Abstract
Proteolysis targeting chimeras (PROTACs) induce intracellular degradation of target proteins. Their bifunctional structure puts degraders in a chemical space where ADME properties often complicate drug discovery. Herein we provide the first structural insight into PROTAC cell permeability obtained by NMR studies of a VHL-based PROTAC (1), which is cell permeable despite having a high molecular weight and polarity and a large number of rotatable bonds. We found that 1 populates elongated and polar conformations in solutions that mimic extra- and intracellular compartments. Conformations were folded and had a smaller polar surface area in chloroform, mimicking a cell membrane interior. Formation of intramolecular and nonclassical hydrogen bonds, π–π interactions, and shielding of amide groups from solvent all facilitate cell permeability by minimization of size and polarity. We conclude that molecular chameleonicity appears to be of major importance for 1 to enter into target cells.
Keywords: PROTAC, cell permeability, NMR spectroscopy, conformation, molecular chameleon
Targeted protein degradation is a new concept in drug discovery which is of particular interest for intracellular targets that are difficult to modulate with small molecule drugs.1,2 This concept is based on proteolysis targeting chimeras (PROTACs), a chemical modality consisting of a ligand that binds to the target protein, a linker, and an E3 ubiquitin ligase ligand. PROTAC induced formation of a ternary complex with the target protein and the E3 ubiquitin leads to ubiquitination and subsequent degradation of the target protein. As a result of this mode of action PROTACs are catalytic.3
Structural information on the ternary complexes between target proteins, PROTACs, and E3 ubiquitin ligases is now beginning to allow rational, structure-based design of PROTACs.4 Design of PROTACs also requires knowledge about how their structures influence their ADME properties, with cell permeability being required for their mode of action. A few recent studies have begun to provide some, albeit sometimes conflicting, insight into how the ADME properties of PROTACs could be optimized. For instance, a study of a small set of seven PROTACs found that their permeabilities across Caco-2 cell monolayers correlated to chromatographically determined descriptors of lipophilicity and polarity.5 Another study of 11 PROTACs highlighted how the combined use of the parallel artificial membrane permeability assay (PAMPA) and lipophilic permeability efficiency (LPE) can be used to provide insight into PROTAC cell permeability.6 This study also suggested that AlogP should be kept below 5 to increase the likelihood for PROTACs to be cell permeable. However, other studies found no correlations between cell permeability and chromatographic lipophilicity or PAMPA permeabilities,7,8 pointing to the need for increased understanding of how in vitro assays may be used for optimization of the ADME properties of PROTACs.8
PROTACs reside in the chemical space close to, or beyond,9,10 the outer limits for oral absorption deduced from analysis of drugs, clinical candidates, and optimized leads in the beyond rule of 5 chemical space.11,12 For instance, the molecular weights of PROTACs typically range from 900 to 1100 Da, while their number of rotatable bonds falls between 20 and 25.9,10 This puts PROTACs at pharmacokinetic risk, where low oral absorption and/or cell permeability may prevent them from reaching their intracellular targets. Currently, efforts to improve these potential shortcomings are hampered by the lack of understanding of PROTAC structure–property relationships, for instance regarding how they cross cell membranes.
For the first time, we provide structural and property based insight into how PROTACs may permeate cell membranes by behaving as molecular chameleons. We used NMR spectroscopy to determine the conformational ensembles of PROTAC 1 (Figure 1A) in solutions having different polarity and hydrogen bonding properties. Chloroform was chosen to mimic the interior of a cell membrane as it has a dielectric constant (ε = 4.8) close to that of a lipid bilayer (ε = 3.0).13 As PROTAC 1 has low aqueous solubility (Table 1), DMSO alone and in a 10:1 mixture with water was used to resemble the aqueous extra- and intracellular environments.
Figure 1.
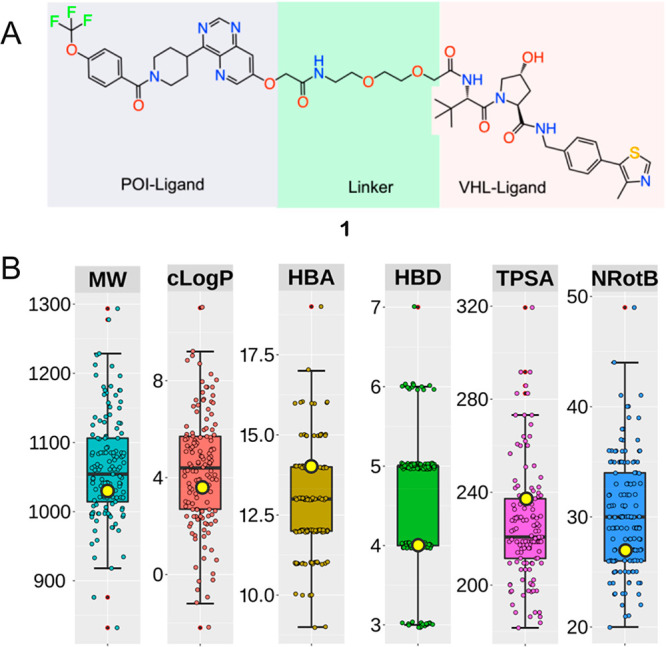
(A) Structure of PROTAC 1. The ligand for the protein target of interest (POI), ERK5, the linker, and the ligand for binding to the E3 ligase (VHL) are indicated by gray, green, and pink shading, respectively. (B) Molecular weight (MW, Da), lipophilicity (cLogP), hydrogen bond acceptors and donors (HBA and HBD), topological polar surface area (TPSA, Å2), and number of rotatable bonds (NRotB) calculated for PROTAC 1 and the subset of 135 VHL PROTACs that have PEG-based linkers reported by Maple et al.10 The values of the six descriptors calculated for 1 are shown as yellow circles (MW = 1034 Da, cLogP = 3.55, HBA = 14, HBD = 4, TPSA = 237 Å2, NRotB = 27). Box plots show the 50th percentiles as horizontal bars, the 25th and 75th percentiles as boxes, the 25th percentile minus 1.5× the interquartile range, and the 75th percentile plus 1.5× the interquartile range as whiskers for the subset of PROTACs having PEG-based linkers. Outliers are shown both as red dots and as circles in the color of the appropriate descriptor.
