Abstract
We have generated a WEE-1.3 strain in C. elegans wherewe have endogenously tagged the C-terminus with GFP. In this publication we demonstrate that this new strain exhibits the same expression localization pattern as the WEE-1.3 antibody and N-terminally endogenously GFP-tagged WEE-1.3 strain that have been previously published. We also show for the first time that endogenously tagging WEE-1.3 at either termini does not affect the reproductive function of the worms.
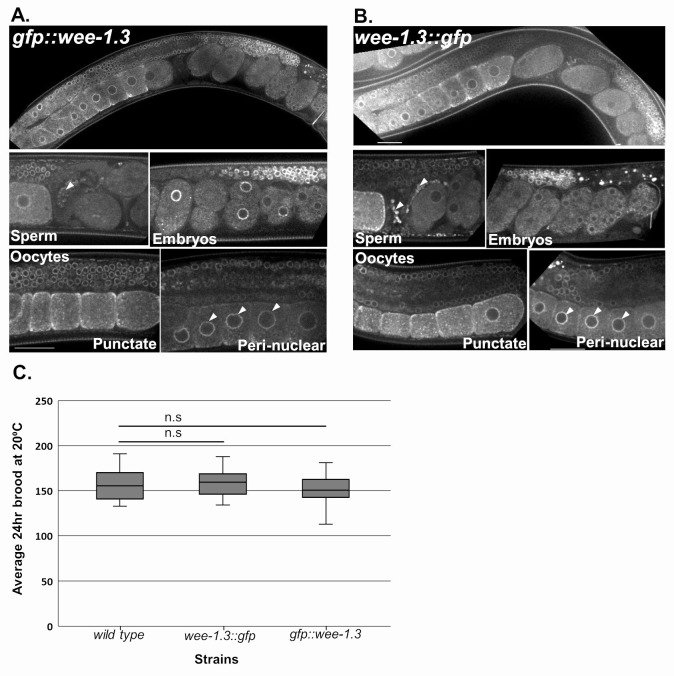
Figure 1. Comparison of N- and C-terminally endogenously GFP-tagged WEE-1.3 strains.
(A-B) Live confocal imaging of germ lines in wee-1.3(ana2[gfp::wee-1.3]) (A) and wee-1.3(ana8[wee-1.3::gfp]) (B) animals. Scale bars indicate 25µm. (C) 24-hour brood analysis comparison of endogenously tagged gfp::wee-1.3 and wee-1.3::gfp strains compared to wild-type (N2) strain. n = 22-28 animals. Statistics were conducted by Student’s T-test, and n.s indicates not significant. Error bars represent the minimum and maximum values.
Description
The Wee1/Myt1 family of kinases are important cell cycle regulators during mitosis and meiosis, and act by specifically phosphorylating certain amino acid residues and inhibiting their target genes. The C. elegans WEE-1.3 protein is a member of this family and has been shown to play an essential role in maintaining oocyte meiotic arrest in the nematode(Allen et al., 2014; Burrows et al., 2006). A complete absence of WEE-1.3 results in embryonic lethality, while depletion of WEE-1.3 starting at the L4 larval stage results in precocious oocyte maturation and sterility (Allen et al., 2014; Burrows et al., 2006). Previously we have shown that WEE-1.3 is expressed throughout the soma and germ line of adult hermaphrodites and developing embryos via antibody staining, transgenic animals tagged with GFP at the N-terminus and C-terminus of WEE-1.3, and endogenously tagging the N-terminus of WEE-1.3 via CRISPR (Allen et al., 2014; Fernando et al., 2020). In these situations, the expression of WEE-1.3 is strongly perinuclear, but with cytoplasmic punctae that co-localizes with ER proteins in oocytes and embryos (Allen et al., 2014; Fernando et al., 2020). Here we have generated a homozygous wee-1.3::gfp strain (WDC8) where GFP is inserted at the 3′ end of the wee-1.3 genomic locus via CRISPR/Cas9 endogenous genome editing. The endogenous C-terminal WEE-1.3::GFPstrain (WDC8) exhibits a perinuclear and cytoplasmic punctate localization expression pattern that is identical to that observed in the WEE-1.3 antibody stained germ lines, transgenic WEE-1.3::GFP animals and in endogenous gfp::wee-1.3 (WDC2) strains (Figure 1A-B). This expression is ubiquitous throughout the soma, in the germ line from the distal tip to the proximal oocytes, in developing embryos, and in sperm stored in the spermatheca. Importantly, both N- and C-terminally endogenously GFP tagged WEE-1.3 strains have a normal 24-hour brood size compared to wild-type control animals implying that we have not disrupted the function of this important reproductive protein kinase by fusing the GFP protein directly to it (Figure 1C). Our data suggest that the new C-terminally endogenously GFP tagged WEE-1.3 strain represents wild-type WEE-1.3 expression and activity, and can be used to monitor endogenous WEE-1.3 levels and localization appropriately.
