Abstract
The pandemics have always been a destructive carrier to living organisms. Humans are the ultimate victims, as now we are facing the SARS CoV-2 virus caused COVID-19 since its emergence in Dec 2019, at Wuhan (China). Due to the new coronavirus’ unexplored nature, we shed light on curcumin for its potential role against the disease. The Nsp9 replicase protein, which plays an essential role in virus replication, was extracted online, followed by 3D PDB model prediction with its validation. The in silico molecular docking of curcumin with the replicase enzyme gave insights into the preventive measures against the virus as curcumin showed multiple interactions with Nsp9 replicase. The current study showed the use of curcumin against the coronavirus and its possible role in developing medicine against it.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00203-020-02163-9.
Keywords: Coronavirus, Nsp9 replicase, Curcumin, In silico, Molecular docking
Introduction
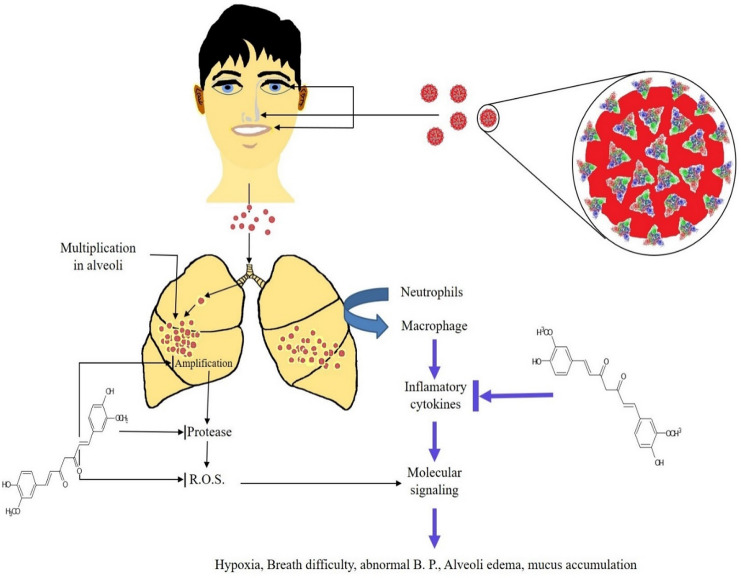
The SARS CoV-2 is responsible for the severe acute respiratory syndrome in humans and is now popularly known as COVID-19 because of the 2019–20 pandemic of coronavirus disease-2019 (Wan et al. 2020). The existing virus is contagious to human health and originated in Wuhan, China, hence sometimes called the “Wuhan virus” (Ralph et al. 2020). It mainly enters the human body through the nasal opening, mouth, and eyes, where it goes to the lungs and multiplies themselves using host cellular machinery. In response to virus attack, host cells start secreting the signaling molecules, which may be the critical markers of viral infection, e.g., difficult breathing (Fig. 1). The coronavirus contains positive-sense single-stranded RNA as genetic material. Structurally, coronavirus contains three major surface proteins called, spike (S) protein (Gallagher and Buchmeier 2001), membrane (M) protein (Neuman et al. 2011), and envelope (E) protein (Ruch et al. 2012). The S-protein makes the crown-like appearance of each virus particle and helps attach the virus to the host protein/glycoproteins present on the host cell surface. Finally, they help in the invasion of virus particles into host cells via the specific receptors called ACE2 receptors. The M-protein is a membrane protein and helps in the assembly of the virus membrane. The E-protein is also a structural protein and helps in the formation of the virion envelope.
Fig. 1.
The common infection pathway of Coronavirus, causes COVID-19 in the human system, with the entry roots; oral, nasal, and mucus membranes from where they reach to the body system to replicate and amplify themselves. Their amplification caused the immune cells to secrete the cytokines (Velazquez et al. 2019), leads to the molecular signaling pathways, cause abnormal breath, hypoxia, etc. Curcumin has the potential to inhibit the virus operated pathways and can lower down the pathological consequences after the virus infection (Qin et al. 2014)
The coronavirus caused severe respiratory illness to the global human population (Su et al. 2016); hence, the WHO declared it a worldwide pandemic (WHO 2020a, b). The virus is non-curable till now. The virus replicates and spreads at a high-speed rate in the human population, depending on their immunity response (The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team 2020). The replicase proteins help the virus to replicate in its host (Littler et al. 2020). These proteins are majorly RNA-dependent RNA polymerase, which binds to the RNA and helps in its replication (Kim et al. 2020). The replicase protein used in the current study is a chain B, Nsp9 replicase from SARS CoV-2 under family Coronaviridae and genus Betacoronavirus.