Table 1. Physicochemical Properties and in Vitro Potencies of 1.
| property | in vitro potency | ||
|---|---|---|---|
| solubility (PBS, pH 6.5, mg/L) | 7.0 | ERK5 IC50, biochemical (μM) | 1.23 |
| logD (pH 7.5) | 2.68 | VHL IC50, biochemical (μM) | 0.307 |
| PAMPA (−logPe, cm/s) | 5.85 | VHL IC50, cellular (μM) | 4.31 |
| VHL cell/biochem. ratio | 14 | ||
PROTAC 1 targets the extracellular signal-regulated kinase 5 (ERK5),14 a potential cancer target, by recruitment of the Von Hippel–Lindau (VHL) tumor suppressor E3 ligase. Thus, the E3 ligase ligand of 1 belongs to one of the two ligand classes that are used in the majority of PROTACs, with the other class being based on cereblon (CRBN) E3 ligase ligands.9,10 Maple et al. recently reported a set of 217 VHL PROTACs,10 the majority of which have flexible linkers that can be classified as either aliphatic (alkyl, n = 63) or based on ethylene glycol (PEG, n = 135). A third, smaller subset has rigid linkers (n = 19) based on alkynes combined with piperidine, pyridine, and/or piperazine moieties. Comparison of the descriptors of Lipinski’s15 and Veber’s16 rules calculated for 1 to those of the subset of PEG-based PROTACs revealed 1 to be a good representative of this subset (Figure 1B). All six descriptors for 1 fall within the 25th to 75th percentile of the corresponding descriptor for the subset. The subsets that have alkyl or rigid linkers differ from the PEG subset by having a higher calculated lipophilicity (cLogP), a lower topological polar surface area (TPSA) and fewer hydrogen bond acceptors (HBA) (Figure S3). As expected, the rigid PROTAC subset has a lower number of rotatable bonds (NRotB) than the two other subsets. It is notable that two or more of the hydrogen bond donors (HBDs) in most VHL PROTACs originate from amide bonds; a feature found to be particularly detrimental for the Caco-2 cell permeability of a collection of macrocycles residing in beyond rule of 5 chemical space.17
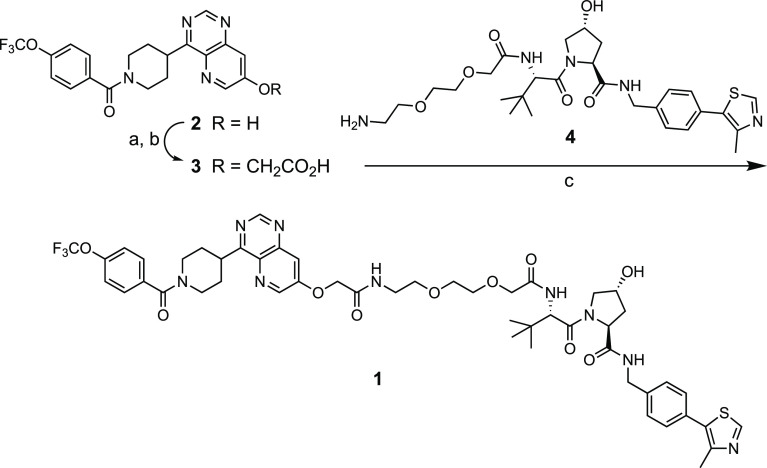
PROTAC 1 was synthesized from phenol 2(14) by alkylation with ethyl bromoacetate followed by saponification of the ethyl ester to give carboxylic acid 3 (Scheme 1). Acid 3 was then coupled with amine 4(18) using HATU as the promoter to give 1 after purification by reversed phase HPLC. Similar to many PROTACs, the aqueous solubility of 1 is low,8 whereas its lipophilicity is in the drug-like range and the permeability in the PAMPA assay is medium to high (Table 1). PROTAC 1 binds to its target protein, ERK5,14 and to the VHL E3 ligase in biochemical assays with IC50 values just above and below 1 μM, respectively (Table 1). The VHL potency drops off 14-fold in a cell-based assay, revealing 1 to have a medium permeability into target cells. Despite residing in chemical space far beyond the rule of 515 and Veber’s rule,16 where cell permeability is expected to be low, PROTAC 1 surprisingly displays medium to high permeability. We therefore considered 1 to be of great interest for investigating how structural and conformational properties, such as molecular chameleonicity, may enable PROTACs to be cell permeable.
