Abstract
Background
Kinase domain duplication (KDD) is a special type of large genomic rearrangement (LGR), occurring in the kinase domain of protein kinase genes. KDD of some lung cancer driver genes, such as
EGFR
KDD, has been identified and implicated to be oncogenic in non-small cell lung cancer (NSCLC). The present study aims to interrogate the spectrum of KDD occurring on classic driver genes in Chinese lung cancer patients without the presence of classic lung cancer driver mutations.
Methods
We retrospectively enrolled 10,525 Chinese lung cancer patients who met the following inclusion criteria; (I) do not carry classic lung cancer driver mutations in any of the 8 driver genes and (II) tyrosine kinase inhibitor (TKI)-naïve. Capture-based targeted sequencing was performed on tissue or plasma samples. LGR and KDD were identified by using in-house analysis scripts. The prevalence and distribution of LGR and KDD in our cohort were analyzed.
Results
The median age of the cohort was 64 years with 68.7% being male. Among all patients, 23.2% and 51.8% were diagnosed with stage III and IV disease respectively. We identified 43 cases (0.41%) harboring LGR in one of the driver genes (EGFR/ERBB2/ALK/RET/ROS1/MET/BRAF), with 24 (0.23%) patients harboring KDD. Of the patients harboring KDD, a majority (n=19) harbored canonical EGFR-KDD involving exons 18–25, whilst one patient harbored duplications of EGFR exons 18–26. There were three MET-KDD patients; in two, the alteration occurred in exons 15–21 and in one, the alteration occurred in exons 3–21. One patient harbored RET-KDD involving exons 12–18. KDD showed a comparable prevalence in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) (0.33% vs. 0.11%, P=0.118). Nineteen non-KDD LGRs, spanning six genes including EGFR (n=6), MET (n=3), ALK (n=4), ROS1 (n=2), ERBB2 (n=2) and BRAF (n=2), were found, each occurring in one patient. The prevalence of LGR in LUADs and LUSCs was comparable (0.55% vs. 0.38%, P=0.452).
Conclusions
We observed a prevalence of 0.41% and 0.23% for LGR and KDD, respectively. Twenty-four different LGR alterations, including 5 KDDs and 19 non-KDD LGRs, were observed. KDDs mainly occurred in EGFR involving exons 18–25 and non-KDD LGRs were distributed more randomly. The prevalence of LGR/KDD in LUSCs and LUADs was comparable.
Keywords: Kinase domain duplication (KDD), lung cancer, large genomic rearrangement (LGR), EGFR-KDD
Introduction
Protein kinases catalyze the transfer of the γ-phosphate group of adenosine triphosphate (ATP) to the hydroxyl group of serine, threonine or tyrosine residues of protein substrates (1-3). These kinases mediate most signal transductions and control various cellular processes including, but not limited to, cell proliferation, movement, apoptosis, metabolism and cell cycle progression (4-7). Mutations, overexpression, and the dysregulation of protein kinases play central roles in the initiation and development of human cancers, including lung cancers, the leading cause of cancer-related deaths worldwide (8-10).
Over the past two decades, the protein kinase family has become one of the most important drug targets for lung cancer therapy (11,12). The development of tyrosine kinase inhibitors (TKIs) has revolutionized the treatment of patients with advanced refractory lung cancer harboring actionable mutations, such as EGFR (exon 19 deletion, G719, L858R, and L861Q), MET (amplification or exon 14 skipping mutation), and rearrangements of ALK, ROS1 and RET (13-19). With advancements in sequencing technologies and our understanding of lung cancer, the list for actionable mutations is expanding.
