Abstract
Most animal species on Earth are insects, and recent reports suggest that their abundance is in drastic decline. Although these reports come from a wide range of insect taxa and regions, the evidence to assess the extent of the phenomenon is sparse. Insect populations are challenging to study, and most monitoring methods are labor intensive and inefficient. Advances in computer vision and deep learning provide potential new solutions to this global challenge. Cameras and other sensors can effectively, continuously, and noninvasively perform entomological observations throughout diurnal and seasonal cycles. The physical appearance of specimens can also be captured by automated imaging in the laboratory. When trained on these data, deep learning models can provide estimates of insect abundance, biomass, and diversity. Further, deep learning models can quantify variation in phenotypic traits, behavior, and interactions. Here, we connect recent developments in deep learning and computer vision to the urgent demand for more cost-efficient monitoring of insects and other invertebrates. We present examples of sensor-based monitoring of insects. We show how deep learning tools can be applied to exceptionally large datasets to derive ecological information and discuss the challenges that lie ahead for the implementation of such solutions in entomology. We identify four focal areas, which will facilitate this transformation: 1) validation of image-based taxonomic identification; 2) generation of sufficient training data; 3) development of public, curated reference databases; and 4) solutions to integrate deep learning and molecular tools.
Keywords: automated monitoring, ecology, insects, image-based identification, machine learning
We are experiencing a mass extinction of species (1), but data on changes in species diversity and abundance have substantial taxonomic, spatial, and temporal biases and gaps (2, 3). The lack of data holds especially true for insects despite the fact that they represent the vast majority of animal species. A major reason for these shortfalls for insects and other invertebrates is that available methods to study and monitor species and their population trends are antiquated and inefficient (4). Nevertheless, some recent studies have demonstrated alarming rates of insect diversity and abundance loss (5–7). To further explore the extent and causes of these changes, we need efficient, rigorous, and reliable methods to study and monitor insects (4, 8).
Data to derive insect population trends are already generated as part of ongoing biomonitoring programs. However, legislative terrestrial biomonitoring (e.g., in the context of the European Union [EU] Habitats Directive) focuses on a very small subset of individual insect species such as rare butterflies and beetles because the majority of insect taxa are too difficult or too costly to monitor (9). In current legislative aquatic monitoring, invertebrates are commonly used in assessments of ecological status (e.g., the US Clean Water Act, the EU Water Framework Directive, and the EU Marine Strategy Framework Directive). Still, spatiotemporal and taxonomic extent and resolution in ongoing biomonitoring programs are coarse and do not provide information on the status of the vast majority of insect populations.
Molecular techniques such as DNA barcoding and metabarcoding will likely become valuable tools for future insect monitoring based on field-collected samples (10, 11), but at the moment, high-throughput methods cannot provide reliable abundance estimates (12, 13), leaving a critical need for other methodological approaches. The state of the art in deep learning and computer vision methods and image processing has matured to the point where it can aid or even replace manual observation in situ (14) as well as in routine laboratory sample processing tasks (15). Image-based observational methods for monitoring of vertebrates using camera traps have undergone rapid development in the past decade (14, 16–18). Similar approaches using cameras and other sensors for investigating diversity and abundance of insects are underway (19, 20). However, despite huge attention in other domains, deep learning is only very slowly beginning to be applied in invertebrate monitoring and biodiversity research (21–25).
Deep learning models learn features of a dataset by iteratively training on example data without the need for manual feature extraction (26). In this way, deep learning is qualitatively different from traditional statistical approaches to prediction (27). Deep learning models specifically designed for dealing with images, so-called convolutional neural networks (CNNs), can extract features from images or objects within them and learn to differentiate among them. There is great potential in automatic detection and classification of insects in video or time-lapse images with trained CNNs for monitoring purposes (20). As the methods become more refined, they will bring exciting new opportunities for understanding insect ecology and for monitoring (19, 28–31).
Here, we argue that deep learning and computer vision can be used to develop novel high-throughput systems for detection, enumeration, classification, and discovery of species as well as for deriving functional traits such as biomass for biomonitoring purposes. These approaches can help solve long standing challenges in ecology and biodiversity research and also address pressing issues in insect population monitoring (32, 33). This article has three goals. First, we present sensor-based solutions for observation of invertebrates in situ and for specimen-based research in the laboratory. We focus on solutions, which either already use or could benefit from deep learning models to analyze the large volume of data involved. Second, we show how deep learning models can be applied to obtained data streams to derive ecologically relevant information. Last, we outline and discuss four main challenges that lie ahead in the implementation of such solutions for invertebrate monitoring, ecology, and biodiversity research.
Sensor-Based Insect Monitoring
Sensors are widely used in ecology for gathering peripheral data such as temperature, precipitation, and light intensity. However, solutions for sensor-based monitoring of insects and other invertebrates in their natural environment are only just emerging (34). The innovation and development are primarily driven by agricultural research to predict occurrence and abundance of beneficial and pest insect species of economic importance (35–37), to provide more efficient screening of natural products for invasive insect species (38), or to monitor disease vectors such as mosquitos (39, 40). The most commonly used sensors are cameras, radars, and microphones. Such sensor-based monitoring is likely to generate datasets that are orders of magnitude larger than those commonly studied in ecology (i.e., big data), which require efficient solutions for extracting relevant biological information. Deep learning could be a critical tool in this respect. Below, we give examples of image-based approaches to insect monitoring, which we argue have the greatest potential for integration with deep learning. We also describe approaches using other types of sensors, where the integration with deep learning is less developed but still could be relevant for detecting and classifying entomological information. We further describe the ongoing efforts in the digitization of natural history collections, which could generate valuable reference data for training and validating deep learning models.
Image-Based Solutions for In Situ Monitoring.
Some case studies have already used cameras and deep learning methods for detecting single species, such as the pest of the fruits of olive trees Bactrocera oleae (41) or for more generic pest detection (42). Here, the pest detection is based on images of insects that have been trapped with either a McPhail-type trap or a trap with pheromone lure and adhesive liner. The images are collected by a microcomputer and transmitted to a remote server where they are analyzed. Other solutions have embedded a digital camera and a microprocessor that can count trapped individuals in real time using object detection based on a deep learning model (37). In both these cases, deep learning networks are trained to recognize and count the number of individuals. However, there are very few examples of invertebrate biodiversity-related field studies applying deep learning models (23). Early attempts used feature vectors extracted from single perspective images and yielded modest accuracy for 35 species of moths (43) or used mostly coarse taxonomic resolution (44). We have recently demonstrated that our custom-built time-lapse cameras can record image data from which a deep learning model can accurately estimate local spatial, diurnal, and seasonal dynamics of honeybees and other flower-visiting insects (45) (Fig. 1). Time-lapse cameras are less likely to create observer bias than direct observation, and data collection can extend across full diurnal and even seasonal timescales. Cameras can be baited just as traditional light and pheromone traps or placed over ephemeral natural resources such as flowers, fruits, dung, fungi, or carrion. Bjerge et al. (46) propose to use an automated light trap to monitor the abundance of moths and other insects attracted to light. As the system is powered by a solar panel, it can be installed in remote locations (Fig. 2). Ultimately, true “Internet of Things”-enabled hardware will make it possible to implement classification algorithms directly on the camera units to provide fully autonomous systems in the field to monitor insects and report detection and classification data back to the user or to online portals in real time (34).