Scheme 1. Synthesis of PROTAC 1.
Reagents and conditions: (a) Ethyl bromoacetate, K2CO3, THF, 100 °C, 8 h; (b) aq. NaOH, EtOH, rt, overnight, 38% from 2; (c) HATU, N,N-diisopropylethylamine, DMF, rt, overnight, 48%.
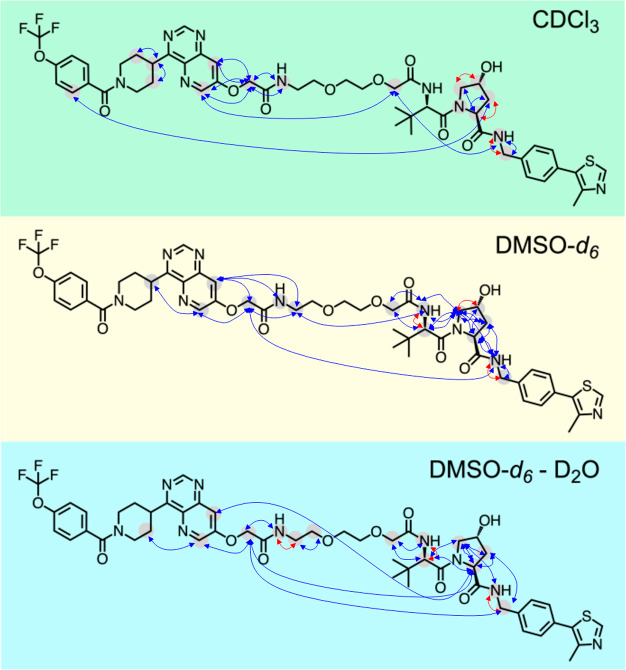
The solution conformational ensembles of 1 were determined by deconvolution of time-averaged NMR data into individual conformations using the NMR analysis of molecular flexibility in solution (NAMFIS) algorithm.19 NAMFIS has previously been used to determine the solution conformations of both flexible, linear compounds and of macrocycles having different flexibility.20−25 Proton–proton distances obtained from nuclear Overhauser effect (NOE) build-up measurements and dihedral angles calculated from vicinal scalar couplings are used as experimental input to NAMFIS. This data was generated from NMR spectra recorded for 1 in CDCl3, DMSO-d6, and DMSO-d6–D2O (ratio of 10:1) at room temperature (Figure 2). Theoretical conformational ensembles that provide a comprehensive coverage of conformational space are also required as input to NAMFIS. The required ensembles were generated by unrestrained Monte Carlo conformational searches using a variety of force fields and implicit solvent models within a 42 kJ/mol energy window.20 Based on these inputs, the NAMFIS algorithm varies the probability of each conformer in the theoretical ensembles to find the best fit of the probability weighted, back-calculated distances and dihedral angles to the corresponding experimental values determined for 1. The three conformational ensembles were validated as described earlier,20 i.e., by the addition of random noise to the experimental data, by the random removal of individual experimental restraints, and by comparison of the experimentally observed and back-calculated distances and scalar coupling constants.
Figure 2.
Overview of experimentally determined proton–proton distances and dihedral angles that were used to determine the solution conformational ensembles of 1. Blue arrows indicate proton–proton distances, while dihedral angles are indicated by red arrows.
Just as most of the reported VHL-based PROTACs,9,101 has a large number of rotatable bonds (NRotB = 27, Figure 1B) indicating a high flexibility. In spite of the potentially high flexibility, inspection of the proton–proton distances and dihedral angles revealed that the two domains of 1 were well constrained in all three solvents (Figure 2). Both the conformation of the VHL ligand domain and of the region linking the ERK5 binding protein of interest (POI) ligand to the first part of the flexible linker were constrained by multiple proton–proton distances and dihedral angles. In addition, a few long-range NOEs were observed for 1 in each of the three solvents between protons located in the POI and VHL ligands, or in either of the two ligands and a distant part of the linker. The distances that originated from these long-range NOEs indicated that folded conformations were found in the ensembles, which brought protons distant from each other in the structure of 1 into close spatial contact. To ensure that the long-range NOEs did not enforce distorted conformations of the POI-linker and VHL ligand domains, the ensembles obtained by NAMFIS analyses using all experimental restraints were validated against ensembles using restraints only from the two well-defined domains of 1, in addition to the standard validation described above.