Large genomic rearrangement (LGR), including deletions or duplications of single or multi-exons (20,21), are rare alterations in lung cancer. Kinase domain duplication (KDD) is a special type of LGR occurring in the kinase domain of protein kinase genes, which results in a novel mechanism for protein kinase activation in tumor cells. A recent study revealed a KDD frequency of 0.62% in pan-cancers in a Caucasian population (22). The most well-studied KDD is EGFR-KDD, which often results from the in-frame tandem duplication of EGFR exons 18–25, causing subsequent constitutive activation of EGFR signaling. Some studies have demonstrated the oncogenic properties of EGFR-KDD from in silico, in vitro and in vivo perspectives (23-25). In silico analysis has shown that EGFR-KDD can form intramolecular dimers and in vitro studies have demonstrated that EGFR-KDD can be targeted by EGFR-TKIs, including erlotinib, afatinib and osimertinib (23). In vivo studies have revealed that increased EGFR-KDD expression promotes tumor growth in nude mice (26). Evidence suggests that KDD might be an oncogenic driver and may also act as a resistance mechanism of TKIs in lung cancer patients (27). EGFR-KDD has been recognized as a therapeutically actionable alteration in lung cancer (23,28). A series of case reports revealed that NSCLC patients with EGFR-KDD responded to EGFR-TKIs including gefitinib, erlotinib, icotinib and afatinib (23,28). A recent work indicates MET-KDD can mediate resistance to ALK inhibitor ceritinib and respond to crizotinib in an ALK-rearranged lung adenocarcinoma (LUAD) patient (27). Recently, great interest has arisen in the role of KDD alone as a primary oncogenic driver in lung cancer. However, most studies interrogating KDD have been conducted in Western populations (22,23). The prevalence of LGR, and specifically KDD in Chinese lung cancer patients remains elusive. In this study, we interrogated the genomic profiling data of 10,525 TKI-naïve Chinese lung cancer patients without the presence of driver mutations to determine the prevalence and distribution of LGR/KDD occurring on classic driver genes.
We present the following article in accordance with the Materials Design Analysis Reporting (MDAR) reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7408).
Methods
Patients and samples
Between January 2017 and October 2019, the genomic profiling records of 31,942 Chinese lung cancer patients from the Burning Rock databank were screened. A total of 10,525 patients were selected for subsequent analysis according to the following inclusion criteria: (I) without the presence of classic lung cancer driver mutations in any of the eight genes (EGFR/ERBB2/ALK/RET/ROS1/MET/BRAF/KRAS); classic lung cancer driver mutations were summarized in Table S1; (II) TKI-naïve. This study was approved by the Ethical Committee of Shenzhen People’s Hospital (No. SYL-201919-01). Informed consent was obtained from each patient for the use of their plasma or tumor tissue samples. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
DNA extraction and library construction
Tissue and plasma DNA were extracted with a QIAamp DNA formalin fixed paraffin embedded (FFPE) tissue kit and the QIAamp Circulating Nucleic Acid Kit, (Qiagen, Valencia, CA, USA), respectively, followed by end repair, phosphorylation, dA addition, and adaptor ligation for library construction. The DNA library was purified by using Agencourt AMPure beads (Beckman Coulter, Fullerton, CA, USA). Capture-based targeted sequencing was performed using a panel consisting of eight lung cancer classic drivers (Burning Rock Biotech, Guangzhou, China).
Sequencing data analysis
The raw sequencing data were preprocessed by using Trimmomatic 0.36 for trimming adaptor, low-quality reads and reads less than 50 base pairs. Preprocessed sequencing data were mapped to the human genome (hg19) by using Burrows-Wheeler Aligner 0.7.10 and the Genome Analysis Toolkit 3.2 (Broad Institute, Cambridge, MA, USA). Variant calling was performed by using VarScan and variants were annotated with ANNOVAR and SnpEff v3.6. Mutations were then filtered against common single nucleotide polymorphisms (SNP) found in 1,000 Genomes ExAC, dbSNP, and ESP6500SI-V2, ClinVar databases. LGR was detected by calculating coverage depth of capture intervals. The coverage depth data of each capture interval in EGFR/ERBB2/ALK/RET/ROS1/MET/BRAF was calculated. The average coverage of all capture regions was calculated as internal control, which was utilized to normalize the coverage of different samples to comparable scales. Those capture intervals with an obvious less (for deletions of single or multi-exons) or larger (for duplication of single or multi-exons) average coverage data than the internal control was identified as LGR. Furthermore, the certain capture intervals with an obvious larger average coverage data than others in the identical gene occurring in kinase domain of EGFR/ERBB2/ALK/RET/ROS1/MET/BRAF was identified as KDD.