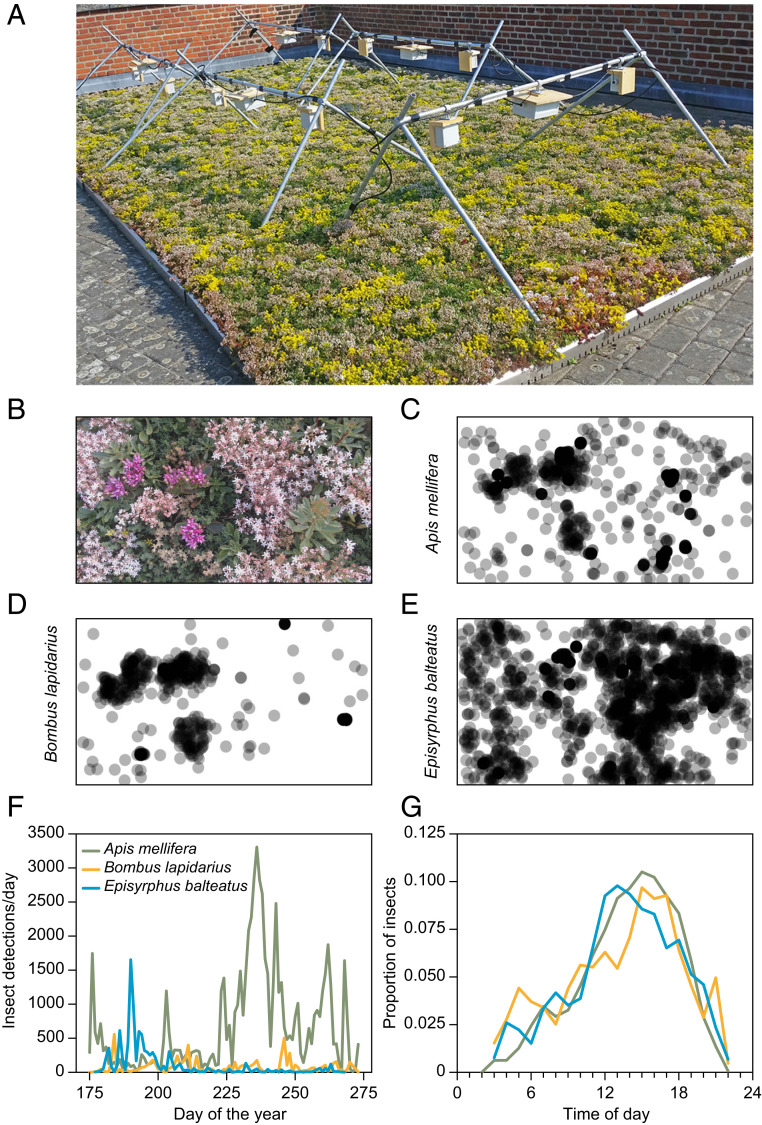
Fig. 1.
We developed and tested a camera trap for monitoring flower-visiting insects, which records images at fixed intervals (45). (A) The setup consists of two web cameras connected to a control unit containing a Raspberry Pi computer and a hard drive. In our test, 10 camera traps were mounted on custom-built steel rod mounts 30 cm above a green roof mix of plants in the genus Sedum. Images were recorded every 30 s during the entire flowering season. After training a CNN (Yolo3), we detected >100,000 instances of pollinators over the course of an entire growing season. (B) An example image from one of the cameras showing a scene consisting of different flowering plant species. The locations of the insect detections varied greatly among three common flower-visiting species: (C) the European honeybee (Apis mellifera), (D) the red-tailed bumblebee (Bombus lapidarius), and (E) the marmalade hoverfly (Episyrphus balteatus). Across the 10 image series, the deep learning model detected detailed variation in (F) seasonal and (G) diurnal variation in the occurrence frequency among the same three species. Adapted with permission from ref. 45.
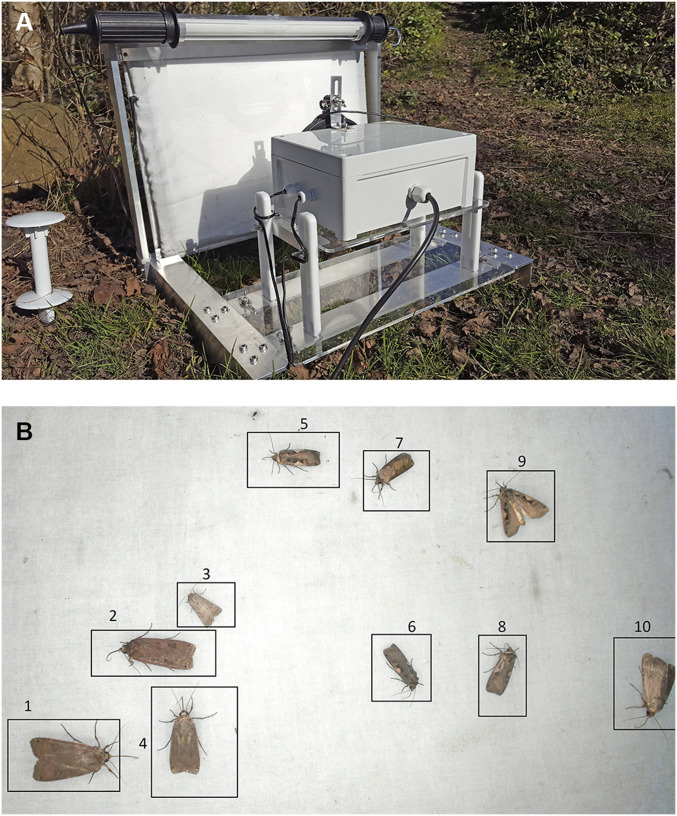
Fig. 2.
(A) To automatically monitor nocturnal moth species, we designed a light trap with an onboard computer vision system (46). The light trap is equipped with three different light sources: a fluorescent tube to attract moths, a light table covered by a white sheet to provide a diffuse background illumination for the resting insects, and a light ring to illuminate the specimens. The system is able to attract moths and automatically capture images based on motion detection. The trap is designed using standard components such as a high-resolution universal serial bus web camera and a Raspberry Pi computer. (B) We have proposed a computer vision algorithm that can track and count individual moths. A customized CNN was trained to detect and classify eight different moth species. Ten (1–10) individuals were automatically detected in this example photo recorded by the trap. The algorithm can run on the onboard computer to allow the system to automatically process and submit species data via a modem to a server. The system works off grid due to a battery and solar panel. Reprinted with permission from ref. 46.
Radar, Acoustic, and Other Solutions for In Situ Monitoring.
The use of radar technology in entomology has allowed for the study of insects at scales not possible with traditional methods, specifically related to both migratory and nonmigratory insects flying at high altitudes (47). Utilizing data from established weather radar networks can provide information at the level of continents (48), while specialized radar technology such as vertical-looking radars (VLRs) can provide finer-grained data albeit at a local scale (49). The VLRs can give estimates of biomass and body shape of the detected object, and direction of flight, speed, and body orientation can be extracted from the return radar signal (50). However, VLR data provide little information on community structure, and conclusive species identification requires aerial trapping (51, 52). Harmonic scanning radars can detect insects flying at low altitudes at a range of several hundred meters, but insects need to be tagged with a radar transponder and must be within line of sight (53, 54). Collectively, the use of radar technology in entomology can provide valuable information in insect monitoring [for example, on the magnitude of biomass flux stemming from insect migrations (55)] but requires validation with other methods (e.g., ref. 56).