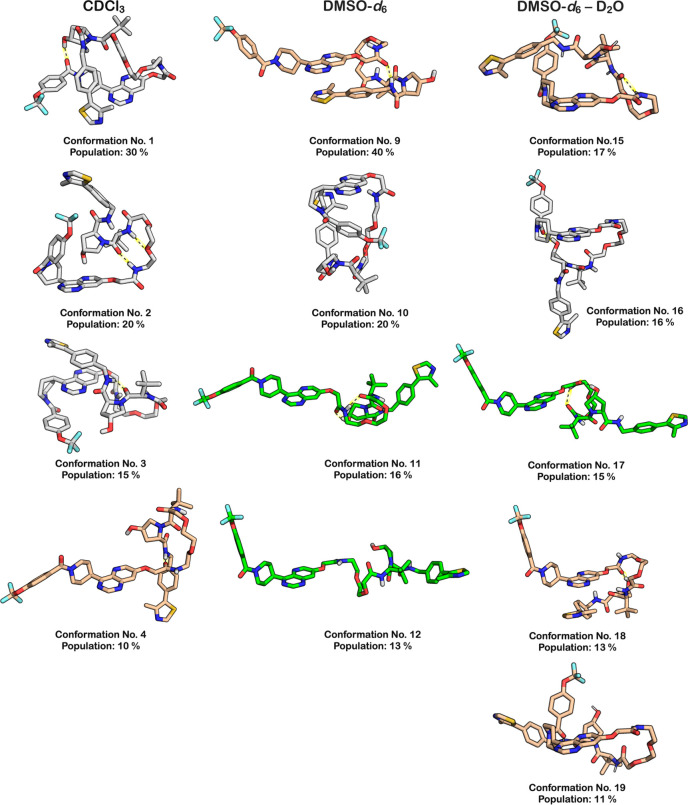
The NAMFIS analysis revealed that each of the three solution ensembles of 1 consisted of a limited number of conformations, ranging from 6 to 10 in each of the three solutions (Table S10). The conformations in the DMSO and DMSO–water ensembles were structurally more diverse than those in chloroform (Table S12). In the two polar solutions, the maximum pairwise RMSD values between the most different conformations were 8.9 and 8.4 Å, while it was 6.6 Å in chloroform. No single (congruent) conformation was adopted in two solutions; the most similar conformations were number 2 (chloroform) and number 13 (DMSO) that had a pairwise RMSD value of 3.2 Å. Four or five major conformations represented approximately 70–80% of each ensemble (a major conformation was defined as having a population ≥10%). In chloroform, PROTAC 1 adopted highly folded conformations. In three of the four major conformations, the backbone of 1 made two turns, while the remaining major conformation had one turn (Figure 3). The four minor conformations in chloroform were all folded with two turns (Figure S2). In the polar solvents, more elongated conformations were observed, with two of the major ones in DMSO being linear and one being folded with one turn. In the DMSO–water solution, one linear conformation and three folded conformations with one turn were found. The minor conformations in the two polar solutions were also more elongated than those in chloroform (Figure S2).
Figure 3.
Structures of the major conformations (population ≥ 10%) in the ensembles adopted by 1 in chloroform, DMSO, and DMSO–water (10:1). Conformations that are predominantly linear are in green, those that are folded with one turn are in tan, while folded conformations with two turns are in gray. The number and population in percent is given below each conformation. Intramolecular hydrogen bonds are indicated with black dotted lines on a yellow background.
Passive cell permeability occurs via desolvation of the compound as it leaves the extracellular aqueous environment, followed by interactions with the negatively charged phospholipid head groups and subsequent passage across the hydrophobic membrane interior. The polarity and the size of the compound, when adopting the permeating conformation(s), are the key properties that determine the kinetics of the desolvation step and the diffusion across the membrane.26 The compound’s polarity is well described by its solvent accessible 3D polar surface area (SA 3D PSA) while its size is approximated by the radius of gyration (Rgyr). The Rgyr is calculated as the root-mean-square distance between the atoms of compound and its center of mass,27 and we calculated the SA 3D PSA of 1 based on its polar atoms (O, N, and attached H) as well as on additional partial charges as recently reported.20
With the exception of one conformation (no. 4, 10%), the conformations adopted by PROTAC 1 in chloroform had a Rgyr in the interval of 5.4–6.2 Å, i.e., at the very low end of the range (5.2–9.7 Å) displayed by all conformations in the three solutions (Figure 4A and B, Table S11). The conformations in DMSO spanned a wider size (Rgyr) range than that in chloroform, whereas the size range in DMSO–water was closer to that in chloroform (Figure 4B). As revealed by the population weighted mean values for Rgyr, conformations were less compact in DMSO and DMSO–water than in chloroform. The SA 3D PSA of six of the conformations which represent 87% of the ensemble in chloroform were ≤207 Å2, i.e., in the lower third of the range of all 24 conformations (172–259 Å2). The ensembles in the three solutions populated similar ranges for SA 3D PSA, but the ensembles in DMSO and DMSO–water were shifted toward higher polarity. Population weighted mean values for SA 3D PSA increased significantly, i.e., by just over 35 Å2, between the ensembles adopted in chloroform and DMSO–water, with the ensemble in DMSO having an intermediate value (Figure 4C).
Figure 4.