Statistical analysis
Differences in two-groups were accessed by two-tailed, unpaired t-test for continuous variables with normal distribution or by Chi-square test for discontinuous variables. P<0.05 was considered statistically significant. All statistical analyses were performed in R 3.3.3 and SPSS 22.0 (IBM Corporation, Armonk, NY, USA).
Results
Patient characteristics
A total of 31,942 Chinese lung cancer patients were screened. A total of 10,525 TKI-naïve patients without the presence of classic lung cancer driver mutations were enrolled in the study. Schematic design of the study is shown in Figure S1. The median age of the cohort was 64 years, ranging from 6 to 98 years. The majority of patients (n=5,459, 51.9%) had lung adenocarcinoma (LUAD), whilst 1,836 (17.4%) had lung squamous cell carcinoma (LUSC). The histological subtype of the remaining 3,230 patients was unknown. The distribution for stage was; stage I (544/10,525, 5.2%), stage II (617/10,525, 5.9%), stage III (2,444/10,525, 23.2%) and stage IV (5,457/10,525, 51.8%). The status of stage in the remaining 1,463 cases (13.9%) was unknown. Patient characteristics are summarized in Table 1.
Table 1. The clinical information of our cohort.
| Characteristics | Enrolled patients (n=10,525), n (%) | LGR patients (n=43), n (%) | KDD patients (n=24), n (%) | P value (LGR vs. enrolled) |
P value (KDD vs. enrolled) |
|---|---|---|---|---|---|
| Median age [range], years | 64 [6–98] | 60.5 [42–80] | 59 [42–80] | 0.055 | 0.044 |
| Sex | 0.122 | 0.648 | |||
| Female | 2,762 (26.2) | 7 (16.3) | 5 (20.8) | ||
| Male | 7,226 (68.7) | 35 (81.4) | 19 (79.2) | ||
| Unknown | 537 (5.1) | 1 (2.3) | 0 | ||
| Histological type | 0.452 | 0.075 | |||
| LUAD | 5,459 (51.9) | 30 (69.8) | 18 (75.0) | ||
| LUSC | 1,836 (17.4) | 7 (16.3) | 2 (8.3) | ||
| Unknown | 3,230 (30.7) | 6 (14.0) | 4 (16.7) | ||
| Stage | 0.346 | 0.404 | |||
| I | 544 (5.2) | 0 (0.0) | 0 (0.0) | ||
| II | 617 (5.9) | 4 (9.3) | 3 (12) | ||
| III | 2,444 (23.2) | 10 (23.3) | 5 (24) | ||
| IV | 5,457 (51.8) | 26 (60.5) | 15 (60.0) | ||
| Unknown | 1,463 (13.9) | 3 (6.9) | 1 (4.0) | ||
| Total | 10,525 patients | 43 patients | 24 patients |
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; LGR, large genomic rearrangement; KDD, kinase domain duplication.
