Abstract
The novel SARS-CoV-2 is responsible for causing the ongoing outbreak of coronavirus disease 19 (COVID-19), a systemic infection in humans. Ever since it was first detected in December 2019, the number of confirmed cases has continued to increase. Within a short period, this disease has become a global issue, and therefore it is characterized as a pandemic. The current understanding and explanations are based on epidemiological, clinical and physiological observations. Besides, it remains a great challenge, as much remains to be understood about this new disease-causing virus. Therefore, we seek to provide an overview of SARS-CoV-2, including its classification, origin, genomic structure, replication cycle, transmission, pathogenesis, clinical aspects, diagnosis, treatments, prevention and vaccine options. We conducted a literature search for the articles published up to August 2020 using the keywords ‘SAR-CoV-2’ and ‘COVID19’ in medical databases; PubMed, google scholar, EMBASE, and web of science. Based on the information collected, the emerging COVID-19, caused by SARS-CoV-2, exhibits strong infectivity but less virulence in terms of severity of disease and mortality rates in certain age groups. It inflicts more damage in terms of peoples’ health and well-being, social life, and global economic impacts. Unfortunately, there is no adequate global and standard response to this pandemic to date, and each country is facing a crisis based on its situation, expertise, and hypotheses. While there is no effective therapy and vaccine against the novel SARS-CoV-2 yet, preventive measures are the only tool available to our disposal to control the spread of the COVID-19 pandemic. Ongoing and future research is focused more on developing standard treatment strategies, and efficacious vaccines, which would be useful to tackle this pandemic globally.
Keywords: Replication, transmission, pathogenesis, SARS-CoV-2, coronavirus disease 19 (COVID-19)
Introduction
Over the past couple of decades, three major international outbreaks of coronaviruses (CoVs) have occurred causing considerable global health concern. The first human CoV outbreak occurred in late 2002. Because the symptoms associated with this CoV outbreak was characterized by severe acute respiratory syndrome (SARS) and the genomic sequence of the causative virus differed significantly from other CoVs previously known to cause human disease, the causative virus was named SARS-CoV (1). A decade later in 2012–2013, another respiratory disease; Middle East Respiratory Syndrome (MERS) broke out and the causative agent of this respiratory disease was named Middle East respiratory syndrome coronavirus (MERS-CoV). Most recently in December 2019, a group of patients presented with pneumonia-like symptoms (2,3). Most of these people had a high fever and some had dyspnea, with chest radiographs revealing invasive lesions in both lungs (4,5). Since the etiology of this pneumonia was unknown, the authorities started conducting an epidemiologic and etiologic investigation. In January 2020, analyses from the lower respiratory tract samples by culture, direct PCR, and whole-genome sequencing identified a novel CoV (2019-nCoV), the causative agent for the observed pneumonia cluster (6). WHO renamed the disease as COVID-19 while the causative agents were named as SARS-CoV-2. The number of positive COVID-19 cases is increasing worldwide (7) and to date, it has now been confirmed on six continents and in more than 100 countries. Being a novel CoV, information is just beginning to emerge regarding biochemical properties, molecular etiology, and clinical aspects of the virus.
Indeed, many issues about COVID-19 still remain unresolved as the current immunological concepts are insufficient to explain the causes for different clinical phenotypes, incubation periods among individuals, cytokine storm with lymphopenia and the damage to organ cells. Accordingly, the current understanding and explanations about COVID-19 are based on epidemiological, clinical and physiological observations (8). Epidemiological observations are supported by the hypothesis of virus transmission from person-to-person within a distance of 2 m from an infected person. However, high aerosol and surface stability of SARS-CoV-2 whereby, the virus remains viable and infectious in aerosol for hours, and the airborne transmission can occur besides close-distance contacts (9). On the other hand, early clinical evidence suggests that severe cases of COVID-19 are frequently characterized by hyper inflammation, renin-angiotensin-aldosterone system imbalance, vasculopathy thrombine microangiopathy and intravascular coagulopathy (10). Meanwhile, other studies have identified D-dimer (11), prothrombin time (12) and thrombocytopenia (13) as prognostic markers for severe COVID-19. In terms of physiological observations, COVID-19 affects multiple organ systems particularly the respiratory system with some patients rapidly progressing to acute respiratory distress syndrome (ARDS) (14). Similarly, cardiac injury is also a common complication in severe COVID-19. As a result of such complications, severe COVID-19 patients are usually admitted to intensive care units (ICUs) and are likely to die. The elderly and particularly those with comorbidities are at the greatest risk of death particularly due to lung injury (14).
In this review, we aim to update the current knowledge about SARS-CoV-2, including its classification, origin, genomic structure, replication cycle, transmission, pathogenesis, clinical symptoms, diagnosis, and available treatment options. We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5272).
Methods
We conducted a comprehensive literature search for the articles published up to August 2020, using keywords ‘SAR-CoV-2’ and ‘COVID19’ in medical databases; PubMed (https://www.ncbi.nlm.nih.gov/pubmed), a search engine accessing the MEDLINE database of references and abstracts on biomedical topics and life sciences. We also searched coronavirus-related literature using EMBASE, google scholar, and web of science. Study selection was based on three medical databases; Web of Science PubMed, and EMBASE. Based on the keywords mention above, qualitative and quantitative data were extracted through the interpretation of published articles. Eligibility criteria included articles indexed in PubMed. Exclusion criteria were non-peer reviewed or unpublished reports. English language restriction was also applied. No clinical trials and/or guidelines were included in the review. Overall, a total of 26,611 articles were retrieved, out of which 130 were selected and used in this review.
Discussion
Coronaviruses
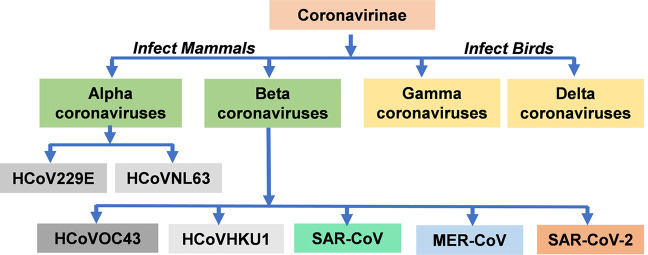
Coronaviruses belong to subfamily orthocoronovirinae of the family Coronaviridae and order Nidovirales. Based on the phylogenetic analysis, the Coronavirinae subfamily is classified into four groups, namely; Alphacoronaviruses (α-CoVs), Betacoronaviruses (β-CoVs), Gammacoronaviruses (γ-CoVs), and Deltacoronaviruses (δ-CoVs) (15). Studies have revealed that γ-CoVs and δ-CoVs usually infect birds and possibly mammals (16). However, they are never known to cause illness in humans. On the contrary, α-CoVs and β-CoVs are known to infect mammals and are capable of causing illnesses to humans (17). Human coronavirus (HCoV) was isolated for the first time in the year 1965 from the nasal discharge of patients with the common cold (18). Reports based on the phylogenetic tree analysis revealed that there are seven known strains of HCoVs. These include HCoV-229E (229E), HCoV-OC43 (OC43), HCoV-NL63 (NL63), HCoV-HKU1 (HKU1), severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and the recently identified SARS-CoV-2 (19). The analysis further suggested that the viral strain; 229E, and NL63 belong to the α-CoVs while OC43, HKU1, SARS-CoV, MERS-CoV, and SARS-CoV-2 belong to the β-CoVs (20) (Figure 1). These suggest that HCoVs belong to either the alpha or beta coronavirus genera. Based on the evidence, viral strains; 229E, OC43, NL63, and HKU1 are already adapted to humans and widely circulated among the human population (21). Therefore, these viruses have been identified to infect humans although with low pathogenicity and mild respiratory syndrome (22). Coronavirus strains; 229E, OC43, and NL63 are distributed globally and cause 15–29% of all common colds (23). Although, SARS-CoV, MERS-CoV, and SARS-CoV-2 were likely acquired by zoonotic transmission through an intermediate host species (20). SARS-CoV-2, unlike SARS-CoV and MERS-CoV, may be an adapted virus to human species for the reason that, there are many asymptomatic carriers and its fatality is mainly observed in older patients with comorbidities (6,24-26).
Figure 1.
Schematic representation of the type of Coronaviruses. HCoVs belong to either the Alphacoronaviruses or Betacoronaviruses. HCoV229E, human coronavirus 229E, HCoVOC43, human coronavirus OC43; HCoVNL63, human coronavirus NL63; HCoVHKU1, human coronavirus HKU1; SARS-COV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome-related coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Origin of coronavirus and SAR-CoV-2
The SARS-CoV and MERS-CoV are known to have originated from bats, which are their natural reservoirs, and then transmitted to humans through an intermediate host; civets, and dromedary camels, respectively (27). Based on some evidence, various proposals have been made regarding the origin of SARS-CoV-2 (28). Anderson KG et.al proposed that the origin may have been from (I) natural selection of the virus in a natural host/intermediate reservoir before zoonotic transfer to humans (II) natural selection of the virus in humans following the zoonotic transfer and (III) adaptation of the virus during the passage in culture/animal model. The first case scenario purports that SARS-CoV-2 originated from bats as the natural host; however, it must have been transmitted to humans via an intermediate host in the Huanan seafood market (29-33). To date, SARS-CoV has been isolated from other animals including the pangolins. Phylogenetic sequence analysis of the virus from pangolins revealed 91% sequence similarity to SARS-CoV-2, which infects humans suggesting that SARS-CoV-2 originated in bats as the natural reservoirs and transmitted to humans through pangolins as the intermediate hosts (Figure 2) (34,35). However, it is worth noting that the Huanan market was not trading in bats but other animals including a hedgehog, badger, snake, pangolin, and bird. Thus, the direct transmission of SARS-CoV-2 from bats to humans appears to be highly unlikely (27). The second case scenario suggests that the progenitor of SARS-CoV-2 was selected and became adapted to the human host after acquiring the genomic features during asymptomatic human-to-human transmission. However, it would not be wise to rule out the third case scenario involving the selection of the virus during the passage in the culture/animal model. Since the research involving CoVs has been performed for many years using the in vitro/or animal models in biosafety level 2 laboratories, SARS-CoV-2 probably may have acquired adaptation mutations during the passage in cell culture/animal model. Nevertheless, in-depth studies to elucidate the actual origin of SARS-CoV-2 are important in order to take precautionary measures against similar future outbreaks.
