Abstract
Oxidative stress-induced vascular endothelial cell dysfunction serves an essential role in the initiation and development of atherosclerosis. Sulforaphane (SFN), a naturally occurring antioxidant, has previously demonstrated to exert protective effects on the endothelium against oxidative stress. However, further studies are required to determine its underlying molecular mechanism prior to clinical application. Accumulating evidence suggests that alterations in the microRNA (miRNA/miR)-34a/sirtuin-1 (SIRT1) axis occur with oxidative stress. Therefore, the present study aimed to investigate if SFN exerts a protective role against oxidative stress in vascular endothelial cells through regulation of the miR-34a/SIRT1 axis. Human umbilical vein endothelial cells (HUVECs) were treated with H2O2 in the presence or absence of SFN pretreatment. Cell viability and apoptosis were analyzed using CellTiter-Blue and flow cytometry, respectively. Reverse transcription-quantitative PCR and western blot analyses were performed to determine changes in the expression levels of miR-34a and SIRT1. The expression levels of miR-34a and SIRT1 were artificially regulated following transfection with miR-34a mimic and inhibitor or SIRT1expression plasmid and small interfering RNA, respectively. Subsequently, the effect of the expression changes of miR-34 and SIRT1 on oxidative stress-induced cell injury was investigated. Dual-luciferase reporter assay was used to confirm the targeted binding of miR-34a to SIRT1. SFN was found to ameliorate cellular damage caused by H2O2 and inhibited intracellular reactive oxygen species production. In addition, miR-34a upregulation was accompanied with reduced SIRT1 expression in HUVECs, following H2O2 treatment. miR-34a was revealed to directly target SIRT1 by binding to its 3'-untranslated region. Down-regulation of miR-34a and up-regulation of SIRT1 increased the survival of HUVECs under oxidative stress. Taken together, the results of the present study suggest that SFN may protect HUVECs from oxidative stress by inducing changes in the miR-34a/SIRT1 axis via upregulation of nuclear factor erythroid-2-related factor 2 expression.
Keywords: atherosclerosis, sulforaphane, microRNA-34a, sirtuin-1, oxidative stress
Introduction
Atherosclerosis is a chronic arterial disease characterized by the gradual formation of atherosclerotic plaques in the arterial walls (1). It is considered to be one of the leading causes (with 2.4 million deaths and 61% of cardiovascular deaths in 2016) of vascular-associated mortality worldwide (2), which can result in ischemic stroke, coronary heart disease and peripheral arterial disease (3). Despite advancements in technology and secondary prevention measures (e.g. percutaneous coronary intervention and oral anticoagulants), the burden of atherosclerotic cardiovascular disease continues to increase, posing a significant obstacle to the clinical prevention and treatment of atherosclerosis (4). Research over the past number of decades has continued to unravel the pathophysiological molecular mechanism of atherosclerotic plaque formation (5). Advancements in understanding the cellular or molecular pathogenesis of atherosclerosis have allowed the development of novel therapeutic strategies (6). A therapeutic strategy that has been frequently reported is by protecting vascular endothelial cells from injury since endothelial cell damage and dysfunction serves a key role in the formation and progression of atherosclerotic plaques, leading to subsequent complications (7). Cardiovascular risk factors (e.g. smoking and alcohol) that induce oxidative stress are the principle drivers of endothelial dysfunction. Under oxidative stress, excess reactive oxygen species (ROS) can inhibit the proliferation of vascular endothelial cells, disrupting repair and induce apoptosis (8) Therefore, preventing oxidative stress in the vascular endothelium may protect against or reverse the development of atherosclerosis (9). Therapeutic approaches designed to inhibit NAD(P)H oxidase, a major source of ROS, have been demonstrated to alleviate cardiovascular oxidative stress and prevent endothelial cell damage (10,11). Development of novel strategies (e.g. administration of anti-radical agents and antioxidants) aimed at attenuating oxidative stress hold significant promise for the prevention and treatment of atherosclerosis.
Sulforaphane (SFN) is a phytochemical antioxidant that can be extracted from cruciferous plants, including broccoli, Brussel sprouts and kale (12). SFN exerts protective effects by activating the nuclear factor erythroid-2-related factor 2 (Nrf2)/antioxidant response element (ARE) pathway, which increases the transcription and activity of antioxidant enzymes (13). Cytoprotective properties of SFN against oxidative stress have been reported both in vivo and in vitro. Yoon et al (14) previously demonstrated that SFN pretreatment protects against ischemia and reperfusion-induced acute renal failure in rats by suppressing oxidative stress in renal tissues. Furthermore, Chen et al (15) reported that SFN reduces ROS levels and protects osteoblasts from apoptosis in vitro, whilst Zhu et al (16) suggested that SFN prevents rat aortic smooth muscle cells death by inhibiting ROS production and oxidative cytotoxicity induced by xanthine oxidase. However, to the best of our knowledge, the potential effects of SFN on vascular endothelial cells under oxidative stress have not been previously reported. Further investigation on its underlying molecular mechanism may facilitate the identification of SFN as a novel therapeutic agent for atherosclerosis.
The association between microRNAs (miRNAs/miRs) and oxidative stress has attracted considerable interest over recent decades. A number of miRNAs (e.g. miR-146a, miR-92 and miR-143) that are responsive to oxidative stress have been previously identified constituting a complex regulatory network in pathological conditions such as atherosclerosis (17). miR-34a is one of the more extensively studied oxidative stress-responsive miRNAs that has been previously reported to be upregulated during oxidative stress and mediate apoptosis in mesenchymal stromal/stem cells, HLE-B3 and HUVECs (18-20). There is increasing consensus suggesting that sirtuin-1 (SIRT1), which regulates cell death/survival balance under oxidative stress expression, is impaired by ROS (21). Down-regulation SIRT1 under oxidative stress promoting endothelial dysfunction and vascular disease progression has also been reported (22). In particular, a previous study has documented that SIRT1 may also be under the regulation of oxidative stress-responsive miRNAs, including miR-34a (23).
The present study hypothesized that SFN may alleviate oxidative stress-induced endothelial cell injury via regulation of the miR-34a/SIRT1 axis, since a number of previous studies have also demonstrated that SFN mediates its function by regulating miRNA expression (24,25).