The prevalence of LGR/KDD
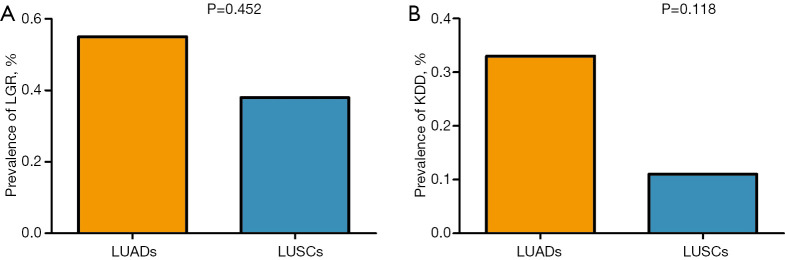
A total of 43 cases (0.41%, 43/10,525) carrying LGR alterations were identified. The median age of this group was 60.5 years, being younger in comparison to that of all enrolled patients (64 years, P=0.055, Table 1). With the exception of one lung cancer harboring BRAF exon 8 deletion and another harboring BRAF exon 14-16 deletion, 41 cases (0.39%) harbored 22 LGR alterations in one of the receptor tyrosine kinase (RTK) genes (EGFR/ERBB2/ALK/RET/ROS1/MET). The distribution of LGR is shown in Figure 1. Of 43 patients with LGR, 24 (0.23%, 24/10,525) carried KDD and 19 (0.18%, 19/10,525) carried non-KDD LGR. Twenty-four patients harbored KDD, spanning in three genes; EGFR (n=20), MET (n=3) and RET (n=1). Five different KDD alterations were observed. For patients with EGFR-KDD, all harbored canonical EGFR-KDD (19/20) involving exons 18–25 except for one, which involved exons 18–26 (Figure 2A). There were three MET-KDD patients; in two, the alteration occurred in exons 15–21 and in one, this occurred in exons 3–21 (Figure 2B). One patient harbored RET-KDD involving exons 12–18 (Figure 2C). The prevalence of LGR in LUAD and LUSC was comparable (0.55% vs. 0.38%, P=0.452, Figure 3A). The median age of the 24 KDD patients was significantly younger than that of enrolled patients (59 vs. 64 years, P=0.044, Table 1). A comparable prevalence of KDD in LUAD and LUSC was observed (0.33% vs. 0.11%, P=0.118, Figure 3B). Nineteen different non-KDD LGR alterations were found, each occurring in one patient. Non-KDD LGR were identified in EGFR (n=6), MET (n=3), ALK (n=4), ROS1 (n=2), ERBB2 (n=2) and BRAF (n=2). Collectively, the prevalence of KDD occurring on driver genes in Chinese lung cancer patients without the presence of classic lung cancer driver mutations was 0.23%. LUAD and LUSC had comparable prevalence of KDD.
Figure 1.
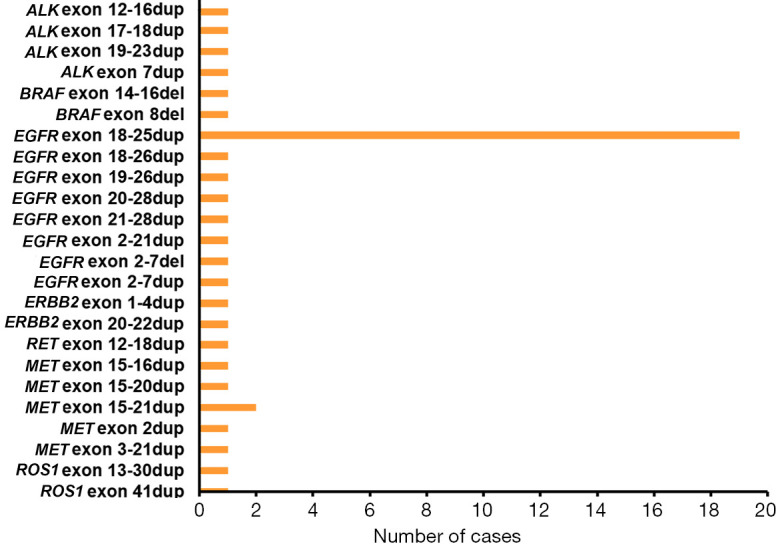
The LGR alterations in our study. The X-coordinate and the Y-coordinate indicate the number of cases and the LGR occurring on classic driver genes, respectively. dup: duplication; del: deletion. LGR, large genomic rearrangement. EGFR, epidermal growth factor receptor; KRAS, KRAS proto-oncogene, GTPase; BRAF, B-Raf proto-oncogene, serine/threonine kinase; ERBB2, erb-b2 receptor tyrosine kinase 2; ALK, ALK (anaplastic lymphoma kinase) receptor tyrosine kinase; RET, ret proto-oncogene; ROS1, ROS proto-oncogene 1, receptor tyrosine kinase; MET, MET (hepatocyte growth factor receptor gene) proto-oncogene, receptor tyrosine kinase.
