Abstract
Porcine astroviruses (PAstVs) have wide distribution in swine herds worldwide. At present, five porcine astrovirus genotypes have been identified. In this study, using viral metagenomics, a novel PAstV strain (designated as Ahast) was identified in fecal samples from pigs in Anhui of China, and the complete genomic sequence of Ahast was obtained by assembling and PCR amplification. Genomic structural analysis indicated that Ahast had a typical ribosomal frameshifting signal, and some conserve amino acid motifs were also found in virally encoded proteins. Phylogenetic analysis and sequence comparison indicated that this virus belonged to porcine astrovirus genotype 4 (PAstV4), which formed a clade clustered with other PAstV4. Multiple recombinant events were confirmed by recombination analysis and indicated that Ahast was a potential recombinant. Epidemiological investigation indicated that PAstV4 has a 10.7% prevalence in this pig farm. The new recombinant identified in this study will be beneficial to comprehend the origin, genetic diversity, and evolution of porcine astroviruses in Anhui of China.
Key words: porcine astroviruses, viral metagenomics, genome recombination
Introduction
Astroviruses belong to the family Astroviridae. The virions are small, about 28–30 nm in diameter, nonenveloped, and contain a + ssRNA genome about 6.4–7.7 kb in length (Rivera et al. 2010). The family Astroviridae is divided into two genera, Mamastrovirus (19 species) and Avastrovirus (3 species) (De Benedictis et al. 2011). Members of the genus Mamastrovirus infect mammals, including human (Vu et al. 2017), bovine (Bouzalas et al. 2014), feline (Yi et al. 2018), porcine (Arruda et al. 2017), mink (Blomstrom et al. 2010), but members of the genus Avastrovirus mainly infect birds such as chicken, turkey, and duck (Bidin et al. 2012a; Bidin et al. 2012b; Sajewicz-Krukowska and Domanska-Blicharz 2016). Infection with Avastroviruses often involves intestinal or extra-intestinal manifestations, while gastroenteritis is the predominant feature of infecting Mamastroviruses.
Porcine astroviruses (PAstVs) belong to the Mamastrovirus genus. PAstV was first identified from the feces sample of diarrheal pigs in 1980 using electron microscopy (Bridger 1980). At present, five genotypes of PAstVs were identified (Brnic et al. 2014). The genome of PAstV encodes for three open reading frames (ORF) including ORF1a, ORF1b, and ORF2 (Bosch et al. 2011). The non-structural proteins and an RNA-dependent RNA polymerase (RdRp) were encoded by ORF1a and ORF1b separately, while ORF2 encodes for the viral capsid structural proteins (De Benedictis et al. 2011). PAstVs have been detected in the intestines and faeces of pigs and are responsible for gastrointestinal disorders, mainly in young individuals. However, some porcine astroviruses infections have been described in sick pigs with extra-intestinal manifestations, including respiratory and neurological signs (Padmanabhan and Hause 2016). Additionally, porcine astroviruses still have been detected in clinically healthy pigs (Lv et al. 2019). The exact relationship between PAstV and diseases is unclear.
In the present study, using the viral metagenomics method, we obtained one novel PAstV4 from pig feces samples collected in Anhui province, China. Further, the complete genome was obtained and analysed for genetic evolution and recombination. The epidemiological investigation indicated that a comparable lower prevalence (10.7%) of PAstV4 in this pig farm of Anhui province.
Experimental
Materials and Methods
Sample collection. In 2018, 215 pig feces samples were collected from a pig farm in Anhui province of China, using disposable 1.5 ml Eppendorf (EP) tubes and shipped on dry ice. Each sample was resuspended in 1 ml of phosphate-buffered saline (PBS), then violently vortex for 10 min. After centrifugation at 12,000 × g for 10 min, the supernatants were collected and were stored at –80°C for the following research.