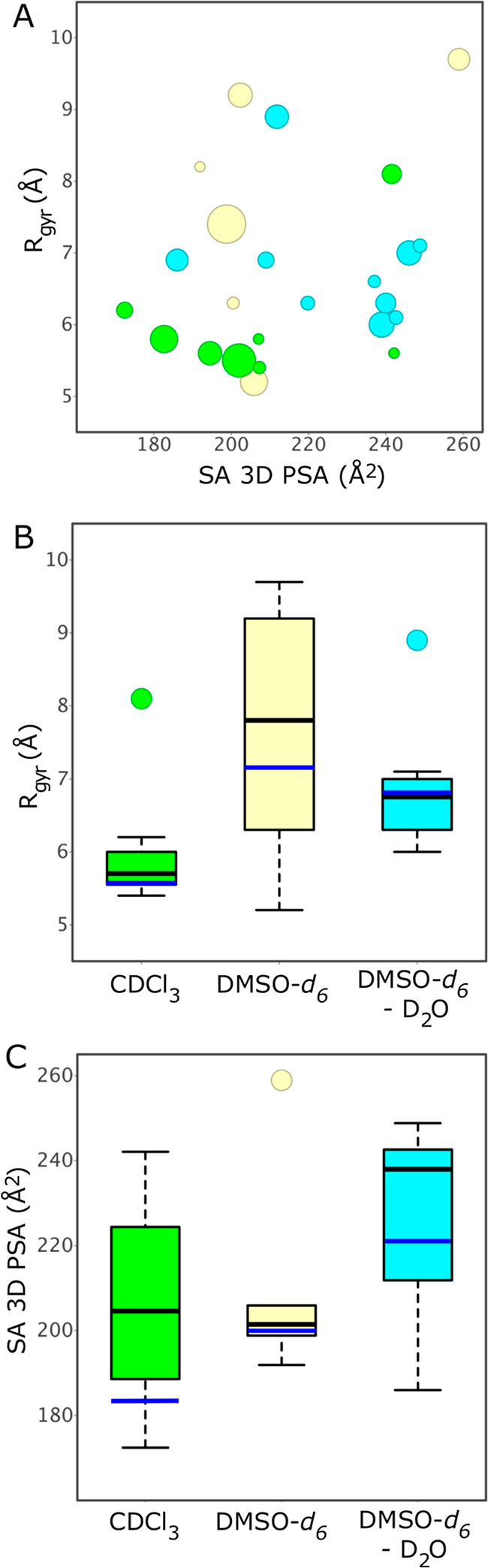
(A) Radius of gyration (Rgyr) versus solvent accessible 3D polar surface area (SA 3D PSA) for all solution conformations populated by 1. The area of each circle is proportional to the population of the corresponding conformation (in %). Conformations in CDCl3 are in green, those in DMSO-d6 are in yellow, while those in DMSO-d6–D2O are in cyan. (B) Radius of gyration (Rgyr) and (C) solvent accessible 3D polar surface area (SA 3D PSA) for the conformations populated by 1 in CDCl3, DMSO-d6, and DMSO-d6–D2O. Box plots show the 50th percentiles as black horizontal bars, the 25th and 75th percentiles as boxes, the 25th percentile minus 1.5× the interquartile range, and the 75th percentile plus 1.5× the interquartile range as whiskers, and outliers as colored circles. Population weighted mean values are shown as blue horizontal bars.
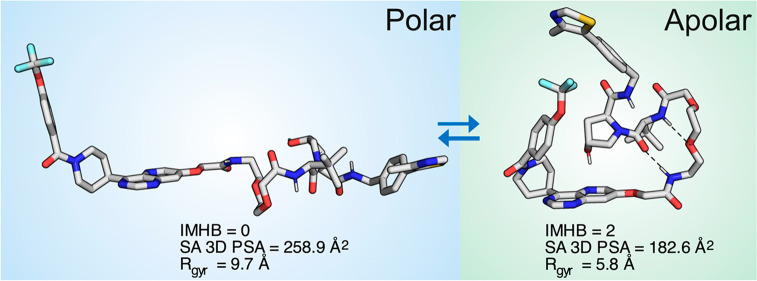
The solution conformations of 1 show a weak correlation between their Rgyr and SA 3D PSA (Figure 4A) and also a weak inverse correlation between the number of intramolecular hydrogen bonds (IMHBs) and the SA 3D PSA (Figure S4). Thus, compact conformations (low Rgyr) tend to have a lower SA 3D PSA and more IMHBs, all of which contribute to a higher permeability than if less compact and more polar conformations were adopted. Importantly, all but one of the conformations in chloroform, i.e., 90% of the ensemble, have a Rgyr below 7 Å, the proposed upper cutoff for passive cell permeability of compounds in bRo5 space.26 Similarly, the majority (87%) of the ensemble in chloroform has a SA 3D PSA in the range were cell permeability may be low-moderate (up to 200–210 Å2), as indicated by a recent study.20 In contrast, conformations in DMSO and DMSO–water were significantly larger, and those in DMSO–water were also more polar. We therefore conclude that most conformations populated in chloroform have values for Rgyr and SA 3D PSA that should allow 1 to permeate cells and that folded conformations having two turns such as conformations 1–3 are likely be the permeating species.
Inspection of the proposed permeating conformations 1–3 provided detailed insight into what intramolecular interactions allow an environment dependent reduction of size and polarity for 1 (Figure 5). In conformation 1, PROTAC 1 forms an IMHB between the hydroxyl group of hydroxyproline and the tertiary amide of the ERK5 ligand. The NH of two of the secondary amide bonds of 1 point toward the center of the folded conformation, thereby shielding them from the surrounding apolar environment, while the third amide NH may be partially shielded by the pyrimidine ring. In addition to the IMHB, a π–π interaction between the pyrimidine and thiazole rings may contribute to minimizing the Rgyr of 1 in this conformation. Two IMHBs, which involve the NH of two of the three secondary amides of 1, are formed in conformation 2 that has a low Rgyr and a very low SA 3D PSA. The third amide NH is partially shielded from the surrounding as it points to the center of the conformation. In addition, the hydroxyl group of the hydroxyproline moiety of 1 is shielded from solvent by the pyrimidine of the ERK5 ligand, potentially by formation of a nonclassical hydrogen bond to the pyrimidine. The SA 3D PSA of conformation 3 is reduced by IMHBs that involve the NH of two of the three secondary amides of 1 and by shielding of the remaining amide NH by the adjacent tert-butyl group. In this case, a π–π interaction between the pyrimidine ring and phenyl ring in the VHL domain could contribute to keeping the Rgyr of conformation 3 low, similar to the π–π interaction of conformation 1. In summary, inspection of conformations 1–3 reveal that the SA 3D PSA of PROTAC 1 can be reduced in an apolar environment by the formation of IMHBs, shielding of amide NH groups from solvent and by formation of nonclassical hydrogen bonds. In addition to these intramolecular interactions, π–π interactions appear to contribute to minimization of Rgyr. We suggest that the ability to adopt folded conformations stabilized by similar intramolecular interactions that minimize Rgyr and SA 3D PSA is likely to be required also for other VHL PROTACs that have flexible PEG-based linkers to enter cells and induce target degradation.