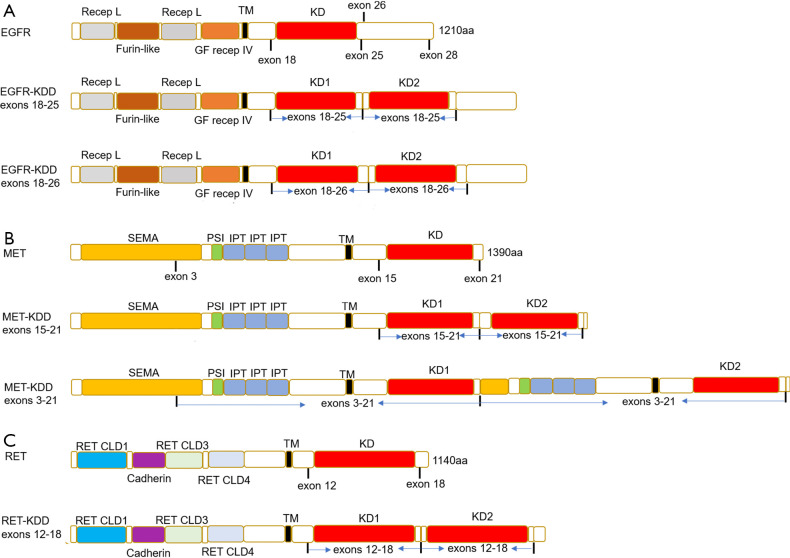
Figure 2.
The schematic representation of proteins. (A) Schematic representation of native EGFR and EGFR-KDD proteins; (B) schematic representation of native MET and MET-KDD proteins; (C) schematic representation of native RET and RET-KDD proteins. Recep L, receptor L domain; Furin-like, Furin-like cysteine rich region; GF recep IV, growth factor receptor domain IV; TM, transmembrane; KD, tyrosine kinase domain; SEMA, semaphorins; IPT, immunoglobin-like fold shared by plexins and transcription factors; PSI, plexin-semaphorin-integrin domain; RET, ret proto-oncogene; RET CLD1, RET Cadherin like domain 1; Cadherin, Cadherin domain; RET CLD3, RET Cadherin like domain 3; RET CLD4, RET Cadherin like domain 4; aa, amino acids; EGFR, epidermal growth factor receptor; MET, MET (hepatocyte growth factor receptor gene) proto-oncogene, receptor tyrosine kinase.
Figure 3.
The prevalence of LGR and KDD in our study. (A) The prevalence of LGR in LUAD and LUSC; (B) the prevalence of KDD in LUAD and LUSC; LGR, large genomic rearrangement; KDD, kinase domain duplication; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Discussion
Over the past few decades, the development of TKIs has revolutionized the management of lung cancer. With advancements in next-generation sequencing technologies and our increased understanding of lung cancer, the list for actionable mutations is expanding. It has been reported that LGR occurring on oncogenic drivers may play an important role in tumorigenesis. The significance of LGR in some cancer types has been elucidated, but remains elusive in lung cancer. For example, oncogenic LGR in EGFR has been reported in human malignant glioma (29). The distribution of LGR in oncogenic gene in transitional cell carcinoma of urinary bladder has been reported (30). As a special type of LGR alteration, KDD occurring in a few classic lung cancer driver genes, such as EGFR-KDD and MET-KDD, have been identified and implicated to be oncogenic in non-small cell lung cancer (NSCLC). KDD occurring on ERBB2, ALK, RET, ROS1 and BRAF as an oncogenic driver has not been reported. The prevalence of KDD has been interrogated in a Western population, however, the prevalence of LGR and KDD in Chinese lung cancer patients remains elusive. A previous study has demonstrated that KDD can act as an acquired resistance against TKIs in lung cancer (27). In order to investigate the prevalence and distribution of de novo KDD in Chinese lung cancer patients without the presence of driver mutations, only TKI-naïve patients were selected in our work. Herein, we conducted a retrospective study to investigate the prevalence and distribution of LGR and KDD occurring on classic driver genes in TKI-naïve Chinese lung cancer patients without the presence of classic lung cancer driver mutations.