Figure 2.
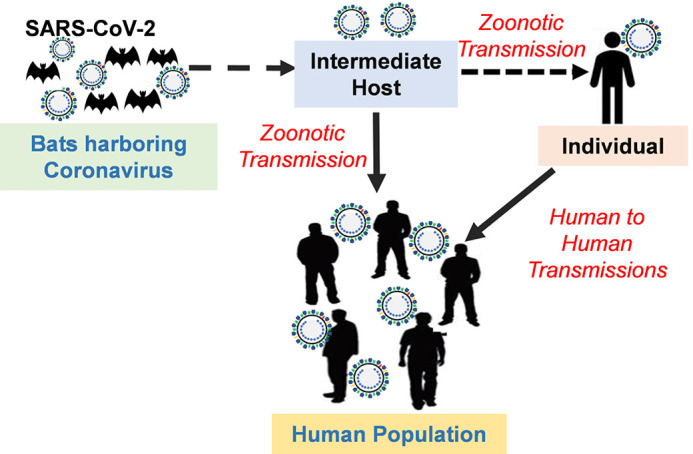
Schematic representation of the zoonotic transmission of SARS-CoV-2: the novel SARS-CoV-2 is believed to have originated from bats, then passed through an intermediate host possibly pangolin, to an individual (across the species barrier into humans) which then resulted in the human to human transmission within populations. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Genomic organization of SAR-CoV-2
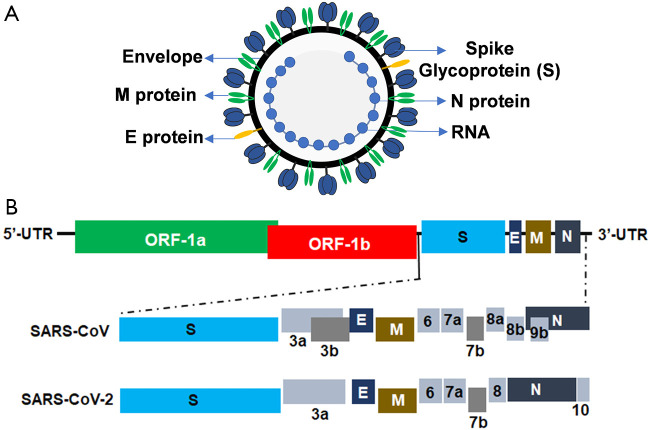
The SARS-CoV-2 is a spherical, enveloped, and positive-sense RNA virus (Figure 3A). The complete genome size of SARS-CoV-2, WuhanHu1 strain is 29.9 kb (36). Studies have shown that CoV genomes contain a variable number of open reading frames (ORFs) (Figure 3B) (37). The first ORF (ORF-1a and ORF-1b) accounts for two-thirds of the viral RNA genome and encodes two polyproteins namely, pp1a and pp1ab and 16 non-structural proteins (NSP). The remaining ORFs encode accessory and structural proteins including four essential proteins, namely, the spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein (17). The S protein, a heavily glycosylated protein forms homotrimeric spikes on the viral surface that mediates viral entry into host cells (Figure 3). M protein, the most abundant protein along with E protein gives the virus its shape. The main function of E protein is to release the viral particles from host cells while the N protein is required for the packaging of the viral RNA into the viral particle during viral assembly.
Figure 3.
Virion and genomic structure of SARS-CoV-2. (A) Schematic structure of the SARS-CoV-2 virion, (B) structural organization of the SARS-CoV-2 genome. Structural proteins, Spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins, non-structural proteins translated from ORF 1a and ORF 1b and accessory proteins, including 3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b (for SARS-CoV), and 3a, 6, 7a, 7b, 8, and 10 (for SARS-CoV-2) are indicated. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ORF, open reading frame.
Mutations and genetic recombination of coronaviruses and SARS-CoV-2
Viruses, particularly RNA viruses are characterized by a high rate of mutations. Recently, Pachetti et al. (38) observed that some mutations were either close to or within RNA -dependent RNA polymerase (RdRp) hydrophobic cleft which is the target for several drug candidates in the pipeline suggesting that such drugs would either fail in preclinical/clinical studies or soon develop drug resistance due to mutations. Similarly, Stefanelli et al. (39) compared the full-length SARS-CoV-2 genome sequences isolated from two patients, (I) a Chinese tourist who visited Italy and (II) Italian patients, with other European sequences and other places around the world. The phylogenetic analysis suggest that multiple SARS-CoV-2 strains were introduced to Europe or that the initial virus that was introduced to Europe continue to evolve through mutations. Consistent with these observations, partial sequence results of SARS-CoV-2 obtained by the Uganda Virus Research Institute (UVRI) in Entebbe, Uganda demonstrates that different SARS-CoV-2 sequences from Ugandan patients’ cluster with viral sequences from the origin where the patient contracted the virus. Besides, another study has identified 116 important mutations out of which three mutations are most common; 29095C>T in the N gene, 28144T>C in ORF8 gene, and 8782C>T in ORF1ab gene (40). There is a high possibility that these mutations might affect the pathogenicity and spread of the SARS-CoV-2 (40).
CoVs have a moderate recombination frequency (41). The recombination and the mutation favor them to adapt to new hosts (42,43). Reports have started to emerge for genomic recombination of HCoVs including; OC43, HKU1, NL63, SARS-CoV (44), and MERS-CoV. Studies have shown that HCoVs; OC43 has A to E five distinct genotypes. Genotype A is exemplified by the prototype VR759 strain that was isolated in 1967 (45). Studies have further shown that genotype D has emerged because of the recombination between naturally occurring genotypes B and C viruses. Moreover, genotype E was also known to have emerged from genotype B, C, and D viruses recombination at different breakpoints in the genes compared to the generation of genotype D (46). The other HCoVs strain; NL63 has the possibility for genetic recombination, for example, the Amsterdam 1 and 496 strains have recombination breakpoints site in S gene (47). HKU1 strain was also reported of having recombinant forms between genotype B and C in some nonstructural proteins and haemagglutinin esterase genes (43). A history of possible recombination with lineages of alpha- and gamma-CoVs has been reported for SAR-CoVs (48). A large number of the recombinant region and many specific breakpoints had been identified especially in the RdRp gene (48). Other studies have demonstrated that the MERS-CoV had recombination events between the different lineages (49). It caused human infections in Riyadh, Saudi Arabia as well as an outbreak in South Korea in 2015 (49,50). Therefore, the foregoing paragraph reveals that genetic recombination events in CoVs results in the emergence of novel viruses with unique genetic diversity and possibly unpredictable virulence following transmission to humans. As the multiple recombinant strains of CoVs are circulating among different animal species, it is just a matter of time until the emergence of new recombinant CoVs and cause another outbreak.
SARS-CoV-2 and its replicative cycle
The CoVs are thought to enter cells via two distinct pathways, namely, one mediated by type II, transmembrane protease, serine 2 (TMPRSS2) present at the cell surface and the other mediated by cathepsin L in the endosomes. Angiotensin-converting enzyme 2 (ACE-2) which is found in cells of the lower respiratory tract of humans is the cellular receptor for SARS-CoVs (51). SARS-CoV-2 also utilizes ACE-2 as a receptor to gain entry into the target cells (52). The viral spike (S) glycoprotein of the SARS-CoV-2 envelope surface attaches to ACE-2 of the target cells to initiate viral entry. Bertram et al. (53) analyzed the role of type II transmembrane serine proteases (TTSPs), human airway trypsin-like protease (HAT) and TMPRSS2 in SARS CoV2 activation and found that HAT cleaves and activates the viral (S) glycoprotein in order to initiate viral entry and it is coexpressed with the viral receptor, ACE-2 in bronchial epithelial cells and pneumocytes. As determined by mutagenesis and mass spectrometry analyses, HAT cleaved the (S) glycoprotein at R667 for cell-cell fusion while TMPRSS2 cleaved the (S) glycoprotein at multiple sites. The observation that TMPRSS2 activates the (S) glycoprotein for viral entry were also confirmed by other authors (54-56). The binding of S glycoprotein of SARS-CoV-2 and ACE-2 receptor is the most critical step for the entry of the virus to the target cells. One study demonstrated that human cells expressing ACE-2 enhanced the entry of SARS-CoV-2 (57). The S protein comprises two functional subunits; S1 subunits and S2 Subunits. S1 subunit binds to the host cell receptor while the S2 subunit is responsible for the fusion of the viral and cellular membranes. Studies have revealed that the distal S1 subunit comprises the receptor-binding domain (RBD) that contributes to the stabilization of the prefusion state of the membrane-anchored S2 subunit that contains the fusion machinery (58). Nevertheless, different CoVs use distinct domains within the S1 subunit to recognize a variety of attachment and entry receptors, SARS-CoV and several SARS-related coronaviruses (SARSr-CoV) interact directly with ACE-2 to enter target cells (31,59). S1 can be further divided into a C-terminal domain (CTD) and an N-terminal domain (NTD), both of which can function as a receptor-binding entity (60,61), whereas mouse hepatitis CoV engages the receptor with its S1 NTD (62). In the case of SARS-CoV-2, the region of the S protein that is responsible for ACE-2 interaction has been identified as the S1 CTD (SARS-CoV-2-CTD).