Materials and methods
Cell culture
Human umbilical vein endothelial cells (HUVECs) were obtained from the American Type Culture Collection (ATCC) and cultured in F-12K medium (ATCC) supplemented with 10% FBS (Thermo Fisher Scientific, Inc.), endothelial cell growth supplements (cat. no. 354006; BD Biosciences), 0.1 mg/ml heparin and 1% penicillin/streptomycin (Merck KGaA) at 37˚C in 5% CO2.
H2O2 treatment and SFN pretreatment
The concentrations of H2O2 and SFN used in the present study were determined following a preliminary study, where treatment with SFN for 4, 8 or 12 h prior to the addition of H2O2 exhibited similar protective effects. HUVECs were divided into four groups as follows: i) Control; ii) H2O2 (cat. no. H1009; Merck KgaA); iii) H2O2 + SFN; and iv) SFN (cat. no. S4441; Merck KgaA). The cells cultured at 37˚C and other culture conditions for each group were as follows: i) Cells in the control group were treated with PBS; ii) Cells in the H2O2 group were treated with 200 µmol/l H2O2 to simulate oxidative stress-induced cell injury; iii) Cells in the H2O2 + SFN group were treated with 1.0 µmol/l SFN for 4 h, prior to the addition of 200 µmol/l H2O2; and iv) Cells in the SFN group were treated with 1.0 µmol/l SFN.
Cell transfection
pcDNA3.1-SIRT1 and pcDNA3.1-NRF2 (Vi-gene Co., Ltd.) expression vectors were constructed and transfected (1,000 ng/ml) into HUVECs to overexpress SIRT1 and NRF2, respectively. Empty pcDNA3.1 was used as the negative control. A total of 50 nM siRNA-SIRT1 (Shanghai GenePharma Co., Ltd.) was transfected into cells to downregulate SIRT1 expression, whilst 50 nM miR-34a mimics and inhibitors (Shanghai GenePharma Co., Ltd.) were used to upregulate and downregulate miR-34a expression in cells, respectively. The Allstar negative control sequence (Qiagen AB) served as a control for transfection with siRNA, mimics and inhibitors. Cells were transfected with pcDNA3.1 vectors using TransPass™ HUVEC transfection reagent (New England Biolabs, Inc.) according to the manufacturer's protocol. Cells were transfected with siRNA, mimics and inhibitors using Lipofectamine® RNAiMAX reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Subsequent experiments were performed 48 h after transfection.
Cell viability assay
The CellTiter-Blue® kit (Promega Corporation) was used to measure the viability of HUVECs in the control, H2O2, H2O2 + SFN and SFN groups in accordance with manufacturer's protocol. The respective cells were seeded into 96-well plates at a density of 6,000 cells/well. After 0, 12, 24 and 48 h, cells were incubated with 10 µl CellTiter-Blue® reagent at 37˚C, following which fluorescence was measured using a FLx800™ microplate fluorescence reader at 560 nm (BioTek Instruments Inc.). Cell viability was analyzed and normalized to the value at 0 h.
Flow cytometric analysis of apoptosis
After 24 h treatment, 1x106/ml cells in the control, H2O2, H2O2+SFN and SFN groups were collected, harvested and subsequently resuspended in 500 µl annexin V binding buffer prior to staining with 5 µl Annexin V-FITC and 10 µl propidium iodide (Thermo Fisher Scientific, Inc.) at 25˚C for 15 min in the dark. Apoptotic cells were measured using an Epics XL™ flow cytometer (Beckman Coulter, Inc.) and analyzed using FlowJo™ v9.0 (FlowJo LLC).
ROS measurement
After 24 h treatment, intracellular ROS levels in HUVECs in the control, H2O2, H2O2 + SFN and SFN groups were measured using the Reactive Oxygen Species Assay kit (cat. no. S0033; Beyotime Institute of Biotechnology). Briefly, HUVECs treated with H2O2 were collected and incubated with 2',7'-dichlorofluorescin diacetate (DCFH-DA) probes (1:1,000) for 30 min at 37˚C. Fluorescence was subsequently measured at the emission wavelength of 523 nm, using an excitation wavelength of 488 nm in a FLx800 microplate fluorescence reader.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from the treated HUVECs 24 h after treatment with H2O2 and/or SFN or 48 h post-transfection, using TRIzol® reagent (Thermo Fisher Scientific, Inc.). To measure SIRT1 mRNA expression, total RNA was reverse transcribed into cDNA using RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Inc.) and amplified using the SYBR™-Green PCR Master Mix (Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's protocols. To measure miR-34a expression, total RNA was reverse transcribed into cDNA using the miScript II RT kit (Qiagen AB). qPCR was subsequently performed using the miScript SYBR®-Green PCR kit (Qiagen AB). The relative expression levels of the target genes were calculated using the 2-ΔΔCq method (26) and normalized to the internal reference genes GAPDH for SIRT1 and SNORD-48(27) for miR-34a. The following primer sequences were used for qPCR: SIRT1 forward, 5'-TAGACACGCTGGAACAGGTTGC-3' and reverse, 5'-CTCCTCGTACAGCTTCACAGTC-3' and GAPDH forward, 5'-GTCTCCTCTGACTTCAACAGCG-3' and reverse, 5'-ACCACCCTGTTGCTGTAGCCAA-3'. miR-34a and SNORD-48 primer sequences were obtained from the miScript Primer Assays kit (Qiagen AB).
Western blotting
Total protein was extracted from treated HUVECs 24 h after treatment with H2O2 and/or SFN or 48 h post-transfection, using a protease inhibitor cocktail added to RIPA buffer (Beyotime Institute of Biotechnology). Total protein was quantified using the BSA Protein Assay kit (Beyotime Institute of Biotechnology) and 30 µg protein/lane was separated by 12% SDS-PAGE. Following electrophoresis, protein samples were transferred onto nitrocellulose membranes and blocked with 5% fat-free milk at room temperature. Membranes were incubated with primary antibodies against SIRT1 (1:1,000; cat. no. ab189494), Nrf2 (1:1,000; cat. no. ab137550) and GAPDH (1:5,000; cat. no. ab9485; all from Abcam) for 12 h at 4˚C. The membranes were subsequently washed and incubated with IRDye®-conjugated secondary antibody (1:2,000; LI-COR Biosciences, cat. no. 926-32211) at room temperature for 2 h. Protein bands were visualized using a Li-Cor Odyssey system v 1.60 (LI-COR Biosciences).