Viral metagenomic analysis. Twenty feces supernatants were randomly chosen to generate one pool. Aspirating 25 μl supernatants per sample and the total 500 μl mixed supernatant was firstly filtered via a 0.45 μm filter (Millipore) to remove bacteria and eukaryotes. Then, the viral particle enrichment filtrate was treated with DNase and RNase to digest free non-viral nucleic acid at 37°C for 60 min and mix once halfway (Zhang et al. 2016). According to the manufacturer’s protocol, the total nucleic acid was extracted using QiaAmp Mini Viral RNA kit (Qiagen). cDNA of viral RNA was synthesized by reverse transcription with six random base primers, then the Klenow enzyme was used to generate the complementary chain of cDNA. Construction of cDNA libraries was done using Nextera XT DNA Sample Preparation Kit (Illumina), and 250 bases paired ends were sequenced using the MiSeq Illumina platform (Ling et al. 2019). The sequenced reads were debarcoded using vendor software (Illumina). Using Phred quality score ten as the threshold, low sequencing quality tails were trimmed. Adaptors and primer sequences were trimmed using VecScreen software (Altschul et al. 1997). The cleaned reads were then de novo assembled by a combination of software including SOAPdenovo2, ABySS, meta-Velvet, and CAP3. The assembled contigs and singlet sequences were compared to the viral proteome database by BLASTx with an E-value cutoff of <10–5 (Deng et al. 2015).
Genomic acquisition and PCR screening. Using Geneious 11.1.2 software, the partial complete porcine astrovirus genome was assembled. Further, PCR primers were designed to amplify 5' terminal and 3' terminal sequences. The primer sequences used for amplification are shown in Table I. The amplification reactions were performed under the following conditions: 95°C for 5 min, 32 cycles of 95°C for 40 sec, 50°C (for the first round) or 55°C (for the second round) for 30 sec and 72°C for 1 min, a final extension at 72°C for 5 min (Zhao et al. 2019). Putative ORFs were analyzed using Geneious 11.1.2 software and ORF finder in NCBI. To investigate the prevalence of PAstV in pigs, a set of primers was designed based on the nucleotide sequences of Ahast ORF2 to perform PCR screening. Primers astWF and astWR were located at 4313–4333 nt and 4818–4799 nt respectively for the first round of PCR, while astNF and astNR were located at 4421–4440 nt and 4725–4706 nt respectively for the second round. The expected length of the amplified fragment is 305 bp.
Table I.
Primers sequences used for screening and amplification for the complete genome of porcine astrovirus.
| Primer ID | Application | Primer sequences (5′-3′) |
|---|---|---|
| astWF astWR |
First round |
ATCACAGCAACCCTAGGCAC AGGTGCAGGTCATTTCAGCA |
| astNF astNR |
Second round |
TGCCTATGGTCCTCTCCAGA ATCCTGCAGTGCACATCTGT |
| 5AstWF 5AstWR |
First round |
TGGTGGCTATGGCCCGTAGG TGCTGCCTCAAGTATGCACA |
| 5AstNF 5AstNR |
Second round |
TGGTGGCTATGGCCCGTAGG TGGGCACAAATGGTTTGCTG |
| 3AstWF 3AstWR |
First round |
GCCCCGATAATGCAGGATGA Oligo(dT)18 |
| 3AstNF 3AstNR |
Second round |
TATTGAAGCCTGGGATGCGG TTTTGCTCAAATTTTTAAATGC |
Phylogenetic analysis. The ORF1b and ORF2 amino acid sequences of Ahast and the representative sequences obtained from GenBank were selected for phylogenetic analysis to classify this virus. All sequences were aligned using CLUSTAL X (version 2.1) with the default settings. The aligned result was saved as a Nexus form file for constructing the phylogenetic tree using Bayes’ theorem in the Mrbayes 3.2.7 program (Liu et al. 2020). The Markov chain was run for a maximum of 1 million generations, in which every 50 generations were sampled, and the first 25% of Markov chain Monte Carlo (mcmc) samples were discarded as burnin. Phylogenetic tree for recombination analysis were constructed using the Maximum Likelihood method in MEGA 5.0. Bootstrap values (based on 500 replicates) for each node were given.
