Abstract
Genetic screens have been used to identify genes involved in the regulation of different biological processes. We identified growth mutants in a Flp/FRT screen using the Drosophila melanogaster eye to identify conditional regulators of cell growth and cell division. One mutant identified from this screen, B.2.16, was mapped and characterized by researchers in undergraduate genetics labs as part of the Fly-CURE. We find that B.2.16 is a non-lethal genetic modifier of the Dark82 mosaic eye phenotype.
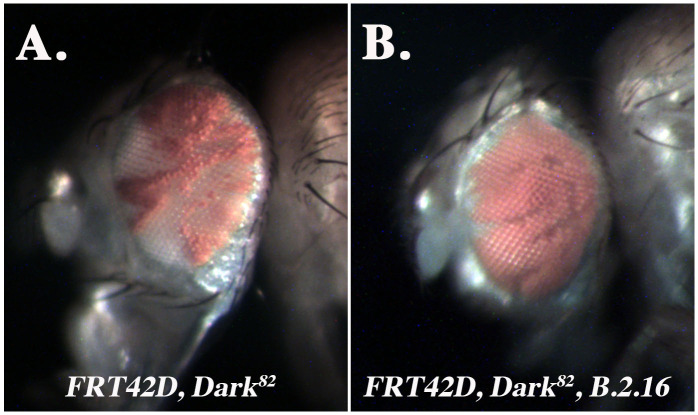
Figure 1. The B.2.16, Dark82 mosaic eye results in an increase in the ratio of mutant pigmented (w+mC) tissue compared to wild type unpigmented tissue.
A. Dark82 mosaic eye (control), mutant tissue is pigmented. B. Dark82, B.2.16 mosaic eye, mutant tissue is pigmented.
Description
Table 1: Complementation tests conducted with mutant B.2.16
| 2R deficiency kit overlapping Dark | |||
| Deficiencies tested within Dark deficiency region | |||
| Deficiency | BDSC Stock # | Region | Complementation test with B.2.16 |
| Df(2R)BSC382 | 24406 | 2R: 16,776,164..16,901,625 | Complement |
| Df(2R)CG15614attP | 84470 | 2R: 16,945,107..16,947,153 | Complement |
| Df(2R)ED1 | 6916 | 2R: 17,026,727..17,097,322 | Complement |
| Df(2R)BSC433 | 24937 | 2R: 17,062,915..17,097,315 | Complement |
| Single genes tested within Dark deficiency region | |||
| Gene | BDSC Stock # | Allele | Complementation test with B.2.16 |
| Ehbp1 | 10931 | Ehbp1k09837 | Complement |
| CG8963 | 12432 | CG8963BG00665 | Complement |
| Pkc53E | 20790 | Pkc53EEY14093 | Complement |
| mute | 22655 | muteEY22147 | Complement |
| PIG-V | 32781 | PIG-VMI01787 | Complement |
| AsnRS-m | 10671 | AsnRS-mk07408 | Complement |
| CG6805 | 78974 | CG6805CR00657-TG4.0 | Complement |
| CG30460 | 59338 | CG30460MI134331 | Complement |
| Genes tested outside of 2R deficiency kit | |||
| Gene | BDSC Stock # | Allele | Complementation test with B.2.16 |
| l(2)41ab | 62050 | l(2)41AbKV00249 | Complement |
| CG40191 | 62029 | CG40191KV00004 | Complement |
In order to identify genes that contribute to the genetic regulation of Drosophila eye development and the regulation of tissue growth, an ethyl methanesulfonate (EMS) mutagenesis screen was performed utilizing the FLP/FRT system, which generates genetically mosaic clones in a tissue specific manner. The screen was performed starting with a copy of chromosome 2R carrying an allele of Death-associated APAF1-Related Killer (Dark), Dark82, which was generated by imprecise P-element excision and therefore retains eye pigmentation due to the presence of an insertion encoding mini-white (Akdemir et al. 2006). When homozygous, the Dark82 allele blocks cell death and allows for the detection of conditional regulators of eye development and tissue growth that might otherwise induce apoptosis (Kagey et al. 2012). Flies that bear a FRT42D, Dark82 chromosome (2R) were mutagenized with EMS and then crossed to ey-Flp; FRT42D flies. The mosaic eyes of resulting offspring were analyzed for phenotypes associated with overall head and eye size, ratio of mutant to wild-type tissue (red over white), or developmental patterning defects (Kagey et al. 2012). One mutant from this screen, B.2.16, was selected for study here. The control mosaic eye, Dark82, has an average ratio of 50-60% mutant (pigmented) tissue (Figure 1A). In comparison the B.2.16, Dark82 mosaic eye ranges from 75-80% mutant tissue (w+mC) demonstrating a consistent increase in the amount of mutant tissue from the Dark82 mosaic ratio (Figure 1B). To understand the mechanism driving this increase in mutant tissue overrepresentation, B.2.16 was mapped via complementation mapping by undergraduate researchers who were part of the Fly-CURE consortium. This type of mapping has been successful for previous mutants from this screen such as Egfr, Ptc, and Shn (Bieser et al. 2019, Kagey et al. 2012, Stamm et al. 2019)