Methods
The wee-1.3(ana8[wee-1.3::gfp]) WDC8 strain was generated via CRISPR/Cas9 genome editing technology following the direct delivery method developed by the Seydoux laboratory (Paix et al., 2017). Superfolder GFP sequence was inserted at the C-terminus immediately upstream of the stop codon (Pédelacq et al., 2006). crRNA30 (5′- atttggatcatcaggcgacg – 3′) (Horizon Discovery Ltd) was used to guide the Cas9 to cut at the 3′ end of the wee-1.3 gene. A PCR repair template was generated using pDONR221 containing Superfolder GFP and wee-1.3 forward (oAKA431) and wee-1.3 reverse (oAKA432) specific primers. oAKA431: 5′- gttttttttccagatgtcatttggatcatcaggcgacgAaGtttccaagggagaggagctctt-3′
oAKA432: 5′ gcaaaaaataaatattcaacaatttttctgatttttgtgcattattacttgtagagctcgtccattc-3′. The bold regions in the primers refer to the Superfolder GFP sequence. The uppercase letters in oAKA431 denote silent mutations that have been introduced to prevent recutting by crRNA30. Screening for successful edits was performed using the co-conversionmethod and unc-58(e665) as a marker (Arribere et al., 2014). Successful edits were confirmed via PCR using forward primer oAKA88 (5′- gaataatgtgatcgacgaggctcc-3′) and reverse primer oAKA61 (5′- agtgcgatatggtcgagagg-3′). Multiple independent strains were generated, and each strain outcrossed five times before experiments were conducted.
Live imaging was conducted by placing 10-15 worms on a slide containing a 3% agar pad and 10uL of anesthetic (0.1% tricane and 0.01% tetramisole in 1x M9 buffer) then lowering a glass coverslip over the sample. Samples were imaged on a Nikon Ti-E-PFS inverted spinning-disk confocal microscope using a 60x 1.4NA Plan Apo Lambda objective, the 488 laser line, and an Andor iXon 897 EMCDD camera. All image editing was done using NIS-elements software.
For the brood assays, L4 hermaphrodites from each strain (N2, gfp::wee-1.3, wee-1.3::gfp) were placed onto E. coli-seeded MYOB plates and then 16 hours after the L4 stage were singled out (1 worm/plate). The worms were grown at 20ºC, and after 24 hours, the adult mothers were discarded. For all plates, the progeny were counted 1 day after transferring the mother off of the plate. All progeny laid on the plate were counted when determining the brood; this includes eggs and developing larvae. Three independent trials were conducted for a total n ranging from 22-28.
Reagents
N2
WDC2 wee-1.3(ana2[gfp::wee-1.3])
WDC8 wee-1.3(ana8[wee-1.3::gfp])
N2 is available from the CGC, WDC strains are available from the Allen Lab.
Acknowledgments
Funding
This work was supported, in part, by a grant W911NF-18-1-0465 from the Department of Defense to A.K.A. (L.M.F. and A.K.A.) and by grant 1R15HD084253-01A1 from the National Institutes of Health to A.K.A (K.G., R.B. and A.K.A). The spinning disk confocal microscope was acquired through a Department of Defense HBCU/MI Equipment/Instrumentation Grant (#64684-RT-REP) to A.K.A.
References
- Allen AK, Nesmith JE, Golden A. An RNAi-based suppressor screen identifies interactors of the Myt1 ortholog of Caenorhabditis elegans. G3 (Bethesda) 2014 Oct 01;4(12):2329–2343. doi: 10.1534/g3.114.013649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 2014 Aug 26;198(3):837–846. doi: 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows AE, Sceurman BK, Kosinski ME, Richie CT, Sadler PL, Schumacher JM, Golden A. The C. elegans Myt1 ortholog is required for the proper timing of oocyte maturation. Development. 2006 Jan 18;133(4):697–709. doi: 10.1242/dev.02241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando LM, Elliot J, Allen AK. The Caenorhabditis elegans proteasome subunit RPN-12 is required for hermaphrodite germline sex determination and oocyte quality. Dev Dyn. 2020 Aug 01; doi: 10.1002/dvdy.235. [DOI] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Seydoux G. Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods. 2017 Apr 01;121-122:86–93. doi: 10.1016/j.ymeth.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2005 Dec 20;24(1):79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]