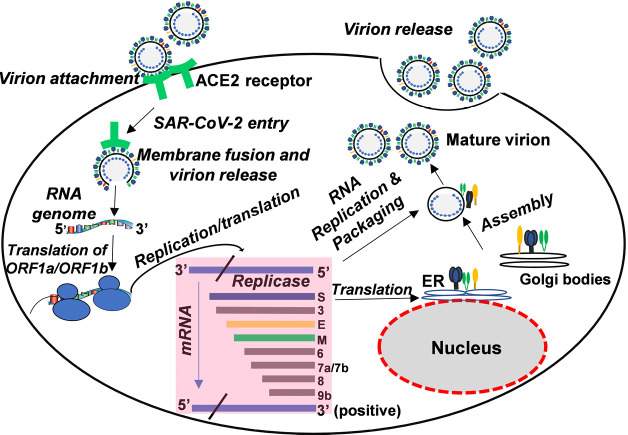
Upon the entry and uncoating of coronavirus into the target cell, the positive sense genomic RNA is translated from open reading frame 1a/b (ORF1a/b) generating the replicase proteins. The replicase proteins use the genome as a template to generate full-length negative-sense RNAs, which then serve as templates in generating additional full-length genomes. CoV mRNAs contain a common 5’ leader sequence fused to downstream gene sequences. The RNA’-RNA polymerase recognizes the 3’ end of the positive strand and full-length template and further proceed to synthesize positive-sense RNA. It continues to synthesize until it encounters the transcriptional regulatory sequences (TRS) located between the coding regions. The translation of sub-genomic mRNAs leads to the production of structural and non-structural viral proteins. Once sufficient structural proteins and genomic viral RNA are formed, viral RNA is then assembled with viral structural proteins into virions (Figure 4). Viral assembly and budding then occur via the smooth-walled vesicles in the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) (63). Through evolution, most viruses are unable to replicate on their own but relay on the host cell’s machinery for effective replication. It is therefore, conceivable that adapted viruses can easily replicate to produce intact virions whereas non-adapted viruses cannot such that the products from their replication process in the infected cells can induce inflammation when released.
Figure 4.
The life cycle of coronavirus in host cells; the virion lifecycle begins when the spike (S) protein binds to ACE2, the cellular receptor. Then the virus releases its plus sense genomic RNA into the host cell. The minus strand is then formed from the plus strand genomic RNA, from which subgenomic mRNAs are formed by transcription and then translated into relevant viral proteins. Genome RNA and viral protein are assembled and then released out of the cell. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; ER, endoplasmic recticulum; ACE2 receptor, angiotensin-converting enzyme 2 receptor.
Transmission of SARS-CoV-2
The SARS-CoV-2 is transmitted from person-to-person at close contact through droplets, aerosol, and possibly through fecal-oral transmission. Liquid droplets/aerosol particles exhaled from the infected person during speech or sneezing can linger in the air (64). If it is inhaled by an uninfected person, then inhaled aerosolized particles can penetrate to the depths of the lungs and deposited in the alveoli causing serious respiratory tract infection (65,66). Indeed, data obtained from studies conducted in Wuhan Hospital demonstrated the presence of SARS-CoV-2 RNA in air samples collected in the Hospital and also in the surroundings leading to the conclusion that the airborne route should be considered a plausible pathway for virus transmission (9). Furthermore, accumulating evidence suggests that SARS-CoV-2, just like many of its related viruses, may also be an enteric virus which can be transmitted via the fecal-oral route (67,68). There are reported cases of COVID-19 infection occurring during pregnancy and the due concern for mother-to-fetal vertical transmission. However, presently, there is little evidence on vertical transmission of SARS-CoV-2, prevalence and clinical features of COVID-19 during pregnancy, regardless to say, infected mothers are at greater risk for more severe respiratory complications from COVID-19 infection (69). Nevertheless, it should be noted that an infected mother is capable of transmitting SARS-CoV-2 through respiratory droplets during breastfeeding. A study conducted to investigate the stability of SARS-CoV-2 through five different environmental conditions (aerosols, stainless steel, plastic, copper, and cardboard) (70) showed that SARS-CoV-2 remained viable for 3hrs and infectious viral titers decrease gradually after 3hrs (70). It further suggested that SARS-CoV-2 was more stable on stainless steel and plastic than on cardboard and copper and the viable virus could be detected on these surfaces even after 72 h (70). Therefore, SARS-CoV-2 can also be contracted by an uninfected person through touching contaminated surfaces or objects and then touching the face. The COVID-19 patients who are asymptomatic or at the incubation period can even shed and transmit the virus to other persons (71).
Pathogenesis of SARS-CoV and SARS-CoV-2 infection
The mechanisms of pathogenicity of SARS-CoV-2 are not fully understood. However, several studies have been done on SARS-CoV and it’s believed that SARS-CoV-2 mechanisms of disease causation are somewhat similar to that of SARS-CoV. In the case of SARS-CoV-infected patients, high virus titers (25), elevated macrophage, monocyte, and neutrophil infiltration in the lung (72), and increased proinflammatory chemokines and cytokines in the serum (73) causes extensive lung damage. This indicates that the viral-induced cytopathic effect and/or immunopathology induced by hyper-cytokinemia are responsible for the clinical deterioration of SARS-CoV infection. Cytokine/chemokine profile analysis showed that the level of cytokines, including CXCL-10, tumor necrosis factor α (TNF-α), interleukin-6 (IL-6), and IL-8 are high that results to the poor prognosis in SARS-CoV infections (74). The level of proinflammatory cytokines in the serum (Interferon γ [IFN-γ], transforming growth factor-β, IL-1, IL-6, and IL-12) and chemokines (CXCL9, CXCL10, CCL2, and IL-8) were found to be high in patients with the severe disease when compared to patients with uncomplicated SARS (73,75-77). Besides, immuno-pathological processes involved in lung injury during SARS-CoV infection are thought to result from the early induction of IL-2, and CXCL10 as well as the subsequent hyperproduction of pro-inflammatory cytokine IL-6 with a coinciding lack of anti-inflammatory cytokine IL-10 production (76). Furthermore, robust and persistent expression of IFN-α, -γ and IFN-stimulated genes (ISGs) accompanied early SARS sequelae (78). Other studies have demonstrated that SARS-CoV infections result in a delayed expression of type I IFN (79). The delay in type I IFN signaling, accompanied by robust virus replication, has been demonstrated to promote the accumulation of pathogenic inflammatory monocyte/macrophages, which resulted in elevated lung cytokine/chemokine levels, vascular leakage, and impaired virus-specific T cell responses (80). However, particularly in COVID-19 patients, initial plasma IL-1Rα, IL-1β, IL-7, IL-8, IL-9, IL-10, basic GCSF, GMCSF, FGF, IFNγ, IP-10, MCP-1, MIP-1α, MIP-1β, TNF-α, PDGF, and vascular endothelial growth factor concentrations were higher in COVID-19 patients as compared to healthy controls. On the other hand, Zou et al. (81) evaluated the correlation between coagulation functions and COVID-19 status and found that coagulation dysfunction was common in patients with COVID-19. In particular, they observed that there was fibrinogen and D-dimer elevations, and the degree of elevation was related to the severity of the COVID-19 disease. However, as patients recovered from severe disease, fibrinogen and activated partial thromboplastin time also returned to normal. These results suggest that immunopathology also plays an important role in the development of disease severity. It is however, worth noting that the presumed immunopathogenesis of COVID-19 are better explained according to the clinical and pathological characteristics of the disease, as well as the protein-homeostasis-system (PHS) hypothesis (8). During the incubation period of COVID-19, substances are produced by SARS-CoV-2 that can induce lung injury as well as other organs such as the cardiac system, which initiate the focus of disease. These substances that are potentially toxic spread locally and systematically and bind to receptors on target organ cells. Innate immune cells and proteins likely control different sized substances during the early stage of COVID-19 infection such that clinical symptoms of the disease including fever, myalgia and pneumonia begin to appear at this stage (8). However, the cytokine storm in progressive severe disease may occur as a result of excessive immune cell activation against large amounts of substances that are produced at this stage of the disease. The etiological substances that induce inflammation, as well as various organ injuries are the smaller substances derived from the virus infected cells where the cytokine imbalance may be responsible for target cell injury. Furthermore, substances emanating from initial tissue injury, as well as secondary bacterial infection can induce further inflammation when released locally or systematically (8).