Recombination analysis. To test whether this virus is a recombinant, the complete genome sequence of all 24 PAstV4 available in the GenBank database were downloaded, then aligned using the MUSCLE algorithm in the MEGA-X program, together with this virus. The alignment results were manually check using Geneious 11.1.2 software. Using Recombination Detection Program 5.0 (RDP 5.0), the potential parental sequences and possible recombination sites were identified through eight different test methods, including RDP, Chimaera, MaxChi, SiSCan, GENECONV, BootScan, LARD, and 3Seq.
GenBank accession number. The two nearly complete genomic sequences of Ahast and Ahast-2 were submitted to the GenBank database under accession number MT470220 and MW082586. The raw sequence reads from the metagenomic library were deposited in the Sequence Read Archive of GenBank database under accession number: SRX9247601.
Results
The libraries from pig fecal samples that were sequenced on the MiSeq platform generated 2,476,530 unique sequence reads. Among them, 173,248 sequence reads were viral sequences. Putative mammalian viruses belong to the families Picornaviridae (60,763 reads), Astroviridae (34,279 reads), Caliciviridae (22,628 reads), Picobirnaviridae (17,569 reads), Parvoviridae (4,434 reads), Coronaviridae (2,273 reads), and Circoviridae (1,301 reads), etc. This study mainly focus on astrovirus.
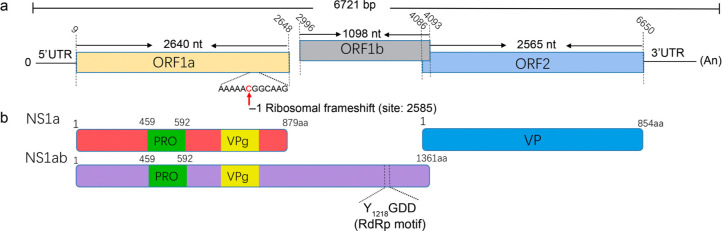
Genomic structure. By assembling the 34,279 reads of astrovirus, two nearly complete genomes of PAstVs were obtained and named Ahast and Ahast-2, respectively. Further, two sets of primers were designed for amplifying the missing 5' and 3' termini sequences of the genome. The complete genome of Ahast is 6,721 bp in length, with the untranslated regions (UTRs) situated in positions 1–9 and 6651–6721. The base composition of A, G, T, and C is 30.23%, 22.38%, 25.17%, and 22.21%, respectively.
The viral genome of Ahast encodes three ORFs, including ORF1a, ORF1b, and ORF2. There is a 8 nt overlap between ORF1b and ORF2. The conserve Kozak sequences of RNNAUGG were identified near the start codon of both three ORFs (GCTATGG for ORF1a, AAAATGT for ORF1b, and CTAATGG for ORF2). A translational ribosomal frameshifting signal existed in the 3' end of Ahast ORF1a Fig. 1a. The Conserved Domains analysis (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) indicated that Ahast NS1a has periplasmic serine protease domain at position 459–592 aa. A conserved RdRp amino acid motif (YGDD), was situated in the N termini of NS1ab. In addition, using online analysis of the FoldIndex software program (http://bip.weizm ann.ac.il/fldbin/findex), a genome-linked viral protein (VPg) with conserved KGKSK and TEEEY was identified (data not shown), at both ends of VPg, potent restriction sites of protease QKKK were found Fig. 1b.
Fig. 1.
Genomic structure and conserve amino acid motif of Ahast. (a) Genomic organization of Ahast. The three ORFs (ORF1a, ORF1b, and ORF2) are shown. The conserve ribosomal frameshift site is marked. (b) The conserve amino acid motif of Ahast. The viral encode polyproteins are marked with different colours, the conserve motif of protease (PRO), RNA-dependent RNA polymerse (RdRp), and genome-linked viral protein (VPg) are shown.
