Abstract
Plasmalogens are vinyl ether-containing lipids produced by mammals and bacteria. The aerobic biosynthetic pathway in eukaryotes and bacteria is known, but the anaerobic pathway has remained a mystery. Here, we describe a two-gene operon (plasmalogen synthase, pls) responsible for plasmalogen production in the anaerobic bacterium Clostridium perfringens. While aerobic plasmalogen biosynthesis involves an oxidative conversion of an ether to a vinyl ether, anaerobic plasmalogen biosynthesis uses the reductive conversion of an ester to an aldehyde equivalent. Heterologous expression of the C. perfringens pls operon in E. coli conferred the ability to produce plasmalogens. The pls operon is predicted to encode a multidomain complex similar to benzoyl-CoA reductase/hydroxylacyl-CoA dehydratase (BCR/HAD) enzymes. Versions of this operon can be found in a wide range of obligate and facultative anaerobic bacteria, including many human gut microbes.
Introduction
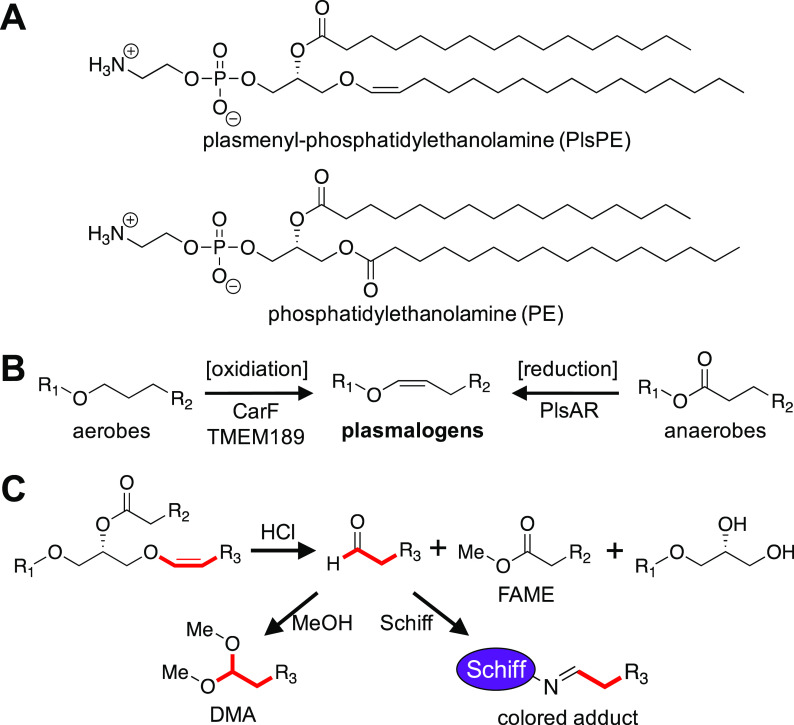
During a systematic screen of bacteria from the human gut microbiota for immunomodulatory compounds, we became interested in the plasmalogen family of lipids. The characteristic feature of plasmalogens is a long-chain vinyl ether linked to a glycerol (Figure 1A).1 Plasmalogens are widely distributed in anaerobic bacteria and animals.2 In humans, plasmalogens are major constituents of most tissues and have their highest concentrations in the brain, heart, and immune cell membranes.3 For example, phosphatidylethanolamines (PEs) in parts of the brain can be up to 60% plasmalogen and only 40% in the more familiar diacyl form (Figure 1A).4 Plasmalogens have roles in signaling, membrane structure, and protection against reactive oxygen species (ROS).5 Mutations in human plasmalogen biosynthesis can lead to rare terminal diseases, including rhizomelic chondrodysplasia punctata (RCDP), which impairs normal growth and development.5 More recently, plasmalogens have been studied for their role (along with other unsaturated lipids) in ferroptosis and cancer.6 In humans and other animals, the characteristic vinyl ether is formed in an O2-dependent oxidation (Figure 1B).3,7 In 2019, Gallego-Garcia and co-workers identified an integral membrane enzyme, CarF, in the aerobic bacterium Myxococcus xanthus as the desaturase necessary for plasmalogen formation, and used that finding to identify the human CarF homologue, TMEM189, as the corresponding human desaturase.8 Subsequently, in 2020, Werner and co-workers used an omics-driven approach to identify TMEM189 as a plasmalogen-forming lipid desaturase in mice and humans.9
Figure 1.
(A) The common polar lipid phosphatidylethanolamine shown in both plasmalogen (top) and diacyl form (bottom). (B) Plasmalogens are found in both aerobic and anaerobic organisms. In aerobes, O2 is required to form plasmalogens from alkyl ethers. In anaerobes, we propose a reductive pathway involving a BCR/HAD-like enzyme. (C) When treated with acid, a typical plasmalogen decomposes to form an aldehyde and a fatty acid methyl ester (FAME). The aldehyde can be converted to a dimethyl-acetal (DMA) in the presence of methanol, or it can react with a Schiff stain to form a purple adduct.
Characterizing CarF and TMEM189 completed the identification of the biosynthetic steps by which aerobic organisms produce plasmalogens, but the way in which anaerobic bacteria biosynthesize plasmalogens remained unknown. Previous studies have shown that the anaerobic pathway in bacteria differs profoundly from the aerobic pathway (see Figure 1B, as well as Figure S1 in the Supporting Information).3,10−12 Aerobic plasmalogen biosynthesis in animals begins with dihydroxyacetone phosphate (DHAP), and feeding studies have shown this is not the case for the anaerobic pathway in bacteria.12 In addition, pulse labeling studies suggest that anaerobic bacteria may directly convert a diacyl precursor to a plasmalogen product.11 Perhaps the starkest difference lies in the creation of the vinyl ether, as the oxygen-dependent aerobic mechanism is not possible for anaerobes (Figure 1B). As part of an ongoing effort in our laboratory to study the chemistry and biology of the human microbiota, we set out to determine the anaerobic plasmalogen biosynthetic pathway. We began by adopting an observation by Robert Feulgen, who originally discovered plasmalogens in 1924 when he stained acidified blood plasma with a Schiff stain (fuschin-sulfurous acid), which is an aldehyde-sensitive dye. Feulgen observed the characteristic purple color of aldehydes, which were formed from plasmalogens upon acid treatment.13 Our pursuit and characterization of the pathway relied on his observation.