Antibody-dependent enhancement of CoVs infection
Some vaccine strategies induce neutralizing and protective antibodies against the target S protein of SARS-CoV. However, the main concern is that vaccinations against different kinds of coronaviruses have different outcomes (82). Studies have shown that anti-spike antibodies offer protection against transmissible gastroenteritis and mouse hepatitis. However, this particular antibody boosts feline-coronavirus (FCoV) infections (83). Moreover, antibody-dependent enhancement (ADE) of virus infection has been observed in viruses including yellow fever, dengue, HIV, and FCoV (82). Even though the mechanism of ADE is poorly understood, it is believed that the virus-specific antibodies enhance the entry of the virus and replication into macrophages/monocytes and granulocytes cells through the interaction with complement/Fc receptors (82). Using SARS-CoV clinical isolates and pseudotyped viruses, Jaume et al. (84) demonstrated that SARS-CoV anti-spike antibodies potentiate ADE infection of both monocytic and lymphoid human immune cell lines. Similarly, Yip et al. (85) suggested the occurrence of ADE in SARS-CoV infected immune cell types, particularly monocytic lineage. However, in addition to monocytes, the presence of anti-viral antibodies enhanced the infection of human macrophages. Nevertheless, macrophages did not support productive viral replication or modify the expression of some pro-inflammatory cytokines. These findings suggest the occurrence of ADE infection of immune cells by SARS-CoV, however, the outcomes of such an alternative infection pathway on the cell functionality/homeostasis remain unclear. The ADE is likely to complicate SARS-CoV-2 vaccine research and development efforts.
Clinical aspects of SARS-CoV-2
Clinical presentation of SARS-CoV-2 infection is similar to those of SARS-CoV and MERS-CoV infections (Table 1). Table 1 summarizes the disease-causing human CoVs and their clinical symptom. The clinical symptom of COVID-19 usually appears 2 to 14 days after exposure. COVID-19 patients commonly experience fever, dry cough, shortness of breath or difficulty breathing, and runny nose as the symptoms. However, other common symptoms include muscle pain, chills, headache, sore throat, loss of taste, or smell (27). Furthermore, approximately 25% of COVID-19 patients experience diarrhea as is the case with SARS-CoV and MERS-CoV infections (5). However, in severe COVID-19 cases, symptoms include acute respiratory distress syndrome, metabolic acidosis, septic shock, and bleeding and coagulation dysfunctions. During the disease, severe and critically sick patients may show moderate to low fever. It’s however, worth noting that some COVID-19 patients may remain asymptomatic though capable of transmitting the virus. Initial reports of COVID-19 indicated infrequent occurrence of disease in children. However, subsequently, multiple reports described children affected by COVID-19 with varying degrees of severity (86-88). Accordingly, during this COVID-19 pandemic, cases of severe Kawasaki disease (KD)-like disease referred to as Multisystem inflammatory syndrome in children (MIS-C) have been reported in large cities in Western countries. Children affected by MIS-C are usually older and manifest shock syndrome with acute heart failure, gastroenteric symptoms and other symptoms such as KD (8). Based on currently available information from clinical expertise personal, there are four higher risk groups for COVID-19: (I) over-70s, regardless of any medical conditions, (II) under 70s who have the underlying health conditions like respiratory diseases (Asthma or chronic obstructive pulmonary disease), diabetes, heart failure, hepatitis, kidney disease, long term neurological problem, and other spleen problem, (III) pregnant women (IV) people with complex health problem like HIV/AIDS patients, organ transplant patients and cancer patients with active chemo/radiotherapy.
Table 1. Human coronavirus based on their types, clinical symptom and incubation time.
| HCoVs | Genus and lineage | Symptom | Incubation time | International outbreak |
|---|---|---|---|---|
| 229E | Alphacoronavirus | General malaise, nasal discharge, fever, sneezing, headache, sore throat and cough | 2–5 days approx. | NA |
| OC43 | Betacoronavirus, lineage A | General malaise, fever, headache, sneezing, nasal discharge, sore throat, and cough | 2–5 days approx. | |
| NL63 | Alphacoronavirus | Rhinorrhea, cough, hypoxia, fever, tachypnea, obstructive laryngitis | 2–5 days approx. | |
| HKU1 | Betacoronavirus, lineage A | Fever, cough, dyspnea, and running nose | 2–5 days approx. | |
| SARS-CoV | Betacoronavirus, lineage B | Fever, myalgia, malaise, headache, chills, dyspnea, nonproductive cough, respiratory distress, and diarrhea | 2–11 days approx. | SARS in 2002 |
| MERS-CoV | Betacoronavirus, lineage C | Cough, fever, sore throat, myalgia, chills, arthralgia, dyspnea, diarrhea, pneumonia, and vomiting | 2–13 days approx. | MERS in late 2012 |
| SARS-CoV-2 | Betacoronavirus, lineage B | Fever, cough, shortness of breath or difficulty breathing, chills, muscle pain, headache, loss of taste or smell, sore throat | 2-14 days approx. | COVID19 in late 2019 |
HCoVs, human coronaviruses; 229E, human coronavirus 229E; OC43, human coronavirus OC43; NL63, human coronavirus NL63; HKU1, human coronavirus HKU1; SARS-COV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome-related coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID19, coronavirus disease 2019.
Just as much as there are differences in clinical manifestations, there are also differing incubation periods among the infected individuals. The observed differences in incubation periods are likely due to different substances that are produced according to the infected cells such as the upper respiratory epithelial cells or the lower respiratory endothelial cells (8). Due to the differences in the intracellular environments of the infected cells, the timing as well as the duration of release of viruses and related substances can also be different. Therefore, viruses and substances in the upper respiratory epithelial cells may be easily discharged outside the host whereas those in regional lymph nodes and lower respiratory endothelial cells are discharged inside the host.
There is evidence to suggest that viruses become attenuated and less virulent once passaged multiple times. It is therefore, possible that people who become infected with SARS-CoV-2 after it has infected several individuals later in the pandemic may be asymptomatic compared to those individuals infected earlier in the pandemic. Similarly, herd immunity can also be achieved as a result of cross-immunity from previous CoVs infections such as those that cause common cold. At the beginning of COVID-19 pandemic, the mortality rate was higher among the elderly although this later varied among populations. Currently, it is unknown if the age distribution of all infected persons will change over time due to herd immunity obtained from other CoVs infections.
Diagnosis of SARS-CoV-2 infection
The WHO recommends mass testing of all suspected cases, isolation, and treatment of all confirmed cases and tracing of all contacts of confirmed cases. However, the high and efficient rates of SARS-CoV-2 transmission among persons have caused global health concern and have overwhelmed healthcare systems of even the developed nations in Europe and the United States of America. Although there are much lower confirmed cases in Africa compared to the developed nations, apparently due to low mass testing capability, there is grave concern regarding how developing countries of Africa and elsewhere will be able to cope with the COVID-19 pandemic, given the limping state of the healthcare systems in these countries. Consequently, as a result of the above, most African countries were quick to close off their borders to the outside world to curtail imported cases of COVID-19 that would subsequently result in local community transmission. Interestingly, although the WHO had projected that Africa could become the next epicenter of COVID-19 pandemic after Europe, confirmed cases of COVID-19 in many African countries including Uganda has been significantly low even months following the first reported case. Many African countries were reasonably prepared to handle the COVID-19 outbreak given the lessons learned from the previous handling of the Ebola and Marburg outbreaks in the continent. Even though commercial diagnosis kits for SARS-COV-2 are available, the antigen detection kits preparation process is complicated and time-consuming as it requires the preparation of monoclonal antibodies and polyclonal antibodies. Therefore, detection of viral RNA using RT-PCR (89) remains a gold standard and it is currently the only diagnostic test approved and available for SARS-CoV-2 infection. Although SARS-CoV-2 can be detected in various human samples including respiratory and fecal samples, testing has been validated for RT-PCR primarily for nasal, nasopharyngeal and oropharyngeal specimens. The rapid spread of SARS-CoV-2 infection in the United States of America and around the world led to Emergency Use Authorization (EUA) which facilitated the development and availability of molecular diagnostic kits without vigorous examination prior to the approval by the Food and Drug Administration (FDA). To these effects, the following RT-PCR platforms have been made available for molecular diagnosis of SARS-CoV-2 infection, namely, the Roche Cobas SARS-CoV-2, the Cepheid Xpert Xpress SARS-CoV-2 and the Abbott ID Now COVID-19 assays. The run time for each assay are 3.5 h, 45 min and 5 min, respectively. Basu et al. (90), analyzed the performance of these three test kits using nasopharyngeal swabs transported in viral transport media and dry nasal swabs. Basu et al. (90) found out that regardless of the method of collection and sample type, Abbott ID Now COVID-19 gave negative results in a third of the samples that tested positive by Cepheid Xpert Xpress. This observation underscores the high level of unreliability associated with the molecular tests currently in use for COVID-19 diagnosis.
On the other hand, Gao et al. (91) investigated the role of clinical laboratory markers in the differential diagnosis of the severe forms of COVID-19 using 43 adult patients. The patients were classified into mild and severe groups. A comparison of the hematological parameters between the mild and the severe groups showed significant differences in IL-6, D-dimer, glucose, thrombin time, fibrinogen and C-reactive protein levels. In particular, high levels of IL-6 and D-dimer were closely related to occurrence of severe COVID-19 in adult patients. Furthermore, the different cardiac manifestations and the use of cardiac-specific biomarkers in terms of their prognostic use in determining the clinical outcomes and disease severity are reviewed in Shafi et al. (92).