Bioacoustics is a well-established scientific discipline, and acoustic signals have been widely used in the field of ecology. Although most commonly used for birds and mammals, bioacoustic techniques have merits in entomological monitoring. For example, Jeliazkov et al. (57) used audio recordings to study population trends of Orthoptera at large spatial and temporal scales, and Kiskin et al. (58) demonstrated the use of a CNN to detect the presence of mosquitoes by identifying the acoustic signal of their wingbeats. Other studies have shown that even species classification for groups such as grasshoppers (59) and bees (60) is possible using machine learning on audio data. It has been argued that the use of pseudoacoustic optical sensors rather than actual acoustic sensors is a more promising technology because of the much improved signal-to-noise ratio in these systems (61). Nevertheless, deep learning methods could be a valuable tool for acoustic entomological monitoring.
Other types of sensor technology are used to automate the recording of insect activity or even body mass, typically without actual consideration of the subsequent processing of the data with deep learning methods (62, 63). In one of these recent studies, researchers used a sensor ring of photodiodes and infrared light-emitting diodes to detect large- and small-sized arthropods, including pollinators and pests, and achieved a 95% detection accuracy for live microarthropods of three different species in the size range from 0.5 to 1.1 mm (62). The Edapholog (63) is a low-power monitoring system for detection of soil microarthropods. Probe and sensing are based on detection of change in infrared light intensity similar to ref. 62, and it counts the organisms falling into the trap and estimates their body size. The probe is connected via radio signals to a logging device that transmits the data to a server for real-time monitoring. Similarly, others have augmented traditional low-cost trapping methods by implementing optoelectronic sensors and wireless communication to allow for real-time monitoring and reporting (35). Since such sensors do not produce images that are intuitive to validate, it could be challenging to generate sufficient, validated training data for implementing deep learning models, although such models could still prove useful.
Digitizing Specimens and Natural History Collections.
There are strong efforts to digitize natural history collections for multiple reasons including the potential for applying deep learning methods (64). The need for and benefits of digitizing natural science collections have motivated the foundation of the Distributed System of Scientific Collections Research Infrastructure (DISSCo RI; https://www.dissco.eu/). DISSCo RI strives for the digital unification of all European natural science assets under common curation and access policies and practices. Most existing databases include single-view digitizations of pinned specimens (65), while datasets of insect specimens recorded using multiple sensors, three-dimensional models, and databases on living insect specimens are only just emerging (66, 67). The latter could be particularly relevant for deep learning models. There is also a valuable archive of entomological data in herbarium specimens in the form of signs of herbivory (68). The standard digitization of herbarium collections has proven suitable for extracting herbivory data using machine learning techniques (69). Techniques to automate digitization will accelerate the development of such valuable databases (64). The BIODISCOVER machine (70) is a solution for automated digitization of liquid-preserved specimens such as most field-collected insects. The process consists of four automatized steps: 1) bin picking of individual insects directly from bulk samples; 2) recording the specimen from multiple angles using high-speed imaging; 3) saving the captured data in an optimized way for deep learning algorithm training and further study; and 4) sorting specimens according to size, taxonomic identity, or rarity for potential further molecular processing (Fig. 3). Implementing such tools for the processing of bulk insect samples from large-scale inventories and monitoring studies could rapidly and nondestructively generate population and community data. Digitization efforts should also carefully consider how images of individual specimens can be leveraged to develop deep learning models for in situ monitoring.
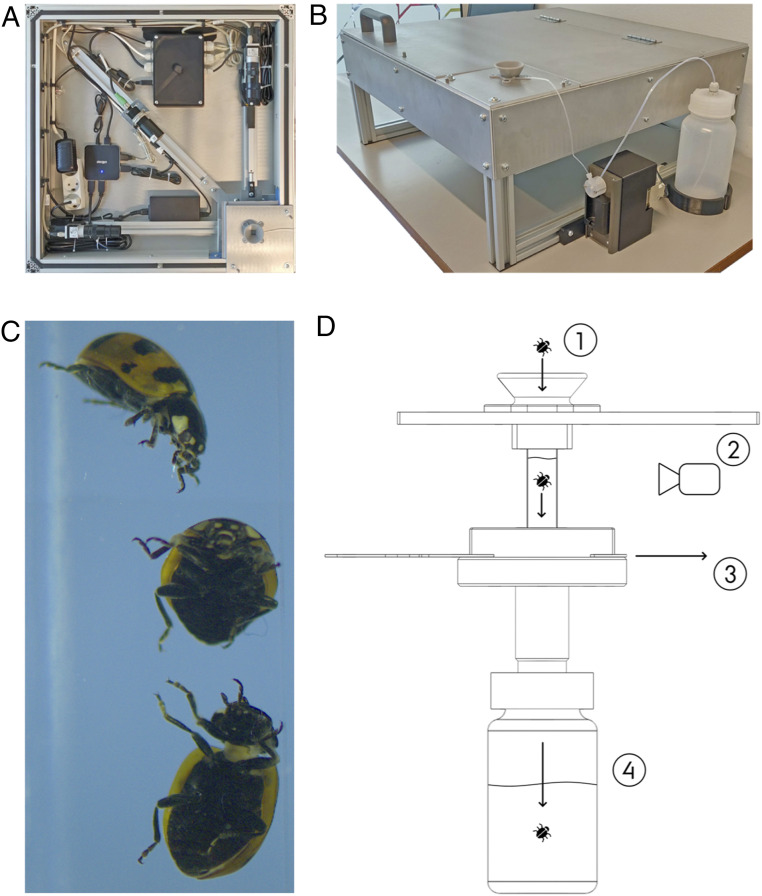
Fig. 3.
The BIODISCOVER machine can automate the process of invertebrate sample sorting, species identification, and biomass estimation (70). (A) The imaging system consists of an ethanol-filled spectroscopic cuvette, a powerful and adjustable light source, and two cameras capable of recording images at 50 frames per second (B) The setup is mounted in a light-proof aluminum box and fitted with a pump for refilling the spectroscopic cuvette. (C) Each specimen is imaged from two angles by the cameras as it is dropped into the ethanol-filled cuvette, and geometric features related to size and biomass are computed automatically. (D) The specimen (1) is imaged by two cameras (2) as it sinks through the ethanol. The system has a built-in flushing mechanism (3) for controlling which specimens should be kept together for subsequent storage or analysis (4). The results for an initial dataset of images of 598 specimens across 12 species of known identity were very promising, with a classification accuracy of 98.0%. Adapted from ref. 70, which is licensed under CC BY 4.0.
Potential Deep Learning Applications in Entomology
The big data collected by sensor-based insect monitoring as described above require efficient solutions for transforming the data into biologically relevant information. Preliminary results suggest that deep learning offers a valuable tool in this respect and could further inspire the collection of new types of data (20, 45). Deep learning software (e.g., for ecological applications) is mostly constructed using open source Python libraries and frameworks such as TensorFlow, Keras, PyTorch, and Scikit-learn (24), and prototype implementations are typically made publicly available (e.g., on https://github.com/). This, in turn, makes the latest advances in other fields related to object detection and fine-grained classification available also for entomological research. As such, the deep learning toolbox is already available to entomologists, but some tools may need to be adapted for specific entomological applications. In the following, we provide a brief description of the transformative potential of deep learning related to entomological data stored in images structured around four main applications.
Detecting and Tracking Individuals In Situ.
Image-based monitoring of insect abundance and diversity could rapidly become globally widespread as countries make efforts to better understand the severity of the global insect decline and identify mitigation measures. Identification of individual insects has recently been facilitated by online portals such as https://www.inaturalist.org/ and https://observation.org/ and their associated smartphone apps. These systems provide instant candidate species when users upload pictures of observed insect species by using deep learning models. While such portals provide powerful tools for rapid generation of biodiversity data, their main purpose is opportunistic recording of species occurrence and not structured ecological monitoring. However, such systems could be adapted for monitoring purposes. For example, a network of time-lapse cameras could generate high-temporal and -spatial resolution image data for monitoring of specific insect species. In some cases, detection of individuals from such data could be achieved by simple modifications to existing models, while in other cases, custom-built solutions may be necessary. Combining image data with acoustic or behavioral data could be a solution for taxa that are harder to detect and identify. In addition to detecting and classifying individuals, object detection models can also pinpoint their exact location within an image. Such models can be applied to time-lapse and video data in order to track the position of individual insects in situ through time. This would add an additional valuable layer of data, which can be derived from image-based observations. For instance, the movement speed of individual insects can be related to the observed microclimatic variation, and more realistic thermal performance curves can be established and contrasted to traditional laboratory-derived thermal performance.