This study revealed 0.41% of lung cancer patients harboring LGR on classic driver genes (EGFR/ERBB2/ALK/RET/ROS1/MET/BRAF) in TKI-naïve Chinese lung cancer patients without the presence of classic lung cancer driver mutations, with 24 (0.23%) patients harboring KDD. The prevalence of EGFR-KDD in Chinese lung cancers in our work was 0.20%. The similar result was observed in a previous study indicating that 0.12% east Asian NSCLC patients harbored EGFR-KDD (31). The prior studies indicated 0.04–0.07% Caucasian lung cancer patients harbored EGFR-KDD (23,25). The higher frequency of EGFR-KDD observed in our study can be explained that East Asians have a higher percentage of having EGFR mutations.
Of the patients with KDD, 24 harbored KDD, including canonical KDD rearrangements involving EGFR exons 18–25, EGFR-KDD exons 18–26, MET-KDD of exons 15–21 and uncommon rearrangements, such as MET-KDD of exons 3–21, and RET-KDD of exons 12–18 that have not previously been described in lung cancer (26,27). Similar to canonical EGFR-KDD involving exons 18–25, EGFR-KDD of exons 18-26 exhibits constitutive autophosphorylation in vitro and induces tumor growth in vivo (26). EGFR-KDD of exons 14–26 and exons 17–25 (31), reported to occur in east Asian population, were not observed in this study. Furthermore, BRAF-KDD, ERBB2-KDD, ROS1-KDD and ALK-KDD occurring in pan-cancers were not found in this study.
The study revealed 19 different non-KDD LGR alterations. Only two non-KDD alterations involving deletion and duplication of EGFR exons 2–7 that have previously been described in human malignant glioma (32,33) were found. The remaining 17 non-KDD alterations appear to have not been previously reported. Evidence suggest that both deletion and duplication of exons 2–7 in EGFR could promote oncogenesis by way of constitutive autophosphorylation in glioblastoma (29).
Multiple studies have demonstrated that KDDs can serve as therapeutic targets. In a computationally constructed three-dimensional structure model of EGFR-KDD, both gefitinib and osimertinib can bind to and inhibit EGFR-KDD, but osimertinib shows higher binding affinity (34). In vitro studies show that erlotinib, afatinib and osimertinib can inhibit EGFR-KDD phosphorylation in a dose-dependent manner (23). The efficacy of EGFR-TKIs in lung cancer patients with EGFR-KDD has been reported, but the treatment response varies. Some case reports show that LUAD patients with EGFR-KDD obtain a substantial clinical response to EGFR-TKIs (including gefitinib, icotinib, afatinib) with a PFS ranging from 5 months to over a year (23,28,31). In contrast, one case report reported an LUAD patient harboring EGFR-KDD refractory to EGFR-TKIs including erlotinib and osimertinib (31). Clinical trials to assess the efficacy of EGFR-TKIs in LUAD patients with EGFR-KDD are needed. Furthermore, the question of whether RET-KDD is the oncogenic driver in lung cancer remains unclear. Research is needed to explore the roles of RET-KDD in lung tumorigenesis.
LGR occurring oncogenic driver might play an important role in tumorigenesis. We revealed the landscape of LGR in Chinese lung cancer to pave the way for identification of possible therapeutic target, which is crucial for development of effective therapeutic strategies for those NSCLC patients who are ineligible for targeted therapy. KDD could serve as an oncogenic driver, therapeutic target, and resistance mechanism of TKIs in lung cancer. We assessed the frequency of KDD to reveal the landscape of oncogenic KDD in Chinese lung cancer patients without the presence of classic lung cancer driver mutations to maximize the identification of all patients that can benefit from precision oncology.
