number of researchers have begun to focus on the development of targeted tumor therapy drugs, such as KRAS (Kristen rat sarcoma viral oncogene, EGFR (epidermal growth factor receptor), and BRAF (V-raf murine sarcoma viral oncogene homolog) (1,16). For these gene targets, certain lung cancer targeted drugs have been developed and put into clinical use. For example, EGFR tyrosine kinase inhibitor (EGFR-TKI) is one of the main molecular targeted drugs. It can effectively inhibit the development of lung cancer, but after the clinical use of EGFR-TKI treatment, some patients develop drug resistance which leads to disease progression (17-19). Therefore, there is still an urgent need to seek new therapeutic targets and targeted therapies for lung cancer.
This study proved that SALL4 is highly expressed in lung cancer tissues and cell lines, and its expression is significantly related to the TNM stage, lymph node metastasis, and prognosis of patients. Knocking down the expression of SALL4 could significantly reduce the proliferation activity, migration, and invasion capabilities of lung cancer cells, and block cell cycle progression. Studies have shown that SALL4 is a transcript with a zinc finger structure, which plays an important regulatory function in maintaining the multidirectional differentiation, self-renewal ability of embryonic stem cells and in promoting early embryonic development (6,20). The results of a number of studies have shown that SALL4 is upregulated in tumor tissues and has a cancer-promoting effect. It is a newly discovered proto-oncogene, and preliminary research suggests that SALL4 can be used as a potential marker for tumor diagnosis or treatment. The 5-year survival rate of early cervical cancer patients with high SALL4 expression has been found to be significantly lower than that of patients with low SALL4 expression, with high SALL4 expression being reported to be an independent prognostic factor for the poor prognosis of cervical cancer (21). SALL4 is also highly expressed in patients with ovarian cancer and has been associated with poor prognosis and the promotion of the invasion and metastasis of ovarian cancer cells. SALL4 may therefore become a new molecular target and prognostic marker for ovarian cancer (22). The present study found that SALL4 expression was high in lung cancer tissues and cell lines, and its expression was significantly related to the patient's TNM stage, lymph node metastasis, and prognosis. The overall survival period and disease-free survival period of patients in the high SALL4 expression group were significantly lower than those in the low SALL4 expression group. The expression level of SALL4 may thus be an independent risk factor influencing the overall survival of lung cancer patients.
Many studies have shown that SALL4 can function as a proto-oncogene to promote tumor progression. For example, knocking down the expression of SALL4 can inhibit the migration and invasion ability of gastric cancer cells, while the overexpression of SALL4 promotes the movement, migration, and invasion ability of gastric cancer cells; furthermore, the overexpression of SALL4 induces the activation of the TGF-β/SMAD signaling pathway and triggers the epithelial–mesenchymal transition (EMT) of gastric cancer cells (23). In breast cancer, knockdown of SALL4 can inhibit the spheroidizing ability of cells. Mechanism studies have shown that SALL4 regulates the alternative splicing of CD44 by up regulating the splicing factor KHDRBS3 of CD44. CD44v overexpression inhibits the reduction of spheroidizing ability caused by KHDRBS3 knockdown (24). In addition, SALL4 can directly bind CTNNB1 promoter to transactivate CTNNB1, upregulate the activity of Wnt/β-catenin signaling pathway, and promote cervical cancer cell proliferation and tumor formation (25). The results of this study are consistent with those of other previous studies, in that the interference with SALL4 expression can significantly inhibit cell proliferation and plate clone formation. In addition, interference with SALL4 expression can block lung cancer cell lines in the G0/G1 phase. Further studies have shown that interference with SALL4 can reduce the expression of cell cycle-related proteins cyclin B, cyclin E, and cyclin D1, further indicating that SALL4 can regulate lung cancer cell growth. The cell cycle contributes to the proliferation of lung cancer cells. Through wound-healing assay, it was found that interference with SALL4 expression significantly reduced the number of cells that migrated and invaded A549 and H358 cells, indicating that inhibition of SALL4 expression could effectively inhibit the migration and invasion of lung cancer cells. Meanwhile, the in vivo experiments demonstrated that inhibiting SALL4 expression can inhibit the growth of transplanted tumors in vivo.
In conclusion, we found that SALL4 is highly expressed in lung cancer cell lines, and confirmed that interfering with SALL4 expression can effectively inhibit the proliferation, migration, and invasion of lung cancer cells, and promote cell cycle arrest and exert the function of tumor suppressor genes. Furthermore, by establishing a subcutaneous xenograft tumor model in nude mice for in vivo experiments, we found that knocking down the expression of SALL4 can also effectively inhibit tumor growth. The occurrence and progression of tumors are regulated by multiple signaling pathways. Therefore, in order to further clarify the regulation mechanism of SALL4, future research will continue to explore the impact of SALL4 on related signaling pathways in lung cancer cells. This may in turn help clarify the mechanism of lung cancer development and progression and improve lung cancer treatment.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by Natural Science Research Project of College and Universities in Anhui Province (KJ2019A0409) and the Young and Middle-aged Project of Wannan Medical College (WK2018F12).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was taken from all individual participants. Experiments involving patients and animals were performed under a project license (NO. 2018-54)granted by the ethics committee of Yijishan Hospital, First Affiliated Hospital of Wannan Medical College. In addition, this study complies with Chinese animal care and use guidelines.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7939
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7939
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7939). The authors have no conflicts of interest to declare.