However, tracking insects in their natural environment is currently a highly challenging task due to issues that include the cluttered scenes and varying lighting conditions. In computer vision, such tasks are termed “detection-based online multiple object tracking” and work under a set of assumptions (71). These assumptions include a precise initial detection (initialization) of the objects to be tracked in a scene; a good ability to visually discriminate between the multiple tracked objects; and smooth motion, velocity, and acceleration patterns of the tracked objects (72). The small visual differences among individual insects and frequent hiding behavior violate the above assumptions. Moreover, current state-of-the-art deep learning models typically use millions of learned parameters and can only run in near real time with low-resolution video, which constrains the visual discrimination of the targeted objects in the scene. Possible solutions to these challenges include the use of nonlinear motion models (73) and the development of compact (74) or compressed (75) deep learning models.
Solving the task of tracking individual insects could open the doors for a new individual-based ecology with profound impacts in such research fields as population, behavioral, and thermal ecology as well as conservation biology. Moreover, considering the recent development in low-cost powerful graphical processing units and dedicated artificial intelligence processors suitable for autonomous and embedded systems (e.g., NVIDIA Jetson Nano, Google Coral Edge TPU, and the Intel AI USB stick), it may soon become feasible to detect, track, and decode behavior of insects in real time and report information back to the user.
Detecting Species Interactions.
Species interactions are critical for the functioning of ecosystems, yet as they are ephemeral and fast, the consequences of a disruption for ecological function are hard to quantify (76). High-temporal resolution image-based monitoring of consumers and resources can allow for a unique quantification of species interactions (77). For instance, insects visiting flowers, defoliation by herbivores, and predation events can be continuously recorded across entire growing seasons with fixed position cameras. To detect such interactions, image recording should be collected at the scales where individuals interact (i.e., by observing interacting individuals at intervals of seconds to minutes), yet they should ideally extend over seasonal and/or multiannual periods (78). Our preliminary results have demonstrated an exciting potential to record plant–insect interactions using time-lapse cameras and deep learning models (28) (Fig. 1).
Taxonomic Identification.
Taxonomic identification can be approached as a deep learning classification problem. Deep learning-based classification accuracies for image-based insect identification of specimens are approaching the accuracy of human experts (79–81). Applications of gradient-weighted class activation mapping can even visualize morphologically important features for CNN classification (81). Classification accuracy is generally much lower when the insects are recorded live in their natural environments (82, 83), but when class confidence is low at the species level, it may still be possible to confidently classify insects to a coarser taxonomic resolution (84). In recent years, impressive results have been obtained by CNNs (85). They can classify huge image datasets, such as the 1,000-class ImageNet dataset, at high accuracy and speed (86). Even with images of >10,000 species of plants, classification accuracy of the best CNNs was close to that of botanical experts. Currently, such performance of CNNs can only be achieved with very large amounts of training data (87), but further improvements are likely, given recent promising results in distributed training of deep neural networks (88) and federated learning (89, 90).
It is common for ecological communities to contain a large fraction of relatively rare species. This often results in highly imbalanced datasets, and the number of specimens representing the rarest species could be insufficient for training neural networks (83, 84). As such, advancing the development of algorithms and approaches for improved identification of rare classes is a key challenge for deep learning-based taxonomic identification (25). Solutions to this challenge could be inspired by class resampling and cost-sensitive training (91) or by multiset feature learning (92). Class resampling aims at balancing the classes by undersampling the larger classes and/or oversampling the smaller classes, while cost-sensitive training assigns a higher loss for errors on the smaller classes. In multiset feature learning, the larger classes are split into smaller subsets, which are combined with the smaller classes to form separate training sets. These methods are all used to learn features that can more robustly distinguish the smaller classes. Species identification performance can vary widely, ranging from species that are correctly identified in most cases to species that are generally difficult to identify (93). Typically, the amount of training data is a key element for successful identification, although recent analyses of images of ∼65,000 specimens in the carabid beetle collection at the Natural History Museum London suggest that imbalances in identification performance are not necessarily related to how well represented a species is in the training data (84). Further work is needed on large datasets to fully understand these challenges.
A related challenge is formed by those species that are completely absent from the reference database on which the deep learning models are trained. Detecting such species requires techniques developed for multiple-class novelty/anomaly detection or open set/world recognition (94, 95). A recent survey introduced various open set recognition methods with the two main approaches being discriminative and generative (96). Discriminative models are based on traditional machine learning techniques or deep neural networks with some additional mechanism to detect outliers, while the main idea of generative models is to generate either positive or negative samples for training. However, the current methods are typically applied to relatively small datasets and do not scale well with the number of classes (96). Insect datasets typically have a high number of classes and a very fine-grained distribution, where the phenotypic differences between species may be minute while intraspecific variation may be large. Such datasets are especially challenging for open set recognition methods. While it will be extremely difficult to overcome this challenge for all species using only phenotype-based identification, combining image-based deep learning and DNA barcoding techniques may help to solve the problem.
Estimating Biomass from Bulk Samples.
Deep learning models can potentially predict biomass of bulk insect samples in a laboratory setting. Legislative aquatic monitoring efforts in the United States and Europe require information about the abundance or biomass of individual taxa from bulk invertebrate samples. Using the BIODISCOVER machine, Ärje et al. (70) were able to estimate biomass variation of individual specimens of Diptera species without destroying specimens. This was achieved from geometric features of the specimen extracted from images recorded by the BIODISCOVER machine and statistically relating such values to subsequently obtained dry mass from the same specimens. To validate such approaches, it is necessary to have accurate information about the dry mass of a large selection of taxa. In the future, deep learning models may provide even more accurate estimates of biomass. Obtaining specimen-specific biomass information nondestructively from bulk samples is a high priority in routine insect monitoring since it will enable more extensive insights into insect population and community dynamics and provide better information for environmental management.
Future Directions
To unlock the full potential of deep learning methods for insect ecology and monitoring, four main challenges need to be addressed with highest priority. We describe each of these challenges below.
Validating Image-Based Taxonomic Identification.
Validation of the detection and identification of species recorded with cameras in the field poses a critical challenge for implementing deep learning tools in entomology. Often, it will not be possible to conclusively identify insects from images, and validation of image-based species classification should be done using independent data. In some cases, it is possible to substantiate claims about species identity from the known occurrence and relative abundance of a species in a particular region or habitat. Independent data can be collected by analyzing DNA traces of insects left (e.g., on flowers) (97) or by directly observing and catching insects visible to the camera. The subsequent identification of specimens can serve as validation of image-based results and can further help in production of training data for optimizing deep learning models.
Generating Training Data.