Sequence comparison and phylogenetic analysis. To investigate the genetic relationships of Ahast, the phylogenetic analysis was performed based on the ORF1b and ORF2 amino acid sequences of the strain in this study, 24 PAstV4 reference strians available in the GenBank database, and other four genotypes reference strains of porcine astrovirus. The result showed that five PAstV genotypes were delineated in the ORF1b phylogenetic tree, the Ahast clustered with other 24 PAstV4 reference strains formed a clade Fig. 2, while genotypes in the ORF2 phylogenetic tree did not ultimately cluster PAstVs. PAstV4 was divided into three clades, respectively, Ahast clustered with KX060809, and the other six PAstV4 strains formed a distinct clade Fig. 3. Sequence analysis using Blastp in NCBI indicated that the ORF1b of Ahast shares the highest amino acid sequence identity (98.63%) with the strain PAstV4/CHN/WG-R2/2017 (MK460231), which was identified in an anal swab of pig from Jiangxi province of China. In contrast, the ORF2 of Ahast shares the highest amino acid sequence identity (68.81%) with the strain CH/JXZS/2014 (KX060809), which was found in the stool of sus scrofa from Hunan province of China. It implied that Ahast might be a recombinant.
Fig. 2.
Phylogenetic analysis of Ahast. Phylogenetic tree based on amino acid sequences of ORF1b of PAstVs. The tree was constructed using MrBayes 3.2.7 software and the average standard deviation of split frequencies were 0.005. The Markov chain was run for a maximum of 1 million generations, in which every 50 generations were sampled and the first 25% of Markov chain Monte Carlo (mcmc) samples were discarded as burn-in.The Ahast strain identified in this study is marked with red dot.
Fig. 3.
Phylogenetic analysis of Ahast. Phylogenetic tree based on amino acid sequences of ORF2 of PAstVs. The tree was constructed using MrBayes 3.2.7 software and the average standard deviation of split frequencies were 0.001 respectively. The Markov chain was run for a maximum of 1 million generations, in which every 50 generations were sampled and the first 25% of Markov chain Monte Carlo (mcmc) samples were discarded as burn-in. The Ahast strain identified in this study is marked with red dot.
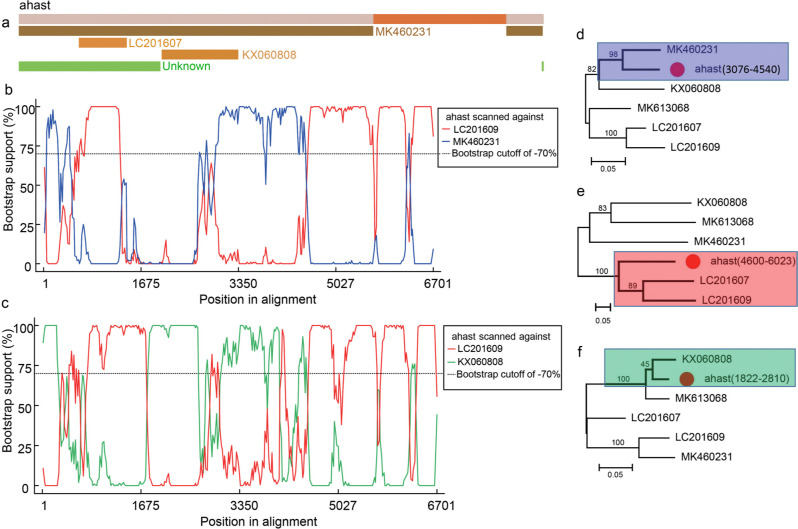
Recombination analysis. To identify potential recombinant events in the Ahast genome, all 24 complete genome sequences of PAstV4, together with that of Ahast, were aligned using the MUSCLE algorithm in the MEGA-X program. Potential parental sequences and possible recombination sites were identified using RDP 5.0. The results showed that Ahast is a novel recombinant resulting from a few recombination events Fig. 4a. Among all those recombination events, two typical recombination events with a high degree of confidence (event 1 with p-value = 3.77 × 10–15, and event 2 with p-value = 2.9 × 10–31) in BootScan method were chosen to further manual bootscan analysis. In both of those two recombination events, The PAstV4/JPN/MoI2-1-1/2015 strain (LC201609) was a major parent, while minor parents were the PAstV4/CHN/WG-R2/2017 strain (MK460231) and the JXJA strain (KX060808), respectively Fig. 4b. Ahast showed higher sequence similarity to the JXJA strain or the PAstV4/CHN/WG-R2/2017 strain in the region of 1822–2810 nt or 3076–4540 nt separately, while in the region of 4600–6023 nt, Ahast displayed higher sequence similarity to the PAstV4/JPN/MoI2-1-1/2015 strain Fig. 4b, and 4c. To confirm this result, phylogenetic trees were constructed basing on the genome region of 1822– 2810 nt, 3076–4540 nt, and 4600–6023 nt, respectively. The trees based on 1822–2810 nt or 3076–4540 nt showed that Ahast clustered with the JXJA strain or the PAstV4/CHN/WG-R2/2017 strain, while the tree based on 3076–4540 nt showed that Ahast clustered with the PAstV4/JPN/MoI2-1-1/2015 strain Fig. 4d, 4e, and 4f. The PAstV4/JPN/MoI2-1-1/2015 strain was first reported in Japan in 2015; the PAstV4/CHN/WG-R2/2017 strain was first found in the Hunan province of China in 2019, while the JXJA strain was first detected in Jiangxi province of China in 2014. From the above data, Ahast might be a potent recombinant form one major parent and two minor parents.