One of the challenges in the management of COVID-19 is the use of an accessible and reliable diagnostic modality that can serve as an alternative to RT-PCR. To this effect, recent studies have demonstrated the usefulness of Chest computed tomography (CT) scan in the diagnosis of COVID-19 (93-95). Indeed, in one instance, the use of CT scan provided an accurate COVID-19 diagnosis even when the molecular RT-PCR was negative (94). On the other hand, in emergency situations, the imaging features of routine admission chest X-ray in patients suspected for COVID-19 infection can be employed for diagnosis. In this case, Ippolito et al. (96) observed that overall, the Chest X-ray sensitivity for SARS-CoV-2 pneumonia was 57% and that sensitivity was higher when COVID-19 symptoms had started more than five days before.
Update on the therapy of SARS-CoV-2 infection
Viral diseases such as COVID-19 are self-limiting whereby, the host’s immune system can potentially control the infection without complications. However, in the case of COVID-19, a small proportion of patients experience disease complications such as progressive pneumonia and ARDS. Therefore, scientists are searching for treatment modalities that can prevent disease progression and complications. Current treatment modalities under consideration include antivirals and antiviral-effecting drugs, as well as immunomodulators and supportive treatment options.
Antivirals and antiviral-effecting drugs
Currently, there is a continuing search for the antiviral drugs that are effective against the SARS-CoV-2 infection. Patients infected with SARS-CoV-2 are required to be kept in bed and closely monitored for vital signs and levels of oxygen saturation (97). Being a novel virus, it will conceivably take several years before effective novel antiviral drugs can become available for COVID-19 treatment. Considering the above challenges, the other solution is to systematically screen existing drugs to determine whether some drugs may have activity against SARS-CoV-2. Accordingly, recent screening of existing drugs revealed that an antiretroviral drug used in the treatment of human immunodeficiency virus (HIV) infection, Kaletra, which comprises of two protease inhibitors; ritonavir and lopinavir may have therapeutic effects on SARS-CoV-2 (98-102).
Another study reported that chloroquine showed inhibitory effects on SARS-CoV-2 in vitro (103) and an early clinical trial showed that chloroquine had a significant effect on clinical outcome and viral clearance (104). As a result of these studies, COVID-19 patients with moderate, and severe cases but without contraindications to chloroquine were recommended to be treated with 500 mg of chloroquine twice a day (97). On the other hand, hydroxychloroquine which is an analog of chloroquine with better pharmacodynamics profiles compared to chloroquine has been shown to have an anti-SARS-CoV-2 activity in vitro (105). Because of having better pharmacodynamics profiles than chloroquine, hydroxychloroquine allows the use of higher daily dose and fewer concerns for drug-to-drug interactions (106,107). Based on these observations, Gautret et al. (108) tested the use of azithromycin and hydroxychloroquine as a treatment for COVID-19 in a non-randomized clinical trial. They reported that hydroxychloroquine effectively cleared viral nasopharyngeal carriage of SARS-CoV-2 in only 3 to 6 days (108). Although small studies have shown a potential benefit of chloroquine/hydroxychloroquine in combination with or without azithromycin for the treatment of COVID-19. Saleh et al. (109) investigated whether the use of these medications alone or in combination can lead to a prolongation of the Quality Time (QT) interval, possibly increasing the risk of Torsade de pointes (TdP) and sudden cardiac death. Paradoxically, in the largest reported cohort of COVID-19 patients treated to date with chloroquine/hydroxychloroquine with or without azithromycin, no instances of TdP or arrhythmogenic death was reported. Similar observations were also made by other authors (110,111), however, Sarayani et al. observed that though treatment with azithromycin resulted in QT prolongation, the use of chloroquine/hydroxychloroquine alone did not produce these effects. In light of all these underlying controversies, the WHO has had to suspend further clinical trials involving the use of chloroquine/hydroxychloroquine for the treatment of COVID-19.
Based on initial studies conducted in China using remdesivir, a new antiviral drug that was originally developed for the treatment of Ebola to treat COVID-19 patients (103), more clinical trials were subsequently conducted in the United States. Indeed, most recently, remdesivir has been approved by the Food and Drug Administration of the United States for emergency treatment of COVID-19 patients based on preliminary results which demonstrated that remdesivir can significantly reduce hospitalization days for severe COVID-19 patients from 15 down to 11 days. Other antiviral drugs that have been tested for COVID-19 treatment but with less success include ribavirin, favipiravir and oseltamivir.
Immunomodulators
COVID-19 is known to induce the release of pro-inflammatory cytokines including IL-1β and IL-6, which mediate lung injury as well as tissue inflammation, fever and fibrosis. Indeed, high levels of IL-6 is the cause of the cytokine storm and therefore, suppression of these pro-inflammatory cytokines may provide treatment options. The immunomodulatory agents used in the treatment of COVID-19 include; Tocilizumab, Interferons and Corticosteroids. Tocilizumab is a recombinant humanized anti-human IL-6 receptor monoclonal antibody that binds to IL-6 receptor with high affinity. Although it is approved for the treatment of cytokine release syndrome (CRS) and rheumatoid arthritis, currently there is not enough data to recommend its use for COVID-19 therapy. On the other hand, interferons are a group of cytokines with non-specific antiviral effects. In this case, interferon α and β have been suggested as candidates for COVID-19 therapy. Interferons act by binding to their receptors on the cell surface membrane, which results in the phosphorylation of STAT1. STAT1 then translocates in to the nucleus where it activates the expression of the interferon-stimulated genes (ISGs). Activated ISGs lead to immunomodulatory effects which interfere with viral replication. However, currently, studies that may support the use of interferons for COVID-19 therapy are inconclusive. Corticosteroids, particularly, Methylprednisolone has been suggested as an adjuvant for COVID-19 treatment. Corticosteroids are widely used in the treatment of severe pneumonia and lung injury because of their ability to suppress systemic inflammation (8). Indeed, studies have shown that ARDS patients treated with Methylprednisolone showed a rapid resolution of pneumonia lesions and clinical improvement and in fact early treatment using corticosteroids as soon as possible may reduce the aberrant immune reactions in the potential stages of ARDS (112,113).
General and supportive treatment
An alternative treatment option is the use of convalescent serum or plasma from COVID-19 recovered patients (114). Recovered patients would have developed specific antibody responses which may help neutralize viruses in newly infected individuals. This treatment approach was effectively and successfully employed in the United States during the 2014/2015 Ebola outbreak (115,116). However, considering the exponential increase in the number of new infections, the use of convalescent sera may be of limited benefit in an outbreak situation since newly infected patients exceed the ability of previous patients to provide donor plasma.
Currently, more than 80 clinical trials are going on for potential SARS-CoV-2 treatments (117). Meanwhile, scientists continue searching for new drugs that would combat multiple coronaviruses. For instance, the surface S protein of the CoVs provides a tantalizing target. The Jiang research group in China and other research groups around the world have found antibodies and compounds binding to the S protein (118), that could prevent CoVs from invading human cells. It should, however, be noted that studies are at an early stage and these compounds need to be verified, developed into drugs, and tested in animals that could take years. It is therefore, advisable that in the meantime, early corticosteroid with preventive antibiotic treatment is very inexpensive and can help to prevent from collapse of medical system in developing countries.
Vaccine research and development efforts
There is an urgent necessity to produce and distribute effective and safe vaccines to immunize a large number of people all over the world from SARS-CoV-2. Owing to the wide geographic diversity of the pandemic and number of people who need the vaccine, more than one vaccine developmental approach is required. Therefore, the collaborative efforts among the biotechnological companies, industry, academicians, and government are essential to design effective vaccine strategies (119,120). For a vaccine to be widely accepted, a high degree of safety is the most primary concern. There is also a theoretical risk that vaccination could even worsen the case of SARS-CoV-2 infection, as we have seen in the case of feline coronaviruses and huge vaccine-challenge animal models of SARS-CoV (119,121). Nonetheless, efficacy trials need to be evaluated considering both benefit and harm. There is also a possibility of controlled human challenge trials, where a small number of young healthy volunteers could be vaccinated and then exposed to SARS-CoV-2 (119). In this trial, as there are no effective therapies for COVID-19 to rescue volunteers if a complication develops, volunteers are at risk of developing severe disease or even death. To nullify these risk factors, the SARS-CoV-2 challenge strain that causes mild illness will be utilized (119). Another concern is that even if it is effective in young healthy adults, it does not necessarily mean to be effective among older adults. Therefore, careful evaluation is needed before conducting the trials.