One of the main challenges with deep learning is the need for large amounts of training data, which is slow, difficult, and expensive to collect and label. Deep learning models typically require hundreds of training instances of a given species to learn to detect its occurrence against the background (83). In a laboratory setting, the collection of data can be eased by automated imaging devices, such as the BIODISCOVER described above, which allows for imaging of large numbers of insects under fixed settings. The imaging of species in situ should be done in a wide range of conditions (e.g., different backgrounds, times of day, and seasons) to avoid the model becoming biased toward specific backgrounds. Approaches to alleviate the challenge of moving from one environment to another include multitask learning (98), style transfer (99), image generation (100), or domain adaptation (101). Multitask learning aims to concurrently learn multiple different tasks (e.g., segmentation, classification, detection) by sharing information leading to better data representations and ultimately, better results. Style transfer methods try to impose properties appearing in one set of data to new data. Image generation can be used to create synthetic training images with, for example, varying backgrounds. Domain adaptation aims at tuning the parameters of a deep learning model trained on data following one distribution (source domain) to adapt so that they can provide high performance on new data following another distribution (target domain).
The motion detection sensors in wildlife cameras are typically not triggered by insects, and species typically only occur in a small fraction of time-lapse images. A key challenge is therefore to detect insects and filter out blank images from images with species of interest (102, 103). Citizen science web portals, such as https://www.zooniverse.org/, can generate data for training and validation of deep learning models, if the organisms of interests are easy to detect and identify (103). When it is difficult to obtain sufficient samples of rare insects, Zhong et al. (104) proposed to use deep learning only to detect all species of flying insects as a single class. Subsequently, the fine-grained species classification can be based on manual feature extraction and support vector machines, a machine learning technique that requires less training data than CNNs.
The issue of scarce training data can also be alleviated with new data synthesis. Data synthesis could be used specifically to augment the training set by creating artificial images of segmented individual insects that are placed randomly in scenes with different backgrounds (105). A promising alternative is to use deep learning models for generating artificial images belonging to the class of interest. The most widely used approach to date is based on generative adversarial networks (106) and has shown promising performance in computer vision problems in general, as well as in ecological problems (107).
Building Reference Databases.
Publicly available reference databases are critical for adapting deep learning tools to entomological research. Initiatives like DISSCO RI and IDigBio (https://www.idigbio.org/) are important for enabling the use of museum collections. However, to enable deep learning-based identification, individual open datasets from entomological research and monitoring are also needed (e.g., refs. 82, 93, and 108). The collation of such datasets will require dedicated projects as well as large, coordinated efforts to promote open access such as the European Open Science Cloud and the Research Data Alliance. Noncollection datasets should also use common approaches and hardware and abide best practices in metadata and data management (109–111). For instance, all of the possible metadata related to the imaging and the specimens should be saved for future analysis, and the corresponding labeling of images of specimens paired to the metadata is critical. Using multiple experts and molecular information about species identity to verify the labeling or performing subsequent validity checks through DNA barcoding will improve the data quality and the performance of the deep learning models. This can be done, for instance, by manually verifying the quality and labeling of images that are repeatedly misclassified by the deep learning methods. Standardized imaging devices such as the BIODISCOVER machine could also play a key role in building reference databases from monitoring programs (70). Training classifiers with species that are currently not encountered in a certain region but can possibly spread there later will naturally help to detect such changes when they occur. Integration of such reference databases with field monitoring methods forms an important future challenge. As a starting point, we provide a list of open-access entomological image databases (SI Appendix).
Integration of Deep Learning and DNA-Based Tools.
For processing insect community samples in the laboratory, molecular methods have gained increasing attention over the past decade, but there are still critical challenges that remain unresolved: specimens are typically destroyed, abundance cannot be accurately estimated, and key specimens cannot be identified in bulk samples. Nevertheless, DNA barcoding is now an established, powerful method to reliably assess biodiversity also in entomology (11). For insects, this works by sequencing a short fragment of the mitochondrial cytochrome-c-oxidase I subunit gene and comparing the DNA sequence with a reference database (112). Even undescribed and morphologically cryptic species can be distinguished with this approach (113), which is unlikely to be possible with deep learning. This is of great importance as morphologically similar species can have distinct ecological preferences (114), and thus, distinguishing them unambiguously is important for monitoring, ecosystem assessment, and conservation biology. However, mass sequencing-based molecular methods cannot provide precise abundance or biomass estimates and assign sequences to individual specimens (12). Therefore, an unparalleled strength lies in combining both image recognition and DNA metabarcoding approaches. When building reference collections for training insect classification models, species identity can be molecularly verified, and potential cryptic species can be separated by the DNA barcode. After image-based species identification of a whole bulk sample, all specimens can be processed via DNA metabarcoding to assess taxonomic resolution at the highest level. A further obvious advantage of linking computer vision and deep learning to DNA is that even in the absence of formal species descriptions, DNA tools can generate distinctly referenced taxonomic assignments via so-called “Barcode-Index-Numbers” (BINs) (115). These BINs provide referenced biodiversity units using the taxonomic backbone of Barcode of Life Data Systems (https://boldsystems.org) and represent a much greater diversity of even yet undescribed species (116). These units can also be directly used as part of ecosystem status assessment despite not yet having Linnean names. BINs can be used for model training. Recent studies convincingly show that with this more holistic approach, which includes cryptic and undescribed species, the predictions of environmental status as required by several legislative monitoring programs actually improve substantially (e.g., ref. 117). For cases of cryptic species with great relevance (e.g., for conservation biology), it is also possible to individually process specimens of a cryptic species complex after automated image-based assignment to further validate their identity and abundance. Combining deep learning with DNA-based approaches could deliver detailed trait information, biomass, and abundance with the best possible taxonomic resolution.
Conclusion
Deep learning is currently influencing a wide range of scientific disciplines (85) but has only just begun to benefit entomology. While there is a vast potential for deep learning of images and other data types to transform insect ecology and monitoring, applying deep learning to entomological research questions brings new technical challenges. The complexity of deep learning models and the challenges of entomological data require substantial investment in interdisciplinary efforts to unleash the potential of deep learning in entomology. However, these challenges also represent ample potential for cross-fertilization among biological and computer sciences. The benefit to entomology is not only more data but also novel kinds of data. As the deep learning tools become widely available and intuitive to use, they can transform field entomology by providing information that is currently intractable to record by human observations (18, 33, 118). Consequently, there is a bright future for entomology. Deep learning and computer vision is opening up new research niches and creates access to unforeseen scales and resolution of data that will benefit future biodiversity assessments.
The shift toward automated methods may raise concerns about the future for taxonomists, much like the debate concerned with developments in molecular species identification (119, 120). We emphasize that the expertise of taxonomists is at the heart of and critical to these developments. Initially, automated techniques will be used in the most routine-like tasks, which in turn, will allow the taxonomic experts to dedicate their focus on the specimens requiring more in-depth studies as well as the plethora of new species that need to be described and studied. To enable this, we need to consider approaches that can pinpoint samples for human expert inspection in a meaningful way [e.g., based on neural network classification confidences (79) or additional rare species detectors (121)]. As deep learning becomes more closely integrated in entomological research, the vision of real-time detection, tracking, and decoding of behavior of insects could be realized for a transformation of insect ecology and monitoring. In turn, efficient tracking of insect biodiversity trends will aid the identification of effective measures to counteract or revert biodiversity loss.
Supplementary Material
Acknowledgments
David Wagner is thanked for convening the session “Insect declines in the Anthropocene” at the Entomological Society of America Annual Meeting 2019 in St. Louis, MO, which brought the group of contributors to the special feature together. Valuable inputs to the manuscript from David Wagner, Matthew L. Forister, and four anonymous reviewers are greatly appreciated. T.T.H. acknowledges funding from Villum Foundation Grant 17523 and Independent Research Fund Denmark Grant 8021-00423B. K.M. acknowledges funding from Nordic Council of Ministers Project 18103 (SCANDNAnet). J.R. acknowledges funding from Academy of Finland Project 324475.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.L.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002545117/-/DCSupplemental.