However, the fact that coronavirus causes COVID-19 is almost non-curable. Still, some molecules/compounds may help prevent or slow down the infection rate by targeting the machinery of viral particles (sampangi et al. 2020). Curcuma longa produces turmeric (diferuloylmethane), named Indian saffron in Europe, with its medicinal uses, including antiviral and anti-inflammatory actions (Araujo et al. 2001). It has shown that curcumin has its inhibitory effects on the virus, including HIV (Hergenhahn et al. 2002), smallpox, measles, and chickenpox are being among its target. It inhibits the integrase and other replication activity needed for viral replication. Figure 1 described coronavirus's entry to the human body and its inhibition by curcumin at multiple steps. In the current study, we showed the possible use of curcumin in the prevention of COVID-19 by targeting the virus replicase protein Nsp9. Turmeric is the principle source of curcumin, and in India it is used as an essential daily ingredient in the food preparation while it has its own antiviral, antifungal, antiallergic properties. Hence, it is preferred over other medicinal compounds in the present study. Nsp9 (non-structural protein 9) RNA binding protein of SARS CoV-2 encoded by ORF1a is supposed to be involved in the viral RNA synthesis (Sutton et al 2004) hence, this protein was targeted in the current study. As, curcumin also showed the antiviral properties, the interaction of curcumin and Nsp9 may be useful in understanding the novel SARS Cov-2.
Material and methods
In silico modeling and molecular docking
The chain B, Nsp9 replicase protein, was found to be a sequence of 117 amino acids and was extracted from NCBI (https://www.ncbi.nlm.nih.gov/protein/6W4B_B) with PDB id; 6W4B. The 3D PDB model of the protein was formed by the SWISS-MODEL (https://swissmodel.expasy.org) and analyzed in PyMOL software (https://pymol.org/2) (Schrodinger 2010). The quality of the predicted protein model was checked by the ProSA web server (https://prosa.services.came.sbg.ac.at/prosa.php) (Wiederstein and Sippl 2007). The active amino acids of chain B, Nsp9 protein were found by the online CASTp server (http://sts.bioe.uic.edu/castp/calculation.html) (Tian et al. 2018) with the default value parameter of 1.4 Å. The structure of curcumin was drawn by chem sketch (http://www.acdlabs.com). The molecular docking of different active amino acids of Nsp9 protein with curcumin was done by Autodock 4.2 software (http://autodock.scripps.edu) (Morris et al. 2009), and the results were analyzed in UCSF chimera software (https://www.cgl.ucsf.edu/chimra) (Pettersen et al. 2004).
Result and discussion
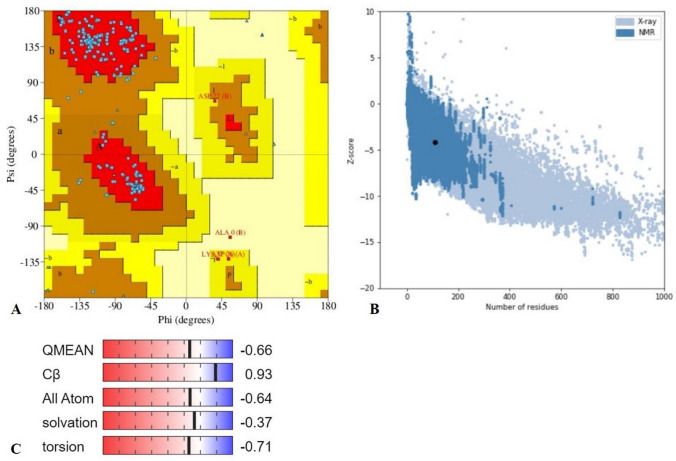
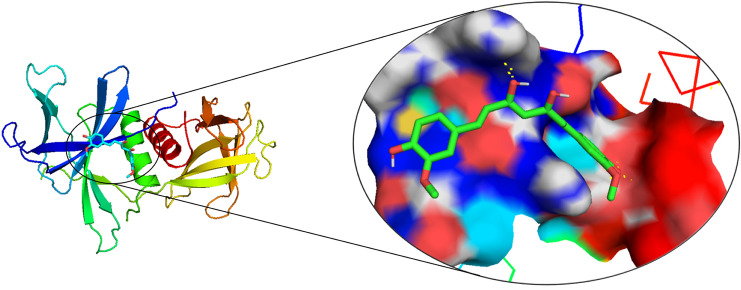
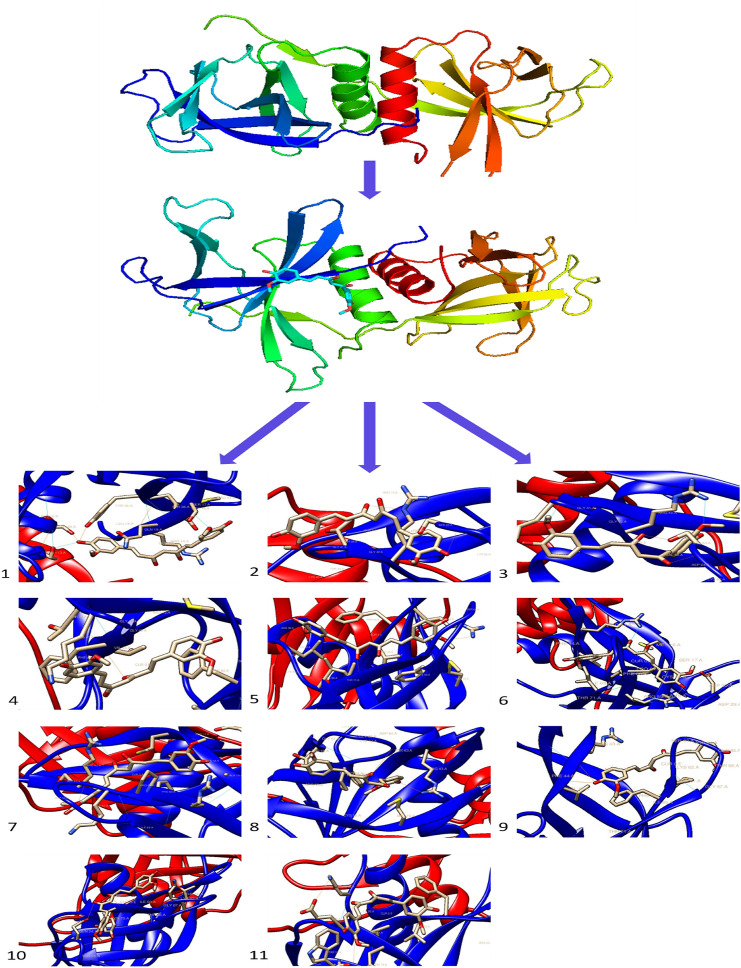