Conclusions
This study revealed the prevalence and distribution of KDD and non-KDD LGR in Chinese lung cancer patients without the presence of classic lung cancer driver mutations, by employing the largest Chinese lung cancer cohort to date. The results advance the understanding of LGR in lung cancer, paving the way for further evaluation of the administration of TKIs in lung cancer patients with KDD.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by the project of Department of Science and Technology of Guangdong Province (2020A1414010024).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethical Committee of Shenzhen People's Hospital (No. SYL-201919-01). Informed consent was obtained from each patient for the use of their plasma or tumor tissue samples. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the Materials Design Analysis Reporting (MDAR) reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7408
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7408). The authors have no conflicts of interest to declare.
References
- 1.Manning G, Whyte DB, Martinez R, et al. The protein kinase complement of the human genome. Science 2002;298:1912-34. 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 2.Knighton DR, Zheng JH, Ten Eyck LF, et al. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 1991;253:414-20. 10.1126/science.1862343 [DOI] [PubMed] [Google Scholar]
- 3.Knighton DR, Zheng JH, Ten Eyck LF, et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 1991;253:407-14. 10.1126/science.1862342 [DOI] [PubMed] [Google Scholar]
- 4.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 2007;26:3291-310. 10.1038/sj.onc.1210422 [DOI] [PubMed] [Google Scholar]
- 5.Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer 2018;17:58. 10.1186/s12943-018-0782-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roskoski R, Jr. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol Res 2015;94:9-25. 10.1016/j.phrs.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 7.Ou SI, Shirai K. Anaplastic Lymphoma Kinase (ALK) Signaling in Lung Cancer. Adv Exp Med Biol 2016;893:179-87. 10.1007/978-3-319-24223-1_9 [DOI] [PubMed] [Google Scholar]
- 8.Golding B, Luu A, Jones R, et al. The function and therapeutic targeting of anaplastic lymphoma kinase (ALK) in non-small cell lung cancer (NSCLC). Mol Cancer 2018;17:52. 10.1186/s12943-018-0810-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 10.Steinfort DP, Kranz S, Dowers A, et al. Sensitive molecular testing methods can demonstrate NSCLC driver mutations in malignant pleural effusion despite non-malignant cytology. Transl Lung Cancer Res 2019;8:513-8. 10.21037/tlcr.2019.07.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roskoski R, Jr. Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol Res 2019;144:19-50. 10.1016/j.phrs.2019.03.006 [DOI] [PubMed] [Google Scholar]
- 12.Wu P, Nielsen TE, Clausen MH. FDA-approved small-molecule kinase inhibitors. Trends Pharmacol Sci 2015;36:422-39. 10.1016/j.tips.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 13.Castellanos E, Feld E, Horn L. Driven by Mutations: The Predictive Value of Mutation Subtype in EGFR-Mutated Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:612-23. 10.1016/j.jtho.2016.12.014 [DOI] [PubMed] [Google Scholar]
- 14.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol 2012;13:e23-31. 10.1016/S1470-2045(11)70129-2 [DOI] [PubMed] [Google Scholar]
- 15.Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. 10.1038/nrc2088 [DOI] [PubMed] [Google Scholar]
- 16.Baik CS, Myall NJ, Wakelee HA. Targeting BRAF-Mutant Non-Small Cell Lung Cancer: From Molecular Profiling to Rationally Designed Therapy. Oncologist 2017;22:786-96. 10.1634/theoncologist.2016-0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dankner M, Rose AAN, Rajkumar S, et al. Classifying BRAF alterations in cancer: new rational therapeutic strategies for actionable mutations. Oncogene 2018;37:3183-99. 10.1038/s41388-018-0171-x [DOI] [PubMed] [Google Scholar]