Data Availability.
There are no data underlying this work.
References
- 1.Ceballos G., Ehrlich P. R., Dirzo R., Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proc. Natl. Acad. Sci. U.S.A. 114, E6089–E6096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dornelas M., et al. , BioTIME: A database of biodiversity time series for the Anthropocene. Glob. Ecol. Biogeogr. 27, 760–786 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blowes S. A., et al. , The geography of biodiversity change in marine and terrestrial assemblages. Science 366, 339–345 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Montgomery G. A., et al. , Is the insect apocalypse upon us? How to find out. Biol. Conserv. 241, 108327 (2020). [Google Scholar]
- 5.Hallmann C. A., et al. , More than 75 percent decline over 27 years in total flying insect biomass in protected areas. PLoS One 12, e0185809 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seibold S., et al. , Arthropod decline in grasslands and forests is associated with landscape-level drivers. Nature 574, 671–674 (2019). [DOI] [PubMed] [Google Scholar]
- 7.van Klink R., et al. , Meta-analysis reveals declines in terrestrial but increases in freshwater insect abundances. Science 368, 417–420 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Wagner D. L., Insect declines in the Anthropocene. Annu. Rev. Entomol. 65, 457–480 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Pawar S., Taxonomic chauvinism and the methodologically challenged. Bioscience 53, 861–864 (2003). [Google Scholar]
- 10.Braukmann T. W. A., et al. , Metabarcoding a diverse arthropod mock community. Mol. Ecol. Resour. 19, 711–727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elbrecht V., et al. , Validation of COI metabarcoding primers for terrestrial arthropods. PeerJ 7, e7745 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elbrecht V., Leese F., Can DNA-based ecosystem assessments quantify species abundance? Testing primer bias and biomass-sequence relationships with an innovative metabarcoding protocol. PLoS One 10, e0130324 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krehenwinkel H., et al. , Estimating and mitigating amplification bias in qualitative and quantitative arthropod metabarcoding. Sci. Rep. 7, 17668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yousif H., Yuan J., Kays R., He Z., Animal scanner: Software for classifying humans, animals, and empty frames in camera trap images. Ecol. Evol. 9, 1578–1589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ärje J., et al. , Human experts vs. machines in taxa recognition. Signal Process. Image Commun. 87, 115917 (2020). [Google Scholar]
- 16.Norouzzadeh M. S., et al. , Automatically identifying, counting, and describing wild animals in camera-trap images with deep learning. Proc. Natl. Acad. Sci. U.S.A. 115, E5716–E5725 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacLeod N., Benfield M., Culverhouse P., Time to automate identification. Nature 467, 154–155 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Steenweg R., et al. , Scaling-up camera traps: Monitoring the planet’s biodiversity with networks of remote sensors. Front. Ecol. Environ. 15, 26–34 (2017). [Google Scholar]
- 19.Steen R., Diel activity, frequency and visit duration of pollinators in focal plants: In situ automatic camera monitoring and data processing. Methods Ecol. Evol. 8, 203–213 (2017). [Google Scholar]
- 20.Pegoraro L., Hidalgo O., Leitch I. J., Pellicer J., Barlow S. E., Automated video monitoring of insect pollinators in the field. Emerg. Top. Life Sci. 4, 87–97 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Wäldchen J., Mäder P., Machine learning for image based species identification. Methods Ecol. Evol. 9, 2216–2225 (2018). [Google Scholar]
- 22.Piechaud N., Hunt C., Culverhouse P. F., Foster N. L., Howell K. L., Automated identification of benthic epifauna with computer vision. Mar. Ecol. Prog. Ser. 615, 15–30 (2019). [Google Scholar]
- 23.Weinstein B. G., A computer vision for animal ecology. J. Anim. Ecol. 87, 533–545 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Christin S., Hervet É., Lecomte N., Applications for deep learning in ecology. Methods Ecol. Evol. 10, 1632–1644 (2019). [Google Scholar]
- 25.Joly A., et al. , Overview of LifeCLEF 2019: Identification of Amazonian Plants, South & North American Birds, and Niche Prediction (Springer International Publishing, Cham, Swtizerland, 2019), pp. 387–401. [Google Scholar]
- 26.Xia D., Chen P., Wang B., Zhang J., Xie C., Insect detection and classification based on an improved convolutional neural network. Sensors (Basel) 18, 4169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bzdok D., Altman N., Krzywinski M., Statistics versus machine learning. Nat. Methods 15, 233–234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran D. T., Høye T. T., Gabbouj M., Iosifidis A., “Automatic flower and visitor detection system” in 2018 26th European Signal Processing Conference (Eusipco) (IEEE, 2018), pp. 405–409. [Google Scholar]
- 29.Collett R. A., Fisher D. O., Time-lapse camera trapping as an alternative to pitfall trapping for estimating activity of leaf litter arthropods. Ecol. Evol. 7, 7527–7533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruczyński I., Hałat Z., Zegarek M., Borowik T., Dechmann D. K. N., Camera transects as a method to monitor high temporal and spatial ephemerality of flying nocturnal insects. Methods Ecol. Evol. 11, 294–302 (2020). [Google Scholar]
- 31.Barlow S. E., O’Neill M. A., Technological advances in field studies of pollinator ecology and the future of e-ecology. Curr. Opin. Insect Sci. 38, 15–25 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Cardoso P., Erwin T. L., Borges P. A. V., New T. R., The seven impediments in invertebrate conservation and how to overcome them. Biol. Conserv. 144, 2647–2655 (2011). [Google Scholar]
- 33.Hortal J., et al. , Seven shortfalls that beset large-scale knowledge of biodiversity. Annu. Rev. Ecol. Evol. Syst. 46, 523–549 (2015). [Google Scholar]
- 34.Potamitis I., Eliopoulos P., Rigakis I., Automated remote insect surveillance at a global scale and the internet of things. Robotics 6, 19 (2017). [Google Scholar]
- 35.Potamitis I., Rigakis I., Vidakis N., Petousis M., Weber M., Affordable bimodal optical sensors to spread the use of automated insect monitoring. J. Sens. 2018, 3949415 (2018). [Google Scholar]
- 36.Rustia D. J. A., Chao J.-J., Chung J.-Y., Lin T.-T., “An online unsupervised deep learning approach for an automated pest insect monitoring system” in 2019 ASABE Annual International Meeting (ASABE, St. Joseph, MI, 2019), pp. 1–13. [Google Scholar]
- 37.Sun Y., et al. , Automatic in-trap pest detection using deep learning for pheromone-based Dendroctonus valens monitoring. Biosyst. Eng. 176, 140–150 (2018). [Google Scholar]
- 38.Poland T. M., Rassati D., Improved biosecurity surveillance of non-native forest insects: A review of current methods. J. Pest. Sci. 92, 37–49 (2019). [Google Scholar]
- 39.Santos D. A. A., Teixeira L. E., Alberti A. M., Furtado V., Rodrigues J. J. P. C., “Sensitivity and noise evaluation of an optoelectronic sensor for mosquitoes monitoring” in 2018 3rd International Conference on Smart and Sustainable Technologies (SpliTech, Split, Croatia, 2018), pp. 1–5. [Google Scholar]
- 40.Park J., Kim D. I., Choi B., Kang W., Kwon H. W., Classification and morphological analysis of vector mosquitoes using deep convolutional neural networks. Sci. Rep. 10, 1012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalamatianos R., Karydis I., Doukakis D., Avlonitis M., DIRT: The dacus image recognition toolkit. J. Imaging 4, 129 (2018). [Google Scholar]
- 42.Ding W., Taylor G., Automatic moth detection from trap images for pest management. Comput. Electron. Agric. 123, 17–28 (2016). [Google Scholar]
- 43.Mayo M., Watson A. T., Automatic species identification of live moths. Knowl. Base. Syst. 20, 195–202 (2007). [Google Scholar]
- 44.Wang J., Lin C., Ji L., Liang A., A new automatic identification system of insect images at the order level. Knowl. Base. Syst. 33, 102–110 (2012). [Google Scholar]
- 45.Høye T. T., Mann H. M. R., Bjerge K., “Camera-based monitoring of insects on green roofs” (Rep. no. 371, Aarhus University DCE–National Centre for Environment and Energy, Aarhus, Denmark, 2020).