Fig. 4.
Recombination analysis of Ahast. (a) Detected the potential recombination events based on complete genome of Ahast. GenBank No. of each putative recombinant is shown on upper right side of the recombinant. (b), (c) BOOTSCAN evidence for the two different recombination origins on the basis of pairwise distance, modeled with a window size 200, step size 20, and 100 Bootstrap replicates. (d), (e), and (f) Phylogenetic trees were constructed using the Maximun Likelihood in MEGA 5.0 based on the region of 3076–4540 nt, 4600–6023 nt, and 1822–2810 nt respectively. Bootstrap values (based on 500 replicates) for each node were given. The Ahast strain identified in this study is marked with red dot.
To computationally verify the boundaries of different regions in the recombinant, the clean raw reads were remapped to the complete genome of Ahast, which revealed evident overlapping read coverage across the boundaries Fig. S1.
Epidemiological analysis. To investigate the prevalence of PAstV4 in this pig farm, viral RNA was extracted from all 215 feces samples. A set of nested primers were designed based on the Ahast ORF2 nucleotide sequence to perform PCR screening. Primers astWF and astWR were used for the first round of PCR, and astNF and astNR for the second round. The expected length of amplified fragment was 305 bp. The results indicated that 23 (10.7%, 23/215) samples were positive for PAstV4.
Discussion
Porcine astroviruses, as widely distributed viruses, have been isolated from many countries, including China, South Korea, Germany, Czech Republic, Canada, Croatia, Italy, Spain, Australia, Hungary, Thailand, India, and USA, which is divided into five genotypes (Indik et al. 2006; Reuter et al. 2011; Lee et al. 2013; Kattoor et al. 2019; Tassoni et al. 2019). The predominant genotypes in domestic pigs were PAstV2 and PAstV4 in China. Both PAstV2 and PAstV4 were identified in this study, which indicates that those two genotypes of porcine astrovirus circulated in the same pig farm. The prevalence of PAstV4 was 16.1% (35/218) in Hunan of China in previous studies (Xiao et al. 2017), while in the present investigation, the prevalence of PAstV4 was 10.7% (23/215), and it was slightly lower than previously reported. The difference in this prevalence may be due to differences in the type or origin of samples. The samples collected in this study were fecal from healthy pigs, but that were fecal swabs or different tissues from diarrhea pigs in previous studies.
Recombination frequently occurs among astroviruses, and it is the major mechanisms contributing to the emergence of novel strains. Our recombination analysis indicates that Ahast is a potent recombinant with multiple recombinant events. The major parent was identified in Japan, while two minor parents were found in Hunan or Jiangxi province of China. The three parental strains were isolated from different countries or districts, which might hint that the pig trade promoted viral recombination.
In conclusion, we identified a novel PAstV4 in healthy pigs and characterized its near-complete genome. Recombinant analysis indicated that it was a recombinant with multiple recombinant events. The epidemiologic study showed that the prevalence rate of Ahast was slightly lower than that of previously reported in China. More work is needed to investigate the association between virus and illness.