- 18.Drilon A, Cappuzzo F, Ou SI, et al. Targeting MET in Lung Cancer: Will Expectations Finally Be MET? J Thorac Oncol 2017;12:15-26. 10.1016/j.jtho.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw AT, Hsu PP, Awad MM, et al. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer 2013;13:772-87. 10.1038/nrc3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodward AM, Davis TA, Silva AG, et al. Large genomic rearrangements of both BRCA2 and BRCA1 are a feature of the inherited breast/ovarian cancer phenotype in selected families. J Med Genet 2005;42:e31. 10.1136/jmg.2004.027961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat 2011;125:325-49. 10.1007/s10549-010-0817-z [DOI] [PubMed] [Google Scholar]
- 22.Ikeda S, Gay L, Pavlick D, et al. Comprehensive Genomic Profiling (CGP) of 114,200 advanced cancers identifies recurrent KDDs (KDD) and novel oncogenic fusions in diverse tumor types. Ann Oncol 2017;28:x1-6. 10.1093/annonc/mdx652 [DOI] [Google Scholar]
- 23.Gallant JN, Sheehan JH, Shaver TM, et al. EGFR KDD (EGFR-KDD) Is a Novel Oncogenic Driver in Lung Cancer That Is Clinically Responsive to Afatinib. Cancer Discov 2015;5:1155-63. 10.1158/2159-8290.CD-15-0654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baik CS, Wu D, Smith C, et al. Durable Response to Tyrosine Kinase Inhibitor Therapy in a Lung Cancer Patient Harboring Epidermal Growth Factor Receptor Tandem KDD. J Thorac Oncol 2015;10:e97-9. 10.1097/JTO.0000000000000586 [DOI] [PubMed] [Google Scholar]
- 25.Costa DB. Kinase inhibitor-responsive genotypes in EGFR mutated lung adenocarcinomas: moving past common point mutations or indels into uncommon KDDs and rearrangements. Transl Lung Cancer Res 2016;5:331-7. 10.21037/tlcr.2016.06.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciesielski MJ, Fenstermaker RA. Oncogenic epidermal growth factor receptor mutants with tandem duplication: gene structure and effects on receptor function. Oncogene 2000;19:810-20. 10.1038/sj.onc.1203409 [DOI] [PubMed] [Google Scholar]
- 27.Plenker D, Bertrand M, de Langen AJ, et al. Structural Alterations of MET Trigger Response to MET Kinase Inhibition in Lung Adenocarcinoma Patients. Clin Cancer Res 2018;24:1337-43. 10.1158/1078-0432.CCR-17-3001 [DOI] [PubMed] [Google Scholar]
- 28.Zhu YC, Wang WX, Xu CW, et al. Lung adenocarcinoma patient with an EGFR KDD (KDD) and the response to icotinib. J Thorac Dis 2018;10:E359-63. 10.21037/jtd.2018.04.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fenstermaker RA, Ciesielski MJ. Deletion and tandem duplication of exons 2 - 7 in the epidermal growth factor receptor gene of a human malignant glioma. Oncogene 2000;19:4542-8. 10.1038/sj.onc.1203802 [DOI] [PubMed] [Google Scholar]
- 30.Bazrafshani MR, Nowshadi PA, Shirian S, et al. Deletion/duplication mutation screening of TP53 gene in patients with transitional cell carcinoma of urinary bladder using multiplex ligation-dependent probe amplification. Cancer Med 2016;5:145-52. 10.1002/cam4.561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Li X, Xue X, et al. Clinical outcomes of EGFR KDD to targeted therapies in NSCLC. Int J Cancer 2019;144:2677-82. 10.1002/ijc.31895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fenstermaker RA, Ciesielski MJ, Castiglia GJ. Tandem duplication of the epidermal growth factor receptor tyrosine kinase and calcium internalization domains in A-172 glioma cells. Oncogene 1998;16:3435-43. 10.1038/sj.onc.1202156 [DOI] [PubMed] [Google Scholar]
- 33.Gan HK, Cvrljevic AN, Johns TG. The epidermal growth factor receptor variant III (EGFRvIII): where wild things are altered. FEBS J 2013;280:5350-70. 10.1111/febs.12393 [DOI] [PubMed] [Google Scholar]
- 34.Jin R, Li J, Jin Z, et al. Osimertinib confers potent binding affinity to EGFR KDD. Int J Cancer 2019;145:2884-5. 10.1002/ijc.32617 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as