Bioinformatics is a successful initiator to explore the systems biology and chemistry at the molecular level while saving time at the critical global pandemic of COVID-19 viral disease. The Nsp9 protein is taking part in viral replication in the host (human) cells (Sutton et al. 2004). Miknis et al. (2009) showed that its dimerization is necessary for efficient viral growth. The 117 amino acid long Nsp9 we have used was extracted from NCBI for the study due to the pandemic of COVID-19. The Nsp9 protein was started from amino acid serine and ended with glutamine, and it contains the initial seven sheets region and one helix region at last. The predicted protein model of Nsp9 replicase was checked and found to be of good quality as more than 90% amino acids were in the favoured region Ramachandran plot (Fig. 2a), and again the X-ray and NMR prediction by ProSA webserver (Fig. 2b) gave a z-score of − 4.2, confirmed the good quality of the protein model and allowed us to use it in the study. The 3D structure of Nsp9 was of good quality homo-dimer with the QMEAN value of − 0.66 (Fig. 2c) and X-ray resolution of 2.95 Å. These quality checks suggest the protein model used by us is an acceptable model. Further, the CASTp server gave 11 active amino acids (MET 16, GLY 41, GLY 42, ARG 43, VAL 45, PHE 60, PRO 61, LYS 62, SER 63, ILE 69, THR 71), which are docked with curcumin, with their confined coordinates. Docking of curcumin with Nsp9 results gave a ligand-binding pocket of the Nsp9 (Fig. 3), and this was probably the confined site where the curcumin showed interaction with other amino acids. Out of 11 docking complexes, six showed direct interaction of amino acids with the curcumin (Fig. 4). They made eight hydrogen bonds with different docking coordinates assigned to them for different active amino acids. All the docking parameters shown in Table 1 and docking coordinates are shown in Suppl Table 1. The hydrogen bonds formed with curcumin involved THR 113 (Fig. 4.1), SER 17 (Fig. 4.1), GLY 41 (Fig. 4.3), ARG 43 (× 3) (Fig. 4.3, 4.6, 4.7), LYS 62 (Fig. 4.10), and VAL 45 (Fig. 4.11) with bond length 2.896, 3.047, 2.916, 3.046, 2.947, 2.912, 2.905 and 2.966 Å, respectively. The supplementary figure file 1 contains all the descriptive images of Fig. 4 (1–11), obtained through molecular docking. Interestingly, ARG 43 was the most common; three times took part in bond formation with different docking coordinates. This suggested the critical role of these amino acids in the interaction with curcumin. Besides the direct interaction and bond formation, there were 21 more possibilities of bond formation between curcumin and amino acids of Nsp9 replicase protein. The six docked complexes showed both actual and possibilities of the bond formation while five complexes were involved in the possible interaction with curcumin. The molecular docking of GLY 41 and LYS 62 gave the same results as the involvement of the same amino acids; AGR 43 and SER63 with different bond length, again showed the importance of ARG 43 of Ns9.