Figure 5.
Structures of conformations 1–3 that are populated in chloroform. The population in percent, the number of intramolecular hydrogen bonds (IMHB), the solvent accessible 3D PSA and the radius of gyration (Rgyr) are given below each conformation. IMHBs are indicated with dashed black lines on a yellow background.
Recent studies of drugs beyond the rule of 5 space,11,28 most of which are semirigid macrocycles, have revealed that they benefit from behaving as molecular chameleons in order to display high aqueous solubility and cell permeability.20,29,30 This study is the first to show that the more flexible PROTACs also can behave as molecular chameleons and that this property is important for their cell permeability. By using a validated NMR technique, we found that the VHL based PROTAC 1 adopts its shape, i.e., its radius of gyration (Rgyr) and its solvent accessible 3D polar surface area (SA 3D PSA) to the surrounding environment. Compound 1 populated conformations that were more elongated and polar in environments used as mimics of polar extra- and intracellular compartments. In contrast, most conformations were folded with a low Rgyr and a low SA 3D PSA in chloroform, which has a polarity close to that of the center of a cell membrane. Importantly, the folded conformations adopted in chloroform had values for Rgyr and SA 3D PSA compatible with satisfactory passive cell permeability,20,26 even though properties such as MW, TPSA, and the number of rotatable bonds place 1 into chemical space far beyond the rule of 5. As the calculated properties of 1 are representative for reported VHL based PROTACs that have flexible, PEG-based linkers, chameleonicity may also be of general importance for the cell permeability of this type of PROTACs. However, this hypothesis needs to be confirmed by studies of additional PROTACs. Design of cell permeable PROTACs could capitalize on recent learnings from other drug candidates, where intramolecular hydrogen bonds31 and shielding of amide bonds32 have been used to improve cell permeability. Design should also benefit from wider analyses of larger PROTAC sets by QSPR and machine learning, just as from methods that allow more accurate sampling of biologically relevant conformational space for this emerging class of drugs.
Acknowledgments
We are grateful for access to an 800 MHz NMR instrument at the Swedish NMR Centre and thank Dieudonné Tshitenge for recording the 13C spectrum of 1. The Monte Carlo conformational searches for the NAMFIS analyses were performed using resources provided by the Swedish National Infrastructure for Computing (SNIC) at NSC (project 2020-5-435).
Glossary
Abbreviations
- CRBN
cereblon
- ERK5
extracellular signal-regulated kinase 5
- IMHB
intramolecular hydrogen bond
- LPE
lipophilic permeability efficiency
- NAMFIS
NMR analysis of molecular flexibility in solution
- NRotB
number of rotatable bonds
- POI
protein of interest
- PROTAC
proteolysis targeting chimera
- Rgyr
radius of gyration
- SA 3D PSA
solvent accessible 3D polar surface area
- TPSA
topological polar surface area
- VHL
Von Hippel–Lindau
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00556.
Synthesis and characterization of 1 and procedures for determination of its physicochemical and in vitro properties; NMR data, Monte Carlo molecular mechanic conformational search and NAMFIS conformational analysis of 1; computational characterization of 1 and reported VHL PROTACs, and NMR spectra of 1 (PDF)
Author Contributions
# Y.A., V.P., and C.S.N. made equal contributions to the manuscript. This manuscript was written through contributions of all authors. All authors have approved the final version of the manuscript.
This work was funded by grants from Bayer AG and the Swedish Research Council (Grant Nos. 2016-05160 to J.K. and 2016-03602 to M.E.).
The authors declare the following competing financial interest(s): D.G. is an employee of Nuvisan Innovation Campus Berlin GmbH. A.G and D.M. are employees of Bayer AG.