Dual-luciferase reporter assay
Both wild-type and mutant SIRT1 3'-untranslated region (3'-UTR) sequences were first cloned into firefly luciferase reporter plasmids pMIR-REPORT™ (Applied Biosystems; Thermo Fisher Scientific, Inc.). Subsequently, 1,000 ng/ml of luciferase reporter vectors (SIRT1 3'-UTR-wild-type or SIRT1 3'-UTR-mutant) were co-transfected with miR-34a mimics (50 nM) or miR-negative controls into HUVECs s. Transfections were performed using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.). Following incubation for 48 h at 37˚C, firefly and Renilla luciferase activities were measured using a Dual-Luciferase® Reporter system (Promega Corporation). Firefly luciferase activity was normalized to that of Renilla luciferase activity.
Statistical analysis
Statistical analysis was performed using the SPSS software (version 19.0; IBM Corp). All experiments were repeated five times. Data are presented as the mean ± standard deviation. Unpaired Student's t-test was used to compare differences between two groups, whilst one-way analysis of variance followed by Tukey's post hoc test was used to compare differences among multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
SFN protects HUVECs from oxidative stress
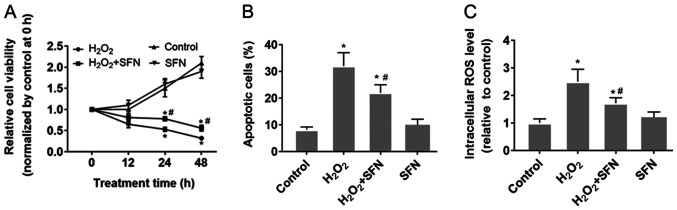
HUVECs were treated with H2O2 to induce oxidative stress. The results demonstrated that H2O2 treatment significantly reduced HUVEC viability whilst significantly increasing apoptosis compared with those under control conditions (P<0.05; Figs. S1A, 1A and B). Furthermore, significantly higher intracellular ROS levels were also observed following H2O2 treatment compared with those in cells in the control group (P<0.05; Fig. 1C). SFN pretreatment was found to relieve the damaging effect of H2O2 in HUVECs compared with that in HUVECs treated with H2O2 alone (Fig. 1). SFN pretreatment was revealed to partially preserve cell viability whilst significantly reducing HUVEC apoptosis induced by H2O2 treatment (P<0.05; Fig. 1A and B). In addition, SFN also alleviated H2O2-induced ROS generation in HUVECs (P<0.05; Fig. 1C). However, HUVECs treated with SFN alone did not exhibit any significant effects in terms of cell viability, apoptosis or ROS production compared with the other three groups (Fig. 1).
Figure 1.
Protective effect of SFN on H2O2-treated HUVECs. (A-C) HUVECs were treated with 200 µmol H2O2, with or without 1.0 µmol/l SFN pretreatment. (A) Cell viability of HUVECs was detected using CellTiter-Blue® assay. (B) Apoptosis was measured by flow cytometry. (C) Intracellular ROS levels were measured using the DCFH-DA probe. *P<0.05 vs. Control and #P<0.05 vs. H2O2. Data are presented as the mean ± standard deviation, n=5. SFN, sulforaphane; HUVECs, human umbilical vein endothelial cells; ROS, reactive oxygen species; DCFH-DA, 2',7'-dichlorofluorescin diacetate.
Effect of SFN on miR-34a and SIRT1 expression in HUVECs under oxidative stress
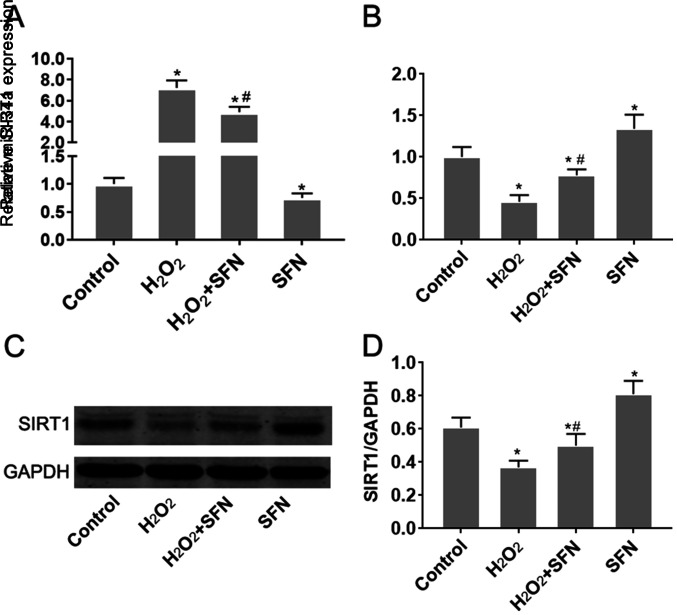
H2O2 treatment was demonstrated to significantly increase miR-34a expression in HUVECs (P<0.05; Fig. 2A). Since TargetScan v7.2 (http://www.targetscan.org/vert_72/) indicated that SIRT1 is a potential target gene of miR-34a, changes in SIRT1 expression were subsequently measured. SIRT1 mRNA and protein expression levels were found to be significantly reduced following treatment with H2O2 (P<0.05; Fig. 2B-D). SFN pretreatment significantly reversed the effects of H2O2 treatment on miR-34a and SIRT1 expression (P<0.05; Fig. 2B-D). Additionally, SFN treatment alone was also found to reduce miR-34a expression whilst increasing SIRT1 expression in HUVECs, compared with those of control cells (Fig. 2). Taken together, these observations suggest that SFN may regulate miR-34a and SIRT1 expression in HUVECs under oxidative stress.
Figure 2.