Fig. 2.
a The Ramachandran plot for assessment of the overall quality of the protein model, indicating up to 90% of the total amino acids in the most favored region of Ramachandran plot (a, b and l). b The Z-score of the protein predicted model was -4.2 indicating the good quality for the study, based on the X-ray and NMR calculations through the ProSA web server. Both quality check program, allowed the predicted model to be used in the current study. c The QMEAN value along with other physical parameters by SWISS MODEL
Fig. 3.
The ligand-binding site of Nsp9 protein holding the curcumin in its pocket
Fig. 4.
Figure showing the in-silico interaction of different active amino acids of Nsp9 replicase protein with the curcumin (1–11). All the active amino acids showed interaction with curcumin with either actual or possibilities of bond formation
Table 1.
The molecular docking parameters used in the study with different active amino acids
| S. no | Active amino acid | Autodock run | Gibbs free energy (ΔG) | Interacting | Hydrogen Bond | ||||
|---|---|---|---|---|---|---|---|---|---|
| AAA’s | AAA’s position | Ligand | Length | Bond nature | |||||
| 1 | MET 16 | 4 | − 6.22 |
THR SER SER TYR |
113 17 109 35 |
CUR CUR CUR CUR |
2.896 Å 3.047 Å 2.938 Å 3.054 Å |
Real Real – – |
– – Possibility Possibility |
| 2 | GLY 41 | 5 | − 5.29 |
ARG SER |
43 63 |
CUR CUR |
3.080 Å 3.024 Å |
– – |
Possibility Possibility |
| 3 | GLY 42 | 4 | − 5.85 |
GLY SER SER ARG |
41 63 63 43 |
CUR CUR CUR CUR |
2.916 Å 2.785 Å 3.151 Å 3.046 Å |
Real – – Real |
Possibility Possibility |
| 4 | ARG 43 | − 7.06 | VAL | 45 | CUR | 2.934 Å | – | Possibility | |
| 5 | VAL 45 | 10 | − 6.70 |
ARG THR |
43 71 |
CUR CUR |
3.433 Å 3.463 Å |
– – |
Possibility Possibility |
| 6 | PHE 60 | 9 | − 7.33 |
ARG ARG VAL |
59 43 45 |
CUR CUR CUR |
3.225 Å 2.947 Å 3.472 Å |
– Real – |
Possibility Possibility |
| 7 | PRO 61 | 6 | − 7.53 |
ARG ARG SER |
59 43 63 |
CUR CUR CUR |
2.969 Å 2.912 Å 3.068 Å |
– Real – |
Possibility Possibility |
| 8 | LYS 62 | 6 | − 5.69 |
ARG SER |
43 63 |
CUR CUR |
3.180 Å 3.538 Å |
– – |
Possibility Possibility |
| 9 | SER 63 | 6 | − 6.25 |
ARG VAL |
43 45 |
CUR CUR |
3.150 Å 3.293 Å |
– – |
Possibility Possibility |
| 10 | ILE 69 | 2 | − 4.86 |
LYS LYS TYR GLN |
62 62 70 24 |
CUR CUR CUR CUR |
2.931 Å 2.905 Å 3.138 Å 3.513 Å |
– Real – – |
Possibility Possibility Possibility |
| 11 | THR 71 | 6 | − 6.54 |
VAL GLU |
45 72 |
CUR CUR |
2.966 Å 2.913 Å |
Real – |
– Possibility |
The autodock 4.2 was programmed for total 10 runs to determine the best fit among them with the minimum binding energy, ΔG (kCal/mol), used for the molecular in silico analysis
As it is well known that in any kind of viral infection, the inflammatory cytokines, IL-1, IL-6, and TNF-α released more actively by immune cells (Velazquez et al. 2019), and they are being the target of curcumin (also diagrammatically represents in Fig. 1). This supports the use of curcumin to reduce the pathological consequences that emerged due to coronavirus infection. So, by targeting the ssRNA of coronavirus at its initial replication stage, through curcumin, when it enters the human is a matter of immediate in-vivo research to possibly overcome the COVID-19 and explore the inhibitory pathways of curcumin to prevent the new coronavirus replication machinery in the human system.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
The author MK would like to thank Hindu college (DU) for providing the infrastructural facility and funds for the ongoing innovation project (SC/2019-2020/05 and SC/2019-2020/06). Kushneet Kaur Sodhi highly acknowledges the University Grant Commission (UGC), Government of India for providing the stipend. DKS particularly acknowledge the NASF (ICAR) for providing the funds for ongoing research, (NASF/CA-6030/2017-18).