Acknowledgments
This work was supported by National Key Research and Development Programs of China No. 2017YFC1200201, Jiangsu Provincial Key Research and Development Projects No. BE2017693 and Independent Project of Chengdu Research Base of Giant Panda Breeding No. 2020CPB-C11.
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Compliance with ethical standards
The experiments were approved and carried out under animal ethics guidelines and approved protocols of Jiangsu University.
Literature
- Altschul S, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997. Sep 1;25(17):3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda B, Arruda P, Hensch M, Chen Q, Zheng Y, Yang C, Gatto IRH, Ferreyra FM, Gauger P, Schwartz K, et al. Porcine Astrovirus Type 3 in central nervous system of swine with Polioencephalomyelitis. Emerg Infect Dis. 2017. Dec;23(12):2097–2100. 10.3201/eid2312.170703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidin M, Bidin Z, Majnaric D, Tisljar M, Lojkic I. Circulation and phylogenetic relationship of chicken and turkey-origin astroviruses detected in domestic ducks (Anas platyrhynchos domesticus). Avian Pathol. 2012a;41(6):555–562. 10.1080/03079457.2012.733340 [DOI] [PubMed] [Google Scholar]
- Bidin M, Lojkic I, Tisljar M, Bidin Z, Majnaric D. Astroviruses associated with stunting and pre-hatching mortality in duck and goose embryos. Avian Pathol. 2012b;41(1):91–97. 10.1080/03079457.2011.642796 [DOI] [PubMed] [Google Scholar]
- Blomström AL, Widén F, Hammer AS, Belák S, Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J Clin Microbiol. 2010. Dec;48(12):4392–4396. 10.1128/JCM.01040-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch AGS, Krishna NK, Méndez E, Monroe SS, Pantin-Jackwood M, Schultz-Cherry S. Family Astroviridae. In: King AMQ, Lefkowitz E, Adams MJ, Carstens EB, edotors. Virus taxonomy: classification and nomenclature of viruses (ninth report of the International Committee on the Taxonomy of Viruses). New York (USA): Elsevier Academic Press; 2011, p. 953–959. [Google Scholar]
- Bouzalas IG, Wüthrich D, Walland J, Drögemüller C, Zurbriggen A, Vandevelde M, Oevermann A, Bruggmann R, Seuberlich T. Neurotropic astrovirus in cattle with nonsuppurative encephalitis in Europe. J Clin Microbiol. 2014. Sep;52(9):3318–3324. 10.1128/JCM.01195-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger JC. Detection by electron microscopy of caliciviruses, astroviruses and rotavirus-like particles in the faeces of piglets with diarrhoea. Vet Rec. 1980. Dec 6;107(23):532–533. [PubMed] [Google Scholar]
- Brnic D, Jemersic L, Keros T, Prpic J, High prevalence and genetic heterogeneity of porcine astroviruses in domestic pigs. Vet J. 2014; 202(2):390–392. https://doi.org/0.1016/j.tvjl.2014.08.015 [DOI] [PubMed] [Google Scholar]
- De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. Astrovirus infections in humans and animals – Molecular biology, genetic diversity, and interspecies transmissions. Infect Genet Evol. 2011b. Oct;11(7):1529–1544. 10.1016/j.meegid.2011.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, Delwart EL. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res. 2015. Apr 20;43(7):e46 10.1093/nar/gkv002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indik S, Valíček L, Šmíd B, Dvořáková H, Rodák L. Isolation and partial characterization of a novel porcine astrovirus. Vet Microbiol. 2006. Oct;117(2–4):276–283. 10.1016/j.vetmic.2006.06.020 [DOI] [PubMed] [Google Scholar]
- Kattoor JJ, Malik YS, Saurabh S, Sircar S, Vinodhkumar OR, Bora DP, Dhama K, Ghosh S, Banyai K, Touil N, et al. First report and genetic characterization of porcine astroviruses of lineage 4 and 2 in diarrhoeic pigs in India. Transbound Emerg Dis. 2019. Jan;66(1): 47–53. 10.1111/tbed.13058 [DOI] [PubMed] [Google Scholar]