Considering some of the facts above, it is likely to take time before a safe, effective, and affordable vaccine becomes available. Nevertheless, most of the vaccine development process is moving toward clinical evaluation. The replicating and nonreplicating viral vectors, traditional recombinant protein and subunit vaccines, and nucleic acid DNA and mRNA approaches are some of the encouraging platforms for vaccine development (119). However, each platform has some advantages as well as demerits. Studies have suggested that the major alterations in the spike protein are not extensive. Therefore, traditional recombinant protein technology can be used to express the spike protein (e.g., Sanofi, Novavax). However, protein vaccines will require a potent adjuvant, but the availability of certain adjuvants is a major concern. The recent progress made in mapping out the SARS-CoV-2 spike protein will pave the way towards the development of vaccines (82). Of particular interest is the use of a relatively new vaccine strategy, RNA vaccines that can elicit potent immune responses against pathogens and certain cancers (122,123). Unlike traditional vaccines that utilize purified proteins from the pathogens, whole deactivated or live attenuated viruses, the RNA-based vaccines use mRNA which upon entering cells, are translated to immunogenic molecules that in turn, stimulate the immune system. This process has been used effectively against some cancers (124,125) and clinical trials are underway for several other cancers (126). Besides, the production of RNA-based vaccines is less expensive and more rapid when compared to traditional vaccines. Therefore, RNA-based vaccine has a major advantage in pandemic situations. Clinical trials for an mRNA-based SARS-CoV-2 vaccine have indeed begun in the United States. Study subjects received the mRNA vaccine in two doses, 28 days apart and the safety and immunogenicity are still in the process to assess.
The other SARS-CoV-2 vaccine relies on producing an immune response against the S protein to block its docking and interaction with the host ACE-2 receptor (127). Based on the recombinant expression of the S protein, Novavax has also generated immunogenic virus-like nanoparticles (128). The Clover Biopharmaceuticals on the other hand is developing a trimerized SARS-CoV-2 S protein as a subunit vaccine using their patented Trimer-Tag technology. Similarly, a consortium comprising of Texas Children’s Hospital Center for Vaccine Development and the University of Texas Medical Branch and New York Blood Center has tested a subunit vaccine comprised of only the receptor-binding domain (RBD) of the SARS-CoV S protein (127,129,130). When administered with alum as an adjuvant, it elicited protective immunity following the homologous virus challenge. The advantage of using an RBD-based vaccine is its ability to minimize host immunopotentiation (127). A safe and effective vaccine needs to be produced in billions of doses for the entire world. Even though new technologies do have the capacity to generate this, there is a need to set up a fund for distributing to every individual’s level. Therefore, it is high time the international agencies collaborated for this noble cause.
Limitations
COVID-19 is wracking havoc globally irrespective of age, gender, sex, ethnic background, lifestyle, and socioeconomic status. Due to the high transmission rate of SARS-Cov-2, it has been a challenge to contain the viral infection. Moreover, the number of trained personnel and health care professionals to conduct testing and taking care of infected individuals has also been a big issue. Despite ongoing research, the precise knowledge about the life cycle of SARS-Cov-2 is very limited, a prerequisite to developing highly specific and more effective therapeutics. Moreover, the current understanding and explanations of this disease are based on epidemiological, clinical and physiological observations (8). A large number of labs worldwide are studying COVID-19, resulting in a huge number of publications in recent months. However, humanity is still waiting for effective anti-SARS-Cov-2 therapy.
Conclusions
The outbreak of COVID-19 rapidly swept across the entire world in a matter of two to three months. As we write this article COVID-19 continues to spread in different parts and corners of the world, scientists have made extraordinary progress in characterizing the novel SARS-CoV-2 and are working incessantly to develop novel therapies and vaccines against the virus. However, many issues about COVID-19 still remain unresolved as the current immunological concepts are insufficient to explain the causes for different clinical phenotypes, incubation periods among individuals, cytokine storm with lymphopenia and the damage to organ cells. Accordingly, the current understanding and explanations about COVID-19 are based on epidemiological, clinical and physiological observations as explained in the introduction section. There is mounting evidence pointing to the fact that there are multiple SARS-CoV-2 strains in global circulation. Compared to SARS and MERS, the emerging COVID-19, caused by SARS-CoV-2, exhibits strong infectivity but less virulence in terms of severity of disease and mortality rates in certain age groups. However, given the rapid and efficient transmission rates of SARS-CoV-2 it has caused in just a few weeks, the COVID-19 appears to inflict more damage in terms of peoples’ health and well-being, social life, and global economic impacts. While there is no effective therapy and vaccine against the novel SARS-CoV-2 yet, preventive measures are the only tool available to our disposal to control the spread of the COVID-19 pandemic. To curtail the transmission, the WHO recommends isolation of suspected cases, testing, and treatment of confirmed cases and tracing of all contacts and quarantining. However, at the individual level, people are advised to avoid becoming infected by practicing social distancing, staying at home, frequently washing hands thoroughly with soap or using alcohol-based sanitizer, avoiding touching of the face particularly the eyes, nose, and mouth with unwashed hands among others. If all these preventive interventions are carefully followed, there is mounting evidence from countries across the world, suggesting that these measures help curtail the epidemic. The recent emergence of COVID-19 pandemic along with other high lethality emerging and re-emerging viruses such as SARS-CoV, MERS-CoV, H5N1, H7N9, Ebola and Marburg should serve as a strong reminder and wake up call to scientists the world over, who should strive to reduce the probability of their re-emergence and the emergence of other novel viruses promptly. For instance, strengthening the investigation of animal etiology, establishing stringent procedures and containment of high-risk pathogens, reducing direct contacts with wildlife while limiting the massive destruction of their habitats, maintaining the barriers between natural reservoirs and human society and completely eradicating wildlife trading, preventing zoonotic transmission will help to achieve these goals.
Supplementary
The article’s supplementary files as
Acknowledgments
We are grateful for the R01 grant (R01DA041746) to MT. We also thank all the lab members of the Center for Translational Medicine, Thomas Jefferson University, who read and commented on the article.
Funding: The research in the Tyagi laboratory is partially funded by the National Institute on Drug Abuse (NIDA), NIH Grant, 1R01DA041746-01. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the U.S. National Institutes of Health. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5272
Conflicts of Interest: All the authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5272). The authors declare no conflict of interest.
References
- 1.Guarner J. Three Emerging Coronaviruses in Two Decades. Am J Clin Pathol 2020;153:420-1. 10.1093/ajcp/aqaa029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565-74. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hui DS, I, Azhar E, Madani TA, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis 2020;91:264-6. 10.1016/j.ijid.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu P, Zhu J, Zhang Z, et al. A familial cluster of infection associated with the 2019 novel coronavirus indicating potential person-to-person transmission during the incubation period. J Infect Dis 2020;221:1757-61. 10.1093/infdis/jiaa077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 2020;382:727-33. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Lancet Infectious Diseases. COVID-19, a pandemic or not? Lancet Infect Dis 2020;20:383. 10.1016/S1473-3099(20)30180-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee KY, Rhim JW, Kang JH. Immunopathogenesis of COVID-19 and early immunomodulators. Clin Exp Pediatr 2020;63:239-50. 10.3345/cep.2020.00759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Setti L, Passarini F, De Gennaro G, et al. Airborne Transmission Route of COVID-19: Why 2 Meters/6 Feet of Inter-Personal Distance Could Not Be Enough. Int J Environ Res Public Health 2020;17:2932. 10.3390/ijerph17082932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henry BM, Vikse J, Benoit S, et al. Hyperinflammation and derangement of renin-angiotensin-aldosterone system in COVID-19: A novel hypothesis for clinically suspected hypercoagulopathy and microvascular immunothrombosis. Clin Chim Acta 2020;507:167-73. 10.1016/j.cca.2020.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lippi G, Favaloro EJ. D-dimer is Associated with Severity of Coronavirus Disease 2019: A Pooled Analysis. Thromb Haemost 2020;120:876-8. 10.1055/s-0040-1709650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry BM, de Oliveira MHS, Benoit S, et al. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med 2020;58:1021-8. 10.1515/cclm-2020-0369 [DOI] [PubMed] [Google Scholar]