Abbreviations
- NCBI
National center for bioinformatics
- CASTp
Computed atlas of surface topography of proteins
- ProSA
Protein structure analysis
- NMR
Nuclear magnetic resonance
- SARS-CoV
Severe acute respiratory syndrome relate corona virus
- ACE
Angiotensin-converting enzyme
- COVID-19
Coronavirus disease-19
- AAA
Active amino acid
- NsP9
Non-structural protein 9
Compliance with ethical standards
Conflict of interest
None of the authors have any kind of conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Araujo CAC, Leon LL. Biological activities of curcuma longa L. Memórias do Instituto Oswaldo Cruz. 2001;96(5):723–728. doi: 10.1590/S0074-02762001000500026. [DOI] [PubMed] [Google Scholar]
- Gallagher TM, Buchmeier MJ. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279(2):371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergenhahn M, Soto U, Weninger A, Polack A, Hsu CH, Cheng AL, Rosl F. The chemopreventive compound curcumin is an efficient inhibitor of Epstein-Barr virus BLZF1 transcription in Raji DR-LUC cells. Mol Carcinogen. 2002;33:137–145. doi: 10.1002/mc.10029. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim D, Lee B (2020) Insufficient Sensitivity of RNA Dependent RNA Polymerase Gene of SARS-CoV-2 Viral Genome as Confirmatory Test using Korean COVID-19 Cases. Preprints 2020, 2020020424 10.20944/preprints202002.0424.v1.
- Littler D, Gully B, Colson RN, Rossjohn J (2020) Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9. bioRxiv. https://www.biorxiv.org/content/10.1101/2020.03.28.013920v1.full.pdf [DOI] [PMC free article] [PubMed]
- Miknis ZJ, Donaldson EF, Umland TC, Rimmer RA, Baric RS, Schultz LW. Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J Virol. 2009;83(7):3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Siddell SG. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Qin Y, Lin L, Chen Y, Wu S, Si X, Wu H, Zhong X. Curcumin inhibits the replication of enterovirus 71 in vitro. Acta Pharmaceutica Sinica B. 2014;4(4):284–294. doi: 10.1016/j.apsb.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph R, Lew J, Zeng T, Francis M, Xue B, Roux M, Kelvin DJ. 2019-nCoV (Wuhan virus), a novel coronavirus: human-to-human transmission, travel-related cases, and vaccine readiness. J Inf Dev Cou. 2020;14(01):3–17. doi: 10.3855/jidc.12425. [DOI] [PubMed] [Google Scholar]
- Ruch TR, Machamer CE. The coronavirus E protein: assembly and beyond. Viruses. 2012;4(3):363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampangi-Ramaiah MH, Vishwakarma R, and Shaanker RU (2020). Molecular docking analysis of selected natural products from plants for inhibition of SARS-CoV-2 main protease. Current Science (00113891), 118(7).
- Schrodinger LLC (2010). The PyMOL molecular graphics system. Version, 1(5), 0.
- Su S, Wong G, Shi W, Liu J, Lai AC, Zhou J, Gao GF. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G, Fry E, Carter L, Sainsbury S, Walter T, Nettleship J, Siddell S. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12(2):341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Chin J Epidemiol. 2020;41(2):145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46(W1):W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Salinas L, Verdugo-Rodriguez A, Rodriguez LL, Borca MV. The role of interleukin 6 during viral infections. Front Microbiol. 2019;10:1057. doi: 10.3389/fmicb.2019.01057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Lepore R. SWISS-model: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35((suppl_2)):W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2020a) Novel coronavirus (2019-nCoV): situation report, 3.
- World Health Organization (2020b) Coronavirus disease 2019 (COVID-19): situation report, 72. https://apps.who.int/iris/bitstream/handle/10665/331685/nCoVsitrep01Apr2020-eng.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.