SFN inhibits miR-34a expression but upregulates SIRT1 expression in HUVECs treated with H2O2. (A-D) HUVECs were treated with H2O2, with or without SFN pretreatment. (A) Changes in miR-34a (B) SIRT1 mRNA expression were measured using RT-qPCR. (C) SIRT1 protein expression was measured by western blot analysis before (D) the protein bands were quantified. *P<0.05 vs. Control; #P<0.05 vs. H2O2. Data are presented as the mean ± standard deviation, n=5. SFN, sulforaphane; miR, microRNA; SIRT1, sirtuin-1; HUVECs, human umbilical vein endothelial cells; RT-qPCR, reverse transcription-quantitative PCR.
miR-34a/SIRT1 axis regulates oxidative stress-induced HUVEC injury and underlies the protective effect of SFN
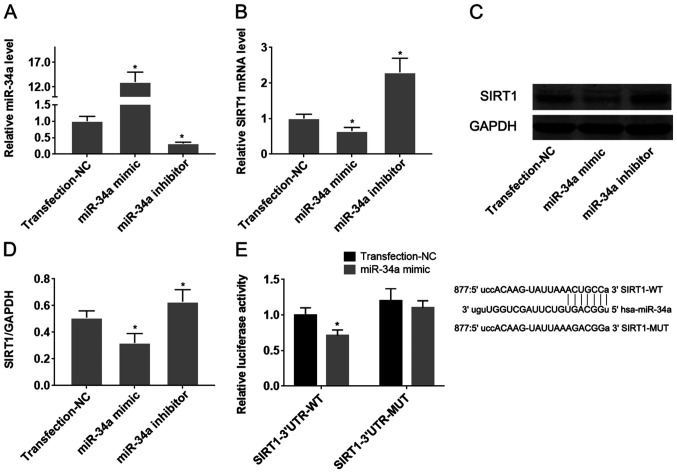
To confirm the target effect of miR-34a on SIRT1 in HUVECs, they were transfected with miR-34a mimics or inhibitors. Transfection with miR-34a mimics and inhibitors significantly was found to significantly increase and reduce miR expression in HUVECs, respectively (Fig. 3A; P<0.05). Overexpression of miR-34a significantly reduced the expression levels of both SIRT1 mRNA and protein (Fig. 3B-D), whilst downregulation of miR-34a mediated the opposite effect on SIRT1 expression, compared with that observed following the upregulation of miR-34a (P<0.05; Fig. 3B-D). Dual-luciferase reporter assay subsequently demonstrated that co-transfection of pMIR-REPORT-SIRT1-3'-UTR-wild-type with the miR-34a mimic significantly reduced luciferase activity, compared with pMIR-REPORT-SIRT1-3'-UTR-mutant-type (P<0.05; Fig. 3E). These results suggest that miR-34a directly targets SIRT1 mRNA by binding to its 3'-UTR.
Figure 3.
miR-34a suppresses SIRT1 mRNA expression by directly binding to its 3'-UTR. miR-34a mimics and inhibitors were transfected into human umbilical vein endothelial cells to change following which the effects of transfection on the level of (A) miR-34a and (B) SIRT1 mRNA expression were verified using reverse transcription-quantitative PCR. (C) Changes in the level of SIRT1 protein expression following transfection with the miR-34a mimic or miR-34a inhibitor were measured using western blot analysis and (D) quantified. (E) The binding capacity of miR-34a to the 3'-UTR of SIRT1 mRNA was verified by dual-luciferase reporter assay. *P<0.05 vs. Transfection-NC. Data are presented as the mean ± standard deviation, n=5. miR, microRNA; SIRT1, sirtuin-1; 3'-UTR, 3'-untranslated region; NC, negative control; WT, wild-type; MUT, mutant.
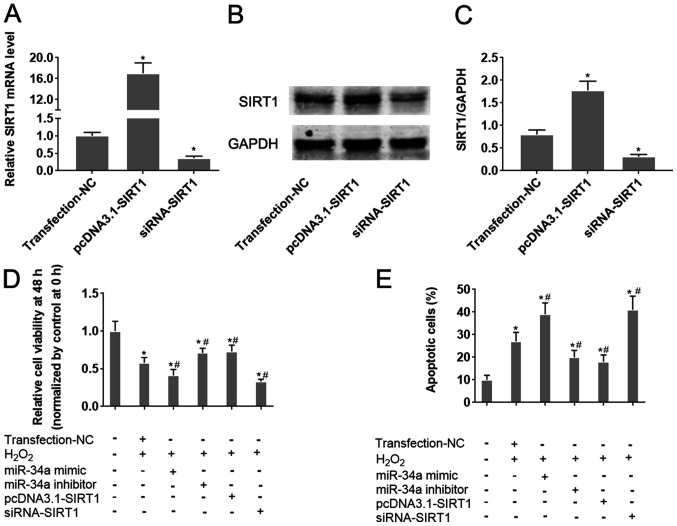
The role of the miR-34a/SIRT1 axis on oxidative stress-induced injury in HUVECs was next analyzed. Transfection of HIUVECs with pcDNA3.1-SIRT1 and siRNA-SIRT1 was demonstrated to significantly upregulate and downregulate SIRT1 expression in HUVECs compared with the respective negative controls (Fig. 4A-C). Overexpression of miR-34a was revealed to significantly aggravate H2O2-induced cell viability inhibition and apoptosis in HUVECs (P<0.05; Fig. 4D and E). Conversely, inhibition of miR-34a antagonized H2O2-induced viability inhibition and apoptosis in HUVECs (P<0.05; Figs. S1B, 4D and E). Following H2O2 treatment, HUVECs transfected the pcDNA3.1-SIRT1 plasmid exhibited significantly increased cell viability and lower apoptotic rates compared with those in cells transfected with negative controls (P<0.05; Figs. S1B, 4D and E). By contrast, opposite effects were observed in HUVECs transfected with siRNA-SIRT1 (P<0.05; Figs. S1B, 4D and E). Taken together, these results collectively suggest that the miR-34a/SIRT1 axis partly underlies the mechanism of oxidative stress-induced injury in HUVECs, where SFN may serve a protective role in HUVECs under oxidative stress by regulating this miR-34a/SIRT1 axis.