Results/Discussion
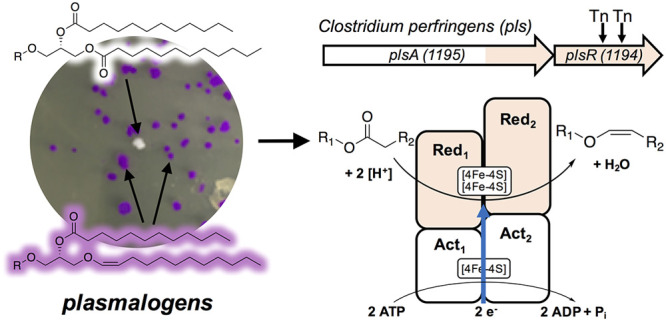
To identify the genes involved in plasmalogen biosynthesis, we employed the genetically tractable and plasmalogen-producing anaerobe Clostridium perfringens and the aldehyde-detecting Schiff stain.14 Upon acid treatment, plasmalogens are converted to aldehydes, which react with Schiff stain to produce a purple color. This procedure allowed us to create a colorimetric high-throughput phenotypic screen to visualize plasmalogen-producing bacterial colonies on agar plates (see Figures 1C and 2A). We used a pre-existing C. perfringens transposon (Tn) library,15 plated ∼12,000 colonies (∼50–100 colonies per plate), and isolated two separate colonies that lacked a positive response to Schiff staining (see Figure 2A, as well as Figure S2 in the Supporting Information). To verify that the loss of Schiff staining was due to the absence of plasmalogens, we analyzed lipids from wild-type C. perfringens and white Tn mutants that lack plasmalogens. Treatment of crude lipid extracts with methanolic HCl liberates aldehydes from plasmalogens and converts them to dimethyl acetals (DMAs) that can be characterized by GC-MS (Figure 1C). No DMAs were observed from the white mutants: only fatty acid methyl esters (FAMEs) were observed, indicating that plasmalogens were absent and replaced by diacyl lipids in the Tn mutants (see Figure S2). The Tn insertion sequence carries an erythromycin resistance gene and lacks HindIII restriction sites, which allowed us to identify the disrupted genes in plasmalogen-deficient mutants. We found that both C. perfringens plasmalogen-deficient mutants contained an insertion site in the same gene, CPE1194, which is part of a two-gene operon with CPE1195 (see Figure 2B).
Figure 2.
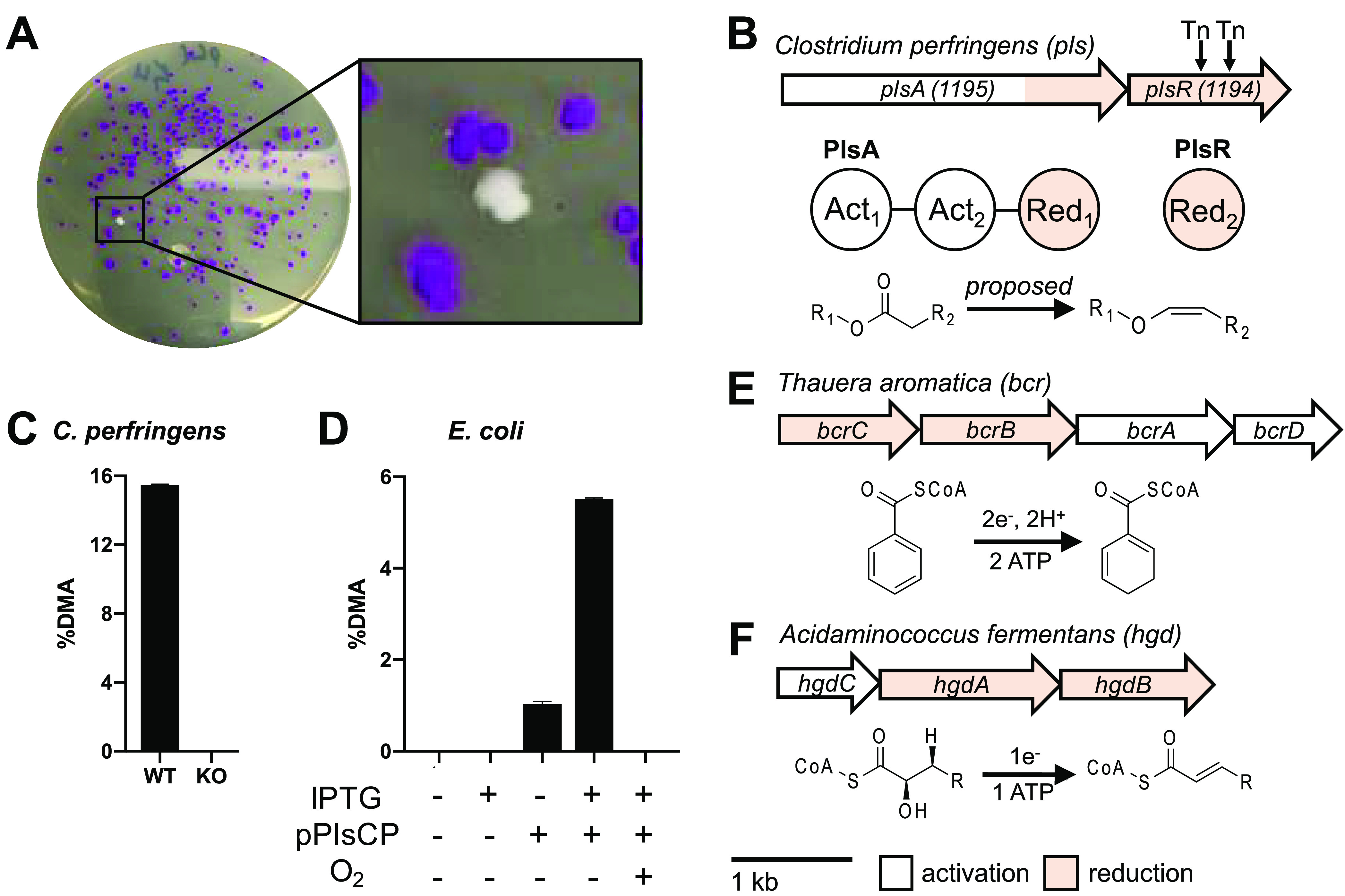
(A) Colonies of a C. perfringens Tn library grown on agar exposed to Schiff stain. The white colony represents a plasmalogen-deficient mutant, and the purple colonies represent plasmalogen producers. (B) Genetic architecture and domain organization of the plasmalogen synthase (pls) operon in C. perfringens (top), protein domain architecture (middle), and proposed reaction to convert diacyl lipids into plasmalogens (bottom). (C) GC-MS analysis of plasmalogen-derived DMA levels in C. perfringens wild-type (WT) and ΔPlsA PlsR. (D) GC-MS analysis of plasmalogen-derived DMA levels E. coli expressing pPlsCP. (E) Genetic architecture of the bcr genes in Thauera aromatica (top) and reaction catalyzed by BCR (bottom). (F) Genetic architecture of the hgd genes in Acidaminococcus fermentans (top) and reaction catalyzed by HGD (bottom).