The genetic mapping of B.2.16 on chromosome 2R was completed by four independent groups of undergraduate researchers at Illinois State University, Nevada State College, the University of Evansville, and the University of Detroit Mercy as a part of the Fly-CURE consortium (Bieser et al. 2018, Bieser et al. 2019, Stamm et al. 2019). Complementation tests were conducted using the 87 deficiency stocks from the Bloomington Stock Center 2R Deficiency Kit that are distal to the FRT42D location (Cook et al. 2012). With the hypothesis that the B.2.16 mutation is homozygous lethal, we mapped homozygous lethality by performing complementation tests using the 87 deficiency stocks. Virgin FRT42D, B.2.16, Dark82/CyO females were crossed to males from each of these deficiency stocks and the F1 progeny were scored for complementation. None of the deficiency stocks failed to complement the B.2.16 mutation. This finding suggests three possible hypotheses: 1) The B.2.16 mutation is linked with the Dark82 mutation and therefore unable to be mapped via complementation due to the homozygous lethal nature of the Dark82 allele. Two deficiencies from the deficiency kit (Df(2R)ED2747 and Df(2R)BSC331) fail to complement Dark82; if the B.2.16 mutation lies within this region of 2R:16,869,330..17,097,303, then a failure to complement for the Dark82 allele would be indistinguishable from a failure to complement for the B.2.16 allele (Gramates et al. 2017). 2) The B.2.16 mutation lies in a region that is not covered by a deficiency stock. There are 46 predicted genes on chromosome 2R that are not covered by the deficiency kit (https://bdsc.indiana.edu/stocks/df/dfkit-info.html, Cook et al. 2012). If the mutation exists in one of these genes, it cannot be mapped using the deficiency kit. 3) Lastly, the B.2.16 mutation is not a homozygous lethal mutation and therefore cannot be mapped via complementation tests.
We conducted additional experiments to investigate these hypotheses. To test the first hypothesis that the B.2.16 mutation is tightly linked to the lethal Dark82,we set up complementation tests with smaller deficiency stocks and all available individual gene lethal alleles within the Dark82 region (see Table 1 for a list of stocks tested). We found that B.2.16 complemented all of these crosses within the Dark82 region, suggesting that B.2.16 is not a lethal mutation tightly linked to Dark. To test the second hypothesis that B.2.16 lies in a region of 2R that is not covered by the deficiency kit, we investigated the 46 predicted genes that are not covered by the Df kit. Of these predicted genes, 33 are considered either pseudo-genes or ‘gene model not supported’ on FlyBase and therefore are unlikely candidates (Thurmond et al. 2019). For the remaining 13 genes, there were two available lethal alleles, which were tested and shown to complement with B.2.16 (Table 1). Since our testing for complementation is limited to available lethal alleles and deficiencies, we also investigated genome-wide sequencing (GWS) data for B.2.16 to look for changes in any gene not directly tested via complementation. We found no mutations within these untested genes (either the Dark overlap region or in areas not covered by the Df kit) that would truncate or alter protein function, per SNP effect predictions by SnpEff software (Cingolani et al. 2012).
Given that the B.2.16 mutation complemented all stocks tested across 2R (Df Kit, additional Df stocks, and single allele stocks) and that GWS provides no clear candidate mutations within the genes not tested by complementation, we conclude that the B.2.16 mutation is a non-lethal mutation that modifies the Dark82 mosaic eye phenotype resulting in an increased ratio of mutant tissue to wild-type tissue.