Figure 4.
Alteration of the miR-34a/SIRT1 axis influences HUVEC cell viability and apoptosis following H2O2 treatment. miR-34a mimics and inhibitors were transfected into HUVECs 24 h before H2O2 treatment. To change SIRT1 expression in cells, SIRT1-siRNA or pcDNA3.1-SIRT1 were transfected into HUVECs. HUVECs were treated with H2O2. (A) The effect of siRNA-SIRT1 and pcDNA3.1-SIRT1 transfection on SIRT1 mRNA expression was verified by reverse transcription-quantitative PCR analysis. (B) The effect of siRNA-SIRT1 and pcDNA3.1-SIRT1 transfection on SIRT1 protein expression was verified by western blot analysis and (C) quantified. (D and E) HUVEC cells were transfected with the indicated combination of miR-34a mimic, miR-34a inhibitor, pcDNA3.1-SIRT1 or siRNA-SIRT1, following which they were treated with H2O2. (D) HUVEC cell viability was measured after 48 h using CellTiter-Blue® assay. (E) HUVEC apoptosis was measured by flow cytometry. *P<0.05 vs. Transfection-NC; #P<0.05 vs. H2O2 + Transfection-NC. Data are presented as the mean ± standard deviation, n=5. miR, microRNA; SIRT1, sirtuin-1; HUVECs, human umbilical vein endothelial cells; siRNA, small interfering RNA; NC, negative control; Transfection-NC, co-transfection with the empty pcDNA3.1 vector and Allstar.
SFN regulates the miR-34a/SIRT1 axis by inducing Nrf2 expression
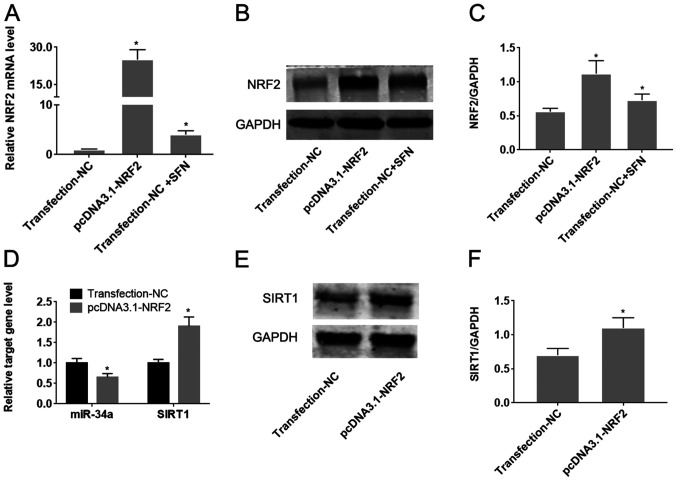
To determine whether Nrf2 regulates the miR-34a/SIRT1 axis in HUVECs, Nrf2 was upregulated by transfection with the pcDNA3.1-Nrf2 plasmid. Transfection with pcDNA3.1-Nrf2 was found to significantly increase Nrf2 expression compared with cells transfected with the empty plasmid (P<0.05; Fig. 5A-C). Treatment of HUVECs with SFN significantly increased the expression of Nrf2 on both mRNA and protein levels (P<0.05; Fig. 5B-D). Overexpression of Nrf2 was also demonstrated to significantly reduce miR-34a expression, whilst increasing SIRT1 expression compared with cells transfected with the empty plasmid (P<0.05; Fig. 5D-F).
Figure 5.
SFN inhibits miR-34a expression whilst enhancing SIRT1 expression by upregulating of Nrf2 in HUVECs. HUVECs were transfected with pcDNA3.1-Nrf2 or NC followed by treatment with SFN. (A) Changes in the level of Nrf2 expression, following treatment with SFN or transfection with pcDNA3.1-Nrf2 were measured using RT-qPCR analysis. (B) The effect of SFN or pcDNA3.1-Nrf2 transfection on Nrf2 protein expression was verified by western blot analysis and (C) the protein bands were quantified. (D) Changes in the expression levels of miR-34a and SIRT1 mRNA following Nrf2 overexpression were measured by RT-qPCR analysis. (E) The effect of pcDNA3.1-Nrf2 transfection on SIRT1 protein expression was verified using western blot analysis and (F) the protein bands were quantified. *P<0.05 vs. Transfection-NC. Data are presented as the mean ± standard deviation, n=5. SFN, sulforaphane; miR, microRNA; Nrf2, nuclear factor erythroid-2-related factor 2; HUVECs, human umbilical vein endothelial cells; RT-qPCR, reverse transcription-quantitative PCR; SIRT1, sirtuin-1; NC, negative control.
Discussion
Vascular endothelial cells are particularly sensitive to oxidative stress, where ROS production results in the induction of inflammatory genes such as vascular cell adhesion protein 1 and monocyte chemoattractant protein-1, oxidation of low density lipoprotein, endothelial cell adhesion and infiltration (28). These alterations disrupt the structure and function of the vascular endothelium, which contributes to the initiation and development of atherosclerosis (29). SFN is a natural antioxidant that can be extracted from cruciferous vegetables (30). The abundant availability of raw materials means that SFN confers certain advantages in clinical applications over other potential naturally occurring agents (31). Previous studies have demonstrated that SFN protects cells against oxidative stress both in vivo and in vitro. Saleh et al (32) reported that SFN has hepatic anti-aging potential through the amelioration of oxidative stress in rats. Another previous in vitro analysis demonstrated that pretreatment of H9c2 rat myoblasts with SFN reduces doxorubicin-induced apoptosis by preventing oxidative stress (33). Although the role of SFN in the protection of endothelial cells through an antioxidant mechanism has been previously reported, information regarding this subject remains insufficient. Campbell et al (34) previously demonstrated that SFN treatment improves the expression of selenoenzymes in the human endothelial cell line EAhy926, protecting the cells from oxidative stress. In brain vascular endothelium, SFN has been found to upregulate antioxidative stress responses, redox signaling and phase 2 drug metabolism/detoxification, thereby exerting neurovascular protective effects (35). Consistent with the aforementioned previous findings, results from the present study demonstrated that SFN inhibited oxidative stress to protect vascular endothelial cells. Pretreatment with SFN was observed to significantly reduce the production of ROS in H2O2-treated HUVECs and suppress apoptosis. SFN has been frequently documented to protects cells against oxidative stress at low-to-moderate doses in cardiomyocytes and nerve cells (36). However, high concentrations of SFN are cytotoxic and confer an acute pro-oxidant effect through the depletion of intracellular glutathione by the formation and export of SFN-glutathione complexes (37). Therefore, the treatment dose of SFN needs to be considered prudently to investigate the potential protective effects of SFN on normal cells.