To verify that the C. perfringensCPE1194/1195 operon is responsible for plasmalogen production, we created an in-frame deletion of the operon in C. perfringensHN13, using homologous recombination, as previously described16 (see Figure S3A in the Supporting Information), and observed a lack of plasmalogen production by Schiff staining and GC-MS analysis of plasmalogen-derived DMAs (see Figure 2C, as well as S3B–S3D in the Supporting Information). These results demonstrate that the CPE1194/1195 operon is necessary for plasmalogen production in C. perfringens; therefore, we designate the operon as the plasmalogen synthase (pls) operon (Figure 2B). Next, we sought to determine if the pls operon was sufficient for plasmalogen production. The pls operon containing plsA and plsR was amplified as a single PCR product and cloned into the plasmid pET-28a to create pPlsCP, allowing for inducible expression of the pls operon in E. coli (see Figure S4A in the Supporting Information), a nonplasmalogen-producing host. Upon anaerobic growth and induction with IPTG, we observed robust plasmalogen production in E. coli harboring the pPlsCP plasmid, but not in the vector control (see Figure 2D, as well as Figures S4B–S4D in the Supporting Information). We also observed low levels of plasmalogen production in uninduced E. coli harboring the pPlsCP, which we attribute to leaky gene expression. The plasmalogen pathway in anaerobic bacteria is proposed to have evolved early in life’s evolution under anaerobic conditions, and it has been speculated that the biosynthetic machinery to make plasmalogens anaerobically is oxygen-sensitive.2,11 To test this hypothesis, we induced expression of the C. perfringenspls operon in aerobically grown E. coli, and we detected no plasmalogens (see Figure 2D, as well as Figure S4B), suggesting that the plasmalogen biosynthetic enzymes PlsA and PlsR (i) may not be expressed, (ii) may be expressed but insoluble, or (iii) may contain oxygen-sensitive components.
The C. perfringenspls operon has two open reading frames (ORFs), plsA and plsR, which are separated by eight base pairs, and have marked homology to enzymes in the BCR/HAD enzyme family (see Figures 2B, 2E, and 2F, as well as Figures S5 and S6 in the Supporting Information).17 Members of this family—benzoyl-CoA reductases (BCRs) and 2-hydroxyacyl-CoA dehydratases (HADs)—catalyze two different reactions: dehydrations for HADs and reductions for BCRs. However, the different reactions share the same first step: a one-electron reduction of a thioester to a ketyl radical anion (see Figure S5).18 The two-electron reduction catalyzed by BCRs is really two successive one-electron reductions. BCR/HAD family members are typically four-protein complexes that consist of two CoA-substrate-specific activation domains (component A), two electron transfer (reduction/dehydration) domains (component D), and three [4Fe-4S] cofactors.17 The pls operon is predicted to encode an enzymatic complex similar to that of HADs and BCRs, including two CoA-substrate-specific activation domains and two reduction/dehydration domains (see Figures 2B, 2E, and 2F, as well as Figure S5).
In BCRs and HADs, a single gene encodes each protein component of the enzymatic complex (see Figures 2E and 2F, as well as Figure S5). In contrast, the pls operon in C. perfringens encodes two proteins, PlsA and PlsR. PlsA is a multidomain protein composed of two activation domains (Act1 and Act2), and one reduction/dehydration domain (Red1). PlsR encodes a second reduction domain (Red2) (see Figure 2B).
To compare activation and reduction domains present in PlsA and PlsR to model HAD/BCR proteins, we split the multidomain protein PlsA into three separate sequences, based on predicted functional domains: Act1 (electron-activating domain, residues 1–288), Act2 (electron-activating domain, residues 289–620), and Red1 (reduction/dehydration domain, residues 621–975) (see Figure S6 and Table S2 in the Supporting Information). The C. perfringens activation domains Act1 and Act2 show low sequence identity to known HAD electron-activation proteins (component A), ranging from 25% to 31%, and a similarly low sequence identity of 30% with each other. Act1 and Act2 also show low sequence identities with BCR electron activation proteins, ranging from 22% to 29%. For comparison, HAD electron-activation domains typically share >50% identity with other HAD activation domains, and this is also generally true for BCR activation domains of the same type within the same families (Bzd-type and Bcr-type) (see Figure S7 in the Supporting Information). Act1 and Act2 also share conserved cysteines for iron–sulfur cluster ligation with HAD and BCR activation domains (see Figure S8 in the Supporting Information). The reduction/dehydration domain Red1 and Red2 show low sequence identity to known HAD reduction/dehydration subunits (component D) ranging from 18% to 24%, and similarly low sequence identities (17% to 23%), when compared to BCR reduction domains subunits. The similar functional annotation, and low sequence identity of PlsAR domains when compared to HAD/BCR proteins suggests that PlsAR is a divergent member of the HAD/BCR family with the capacity to perform reductive chemistry.