Supplementary Material
References
- Lai A. C.; Crews C. M. Induced protein degradation: an emerging drug discovery paradigm. Nat. Rev. Drug Discov. 2017, 16, 101–114. 10.1038/nrd.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira M.; Calabrese M. F.; Bullock A. N.; Crews C. M. Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discov. 2019, 18, 949–963. 10.1038/s41573-019-0047-y. [DOI] [PubMed] [Google Scholar]
- Bondeson D. P.; Mares A.; Smith I. E. D.; Ko E.; Campos S.; Miah A. H.; Mulholland K. E.; Routly N.; Buckley D. L.; Gustafson J. L.; Zinn N.; Grandi P.; Shimamura S.; Bergamini G.; Faelth-Savitski M.; Bantscheff M.; Cox C.; Gordon D. A.; Willard R. R.; Flanagan J. J.; Casillas L. N.; Votta B. J.; den Besten W.; Famm K.; Kruidenier L.; Carter P. S.; Harling J. D.; Churcher I.; Crews C. M. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015, 11, 611–617. 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa A.; Hughes S. J.; Lucas X.; Wright J. E.; Ciulli A. Structure-based design of a macrocyclic PROTAC. Angew. Chem., Int. Ed. 2020, 59, 1727–1734. 10.1002/anie.201914396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermondi G.; Vallaro M.; Caron G. Degraders early developability assessment: face-to-face with molecular properties. Drug Discov. Today 2020, 25, 1585. 10.1016/j.drudis.2020.06.015. [DOI] [PubMed] [Google Scholar]
- Klein V. G.; Townsend C. E.; Testa A.; Zengerle M.; Maniaci C.; Hughes S. J.; Chan K.-H.; Ciulli A.; Lokey R. S. Understanding and improving the membrane permeability of VH032-based PROTACs. ACS Med. Chem. Lett. 2020, 11, 1732. 10.1021/acsmedchemlett.0c00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. E.; Rooney T. P. C.; Bayle E. D.; Mirza T.; Willems H. M. G.; Clarke J. H.; Andrews S. P.; Skidmore J. Systematic investigation of the permeability of androgen receptor PROTACs. ACS Med. Chem. Lett. 2020, 11, 1539–1547. 10.1021/acsmedchemlett.0c00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrill C.; Chaturvedi P.; Rynn C.; Schaffland J. P.; Walter I.; Wittwer M. B. Fundamental aspects of DMPK optimization of targeted protein degraders. Drug Discov. Today 2020, 25, 969–982. 10.1016/j.drudis.2020.03.012. [DOI] [PubMed] [Google Scholar]
- Edmondson S. D.; Yang B.; Fallan C. Proteolysis targeting chimeras (PROTACs) in ‘beyond rule-of-five’ chemical space: Recent progress and future challenges. Bioorg. Med. Chem. Lett. 2019, 29, 1555–1564. 10.1016/j.bmcl.2019.04.030. [DOI] [PubMed] [Google Scholar]
- Maple H. J.; Clayden N.; Baron A.; Stacey C.; Felix R. Developing degraders: principles and perspectives on design and chemical space. MedChemComm 2019, 10, 1755–1764. 10.1039/C9MD00272C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doak B. C.; Over B.; Giordanetto F.; Kihlberg J. Oral druggable space beyond the rule of 5: Insights from drugs and clinical candidates. Chem. Biol. 2014, 21, 1115–1142. 10.1016/j.chembiol.2014.08.013. [DOI] [PubMed] [Google Scholar]
- DeGoey D. A.; Chen H.-J.; Cox P. B.; Wendt M. D. Beyond the rule of 5: Lessons learned from AbbVie’s drugs and compound collection. J. Med. Chem. 2018, 61, 2636–2651. 10.1021/acs.jmedchem.7b00717. [DOI] [PubMed] [Google Scholar]
- Gramse G.; Dols-Perez A.; Edwards M. A.; Fumagalli L.; Gomila G. Nanoscale measurement of the dielectric constant of supported lipid bilayers in aqueous solutions with electrostatic force microscopy. Biophys. J. 2013, 104, 1257–1262. 10.1016/j.bpj.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.; Lemos C.; Wortmann L.; Eis K.; Holton S. J.; Boemer U.; Moosmayer D.; Eberspaecher U.; Weiske J.; Lechner C.; Prechtl S.; Suelzle D.; Siegel F.; Prinz F.; Lesche R.; Nicke B.; Nowak-Reppel K.; Himmel H.; Mumberg D.; von Nussbaum F.; Nising C. F.; Bauser M.; Haegebarth A. Discovery and characterization of the potent and highly selective (piperidin-4-yl)pyrido[3,2-d]pyrimidine based in vitro probe BAY-885 for the kinase ERK5. J. Med. Chem. 2019, 62, 928–940. 10.1021/acs.jmedchem.8b01606. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Veber D. F.; Johnson S. R.; Cheng H.-Y.; Smith B. R.; Ward K. W.; Kopple K. D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Over B.; Matsson P.; Tyrchan C.; Artursson P.; Doak B. C.; Foley M. A.; Hilgendorf C.; Johnston S.; Lee I. M. D.; Lewis R.; McCarren P.; Muncipinto G.; Norinder U.; Perry M.; Duvall J. R.; Kihlberg J. Structural and conformational determinants of macrocycle cell permeability. Nat. Chem. Biol. 2016, 12, 1065–1074. 10.1038/nchembio.2203. [DOI] [PubMed] [Google Scholar]