Prior to its clinical application, it is important to determine the molecular mechanism by which SFN exerts endothelial cell protection. A canonical pathway by which SFN performs a protective role is the Nrf2-ARE pathway (38). SFN can induce the expression or activation of Nrf2 to enhance its binding to ARE (39). Nrf2 is a helix-loop-helix basic leucine zipper transcription factor that serves as a key regulator in the cellular defense system against oxidative stress (40). Through binding to the promoter region of ARE, Nrf2 activation induces the transcription of a series of anti-oxidative stress/detoxifying enzymes, including heme oxygenase-1, NAD(P)H: Quinone oxidoreductase-1, UDP-glucuronosyltransferases and glutathione-S-transferases (41). Accumulating evidence suggests that in addition to inducing the level of antioxidative stress enzymes via the NRF2-ARE pathway, SFN may also alter of expression profile of miRNAs under oxidative stress. Eren et al (42) previously reported that SFN alleviates lipopolysaccharide-induced cell injury and suppresses oxidative stress by inhibiting miR-155. The function of miRNA regulatory networks during oxidative stress in atherosclerosis and arterial remodeling have been discussed in previous studies (43,44). It is therefore possible that SFN may be involved in the regulation of miRNA expression during the protection of endothelial cells from oxidative stress damage.
The present study selected miR-34a as the candidate target gene of SFN in HUVECs under oxidative stress conditions. miR-34a has been previously confirmed as an miRNA that is upregulated in response to oxidative stress. For example, Tong et al (45) demonstrated that miR-34a induction in response to ROS reduced the tolerance of spontaneously arising retinal pigment epithelia cells to oxidative stress. Furthermore, increased miR-34a expression has been previously reported to mediate visfatin-induced apoptosis and oxidative stress in human osteoarthritic chondrocytes, which was reversed by the inhibition of miR-34a (46). However, role of SFN on miR-34a expression remains to be investigated.
SIRT1 is an NAD-dependent deacetylase that regulates apoptosis in response to oxidative and genotoxic stress that has also been found to be a potential target gene of miR-34a (47). The role of the miR-34a/SIRT1 axis in oxidative stress-induced cell injury has been confirmed in endothelial progenitor cells (48). Therefore, the present study hypothesized that SFN may protect endothelial cells partly through regulation of the miR-34a/SIRT1 axis. The results of the present study demonstrated that H2O2 treatment upregulated miR-34a expression, whilst inhibiting SIRT1 expression. Down-regulation of miR-34a and up-regulation of SIRT1 was found to protect cells from oxidative stress. These results are consistent with previous findings, suggesting a role for the miR-34a/SIRT1 axis in HUVECs (48). SFN treatment was demonstrated to reduce the expression levels of miR-34a and induce SIRT1 in HUVECs, indicating that miR-34a/SIRT1 axis counteracted the protective effect of SFN. Taken together, these results suggest that SFN may modulate the miR-34a/SIRT1 axis to protect against oxidative stress-induced HUVEC injury. Furthermore, overexpression of Nrf2 exerted a regulatory effect on the miR-34a/SIRT1 axis, similar to that exhibited by SFN. These results are consistent with previous findings (38,39), which have demonstrated that SFN regulates Nrf2, suggesting that SFN may also regulate the miR-34a/SIRT1 axis via Nrf2 in HUVECs. Nrf2 is considered to be a regulator of miRNA expression during antioxidant responses (35). Nrf2 can directly regulate the expression of miRNAs that contain the functional Nrf2-responsive elements (49). Nrf2 can also indirectly regulate miRNA expression by controlling transcription. For example, miR-125B1 and miR-29B1 were also identified as direct transcriptional targets of NRF2 in acute myeloid leukemia cells (50). However, the present study failed to investigate if Nrf2 directly or indirectly regulates miR-34a. Therefore, prospective studies will aim to perform chromatin immunoprecipitation or luciferase assays to address this.