The two reactions that are performed by BCR/HAD family members are different, yet they share a common first step: the one-electron reduction of a thioester to a ketyl radical anion. The one-electron reduction of a thioester (or, in the case of a plasmalogen synthase (PlsAR), an ester) is a difficult reduction for typical biological cofactors. In both HADs and BCRs, the activation module (component A) accepts electrons from ferredoxin and transfers them in ATP-requiring steps to the reduction/dehydration module (component D), which is a heterodimer that contains at least one [4Fe-4S] cluster.18 BCRs are found in multiple species of anaerobic bacteria and reduce the benzene ring of benzoyl-CoA, the functional equivalent of a Birch reduction.19 HADs play key roles in amino acid fermentation in anaerobic bacteria, but they do not perform redox reactions.20 HAD reactions functionally resemble photoredox-catalyzed transformations.21 In addition, HADs require catalytic ATP, whereas BCRs require stoichiometric ATP (see Figure S5).
While plasmalogen biosynthesis in anaerobic bacteria has never been fully defined, various studies, primarily feeding studies, have outlined the pathway’s early steps.11 The step(s) introducing the vinyl ether remain(s) unknown, but the existing evidence suggests that an ester is converted to the vinyl ether.11 In principle, the conversion of an ester to a vinyl ether through reduction and dehydration is plausible, but reducing an ester with familiar biological reducing agents (e.g., NAD(P)H, flavins, or hydroquinones) is not thermodynamically feasible. BCR/HAD enzymes, as discussed above, can perform challenging reductions such as the reduction of the thioester in the BCR reaction. In analogy with the proposed mechanism of the BCR reaction, likely mechanistic steps for the plasmalogen synthase from C. perfringens can be proposed. The sequence would involve an initial reduction of the sn-1 linked ester to a ketyl radical anion, protonation, a second reduction-protonation sequence, and a final dehydration (see Figure S5F).
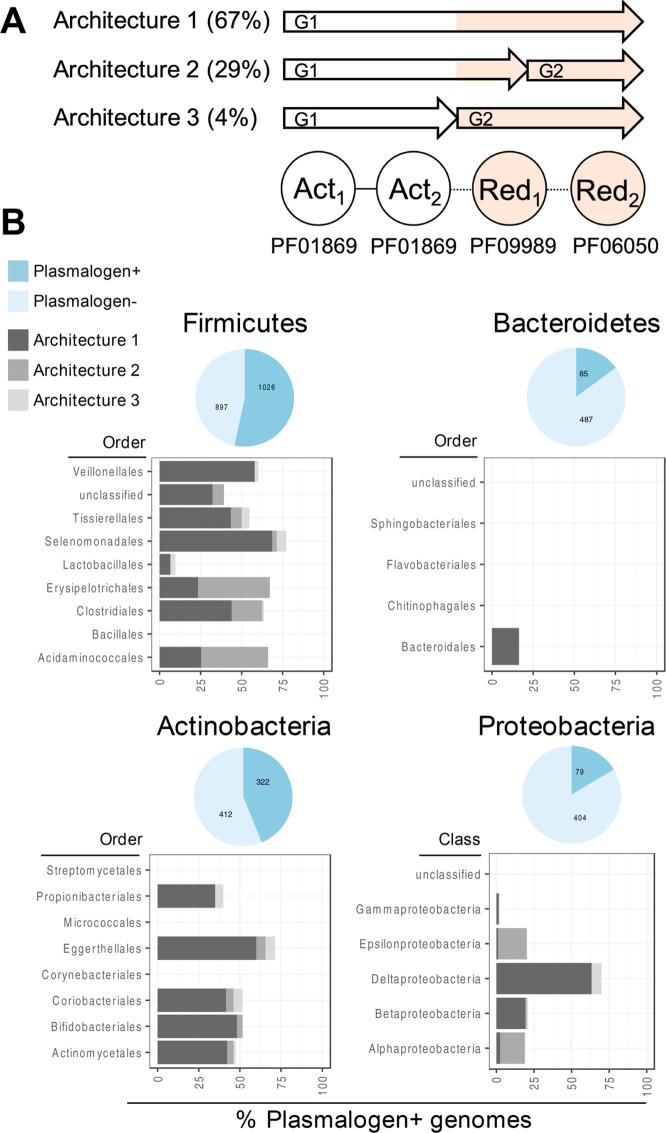
Identifying the pls operon in C. perfringens allowed us to assess the distribution of plasmalogen biosynthesis genes more broadly in bacteria. We focused on the following Pfam domains: PF01896 (BcrAD_BadFG, Benzoyl-CoA reductase, 2-hydroxyglutaryl-CoA dehydratase, which correspond to Act1 and Act2 of PlsA), PF09989 (CoA enzyme activase uncharacterized domain, which corresponds to Red1 of PlsA), and PF06050 (HGD-D, 2-hydroxyglutaryl-CoA dehydratase, which corresponds to Red2 of PlsR). We searched the proteomes from ∼5,000 metagenomic species (MGS) assembled in the human microbiomes for genes homologous to C. perfringens pls operon (BLAST, e-value <1e–4) that contained the prioritized Pfam domains (InterProScan, default settings; see Figure 3).22,23 To classify a species as plasmalogen-positive, we required all four Pfam domains to be detected in the genes returned by a BLAST search, which resulted in 1910 MGS plasmalogen-positive species. Close inspection revealed that plasmalogen-related Pfams exist in three distinct architectures: architecture 1 (single gene: PF01869 + PF01869 + PF09989 + PF06050), architecture 2 (gene 1, G1: PF01869 + PF01869 + PF09989 and gene 2, G2: PF06050), and architecture 3 (gene 1, G1: PF01869 + PF01869 and gene 2, G2: PF09989 + PF06050) (Figure 3A). The most prevalent architecture 1 was observed in 1283 MGS (e.g., Collinsella aerofaciens, Bifidobacterium breve, or Eubacterium rectale), while architecture 2 and architecture 3 were observed in 575 MGS (e.g., Clostridium perfringens, Clostridium innocuum, or Faecalibacterium prausnitzii) and 81 MGS (e.g., Streptococcus lutetiensis, Streptococcus equinus, or Eubacterium callanderi), respectively. We also found that a subset of genomes represented by 29 species contain multiple architectures composed of plasmalogen-related Pfams (see Table S3 in the Supporting Information). Among the microbial phyla most typical for the human gut microbiome, we observed a variable distribution of plasmalogen-positive species that partially corresponded to the Gram staining. Larger proportions of Gram-positive Firmicutes and Actinobacteria were classified as plasmalogen-positive (53% and 44% MGS, respectively), in contrast to only ∼15% of Bacteroidetes and Proteobacteria MGS. In summary, our analysis revealed the presence of the pls operon in a wide variety of obligate and facultative anaerobes (Figure 3B).