- Chan K.-H.; Zengerle M.; Testa A.; Ciulli A. Impact of target warhead and linkage vector on inducing protein degradation: Comparison of bromodomain and extra-terminal (BET) degraders derived from triazolodiazepine (JQ1) and tetrahydroquinoline (I-BET726) BET inhibitor scaffolds. J. Med. Chem. 2018, 61, 504–513. 10.1021/acs.jmedchem.6b01912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicero D. O.; Barbato G.; Bazzo R. NMR analysis of molecular flexibility in solution: A new method for the study of complex distributions of rapidly exchanging conformations. Application to a 13-residue peptide with an 8-residue loop. J. Am. Chem. Soc. 1995, 117, 1027–1033. 10.1021/ja00108a019. [DOI] [Google Scholar]
- Danelius E.; Poongavanam V.; Peintner S.; Wieske L. H. E.; Erdélyi M.; Kihlberg J. Solution conformations explain the chameleonic behavior of macrocyclic drugs. Chem. - Eur. J. 2020, 26, 5231–5244. 10.1002/chem.201905599. [DOI] [PubMed] [Google Scholar]
- Danelius E.; Andersson H.; Jarvoll P.; Lood K.; Gräfenstein J.; Erdélyi M. Halogen bonding: A powerful tool for modulation of peptide conformation. Biochemistry 2017, 56, 3265–3272. 10.1021/acs.biochem.7b00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danelius E.; Ohm R. G.; Ahsanullah; Mulumba M.; Ong H.; Chemtob S.; Erdelyi M.; Lubell W. D. Dynamic chirality in the mechanism of action of allosteric CD36 modulators of macrophage-driven inflammation. J. Med. Chem. 2019, 62, 11071–11079. 10.1021/acs.jmedchem.9b00918. [DOI] [PubMed] [Google Scholar]
- Dickman R.; Danelius E.; Mitchell S. A.; Hansen D. F.; Erdelyi M.; Tabor A. T. A chemical biology approach to understanding molecular recognition of lipid II by nisin(1–12): Synthesis and NMR ensemble analysis of nisin(1–12) and analogues. Chem. - Eur. J. 2019, 25, 14572–14582. 10.1002/chem.201902814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thepchatri P.; Cicero D. O.; Monteagudo E.; Ghosh A. K.; Cornett B.; Weeks E. R.; Snyder J. P. Conformations of laulimalide in DMSO-d6. J. Am. Chem. Soc. 2005, 127, 12838–12846. 10.1021/ja042890e. [DOI] [PubMed] [Google Scholar]
- Koivisto J. J.; Kumpulainen E. T. T.; Koskinen A. M. P. Conformational ensembles of flexible β-turn mimetics in DMSO-d6. Org. Biomol. Chem. 2010, 8, 2103–2116. 10.1039/b921794k. [DOI] [PubMed] [Google Scholar]
- Guimarães C. R. W.; Mathiowetz A. M.; Shalaeva M.; Goetz G.; Liras S. Use of 3D properties to characterize beyond rule-of-5 property space for passive permeation. J. Chem. Inf. Model. 2012, 52, 882–890. 10.1021/ci300010y. [DOI] [PubMed] [Google Scholar]
- Narang P.; Bhushan K.; Bose S.; Jayaram B. A computational pathway for bracketing native-like structures for small alpha helical globular proteins. Phys. Chem. Chem. Phys. 2005, 7, 2364–2375. 10.1039/b502226f. [DOI] [PubMed] [Google Scholar]
- Poongavanam V.; Doak B. C.; Kihlberg J. Opportunities and guidelines for discovery of orally absorbed drugs in beyond rule of 5 space. Curr. Opin. Chem. Biol. 2018, 44, 23–29. 10.1016/j.cbpa.2018.05.010. [DOI] [PubMed] [Google Scholar]
- Whitty A.; Zhong M.; Viarengo L.; Beglov D.; Hall D. R.; Vajda S. Quantifying the chameleonic properties of macrocycles and other high-molecular-weight drugs. Drug Discov. Today 2016, 21, 712–717. 10.1016/j.drudis.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi Sebastiano M.; Doak B. C.; Backlund M.; Poongavanam V.; Over B.; Ermondi G.; Caron G.; Matsson P.; Kihlberg J. Impact of dynamically exposed polarity on permeability and solubility of chameleonic drugs beyond the rule of 5. J. Med. Chem. 2018, 61, 4189–4202. 10.1021/acs.jmedchem.8b00347. [DOI] [PubMed] [Google Scholar]
- Mackman R. L.; Steadman V. A.; Dean D. K.; Jansa P.; Poullennec K. G.; Appleby T.; Austin C.; Blakemore C. A.; Cai R.; Cannizzaro C.; Chin G.; Chiva J.-Y. C.; Dunbar N. A.; Fliri H.; Highton A. J.; Hui H.; Ji M.; Jin H.; Karki K.; Keats A. J.; Lazarides L.; Lee Y.-J.; Liclican A.; Mish M.; Murray B.; Pettit S. B.; Pyun P.; Sangi M.; Santos R.; Sanvoisin J.; Schmitz U.; Schrier A.; Siegel D.; Sperandio D.; Stepan G.; Tian Y.; Watt G. M.; Yang H.; Schultz B. E. Discovery of a potent and orally bioavailable cyclophilin inhibitor derived from the sanglifehrin macrocycle. J. Med. Chem. 2018, 61, 9473–9499. 10.1021/acs.jmedchem.8b00802. [DOI] [PubMed] [Google Scholar]
- Tyagi M.; Poongavanam V.; Lindhagen M.; Pettersen A.; Sjö P.; Schiesser S.; Kihlberg J. Toward the design of molecular chameleons: Flexible shielding of an amide bond enhances macrocycle cell permeability. Org. Lett. 2018, 20, 5737–5742. 10.1021/acs.orglett.8b02447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.