In conclusion, results from the present study suggest that SFN protects HUVECs against oxidative stress by regulating the miR-34a/SIRT1 axis via Nrf2. Furthermore, SFN exhibited cytoprotective effects on vascular endothelial cells under oxidative stress, putting it forward as a potential candidate for the therapeutic intervention of atherosclerosis.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
TL and ZZ designed the study; TL and QP analyzed and interpreted the data; TL, YL and QP drafted the manuscript; MB and YP performed statistical analysis; TL and YP performed data visualization; YL also contributed to the acquisition and analysis part of data. ZZ supervised the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kobiyama K, Ley K. Atherosclerosis. Circ Res. 2018;123:1118–1120. doi: 10.1161/CIRCRESAHA.118.313816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao D, Liu J, Wang M, Zhang X, Zhou M. Epidemiology of cardiovascular disease in China: Current features and implications. Nat Rev Cardiol. 2019;16:203–212. doi: 10.1038/s41569-018-0119-4. [DOI] [PubMed] [Google Scholar]
- 3.Herrington W, Lacey B, Sherliker P, Armitage J, Lewington S. Epidemiology of atherosclerosis and the potential to reduce the global burden of atherothrombotic disease. Circ Res. 2016;118:535–546. doi: 10.1161/CIRCRESAHA.115.307611. [DOI] [PubMed] [Google Scholar]
- 4.Torres N, Guevara-Cruz M, Velázquez-Villegas LA, Tovar AR. Nutrition and atherosclerosis. Arch Med Res. 2015;46:408–426. doi: 10.1016/j.arcmed.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Criqui MH, Aboyans V. Epidemiology of peripheral artery disease. Circ Res. 2015;116:1509–1526. doi: 10.1161/CIRCRESAHA.116.303849. [DOI] [PubMed] [Google Scholar]
- 6.Fredman G, Tabas I. Boosting inflammation resolution in atherosclerosis: The next frontier for therapy. Am J Pathol. 2017;187:1211–1221. doi: 10.1016/j.ajpath.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma H, Su L, He X, Miao J. Loss of HMBOX1 promotes LPS-induced apoptosis and inhibits LPS-induced autophagy of vascular endothelial cells in mouse. Apoptosis. 2019;24:946–957. doi: 10.1007/s10495-019-01572-6. [DOI] [PubMed] [Google Scholar]
- 8.Marchio P, Guerra-Ojeda S, Vila JM, Aldasoro M, Victor VM, Mauricio MD. Targeting early atherosclerosis: A focus on oxidative stress and inflammation. Oxid Med Cell Longev. 2019;2019(8563845) doi: 10.1155/2019/8563845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kattoor AJ, Pothineni NV, Palagiri D, Mehta JL. Oxidative stress in atherosclerosis. Curr Atheroscler Rep. 2017;19(42) doi: 10.1007/s11883-017-0678-6. [DOI] [PubMed] [Google Scholar]
- 10.Libby P, Bornfeldt KE, Tall AR. Atherosclerosis: Successes, surprises, and future challenges. Circ Res. 2016;118:531–534. doi: 10.1161/CIRCRESAHA.116.308334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Hyseni X, Carter JD, Soukup JM, Dailey LA, Huang YC. Pollutant particles enhanced H2O2 production from NAD(P)H oxidase and mitochondria in human pulmonary artery endothelial cells. Am J Physiol Cell Physiol. 2006;291:C357–C365. doi: 10.1152/ajpcell.00365.2005. [DOI] [PubMed] [Google Scholar]
- 12.Sita G, Hrelia P, Graziosi A, Morroni F. Sulforaphane from cruciferous vegetables: Recent advances to improve glioblastoma treatment. Nutrients. 2018;10(1755) doi: 10.3390/nu10111755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C, Wu J, Chen D, Jin J, Wu Y, Chen Z. Effects of sulforaphane in the central nervous system. Eur J Pharmacol. 2019;853:153–168. doi: 10.1016/j.ejphar.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Yoon HY, Kang NI, Lee HK, Jang KY, Park JW, Park BH. Sulforaphane protects kidneys against ischemia-reperfusion injury through induction of the nrf2-dependent phase 2 enzyme. Biochem Pharmacol. 2008;75:2214–2223. doi: 10.1016/j.bcp.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 15.Chen Q, Wu S, Lu T, Chen J, Xu Z, Chen J. The effect of sulforaphane on the activity and mineralization of osteoblasts under oxidative stress. Pharmacology. 2019;104:147–156. doi: 10.1159/000500846. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Jia Z, Strobl JS, Ehrich M, Misra HP, Li Y. Potent induction of total cellular and mitochondrial antioxidants and phase 2 enzymes by cruciferous sulforaphane in rat aortic smooth muscle cells: Cytoprotection against oxidative and electrophilic stress. Cardiovasc Toxicol. 2008;8:115–125. doi: 10.1007/s12012-008-9020-4. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg MW, Moore KJ. MicroRNA regulation of atherosclerosis. Circ Res. 2016;118:703–720. doi: 10.1161/CIRCRESAHA.115.306300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Zhang X, Chen J, Li T. Inhibition of mircoRNA-34a enhances survival of human bone marrow mesenchymal stromal/stem cells under oxidative stress. Med Sci Monit. 2018;24:264–271. doi: 10.12659/msm.904618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Guo C, Yu M, Ning X, Yan B, Zhao J, Yang A, Yan H. Identification of H2O2 induced oxidative stress associated microRNAs in HLE-B3 cells and their clinical relevance to the progression of age-related nuclear cataract. BMC Ophthalmol. 2018;18(93) doi: 10.1186/s12886-018-0766-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong X, Li P, Li J, He R, Cheng G, Li Y. Downregulation of microRNA34a inhibits oxidized lowdensity lipoproteininduced apoptosis and oxidative stress in human umbilical vein endothelial cells. Int J Mol Med. 2018;42:1134–1144. doi: 10.3892/ijmm.2018.3663. [DOI] [PubMed] [Google Scholar]
- 21.Singh P, Hanson PS, Morris CM. SIRT1 ameliorates oxidative stress induced neural cell death and is down-regulated in parkinson's disease. BMC Neurosci. 2017;18(46) doi: 10.1186/s12868-017-0364-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Huang Q, Zeng Z, Wu J, Zhang Y, Chen Z. Sirt1 inhibits oxidative stress in vascular endothelial cells. Oxid Med Cell Longev. 2017;2017(7543973) doi: 10.1155/2017/7543973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z, Shentu TP, Wen L, Johnson DA, Shyy JY. Regulation of SIRT1 by oxidative stress-responsive miRNAs and a systematic approach to identify its role in the endothelium. Antioxid Redox Signal. 2013;19:1522–1538. doi: 10.1089/ars.2012.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dacosta C, Bao Y. The role of microRNAs in the chemopreventive activity of sulforaphane from cruciferous vegetables. Nutrients. 2017;9(902) doi: 10.3390/nu9080902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rafiei H, Ashrafizadeh M, Ahmadi Z. MicroRNAs as novel targets of sulforaphane in cancer therapy: The beginning of a new tale? Phytother Res. 2020;34:721–728. doi: 10.1002/ptr.6572. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Bignotti E, Calza S, Tassi RA, Zanotti L, Bandiera E, Sartori E, Odicino FE, Ravaggi A, Todeschini P, Romani C. Identification of stably expressed reference small non-coding RNAs for microRNA quantification in high-grade serous ovarian carcinoma tissues. J Cell Mol Med. 2016;20:2341–2348. doi: 10.1111/jcmm.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrotta I, Aquila S. The role of oxidative stress and autophagy in atherosclerosis. Oxid Med Cell Longev. 2015;2015(130315) doi: 10.1155/2015/130315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Förstermann U, Xia N, Li H. Roles of vascular oxidative stress and nitric oxide in the pathogenesis of atherosclerosis. Circ Res. 2017;120:713–735. doi: 10.1161/CIRCRESAHA.116.309326. [DOI] [PubMed] [Google Scholar]