Figure 3.
Distribution of plasmalogen encoding genes in the metagenomic species assembled in the human microbiome. (A) Plasmalogen-related Pfam domain architectures identified in the analyzed species (architecture 1 consists of a single gene encoding all four Pfam domains, while architectures 2 and 3 consist of two genes, G1 and G2, with variable number of domains). (B) Distribution of plasmalogen-positive species among the main phyla in the human gut microbiome. Bar plots were stratified further by order or class (Proteobacteria).
Previous reports suggested that plasmalogens are not present in facultative anaerobic bacteria;2 however, we identified the pls operon as a single gene in several facultative anaerobes, including Enterococcus faecalis, a common gut microbe that can also cause opportunistic infections (see Figure 4A).24 The E. faecalis gene EF1327 encodes all activation and reduction/dehydration domains as a single polypeptide (PlsA), with overall sequence identity of 48% when compared to PlsAR from C. perfringens. The relatively high protein sequence identity between PlsA (E. faecalis) and PlsAR (C. perfringens) suggested that E. faecalis may have the biosynthetic capacity to produce plasmalogens. We cultured E. faecalis under anaerobic and aerobic conditions in liquid culture and detected plasmalogen production under anaerobic but not aerobic conditions, as determined by Schiff staining and GC-MS analysis of plasmalogen-derived DMAs (see Figures 4B and 4C, as well as Figure S9). We also cultured E. faecalis under aerobic and anaerobic conditions on agar; we observed robust purple staining of anaerobically grown colonies and, notably, a more complex staining pattern for aerobically grown colonies (Figure 4D). For aerobically grown colonies, it appeared that only the colony interior was stained purple, while the exterior remained unstained and white. This argues that plasmalogen production in aerobically grown colonies is spatially restricted to the colony interior, where oxygen is limited (see Figures 4D and 4E). We identified a TetR/AcrR-like transcriptional regulator (EF1326) directly upstream of the pls gene (EF1327), and this regulator is conserved in other facultative anaerobes, including Enterococcus casseliflavus and Streptococcus mutans (Figure 4A), but absent in obligate anaerobes, where the pls operon is present. The TetR/AcrR-like regulator associated with the pls operon in facultative anaerobes is likely to be an O2-sensing transcriptional regulator. The predicted [4Fe-4S] cluster-containing multidomain PlsA enzyme is likely rendered nonfunctional in the presence of oxygen, and its expression may be repressed under aerobic conditions. In addition, a pET-29b expression vector containing the EF1327 coding sequence was synthesized (pPlsEF), allowing for inducible expression of the E. faecalis pls operon in E. coli. Upon anaerobic growth and induction with IPTG, we observed plasmalogen production in E. coli harboring the pPlsEF plasmid (see Figure S10 in the Supporting Information).
Figure 4.
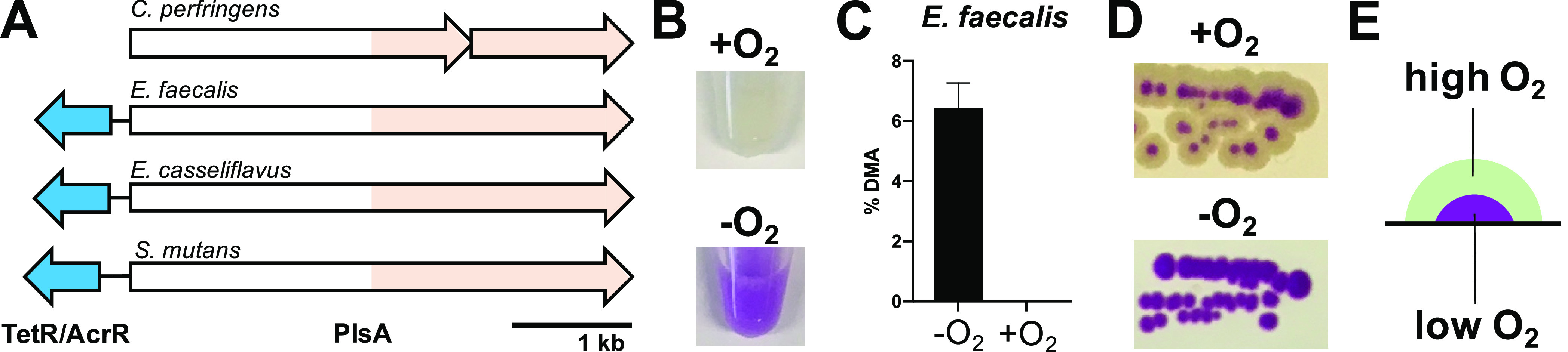
(A) The pls operon from facultative anaerobes and conserved transcription regulator. (B) Schiff-stained pellets from liquid cultures of Enterococcus faecalis grown aerobically (top) and anaerobically (bottom). (C) GC-MS analysis of plasmalogen derived DMAs from E. faecalis liquid culture grown under aerobic and anaerobic conditions. (D) Schiff-stained colonies of E. faecalis grown on agar under aerobic and anaerobic conditions. (E) A model for differential plasmalogen production in a single E. faecalis colony grown under aerobic conditions.