- 46.Bjerge K., Sepstrup M. V., Nielsen J. B., Helsing F., Høye T. T., An automated light trap to monitor moths (Lepidoptera) using computer vision-based tracking and deep learning. bioRxiv:10.1101/2020.03.18.996447 (31 October 2020). [DOI] [PMC free article] [PubMed]
- 47.Chapman J. W., Drake V. A., Reynolds D. R., Recent insights from radar studies of insect flight. Annu. Rev. Entomol. 56, 337–356 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Hueppop O., et al. , Perspectives and challenges for the use of radar in biological conservation. Ecography 42, 912–930 (2019). [Google Scholar]
- 49.Wotton K. R., et al. , Mass seasonal migrations of hoverflies provide extensive pollination and crop protection services. Curr. Biol. 29, 2167–2173.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Chapman J. W., Smith A. D., Woiwod I. P., Reynolds D. R., Riley J. R., Development of vertical-looking radar technology for monitoring insect migration. Comput. Electron. Agric. 35, 95–110 (2002). [Google Scholar]
- 51.Chapman J. W., Reynolds D. R., Smith A. D., Migratory and foraging movements in beneficial insects: A review of radar monitoring and tracking methods. Int. J. Pest Manage. 50, 225–232 (2004). [Google Scholar]
- 52.Chapman J. W., et al. , High-altitude migration of the diamondback moth Plutella xylostella to the UK: A study using radar, aerial netting, and ground trapping. Ecol. Entomol. 27, 641–650 (2002). [Google Scholar]
- 53.Kissling W. D., Pattemore D. E., Hagen M., Challenges and prospects in the telemetry of insects. Biol. Rev. Camb. Philos. Soc. 89, 511–530 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Maggiora R., Saccani M., Milanesio D., Porporato M., An innovative harmonic radar to track flying insects: The case of Vespa velutina. Sci. Rep. 9, 11964 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu G., et al. , Mass seasonal bioflows of high-flying insect migrants. Science 354, 1584–1587 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Stepanian P. M., et al. , Declines in an abundant aquatic insect, the burrowing mayfly, across major North American waterways. Proc. Natl. Acad. Sci. U.S.A. 117, 2987–2992 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeliazkov A., et al. , Large-scale semi-automated acoustic monitoring allows to detect temporal decline of bush-crickets. Glob. Ecol. Conserv. 6, 208–218 (2016). [Google Scholar]
- 58.Kiskin I., et al. , Bioacoustic detection with wavelet-conditioned convolutional neural networks. Neural Comput. Appl. 32, 915–927 (2020). [Google Scholar]
- 59.Chesmore E. D., Ohya E., Automated identification of field-recorded songs of four British grasshoppers using bioacoustic signal recognition. Bull. Entomol. Res. 94, 319–330 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Kawakita S., Ichikawa K., Automated classification of bees and hornet using acoustic analysis of their flight sounds. Apidologie (Celle) 50, 71–79 (2019). [Google Scholar]
- 61.Chen Y., Why A., Batista G., Mafra-Neto A., Keogh E., Flying insect detection and classification with inexpensive sensors. J. Vis. Exp. 27, e52111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balla E., et al. , An opto-electronic sensor-ring to detect arthropods of significantly different body sizes. Sensors (Basel) 20, 982 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dombos M., et al. , EDAPHOLOG monitoring system: Automatic, real-time detection of soil microarthropods. Methods Ecol. Evol. 8, 313–321 (2017). [Google Scholar]
- 64.Hedrick B. P., et al. , Digitization and the future of natural history collections. Bioscience 70, 243–251 (2020). [Google Scholar]
- 65.Blagoderov V., Kitching I. J., Livermore L., Simonsen T. J., Smith V. S., No specimen left behind: Industrial scale digitization of natural history collections. ZooKeys, 133–146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ströbel B., Schmelzle S., Blüthgen N., Heethoff M., An automated device for the digitization and 3D modelling of insects, combining extended-depth-of-field and all-side multi-view imaging. ZooKeys 759, 1–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Short A. E. Z., Dikow T., Moreau C. S., Entomological collections in the age of big data. Annu. Rev. Entomol. 63, 513–530 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Meineke E. K., Davies T. J., Museum specimens provide novel insights into changing plant-herbivore interactions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20170393 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meineke E. K., Tomasi C., Yuan S., Pryer K. M., Applying machine learning to investigate long-term insect-plant interactions preserved on digitized herbarium specimens. Appl. Plant Sci. 8, e11369 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ärje J., et al. , Automatic image-based identication and biomass estimation of invertebrates. Methods Ecol. Evol. 11, 922–931 (2020). [Google Scholar]
- 71.Wang Q., Zhang L., Bertinetto L., Hu W., Torr P. H. S., “Fast online object tracking and segmentation: A unifying approach” in IEEE Conference on Computer Vision and Pattern Recognition (IEEE, 2019), pp. 1328–1338.
- 72.Luo W., Zhao X., Kim T.-K., Multiple object tracking: A review. arXiv:1409.7618 (26 September 2014).
- 73.Yang B., Nevatia R., “Multi-target tracking by online learning of non-linear motion patterns and robust appearance models” in 2012 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2012), pp. 1918–1925. [Google Scholar]
- 74.Tran D. T., Kiranyaz S., Gabbouj M., Iosifidis A., Heterogeneous multilayer generalized operational perceptron. IEEE Trans. Neural Netw. Learn. Syst. 31, 710–724 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Tran D. T., Iosifidis A., Gabbouj M., Improving efficiency in convolutional neural networks with multilinear filters. Neural Netw. 105, 328–339 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Valiente-Banuet A., et al. , Beyond species loss: The extinction of ecological interactions in a changing world. Funct. Ecol. 29, 299–307 (2015). [Google Scholar]
- 77.Hamel S., et al. , Towards good practice guidance in using camera-traps in ecology: Influence of sampling design on validity of ecological inferences. Methods Ecol. Evol. 4, 105–113 (2013). [Google Scholar]
- 78.Estes L., et al. , The spatial and temporal domains of modern ecology. Nat. Ecol. Evol. 2, 819–826 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Raitoharju J., Meissner K., “On confidences and their use in (semi-)automatic multi-image taxa identification” in 2019 IEEE Symposium Series on Computational Intelligence (SSCI) (IEEE, 2019), pp. 1338–1343. [Google Scholar]
- 80.Valan M., Makonyi K., Maki A., Vondráček D., Ronquist F., Automated taxonomic identification of insects with expert-level accuracy using effective feature transfer from convolutional networks. Syst. Biol. 68, 876–895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Milošević D., et al. , Application of deep learning in aquatic bioassessment: Towards automated identification of non-biting midges. Sci. Total Environ. 711, 135160 (2020). [DOI] [PubMed] [Google Scholar]
- 82.Wu X., Zhan C., Lai Y., Cheng M., Yang J., “IP102: A large-scale benchmark dataset for insect pest recognition” in 2019 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2019), pp. 8779–8788. [Google Scholar]
- 83.Horn G. V., et al. , The iNaturalist challenge 2017 dataset. arXiv:1707.06642 (20 July 2017).