Reagents
FRT42D Dark82/CyO (Akdemir et al.., 2006)FRT42D Dark82, B.2.16 /CyO (this manuscript)Ey-Flp; FRT42D (BDSC 8211) Bloomington Drosophila Stock Center 2R Deficiency Kit (Cook et al.,2012)
Additional Bloomington Stocks (See Table 1 for complete list of stock numbers)
Acknowledgments
Acknowledgments
Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Funding
A. Vrailas-Mortimer is funded by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R15AR070505. Fly-CURE (K. Bieser, J. Kagey, J. Stamm, and A Vrailas-Mortimer) is funded by a National Science Foundation IUSE Award (NSF 2021146).
References
- Akdemir F, Farkas R, Chen P, Juhasz G, Medved'ová L, Sass M, Wang L, Wang X, Chittaranjan S, Gorski SM, Rodriguez A, Abrams JM. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development. 2006 Mar 15;133(8):1457–1465. doi: 10.1242/dev.02332. [DOI] [PubMed] [Google Scholar]
- Bieser K, Stamm J, Aldo A, Bhaskara S, Clairborne M, Coronel Gómez J, Dean R, Dowell A, Dowell E, Eissa M, Fawaz A, Fouad-Meshriky M, Godoy D, Gonzalez K, Hachem M, Hammoud M, Huffman A, Ingram H, Jackman A, Karki B, Khalil N, Khalil H, Ha TK, Kharel A, Kobylarz I, Lomprey H, Lonnberg A, Mahbuba S, Massarani H, Minster M, Molina K, Molitor L, Murray T, Patel P, Pechulis S, Raja A, Rastegari G, Reeves S, Sabu N, Salazar R, Schulert D, Senopole M, Sportiello K, Torres C, Villalobos J, Wu J, Zeigler S, Kagey J. The mapping of Drosophila melanogaster mutant A.4.4. MicroPubl Biol. 2018 Dec 17;2018 doi: 10.17912/micropub.biology.000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieser K, Sanford J, Saville K, Arreola K, Ayres Z, Basulto D, Benito S, Breen C, Brix J, Brown N, Burton K, Chadwick T, Chen M, Chu K, Corbett B, Dill Z, Faughender M, Hickey A, Julia J, Kelty S, Jobs B, Krason B, Lam B, McCullough C, McEwen B, McKenzie J, McQuinn K, Moritz C, Myers K, Naugle E, Nutter A, O'Conke D, O'Grondik M, Patel K, Rudowski S, Sberna E, Stall G, Steiner T, Tanriverdi E, Torres Patarroyo N, Traster V, Tsai L, Valenti A, Villegas M, Voors S, Watson K, Wright M, Kagey J. Genetic mapping of shnE.3.2 in Drosophila melanogaster. MicroPubl Biol. 2019 Jun 01;2019 doi: 10.17912/micropub.biology.000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 1970 Jan 01;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, Kaufman TC, Cook KR. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13(3):R21–R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates LS, Marygold SJ, Santos GD, Urbano JM, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, Falls K, Goodman JL, Hu Y, Ponting L, Schroeder AJ, Strelets VB, Thurmond J, Zhou P, the FlyBase Consortium. FlyBase at 25: looking to the future. Nucleic Acids Res. 2016 Oct 30;45(D1):D663–D671. doi: 10.1093/nar/gkw1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey JD, Brown JA, Moberg KH. Regulation of Yorkie activity in Drosophila imaginal discs by the Hedgehog receptor gene patched. Mech Dev. 2012 Jun 15;129(9-12):339–349. doi: 10.1016/j.mod.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm J, Joshi G, Anderson MA, Bussing K, Houchin C, Elinsky A, Flyte J, Husseini N, Jarosz D, Johnson C, Johnson A, Jones C, Kooner T, Myhre D, Rafaill T, Sayed S, Swan K, Toma J, Kagey J. Genetic mapping of EgfrL.3.1 in Drosophila melanogaster. MicroPubl Biol. 2019 Apr 26;2019 doi: 10.17912/micropub.biology.000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, Kaufman TC, Calvi BR, FlyBase Consortium. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019 Jan 01;47(D1):D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]