The defining feature of plasmalogens, the vinyl ether at the sn–1 position of a diacyl glyceride, renders them challenging molecules to study in detail, and they are typically characterized collectively. Plasmalogen-containing lipid mixtures are treated with methanol and HCl to liberate the vinyl ether fragment as a dimethyl acetal that can be characterized by GC-MS analysis. The very reactive vinyl ether moiety is also thought to be responsible for one of the key biological functions of plasmalogens: reacting with reactive oxygen species (ROS) to prevent cellular damage.1 They are the biological equivalent of the sacrificial anodes used to prevent corrosion to metal structures. Plasmalogens have limited phylogenetic distribution, primarily in animals and anaerobic bacteria.2 The formation of plasmalogens in animals is reasonably well understood, and the vinyl ether moiety is created by an oxidation of an ether in an enzyme-catalyzed oxygen-consuming reaction.8,9 The corresponding steps in anaerobic bacteria are not known.2,11
In conclusion, the studies reported here identify a pathway involving an unusual reductive transformation of an ester moiety to an aldehyde equivalent, a vinyl ether, the defining feature of a plasmalogen. The stark difference between aerobic and anaerobic biosynthetic routes is another reminder of Konrad Bloch’s observation that unsaturated lipids, which are widespread in biology, have two independent origins: one in the anaerobic world and one in the aerobic world.25 The reduction of an ester requires a potent reducing agent, and the BCR/HAD family of enzymes initiate the reduction using an ATP-driven electron transfer to create a ketyl intermediate.18 Protonation and a subsequent one-electron reduction, protonation, and dehydration finish the sequence. The identification and deletion studies of the pls operon in C. perfringens, and complementation in E. coli, demonstrate a tight link between plasmalogen biosynthesis and the BCR/HAD enzyme encoded in the pls operon. This expands our understanding of the BCR/HAD enzyme family, and it suggests that, in addition to acyl-CoAs, diacyl lipids may also serve as substrates for reduction/dehydration.
As is often the case, discovering the genes responsible for the biosynthesis of plasmalogens led to the realization that they are widespread in many members of the gut microbiota. In addition, facultative anaerobes, not just obligate anaerobes, can and do make plasmalogens. The characterization of the plasmalogen biosynthetic pathway enables future research on the roles of these highly reactive, widespread, and enigmatic lipids.
Methods
A Transposon Screen in C. perfringens HN13 To Identify Plasmalogen-Deficient Mutants
Bacterial strains, plasmids, and primers used in this study are listed in Table S1 in the Supporting Information. Two types of media were used to culture C. perfringens for transposon screening: PGY (3% proteose peptone #3, 2% glucose, 1% yeast extract, and 0.1% sodium thioglycolate) for plating on agar, and Brain Heart Infusion (BHI) (Bacto) for liquid growth. A previously described transposon library of C. perfringens HN13 was grown overnight in BHI, diluted 10,000-fold with D-PBS, and then ∼120 plates (PGY agar) were inoculated with 15 μL/plate of this dilution using glass beads, resulting in ∼100 colonies per plate for a total of ∼12,000 colonies.15 The plates were incubated for 48 h in an anaerobic chamber (Coy) and replica-plated onto a new set of anaerobic PGY plates, using velvet squares. Original and replica plates were grown for an additional 48 h anaerobically. Original plates were stained with Schiff Reagent (Sigma–Aldrich #84655) by carefully pouring Schiff Reagent over each plate until colonies were completely submerged. After 15–20 min, two colonies (C. perfringens Tn48 and C. perfringens Tn54) were identified, because of the lack of purple staining, suggesting these mutants lack plasmalogens (see Figure S2). The corresponding colonies on the replica plates were restreaked, and subsequent growth was used to inoculate 5 mL overnight cultures in BHI. Pellets from 5 mL of liquid culture were stained with Schiff Reagent to verify the plasmalogen-deficient phenotype and both mutants were verified as C. perfringens by 16S rRNA sequencing. Furthermore, cell pellets from 50 mL cultures were extracted with chloroform:methanol (1:1) to yield crude lipid extracts, which were sent to Microbial ID, Inc. for GC-MS analysis of lipid-derived fatty acid methyl esters (FAMEs) and plasmalogen-derived dimethyl acetals (DMAs) (see Figure S2).
Identifying Tn Insertion Sites in C. perfringens Plasmalogen-Deficient Mutants
The Tn insertion of the C. perfringens Tn mutant library contains an erythromycin resistance (Ermr) gene lacking any internal HindIII restriction sites. To identify Tn insertion sites, gDNA was isolated from C. perfringens Tn48 and Tn54 using a Qiagen Miniprep kit with an additional step: after the addition of buffer P1, 250 μL of 2% sodium dodecyl sulfate (SDS) in water was added to the cell suspension and heated at 55 °C for 20 min, gDNA was then digested with HindIII (ThermoFisher), and the resulting fragments were treated with alkaline phosphatase (FastAP, Thermo Scientific), gel-purified (Qiagen Gel Extraction Kit), and ligated into pUC19 with T4 DNA ligase (16 °C for 16 h). Ligation reactions were transformed into E. coli DH5α by heat shock (42 °C for 45 s), incubated on ice for 2 min, grown in Luria–Bertani (LB) for 1 h at 37 °C, then plated on LB-agar plates containing 300 μg/mL erythromycin (resistance mediated by Tn insertion) and 50 μg/mL ampicillin (resistance-mediated by pUC19). Resulting colonies were used to inoculate overnight cultures for plasmid isolation (Qiagen Miniprep Kit) and sequencing (Genewiz), using previously described primers OHL21 and OHL22 to map insertions.15 Insertions for Tn48 and Tn54 were both mapped to the CPE1194 gene of C. perfringens HN13.