- 84.Hansen O. L. P., et al. , Species-level image classification with convolutional neural network enables insect identification from habitus images. Ecol. Evol. 10, 737–747 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.LeCun Y., Bengio Y., Hinton G., Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 86.He K., Zhang X., Ren S., Sun J., “Delving deep into rectifiers: Surpassing human-level performance on ImageNet classification” in 2015 IEEE International Conference on Computer Vision (ICCV) (IEEE, 2015), pp. 1026–1034. [Google Scholar]
- 87.Joly A., et al. , “Overview of LifeCLEF 2018: A large-scale evaluation of species identification and recommendation algorithms in the Era of AI” in Experimental IR Meets Multilinguality, Multimodality, and Interaction, Bellot P., et al., Eds. (Springer International Publishing, Cham, Switzerland, 2018), pp. 247–266. [Google Scholar]
- 88.Dean J., et al. , “Large scale distributed deep networks” in Proceedings of the 25th International Conference on Neural Information Processing Systems–Volume 1, Pereira F., Burges C. J. C., Bottou L., Weinberger K. Q., Eds. (Curran Associates Inc., Lake Tahoe, NV, 2012), pp. 1223–1231. [Google Scholar]
- 89.McMahan H. B., Moore E., Ramage D., Hampson S., Arcas B. A. y., “Communication-efficient learning of deep networks from decentralized data” in Proceedings of the 20th International Conference on Artificial Intelligence and Statistics (PMLR, 2016), vol. 54, pp. 1273–1282. [Google Scholar]
- 90.Bonawitz K., et al. , Towards federated learning at scale: System design. arXiv:1902.01046 (4 February 2019).
- 91.Huang C., Li Y. N., Loy C. C., Tang X. O., “Learning deep representation for imbalanced classification” in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2016), pp. 5375–5384. [Google Scholar]
- 92.Jing X. Y., et al. , Multiset feature learning for highly imbalanced data classification. IEEE Trans. Pattern Anal. Mach. Intell., 10.1109/TPAMI.2019.2929166 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Raitoharju J., et al. , Benchmark database for fine-grained image classification of benthic macroinvertebrates. Image Vis. Comput. 78, 73–83 (2018). [Google Scholar]
- 94.Turkoz M., Kim S., Son Y., Jeong M. K., Elsayed E. A., Generalized support vector data description for anomaly detection. Pattern Recognit. 100, 107119 (2020). [Google Scholar]
- 95.Perera P., Patel V. M., “Deep transfer learning for multiple class novelty detection” in 2019 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2019), pp. 11536–11544. [Google Scholar]
- 96.Geng C., Huang S. J., Chen S., Recent advances in open set recognition: A survey. IEEE Trans. Pattern Anal. Mach. Intell., 10.1109/TPAMI.2020.2981604 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Thomsen P. F., Sigsgaard E. E., Environmental DNA metabarcoding of wild flowers reveals diverse communities of terrestrial arthropods. Ecol. Evol. 9, 1665–1679 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kendall A., Gal Y., Cipolla R., “Multi-task learning using uncertainty to weigh losses for scene geometry and semantics” in 2018 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2018), pp. 7482–7491. [Google Scholar]
- 99.Gatys L. A., Ecker A. S., Bethge M., “Image style transfer using convolutional neural networks” in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2016), pp. 2414–2423. [Google Scholar]
- 100.Bao J. M., Chen D., Wen F., Li H. Q., Hua G., “CVAE-GAN: Fine-grained image generation through asymmetric training” in 2017 IEEE International Conference on Computer Vision (ICCV) (IEEE, 2017), pp. 2764–2773. [Google Scholar]
- 101.Tzeng E., Hoffman J., Saenko K., Darrell T., “Adversarial discriminative domain adaptation” in 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE, 2017), pp. 2962–2971. [Google Scholar]
- 102.Glover-Kapfer P., Soto-Navarro C. A., Wearn O. R., Camera-trapping version 3.0: Current constraints and future priorities for development. Remote Sens. Ecol. Conserv. 5, 209–223 (2019). [Google Scholar]
- 103.Willi M., et al. , Identifying animal species in camera trap images using deep learning and citizen science. Methods Ecol. Evol. 10, 80–91 (2019). [Google Scholar]
- 104.Zhong Y., Gao J., Lei Q., Zhou Y., A vision-based counting and recognition system for flying insects in intelligent agriculture. Sensors (Basel) 18, 1489 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Inoue H., Data augmentation by pairing samples for images classification. arXiv:1801.02929 (9 January 2018).
- 106.Goodfellow I. J., et al. , Generative adversarial networks. arXiv:1406.2661 (10 June 2014).
- 107.Lu C.-Y., Arcega Rustia D. J., Lin T.-T., Generative adversarial network based image augmentation for insect pest classification enhancement. IFAC-PapersOnLine 52, 1–5 (2019). [Google Scholar]
- 108.Martineau M., et al. , A survey on image-based insect classification. Pattern Recognit. 65, 273–284 (2017). [Google Scholar]
- 109.Forrester T., et al. , An open standard for camera trap data. Biodivers. Data J. 4, e10197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scotson L., et al. , Best practices and software for the management and sharing of camera trap data for small and large scales studies. Remote Sens. Ecol. Conserv. 3, 158–172 (2017). [Google Scholar]
- 111.Nieva de la Hidalga A., van Walsun M., Rosin P., Sun X., Wijers A., Data from “Quality management methodologies for digitisation operations.” Zenodo. 10.5281/zenodo.3469520. Accessed 19 November 2020. [DOI]
- 112.Ratnasingham S., Hebert P. D. N., bold: The Barcode of Life Data System ( http://www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hajibabaei M., Janzen D. H., Burns J. M., Hallwachs W., Hebert P. D. N., DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl. Acad. Sci. U.S.A. 103, 968–971 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Macher J. N., et al. , Multiple-stressor effects on stream invertebrates: DNA barcoding reveals contrasting responses of cryptic mayfly species. Ecol. Indic. 61, 159–169 (2016). [Google Scholar]
- 115.Ratnasingham S., Hebert P. D. N., A DNA-based registry for all animal species: The barcode index number (BIN) system. PLoS One 8, e66213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hebert P. D. N., Penton E. H., Burns J. M., Janzen D. H., Hallwachs W., Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. U.S.A. 101, 14812–14817 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cordier T., et al. , Supervised machine learning outperforms taxonomy-based environmental DNA metabarcoding applied to biomonitoring. Mol. Ecol. Resour. 18, 1381–1391 (2018). [DOI] [PubMed] [Google Scholar]
- 118.Burton A. C., et al. , REVIEW: Wildlife camera trapping: A review and recommendations for linking surveys to ecological processes. J. Appl. Ecol. 52, 675–685 (2015). [Google Scholar]
- 119.Kelly M. G., Schneider S. C., King L., Customs, habits, and traditions: The role of nonscientific factors in the development of ecological assessment methods. Wiley Interdisciplinary Rev.-Water 2, 159–165 (2015). [Google Scholar]
- 120.Leese F., et al. , Why we need sustainable networks bridging countries, disciplines, cultures and generations for aquatic biomonitoring 2.0: A perspective derived from the DNAqua-Net COST action. Next Gener. Biomonitoring. P&T 1, 63–99 (2018). [Google Scholar]
- 121.Sohrab F., Raitoharju J., Boosting rare benthic macroinvertebrates taxa identification with one-class classification. arXiv:2002.10420 (12 February 2020).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
There are no data underlying this work.