- 13.Lippi G, Plebani M, Henry BM. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clin Chim Acta 2020;506:145-8. 10.1016/j.cca.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Ma X. Acute respiratory failure in COVID-19: is it "typical" ARDS? Crit Care 2020;24:198. 10.1186/s13054-020-02911-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Liu Q, Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol 2020;92:418-23. 10.1002/jmv.25681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo PC, Lau SK, Lam CS, et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 2012;86:3995-4008. 10.1128/JVI.06540-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17:181-92. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyrrell DA, Bynoe ML. Cultivation of a Novel Type of Common-Cold Virus in Organ Cultures. Br Med J 1965;1:1467-70. 10.1136/bmj.1.5448.1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kin N, Miszczak F, Lin W, et al. Genomic Analysis of 15 Human Coronaviruses OC43 (HCoV-OC43s) Circulating in France from 2001 to 2013 Reveals a High Intra-Specific Diversity with New Recombinant Genotypes. Viruses 2015;7:2358-77. 10.3390/v7052358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su S, Wong G, Shi W, et al. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol 2016;24:490-502. 10.1016/j.tim.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Li X, Liu W, et al. Discovery of a subgenotype of human coronavirus NL63 associated with severe lower respiratory tract infection in China, 2018. Emerg Microbes Infect 2020;9:246-55. 10.1080/22221751.2020.1717999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye ZW, Yuan S, Yuen KS, et al. Zoonotic origins of human coronaviruses. Int J Biol Sci 2020;16:1686-97. 10.7150/ijbs.45472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monto AS. Medical reviews. Coronaviruses. Yale J Biol Med 1974;47:234-51. [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology 2018;23:130-7. 10.1111/resp.13196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003;361:1319-25. 10.1016/S0140-6736(03)13077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raj VS, Osterhaus AD, Fouchier RA, et al. MERS: emergence of a novel human coronavirus. Curr Opin Virol 2014;5:58-62. 10.1016/j.coviro.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li JY, You Z, Wang Q, et al. The epidemic of 2019-novel-coronavirus (2019-nCoV) pneumonia and insights for emerging infectious diseases in the future. Microbes Infect 2020;22:80-5. 10.1016/j.micinf.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen KG, Rambaut A, Lipkin WI, et al. The proximal origin of SARS-CoV-2. Nat Med 2020;26:450-2. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012;367:1814-20. 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 30.Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003;302:276-8. 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
- 31.Ge XY, Li JL, Yang XL, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013;503:535-8. 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azhar EI, El-Kafrawy SA, Farraj SA, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med 2014;370:2499-505. 10.1056/NEJMoa1401505 [DOI] [PubMed] [Google Scholar]
- 33.Kupferschmidt K. The camel connection. Science 2014;343:1422-3, 5. [DOI] [PubMed]
- 34.Lam TT, Shum MH, Zhu HC, et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature 2020;583:282-5. 10.1038/s41586-020-2169-0 [DOI] [PubMed] [Google Scholar]
- 35.Zhang T, Wu Q, Zhang Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr Biol 2020;30:1346-51.e2. 10.1016/j.cub.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265-9. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song Z, Xu Y, Bao L, et al. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses 2019;11:59. 10.3390/v11010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pachetti M, Marini B, Benedetti F, et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med 2020;18:179. 10.1186/s12967-020-02344-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stefanelli P, Faggioni G, Lo Presti A, et al. Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill 2020;25:2000305. 10.2807/1560-7917.ES.2020.25.13.2000305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep 2020;19:100682. 10.1016/j.genrep.2020.100682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res 1997;48:1-100. 10.1016/S0065-3527(08)60286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herrewegh AA, Smeenk I, Horzinek MC, et al. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol 1998;72:4508-14. 10.1128/JVI.72.5.4508-4514.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woo PC, Lau SK, Yip CC, et al. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J Virol 2006;80:7136-45. 10.1128/JVI.00509-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau SK, Li KS, Huang Y, et al. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J Virol 2010;84:2808-19. 10.1128/JVI.02219-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau SK, Lee P, Tsang AK, et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J Virol 2011;85:11325-37. 10.1128/JVI.05512-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Li J, Xiao Y, et al. Genotype shift in human coronavirus OC43 and emergence of a novel genotype by natural recombination. J Infect 2015;70:641-50. 10.1016/j.jinf.2014.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pyrc K, Dijkman R, Deng L, et al. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J Mol Biol 2006;364:964-73. 10.1016/j.jmb.2006.09.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stanhope MJ, Brown JR, Amrine-Madsen H. Evidence from the evolutionary analysis of nucleotide sequences for a recombinant history of SARS-CoV. Infect Genet Evol 2004;4:15-9. 10.1016/j.meegid.2003.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sabir JS, Lam TT, Ahmed MM, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 2016;351:81-4. 10.1126/science.aac8608 [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Liu D, Shi W, et al. Origin and Possible Genetic Recombination of the Middle East Respiratory Syndrome Coronavirus from the First Imported Case in China: Phylogenetics and Coalescence Analysis. mBio 2015;6:e01280-15. 10.1128/mBio.01280-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia HP, Look DC, Shi L, et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J Virol 2005;79:14614-21. 10.1128/JVI.79.23.14614-14621.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan Y, Shang J, Graham R, et al. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J Virol 2020;94:e00127-20. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bertram S, Glowacka I, Muller MA, et al. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J Virol 2011;85:13363-72. 10.1128/JVI.05300-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsuyama S, Nagata N, Shirato K, et al. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 2010;84:12658-64. 10.1128/JVI.01542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawase M, Shirato K, van der Hoek L, et al. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol 2012;86:6537-45. 10.1128/JVI.00094-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heurich A, Hofmann-Winkler H, Gierer S, et al. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol 2014;88:1293-307. 10.1128/JVI.02202-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 2020;5:562-9. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gui M, Song W, Zhou H, et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res 2017;27:119-29. 10.1038/cr.2016.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirchdoerfer RN, Wang N, Pallesen J, et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep 2018;8:15701. 10.1038/s41598-018-34171-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li F, Li W, Farzan M, et al. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005;309:1864-8. 10.1126/science.1116480 [DOI] [PubMed] [Google Scholar]
- 61.Lu G, Hu Y, Wang Q, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 2013;500:227-31. 10.1038/nature12328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taguchi F, Hirai-Yuki A. Mouse Hepatitis Virus Receptor as a Determinant of the Mouse Susceptibility to MHV Infection. Front Microbiol 2012;3:68. 10.3389/fmicb.2012.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ashour HM, Elkhatib WF, Rahman MM, et al. Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks. Pathogens 2020;9:186. 10.3390/pathogens9030186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anfinrud P, Stadnytskyi V, Bax CE, et al. Visualizing Speech-Generated Oral Fluid Droplets with Laser Light Scattering. N Engl J Med 2020;382:2061-3. 10.1056/NEJMc2007800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meselson M. Droplets and Aerosols in the Transmission of SARS-CoV-2. N Engl J Med 2020;382:2063. 10.1056/NEJMc2009324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arslan M, Xu B, Gamal El-Din M. Transmission of SARS-CoV-2 via fecal-oral and aerosols-borne routes: Environmental dynamics and implications for wastewater management in underprivileged societies. Sci Total Environ 2020;743:140709. 10.1016/j.scitotenv.2020.140709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding S, Liang TJ. Is SARS-CoV-2 Also an Enteric Pathogen With Potential Fecal-Oral Transmission? A COVID-19 Virological and Clinical Review. Gastroenterology 2020;159:53-61. 10.1053/j.gastro.2020.04.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Donà D, Minotti C, Costenaro P, et al. Fecal-Oral Transmission of SARS-CoV-2 In Children: is it Time to Change Our Approach? Pediatr Infect Dis J 2020;39:e133-4. 10.1097/INF.0000000000002704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karimi-Zarchi M, Neamatzadeh H, Dastgheib SA, et al. Vertical Transmission of Coronavirus Disease 19 (COVID-19) from Infected Pregnant Mothers to Neonates: A Review. Fetal Pediatr Pathol 2020;39:246-50. 10.1080/15513815.2020.1747120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N Engl J Med 2020;382:1564-7. 10.1056/NEJMc2004973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang C, Horby PW, Hayden FG, et al. A novel coronavirus outbreak of global health concern. Lancet 2020;395:470-3. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nicholls JM, Poon LL, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet 2003;361:1773-8. 10.1016/S0140-6736(03)13413-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wong CK, Lam CW, Wu AK, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol 2004;136:95-103. 10.1111/j.1365-2249.2004.02415.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kong SL, Chui P, Lim B, et al. Elucidating the molecular physiopathology of acute respiratory distress syndrome in severe acute respiratory syndrome patients. Virus Res 2009;145:260-9. 10.1016/j.virusres.2009.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang KJ, Su IJ, Theron M, et al. An interferon-gamma-related cytokine storm in SARS patients. J Med Virol 2005;75:185-94. 10.1002/jmv.20255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chien JY, Hsueh PR, Cheng WC, et al. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology 2006;11:715-22. 10.1111/j.1440-1843.2006.00942.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Li J, Zhan Y, et al. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect Immun 2004;72:4410-5. 10.1128/IAI.72.8.4410-4415.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cameron MJ, Ran L, Xu L, et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol 2007;81:8692-706. 10.1128/JVI.00527-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Menachery VD, Eisfeld AJ, Schafer A, et al. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio 2014;5:e01174-14. 10.1128/mBio.01174-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Channappanavar R, Fehr AR, Vijay R, et al. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016;19:181-93. 10.1016/j.chom.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zou Y, Guo H, Zhang Y, et al. Analysis of coagulation parameters in patients with COVID-19 in Shanghai, China. Biosci Trends 2020;14:285-9. 10.5582/bst.2020.03086 [DOI] [PubMed] [Google Scholar]