(English Language Editor: J. Gray)
References
- 1.Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. 10.1016/S0140-6736(16)30958-8 [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Canales J, Parra-Cuentas E, Wistuba II. Diagnosis and Molecular Classification of Lung Cancer. Cancer Treat Res 2016;170:25-46. 10.1007/978-3-319-40389-2_2 [DOI] [PubMed] [Google Scholar]
- 3.Duma N, Santana-Davila R, Molina JR. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin Proc 2019;94:1623-40. 10.1016/j.mayocp.2019.01.013 [DOI] [PubMed] [Google Scholar]
- 4.Gregg JP, Li T, Yoneda KY. Molecular testing strategies in non-small cell lung cancer: optimizing the diagnostic journey. Transl Lung Cancer Res 2019;8:286-301. 10.21037/tlcr.2019.04.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matyskiela ME, Couto S, Zheng X, et al. SALL4 mediates teratogenicity as a thalidomide-dependent cereblon substrate. Nat Chem Biol 2018;14:981-7. 10.1038/s41589-018-0129-x [DOI] [PubMed] [Google Scholar]
- 6.Yang J. SALL4 as a transcriptional and epigenetic regulator in normal and leukemic hematopoiesis. Biomark Res 2018;6:1. 10.1186/s40364-017-0115-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang G, Liu CT. Knockdown of SALL4 overcomes cisplatin-resistance through AKT/mTOR signaling in lung cancer cells. Int J Clin Exp Pathol 2018;11:634-41. [PMC free article] [PubMed] [Google Scholar]
- 8.Tatetsu H, Kong NR, Chong G, et al. SALL4, the missing link between stem cells, development and cancer. Gene 2016;584:111-9. 10.1016/j.gene.2016.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheikhrezaei Z, Heydari P, Farsinezhad A, et al. A New Indole Derivative Decreased SALL4 Gene Expression in Acute Promyelocytic Leukemia Cell Line (NB4). Iran Biomed J 2018;22:99-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin F, Han X, Yao SK, et al. Importance of SALL4 in the development and prognosis of hepatocellular carcinoma. World J Gastroenterol 2016;22:2837-43. 10.3748/wjg.v22.i9.2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirican E, Akkiprik M. Functional and clinical significance of SALL4 in breast cancer. Tumour Biol 2016;37:11701-9. 10.1007/s13277-016-5150-7 [DOI] [PubMed] [Google Scholar]
- 12.Grenda TR, Revels SL, Yin H, et al. Lung Cancer Resection at Hospitals With High vs Low Mortality Rates. JAMA Surg 2015;150:1034-40. 10.1001/jamasurg.2015.2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lederman D, Easwar J, Feldman J, et al. Anesthetic considerations for lung resection: preoperative assessment, intraoperative challenges and postoperative analgesia. Ann Transl Med 2019;7:356. 10.21037/atm.2019.03.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shroff GS, de Groot PM, Papadimitrakopoulou VA, et al. Targeted Therapy and Immunotherapy in the Treatment of Non-Small Cell Lung Cancer. Radiol Clin North Am 2018;56:485-95. 10.1016/j.rcl.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 15.Waqar SN, Morgensztern D. Treatment advances in small cell lung cancer (SCLC). Pharmacol Ther 2017;180:16-23. 10.1016/j.pharmthera.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 16.Arbour KC, Riely GJ. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA 2019;322:764-74. 10.1001/jama.2019.11058 [DOI] [PubMed] [Google Scholar]
- 17.Li F, Zhu T, Cao B, et al. Apatinib enhances antitumour activity of EGFR-TKIs in non-small cell lung cancer with EGFR-TKI resistance. Eur J Cancer 2017;84:184-92. 10.1016/j.ejca.2017.07.037 [DOI] [PubMed] [Google Scholar]
- 18.Planchard D, Boyer MJ, Lee JS, et al. Postprogression Outcomes for Osimertinib versus Standard-of-Care EGFR-TKI in Patients with Previously Untreated EGFR-mutated Advanced Non-Small Cell Lung Cancer. Clin Cancer Res 2019;25:2058-63. 10.1158/1078-0432.CCR-18-3325 [DOI] [PubMed] [Google Scholar]
- 19.Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer 2018;17:38. 10.1186/s12943-018-0777-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tahara N, Kawakami H, Chen KQ, et al. Sall4 regulates neuromesodermal progenitors and their descendants during body elongation in mouse embryos. Development 2019;146:dev177659. 10.1242/dev.177659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di C, Sun J, Zhang H, et al. High expression of SALL4 is associated with poor prognosis in squamous cell carcinoma of the uterine cervix. Int J Clin Exp Pathol 2018;11:1391-8. [PMC free article] [PubMed] [Google Scholar]
- 22.Yang M, Xie X, Ding Y. SALL4 is a marker of poor prognosis in serous ovarian carcinoma promoting invasion and metastasis. Oncol Rep 2016;35:1796-806. 10.3892/or.2016.4545 [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Zhang P, Shao M, et al. SALL4 activates TGF-beta/SMAD signaling pathway to induce EMT and promote gastric cancer metastasis. Cancer Manag Res 2018;10:4459-70. 10.2147/CMAR.S177373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto Y, Itou J, Sato F, et al. SALL4 - KHDRBS3 network enhances stemness by modulating CD44 splicing in basal-like breast cancer. Cancer Med 2018;7:454-62. 10.1002/cam4.1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen M, Li L, Zheng PS. SALL4 promotes the tumorigenicity of cervical cancer cells through activation of the Wnt/beta-catenin pathway via CTNNB1. Cancer Sci 2019;110:2794-805. 10.1111/cas.14140 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as