- 30.Vanduchova A, Anzenbacher P, Anzenbacherova E. Isothiocyanate from broccoli, sulforaphane, and its properties. J Med Food. 2019;22:121–126. doi: 10.1089/jmf.2018.0024. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Palliyaguru DL, Kensler TW. Frugal chemoprevention: Targeting nrf2 with foods rich in sulforaphane. Semin Oncol. 2016;43:146–153. doi: 10.1053/j.seminoncol.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saleh DO, Mansour DF, Hashad IM, Bakeer RM. Effects of sulforaphane on D-galactose-induced liver aging in rats: Role of keap-1/nrf-2 pathway. Eur J Pharmacol. 2019;855:40–49. doi: 10.1016/j.ejphar.2019.04.043. [DOI] [PubMed] [Google Scholar]
- 33.Li B, Kim DS, Yadav RK, Kim HR, Chae HJ. Sulforaphane prevents doxorubicin-induced oxidative stress and cell death in rat h9c2 cells. Int J Mol Med. 2015;36:53–64. doi: 10.3892/ijmm.2015.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell L, Howie F, Arthur JR, Nicol F, Beckett G. Selenium and sulforaphane modify the expression of selenoenzymes in the human endothelial cell line EAhy926 and protect cells from oxidative damage. Nutrition. 2007;23:138–144. doi: 10.1016/j.nut.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Sajja RK, Kaisar MA, Vijay V, Desai VG, Prasad S, Cucullo L. In vitro modulation of redox and metabolism interplay at the brain vascular endothelium: Genomic and proteomic profiles of sulforaphane activity. Sci Rep. 2018;8(12708) doi: 10.1038/s41598-018-31137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerrero-Beltrán CE, Calderón-Oliver M, Pedraza-Chaverri J, Chirino YI. Protective effect of sulforaphane against oxidative stress: Recent advances. Exp Toxicol Pathol. 2012;64:503–508. doi: 10.1016/j.etp.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Yanaka A, Zhang S, Tauchi M, Suzuki H, Shibahara T, Matsui H, Nakahara A, Tanaka N, Yamamoto M. Role of the nrf-2 gene in protection and repair of gastric mucosa against oxidative stress. Inflammopharmacology. 2005;13:83–90. doi: 10.1163/156856005774423863. [DOI] [PubMed] [Google Scholar]
- 38.Bai Y, Wang X, Zhao S, Ma C, Cui J, Zheng Y. Sulforaphane protects against cardiovascular disease via nrf2 activation. Oxid Med Cell Longev. 2015;2015(407580) doi: 10.1155/2015/407580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houghton CA, Fassett RG, Coombes JS. Sulforaphane and other nutrigenomic nrf2 activators: Can the clinician's expectation be matched by the reality? Oxid Med Cell Longev. 2016;2016(7857186) doi: 10.1155/2016/7857186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su X, Jiang X, Meng L, Dong X, Shen Y, Xin Y. Anticancer activity of sulforaphane: The epigenetic mechanisms and the nrf2 signaling pathway. Oxid Med Cell Longev. 2018;2018(5438179) doi: 10.1155/2018/5438179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dinkova-Kostova AT, Fahey JW, Kostov RV, Kensler TW. KEAP1 and done? Targeting the NRF2 pathway with sulforaphane. Trends Food Sci Technol. 2017;69:257–269. doi: 10.1016/j.tifs.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eren E, Tufekci KU, Isci KB, Tastan B, Genc K, Genc S. Sulforaphane inhibits lipopolysaccharide-induced inflammation, cytotoxicity, oxidative stress, and miR-155 expression and switches to mox phenotype through activating extracellular signal-regulated kinase 1/2-nuclear factor erythroid 2-related factor 2/antioxidant response element pathway in murine microglial cells. Front Immunol. 2018;9(36) doi: 10.3389/fimmu.2018.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zampetaki A, Dudek K, Mayr M. Oxidative stress in atherosclerosis: The role of microRNAs in arterial remodeling. Free Radic Biol Med. 2013;64:69–77. doi: 10.1016/j.freeradbiomed.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 44.Chen T, Huang Z, Wang L, Wang Y, Wu F, Meng S, Wang C. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 2009;83:131–139. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 45.Tong N, Jin R, Zhou Z, Wu X. Involvement of microRNA-34a in age-related susceptibility to oxidative stress in ARPE-19 cells by targeting the silent mating type information regulation 2 homolog 1/p66shc pathway: Implications for age-related macular degeneration. Front Aging Neurosci. 2019;11(137) doi: 10.3389/fnagi.2019.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheleschi S, Tenti S, Mondanelli N, Corallo C, Barbarino M, Giannotti S, Gallo I, Giordano A, Fioravanti A. MicroRNA-34a and microRNA-181a mediate visfatin-induced apoptosis and oxidative stress via NF-kappaB pathway in human osteoarthritic chondrocytes. Cells. 2019;8(874) doi: 10.3390/cells8080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamakuchi M, Ferlito M, Lowenstein CJ. MiR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo Y, Li P, Gao L, Zhang J, Yang Z, Bledsoe G, Chang E, Chao L, Chao J. Kallistatin reduces vascular senescence and aging by regulating microRNA-34a-SIRT1 pathway. Aging Cell. 2017;16:837–846. doi: 10.1111/acel.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurinna S, Werner S. NRF2 and microRNAs: New but awaited relations. Biochem Soc Trans. 2015;43:595–601. doi: 10.1042/BST20140317. [DOI] [PubMed] [Google Scholar]
- 50.Shah NM, Zaitseva L, Bowles KM, MacEwan DJ, Rushworth SA. NRF2-Driven miR-125B1 and miR-29B1 transcriptional regulation controls a novel anti-apoptotic miRNA regulatory network for AML survival. Cell Death Differ. 2015;22:654–664. doi: 10.1038/cdd.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the present study are available from the corresponding author upon reasonable request.