- 82.Tirado SM, Yoon KJ. Antibody-dependent enhancement of virus infection and disease. Viral Immunol 2003;16:69-86. 10.1089/088282403763635465 [DOI] [PubMed] [Google Scholar]
- 83.Olsen CW, Corapi WV, Ngichabe CK, et al. Monoclonal antibodies to the spike protein of feline infectious peritonitis virus mediate antibody-dependent enhancement of infection of feline macrophages. J Virol 1992;66:956-65. 10.1128/JVI.66.2.956-965.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jaume M, Yip MS, Kam YW, et al. SARS CoV subunit vaccine: antibody-mediated neutralisation and enhancement. Hong Kong Med J 2012;18 Suppl 2:31-6. [PubMed] [Google Scholar]
- 85.Yip MS, Leung HL, Li PH, et al. Antibody-dependent enhancement of SARS coronavirus infection and its role in the pathogenesis of SARS. Hong Kong Med J 2016;22:25-31. [PubMed] [Google Scholar]
- 86.Lu X, Zhang L, Du H, et al. SARS-CoV-2 Infection in Children. N Engl J Med 2020;382:1663-5. 10.1056/NEJMc2005073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu X, Xiang Y, Du H, et al. SARS-CoV-2 infection in children - Understanding the immune responses and controlling the pandemic. Pediatr Allergy Immunol 2020;31:449-53. 10.1111/pai.13267 [DOI] [PubMed] [Google Scholar]
- 88.Chao JY, Derespina KR, Herold BC, et al. Clinical Characteristics and Outcomes of Hospitalized and Critically Ill Children and Adolescents with Coronavirus Disease 2019 at a Tertiary Care Medical Center in New York City. J Pediatr 2020;223:14-9.e2. 10.1016/j.jpeds.2020.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Udugama B, Kadhiresan P, Kozlowski HN, et al. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020;14:3822-35. 10.1021/acsnano.0c02624 [DOI] [PubMed] [Google Scholar]
- 90.Basu A, Zinger T, Inglima K, et al. Performance of Abbott ID Now COVID-19 Rapid Nucleic Acid Amplification Test Using Nasopharyngeal Swabs Transported in Viral Transport Media and Dry Nasal Swabs in a New York City Academic Institution. J Clin Microbiol 2020;58:e01136-20. 10.1128/JCM.01136-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gao Y, Li T, Han M, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol 2020;92:791-6. 10.1002/jmv.25770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shafi AMA, Shaikh SA, Shirke MM, et al. Cardiac manifestations in COVID-19 patients-A systematic review. J Card Surg 2020;35:1988-2008. 10.1111/jocs.14808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tenda ED, Yulianti M, Asaf MM, et al. The Importance of Chest CT Scan in COVID-19. Acta Med Indones 2020;52:68-73. [PubMed] [Google Scholar]
- 94.Burhan E, Prasenohadi P, Rogayah R, et al. Clinical Progression of COVID-19 Patient with Extended Incubation Period, Delayed RT-PCR Time-to-positivity, and Potential Role of Chest CT-scan. Acta Med Indones 2020;52:80-3. [PubMed] [Google Scholar]
- 95.Harahwa TA, Lai Yau TH, Lim-Cooke MS, et al. The optimal diagnostic methods for COVID-19. Diagnosis (Berl) 2020. [Epub ahead of print]. [DOI] [PubMed]
- 96.Ippolito D, Pecorelli A, Maino C, et al. Diagnostic impact of bedside chest X-ray features of 2019 novel coronavirus in the routine admission at the emergency department: case series from Lombardy region. Eur J Radiol 2020;129:109092. 10.1016/j.ejrad.2020.109092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li T. Diagnosis and clinical management of severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) infection: an operational recommendation of Peking Union Medical College Hospital (V2.0). Emerg Microbes Infect 2020;9:582-5. 10.1080/22221751.2020.1735265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tai DY. Pharmacologic treatment of SARS: current knowledge and recommendations. Ann Acad Med Singapore 2007;36:438-43. [PubMed] [Google Scholar]
- 99.Chu CM, Cheng VC, Hung IF, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 2004;59:252-6. 10.1136/thorax.2003.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nukoolkarn V, Lee VS, Malaisree M, et al. Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors. J Theor Biol 2008;254:861-7. 10.1016/j.jtbi.2008.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chong YP, Song JY, Seo YB, et al. Antiviral Treatment Guidelines for Middle East Respiratory Syndrome. Infect Chemother 2015;47:212-22. 10.3947/ic.2015.47.3.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Arabi YM, Alothman A, Balkhy HH, et al. Treatment of Middle East Respiratory Syndrome with a combination of lopinavir-ritonavir and interferon-beta1b (MIRACLE trial): study protocol for a randomized controlled trial. Trials 2018;19:81. 10.1186/s13063-017-2427-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020;30:269-71. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends 2020;14:72-3. 10.5582/bst.2020.01047 [DOI] [PubMed] [Google Scholar]
- 105.Biot C, Daher W, Chavain N, et al. Design and synthesis of hydroxyferroquine derivatives with antimalarial and antiviral activities. J Med Chem 2006;49:2845-9. 10.1021/jm0601856 [DOI] [PubMed] [Google Scholar]
- 106.Marmor MF, Kellner U, Lai TY, et al. Recommendations on Screening for Chloroquine and Hydroxychloroquine Retinopathy (2016 Revision). Ophthalmology 2016;123:1386-94. 10.1016/j.ophtha.2016.01.058 [DOI] [PubMed] [Google Scholar]
- 107.Yao X, Ye F, Zhang M, et al. In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin Infect Dis 2020;71:732-9. 10.1093/cid/ciaa237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gautret P, Lagier JC, Parola P, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 2020;56:105949. 10.1016/j.ijantimicag.2020.105949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Saleh M, Gabriels J, Chang D, et al. Effect of Chloroquine, Hydroxychloroquine, and Azithromycin on the Corrected QT Interval in Patients With SARS-CoV-2 Infection. Circ Arrhythm Electrophysiol 2020;13:e008662. 10.1161/CIRCEP.120.008662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sarayani A, Cicali B, Henriksen CH, et al. Safety signals for QT prolongation or Torsades de Pointes associated with azithromycin with or without chloroquine or hydroxychloroquine. Res Social Adm Pharm 2020. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mercuro NJ, Yen CF, Shim DJ, et al. Risk of QT Interval Prolongation Associated With Use of Hydroxychloroquine With or Without Concomitant Azithromycin Among Hospitalized Patients Testing Positive for Coronavirus Disease 2019 (COVID-19). JAMA Cardiol 2020;5:1036-41. 10.1001/jamacardio.2020.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee KY. Pneumonia, Acute Respiratory Distress Syndrome, and Early Immune-Modulator Therapy. Int J Mol Sci 2017;18:388. 10.3390/ijms18020388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sheianov MV, Udalov YD, Ochkin SS, et al. Pulse Therapy With Corticosteroids and Intravenous Immunoglobulin in the Management of Severe Tocilizumab-Resistant COVID-19: A Report of Three Clinical Cases. Cureus 2020;12:e9038. 10.7759/cureus.9038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jawhara S. Could Intravenous Immunoglobulin Collected from Recovered Coronavirus Patients Protect against COVID-19 and Strengthen the Immune System of New Patients? Int J Mol Sci 2020;21:2272. 10.3390/ijms21072272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kraft CS, Hewlett AL, Koepsell S, et al. The Use of TKM-100802 and Convalescent Plasma in 2 Patients With Ebola Virus Disease in the United States. Clin Infect Dis 2015;61:496-502. 10.1093/cid/civ334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Walker LM, Burton DR. Passive immunotherapy of viral infections: 'super-antibodies' enter the fray. Nat Rev Immunol 2018;18:297-308. 10.1038/nri.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maxmen A. More than 80 clinical trials launch to test coronavirus treatments. Nature 2020;578:347-8. 10.1038/d41586-020-00444-3 [DOI] [PubMed] [Google Scholar]
- 118.Xia S, Yan L, Xu W, et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv 2019;5:eaav4580. 10.1126/sciadv.aav4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Corey L, Mascola JR, Fauci AS, Collins FS. A strategic approach to COVID-19 vaccine R&D. Science 2020;368:948-50. 10.1126/science.abc5312 [DOI] [PubMed] [Google Scholar]
- 120.Lurie N, Saville M, Hatchett R, et al. Developing Covid-19 Vaccines at Pandemic Speed. N Engl J Med 2020;382:1969-73. 10.1056/NEJMp2005630 [DOI] [PubMed] [Google Scholar]
- 121.Bolles M, Deming D, Long K, et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol 2011;85:12201-15. 10.1128/JVI.06048-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wolff JA, Malone RW, Williams P, et al. Direct gene transfer into mouse muscle in vivo. Science 1990;247:1465-8. 10.1126/science.1690918 [DOI] [PubMed] [Google Scholar]
- 123.Sahin U, Kariko K, Tureci O. mRNA-based therapeutics--developing a new class of drugs. Nat Rev Drug Discov 2014;13:759-80. 10.1038/nrd4278 [DOI] [PubMed] [Google Scholar]
- 124.Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017;547:222-6. 10.1038/nature23003 [DOI] [PubMed] [Google Scholar]
- 125.Sebastian M, Schroder A, Scheel B, et al. A phase I/IIa study of the mRNA-based cancer immunotherapy CV9201 in patients with stage IIIB/IV non-small cell lung cancer. Cancer Immunol Immunother 2019;68:799-812. 10.1007/s00262-019-02315-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Diken M, Kranz LM, Kreiter S, et al. mRNA: A Versatile Molecule for Cancer Vaccines. Curr Issues Mol Biol 2017;22:113-28. 10.21775/cimb.022.113 [DOI] [PubMed] [Google Scholar]
- 127.Jiang S, Bottazzi ME, Du L, et al. Roadmap to developing a recombinant coronavirus S protein receptor-binding domain vaccine for severe acute respiratory syndrome. Expert Rev Vaccines 2012;11:1405-13. 10.1586/erv.12.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Coleman CM, Liu YV, Mu H, et al. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine 2014;32:3169-74. 10.1016/j.vaccine.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen WH, Chag SM, Poongavanam MV, et al. Optimization of the Production Process and Characterization of the Yeast-Expressed SARS-CoV Recombinant Receptor-Binding Domain (RBD219-N1), a SARS Vaccine Candidate. J Pharm Sci 2017;106:1961-70. 10.1016/j.xphs.2017.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen WH, Du L, Chag SM, et al. Yeast-expressed recombinant protein of the receptor-binding domain in SARS-CoV spike protein with deglycosylated forms as a SARS vaccine candidate. Hum Vaccin Immunother 2014;10:648-58. 10.4161/hv.27464 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as