Constructing an In-Frame Deletion of CPE1194–1195
An in-frame deletion of the CPE1194/1195 was created using the pCM-GALK plasmid, as described previously by Nariya et al.(16) Briefly, the flanking 5′-region of CPE1195 and 3′-region of CPE1194 were amplified from C. perfringens HN13 gDNA as the template, using primers pairs DJ031/DJ032 (5′-region) and DJ033/DJ034 (3′-region), and fused using a single round of overlap extension PCR with primers DJ031/DJ034. The resulting PCR product was digested with NheI and BamHI and ligated into pCM-GALK to yield pCM-GALK-Δ1194/1195, which was electroporated into C. perfringens HN13, and mutants were selected as previously described.15 Deletion of CPE1194/1195 was confirmed using PCR with primers DJ049 and DJ050. The plasmalogen-deficient phenotype of C. perfringens HN13ΔPlsA PlsR was first verified using Schiff staining of pellets from 5 mL overnight cultures. Furthermore, lipid extracts from cell pellets were prepared (as described above) and sent to Microbial ID, Inc. for GC-MS analysis of lipid-derived fatty acid methyl esters (FAMEs) and plasmalogen-derived dimethyl acetals (DMAs) (see Figures S3C and S3D).
Expression of C. perfringens HN13CPE1194–1195 in E. coli
The 4199 base-pair region encompassing CPE1194/1195 of the C. perfringens HN13 genome was amplified using primers DJ009 and DJ010, digested using NheI and XhoI (ThermoFisher), ligated into pET-28a, and transformed into E. coli DH5α (New England Biolabs) supplemented with 50 μg/mL kanamycin. pPlsCP was mini-prepped (Qiagen Miniprep Kit) and sequenced prior to transformation into E. coli BL21(DE3) (New England Biolabs) in the presence of 50 μg/mL kanamycin. Expression was performed aerobically and anaerobically in Brain Heart Infusion (BHI) media. For aerobic induction, 5 mL of BHI in a 50 mL conical tube was inoculated from a frozen glycerol stock of E. coli BL21(DE3)/pPlsCP and shaken in an aerobic incubator at 220 rpm at 37 °C until the OD600 reached 0.4, then 100 μM IPTG (final concentration) was added and the culture was grown for an additional 12–16 h. For anaerobic induction, overnight cultures of E. coli BL21(DE3)/pPlsCP were grown in a Coy anaerobic chamber and diluted 1:100 in fresh BHI. When the OD600 reached 0.4, 100 μM IPTG (final concentration) was added and the culture was grown for an additional 12–16 h. Plasmalogen production was evaluated by Schiff staining and GC-MS (Microbial ID, Inc.) (see Figure S4).
E. faecalis OG1RF Aerobic/Anaerobic Growth and E. faecalis EF1327 Expression in E. coli
For aerobic growth, 5 mL of BHI in a 50 mL conical tube was inoculated from a frozen glycerol stock and shaken in an aerobic incubator at 220 rpm at 37 °C for 12–16 h. For anaerobic growth, overnight cultures of E. faecalis OG1RF were grown in a Coy anaerobic chamber, diluted 1:100 in fresh BHI, and grown for 12–16 h, and assessment of plasmalogens was performed as described above. To express the E. faecalis gene EF1327 in E. coli, the 4248 base pair coding region corresponding to EF1327 was synthesized and cloned into pET-29b (TWIST Biosciences) to yield the expression vector pPlsEF. E. coli BL21(DE3) was transformed with pPlsEF under kanamycin selection (50 μg/mL), and anaerobic induction and assessment of plasmalogens was performed as described above for E. coli BL21(DE3) harboring pPlsCP (see Figure S10 in the Supporting Information).
Acknowledgments
We thank H. Goldfine (University of Pennsylvania) for kindly providing insights from his laboratory’s work on plasmalogens in anaerobic bacteria, and for inspiring and supporting us early on in our research efforts. We acknowledge funding support from the National Institutes of Health (Grant Nos. R01AT009708 (J.C. and R.J.X.), F32GM122233 (D.R.J.), and F32AT010415 (C.D.C.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.0c00673.
The authors declare no competing financial interest.