- Lee MH, Jeoung HY, Park HR, Lim JA, Song JY, An DJ. Phylogenetic analysis of porcine astrovirus in domestic pigs and wild boars in South Korea. Virus Genes. 2013. Feb;46(1):175–181. 10.1007/s11262-012-0816-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Zhang X, Qi G, Yang S, Jingjiao L, Shen Q, Wang X, Cui L, Hua X, Deng X, et al. Viral metagenomics reveals significant viruses in the genital tract of apparently healthy dairy cows. Arch Virol. 2019. Apr;164(4):1059–1067. 10.1007/s00705-019-04158-4 [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang H, Ling Y, Yang SX, Wang XC, Zhou R, Xiao YQ, Chen X, Yang J, Fu WG, et al. Viral metagenomics revealed diverse CRESS-DNA virus genomes in faeces of forest musk deer. Virol J. 2020. Dec;17(1):61 10.1186/s12985-020-01332-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv SL, Zhang HH, Li JY, Hu WQ, Song YT, Opriessnig T, Xiao CT. High genetic diversity and recombination events of porcine astrovirus strains identified from ill and asymptomatic pigs in 2017, Hunan Province, China. Virus Genes. 2019. Oct;55(5):673–681. 10.1007/s11262-019-01692-w [DOI] [PubMed] [Google Scholar]
- Padmanabhan A, Hause BM. Detection and characterization of a novel genotype of porcine astrovirus 4 from nasal swabs from pigs with acute respiratory disease. Arch Virol. 2016. Sep;161(9):2575–2579. 10.1007/s00705-016-2937-1 [DOI] [PubMed] [Google Scholar]
- Reuter G, Pankovics P, Boros Á. Identification of a novel astrovirus in a domestic pig in Hungary. Arch Virol. 2011. Jan;156(1):125–128. 10.1007/s00705-010-0827-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera R, Nollens HH, Venn-Watson S, Gulland FMD, Wellehan JFX Jr.. Characterization of phylogenetically diverse astroviruses of marine mammals. J Gen Virol. 2010. Jan 01;91(1):166–173. 10.1099/vir.0.015222-0 [DOI] [PubMed] [Google Scholar]
- Sajewicz-Krukowska J, Domanska-Blicharz K. Nearly full-length genome sequence of a novel astrovirus isolated from chickens with ‘white chicks’ condition. Arch Virol. 2016. Sep;161(9):2581–2587. 10.1007/s00705-016-2940-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassoni L, Zamperin G, Schiavon E, Vendramin V, Cavicchio L, Mion M, Tonon FT, Monne I, Beato MS. First whole genome characterization of porcine astrovirus detected in swine faeces in Italy. Vet Ital. 2019. Sep 30;55(3):221–229. 10.12834/VetIt.1873.9956.1 [DOI] [PubMed] [Google Scholar]
- Vu DL, Bosch A, Pintó R, Guix S. Epidemiology of classic and novel human astrovirus: gastroenteritis and beyond. Viruses. 2017. Feb 18;9(2):33 10.3390/v9020033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao CT, Luo Z, Lv SL, Opriessnig T, Li RC, Yu XL. Identification and characterization of multiple porcine astrovirus genotypes in Hunan province, China. Arch Virol. 2017. Apr;162(4):943–952. 10.1007/s00705-016-3185-0 [DOI] [PubMed] [Google Scholar]
- Yi S, Niu J, Wang H, Dong G, Guo Y, Dong H, Wang K, Hu G. Molecular characterization of feline astrovirus in domestic cats from Northeast China. PLoS One. 2018. Oct 9;13(10):e0205441 10.1371/journal.pone.0205441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li L, Deng X, Blümel J, Nübling CM, Hunfeld A, Baylis SA, Delwart E. Viral nucleic acids in human plasma pools. Transfusion. 2016. Sep;56(9):2248–2255. 10.1111/trf.13692 [DOI] [PubMed] [Google Scholar]
- Zhao C, Chen C, Li Y, Dong S, Tan K, Tian Y, Zhang L, Huang J, Zhang L. Genomic characterization of a novel recombinant porcine astrovirus isolated in northeastern China. Arch Virol. 2019. May; 164(5):1469–1473. 10.1007/s00705-019-04162-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.