Supplementary Material
References
- Dean J. M.; Lodhi I. J. (2018) Structural and functional roles of ether lipids. Protein Cell 9, 196–206. 10.1007/s13238-017-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfine H. (2010) The appearance, disappearance and reappearance of plasmalogens in evolution. Prog. Lipid Res. 49, 493–498. 10.1016/j.plipres.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Nagan N.; Zoeller R. A. (2001) Plasmalogens: biosynthesis and functions. Prog. Lipid Res. 40, 199–229. 10.1016/S0163-7827(01)00003-0. [DOI] [PubMed] [Google Scholar]
- Han X.; Holtzman D. M.; McKeel D. W. Jr. (2001) Plasmalogen deficiency in early Alzheimer’s disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J. Neurochem. 77, 1168–1180. 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- Braverman N. E.; Moser A. B. (2012) Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta, Mol. Basis Dis. 1822, 1442–1452. 10.1016/j.bbadis.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Zou Y.; Palte M. J.; Deik A. A.; Li H.; Eaton J. K.; Wang W.; Tseng Y. Y.; Deasy R.; Kost-Alimova M.; Dancik V.; Leshchiner E. S.; Viswanathan V. S.; Signoretti S.; Choueiri T. K.; Boehm J. S.; Wagner B. K.; Doench J. G.; Clish C. B.; Clemons P. A.; Schreiber S. L. (2019) A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat. Commun. 10, 1617. 10.1038/s41467-019-09277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltauf F. (1972) Plasmalogen biosynthesis in a cell-free system. Enzymic desaturation of 1-O-alkyl (2-acyl) glycerophosphoryl ethanolamine. FEBS Lett. 20, 79–82. 10.1016/0014-5793(72)80021-8. [DOI] [PubMed] [Google Scholar]
- Gallego-Garcia A.; Monera-Girona A. J.; Pajares-Martinez E.; Bastida-Martinez E.; Perez-Castano R.; Iniesta A. A.; Fontes M.; Padmanabhan S.; Elias-Arnanz M. (2019) A bacterial light response reveals an orphan desaturase for human plasmalogen synthesis. Science 366, 128–132. 10.1126/science.aay1436. [DOI] [PubMed] [Google Scholar]
- Werner E. R.; Keller M. A.; Sailer S.; Lackner K.; Koch J.; Hermann M.; Coassin S.; Golderer G.; Werner-Felmayer G.; Zoeller R. A.; Hulo N.; Berger J.; Watschinger K. (2020) The TMEM189 gene encodes plasmanylethanolamine desaturase which introduces the characteristic vinyl ether double bond into plasmalogens. Proc. Natl. Acad. Sci. U. S. A. 117 (14), 7792–7798. 10.1073/pnas.1917461117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins R. A.; Van Golde L. M. (1976) Entrance of glycerol into plasmalogens of some strictly anaerobic bacteria and protozoa. FEBS Lett. 63, 107–111. 10.1016/0014-5793(76)80204-9. [DOI] [PubMed] [Google Scholar]
- Goldfine H. (2017) The anaerobic biosynthesis of plasmalogens. FEBS Lett. 591, 2714–2719. 10.1002/1873-3468.12714. [DOI] [PubMed] [Google Scholar]
- Hill E. E.; Lands W. E. (1970) Formation of acyl and alkenyl glycerol derivatives in Clostridium butyricum. Biochim. Biophys. Acta, Lipids Lipid Metab. 202, 209–211. 10.1016/0005-2760(70)90239-0. [DOI] [PubMed] [Google Scholar]
- Feulgen R.; Rossenbeck H. (1924) Micnoskopisch-chemischer Nachweis einer Nucleinsaure vom Typus der Thymonucleinsaure und die- darauf beruhende elektive Farbung von Zellkernen in mikroskopischen Praparaten. Hoppe-Seyler's Z. Physiol. Chem. 135, 203. 10.1515/bchm2.1924.135.5-6.203. [DOI] [Google Scholar]
- Johnston N. C.; Baker J. K.; Goldfine H. (2004) Phospholipids of Clostridium perfringens: a reexamination. FEMS Microbiol. Lett. 233, 65–68. 10.1016/j.femsle.2004.01.048. [DOI] [PubMed] [Google Scholar]
- Liu H.; Bouillaut L.; Sonenshein A. L.; Melville S. B. (2013) Use of a mariner-based transposon mutagenesis system to isolate Clostridium perfringens mutants deficient in gliding motility. J. Bacteriol. 195, 629–636. 10.1128/JB.01288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nariya H.; Miyata S.; Suzuki M.; Tamai E.; Okabe A. (2011) Development and application of a method for counterselectable in-frame deletion in Clostridium perfringens. Appl. Environ. Microbiol. 77, 1375–1382. 10.1128/AEM.01572-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckel W.; Kung J. W.; Boll M. (2014) The benzoyl-coenzyme a reductase and 2-hydroxyacyl-coenzyme a dehydratase radical enzyme family. ChemBioChem 15, 2188–2194. 10.1002/cbic.201402270. [DOI] [PubMed] [Google Scholar]
- Buckel W. (2019) Enzymatic Reactions Involving Ketyls: From a Chemical Curiosity to a General Biochemical Mechanism. Biochemistry 58, 5221–5233. 10.1021/acs.biochem.9b00171. [DOI] [PubMed] [Google Scholar]
- Boll M. (2005) Key enzymes in the anaerobic aromatic metabolism catalysing Birch-like reductions. Biochim. Biophys. Acta, Bioenerg. 1707, 34–50. 10.1016/j.bbabio.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Kim J.; Hetzel M.; Boiangiu C. D.; Buckel W. (2004) Dehydration of (R)-2-hydroxyacyl-CoA to enoyl-CoA in the fermentation of alpha-amino acids by anaerobic bacteria. FEMS Microbiol. Rev. 28, 455–468. 10.1016/j.femsre.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Romero N. A.; Nicewicz D. A. (2016) Organic Photoredox Catalysis. Chem. Rev. 116, 10075–10166. 10.1021/acs.chemrev.6b00057. [DOI] [PubMed] [Google Scholar]
- Jones P.; Binns D.; Chang H. Y.; Fraser M.; Li W.; McAnulla C.; McWilliam H.; Maslen J.; Mitchell A.; Nuka G.; Pesseat S.; Quinn A. F.; Sangrador-Vegas A.; Scheremetjew M.; Yong S. Y.; Lopez R.; Hunter S. (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasolli E.; Asnicar F.; Manara S.; Zolfo M.; Karcher N.; Armanini F.; Beghini F.; Manghi P.; Tett A.; Ghensi P.; Collado M. C.; Rice B. L.; DuLong C.; Morgan X. C.; Golden C. D.; Quince C.; Huttenhower C.; Segata N. (2019) Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 176, 649–662. 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore E., Van Tyne D., and Gilmore M. S. (2019) Pathogenicity of Enterococci. Microbiol. Spectrum 7, 10.1128/microbiolspec.GPP3-0053-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch K. (1969) Enzymatic Synthesis of Monounsaturated Fatty Acids. Acc. Chem. Res. 2, 193–202. 10.1021/ar50019a001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.