Significance
Cryptococcus neoformans is an opportunistic human pathogen, causing cryptococcal meningitis in immunocompromised individuals, with a mortality rate of more than 50%. We previously found that the Prp8 intein was a viable drug target for C. neoformans. In an effort to find new therapies, we developed screening assays and identified a small molecule and its fluoride derivative as potent Prp8 intein-splicing inhibitors. These inhibitors bind covalently to an intein active-site residue and inhibit protein splicing, leading to nonfunctional Prp8, which is essential for fungal growth. These molecules inhibited growth of C. neoformans and showed synergistic or additive effects with Food and Drug Administration-approved antimycotics. Overall, the identification of these intein-splicing inhibitors opens up prospects for a new class of antifungals.
Keywords: small-molecule inhibitor, antifungal, Cryptococcus, Prp8 intein, protein splicing
Abstract
Self-splicing proteins, called inteins, are present in many human pathogens, including the emerging fungal threats Cryptococcus neoformans (Cne) and Cryptococcus gattii (Cga), the causative agents of cryptococcosis. Inhibition of protein splicing in Cryptococcus sp. interferes with activity of the only intein-containing protein, Prp8, an essential intron splicing factor. Here, we screened a small-molecule library to find addititonal, potent inhibitors of the Cne Prp8 intein using a split-GFP splicing assay. This revealed the compound 6G-318S, with IC50 values in the low micromolar range in the split-GFP assay and in a complementary split-luciferase system. A fluoride derivative of the compound 6G-318S displayed improved cytotoxicity in human lung carcinoma cells, although there was a slight reduction in the inhibition of splicing. 6G-318S and its derivative inhibited splicing of the Cne Prp8 intein in vivo in Escherichia coli and in C. neoformans. Moreover, the compounds repressed growth of WT C. neoformans and C. gattii. In contrast, the inhibitors were less potent at inhibiting growth of the inteinless Candida albicans. Drug resistance was observed when the Prp8 intein was overexpressed in C. neoformans, indicating specificity of this molecule toward the target. No off-target activity was observed, such as inhibition of serine/cysteine proteases. The inhibitors bound covalently to the Prp8 intein and binding was reduced when the active-site residue Cys1 was mutated. 6G-318S showed a synergistic effect with amphotericin B and additive to indifferent effects with a few other clinically used antimycotics. Overall, the identification of these small-molecule intein-splicing inhibitors opens up prospects for a new class of antifungals.
Many microbial pathogens contain self-splicing elements called inteins, which are internal proteins that self-excise from their intein-hosting proteins and catalyze ligation of the flanking sequences (exteins) with a natural peptide bond (1–4). Overall, more than 1,700 inteins have been identified (5). Among intein-containing deadly human pathogens is Mycobacterium tuberculosis, which has inteins in three critical genes, involved in replication (dnaB), iron-sulfur cluster assembly (sufB), and recombination (recA). M. tuberculosis infections cause 2 million annual tuberculosis-related deaths worldwide (6). Pathogenic fungi, such as Crypotococcus neoformans (Cne), Crypotococcus gattii (Cga), and Aspergillus fumigatus also encode inteins, in the pre-mRNA processing factor 8 (Prp8) gene (7, 8). Globally, over 300 million people are affected by invasive fungal infections, with estimated deaths of over 1 million people every year (9–12). Moreover, the emergence of severely drug-resistant strains of M. tuberculosis and pathogenic fungi, plus the deadly synergistic association with HIV/AIDS, represent significant public health challenges (13–18).
Since inteins consistently interrupt highly conserved sites of intein-hosting proteins, splicing inhibition can cause a disruption of functions that are essential for the pathogen’s survival. Inteins are therefore attractive targets for drug development (7, 8, 19–21). Additionally, inteins do not occur in multicellular organisms including humans nor in unicellular organisms including bacteria normally associated with the human gut flora, making intein-inhibiting drugs highly selective for intein-containing pathogens, such as M. tuberculosis, C. neoformans, C. gattii, and A. fumigatus.
Previously we found that cisplatin, a Food and Drug Administration (FDA)-approved chemotherapeutic drug, inhibited fungal Prp8 intein splicing in vitro and reduced fungal burden in vivo (8). The action of cisplatin is by the platinum ion being coordinated by the catalytic Cys1 of the inetin, thereby inhibiting the first step of splicing and subsequent branched intermediate formation and extein ligation. However, the high cytotoxicity of cisplatin and its derivatives (22–24) may limit their use in immunocompromised patients. In the present study, we performed a pilot screening of a small-molecule library and found a compound and its derivative that impede fungal Prp8 intein splicing in a dose-dependent manner. In addition, these molecules inhibited the Prp8 intein-containing fungi C. neoformans and C. gattii, but not the yeast Candida albicans, a major human pathogen that does not encode a Prp8 intein. Furthermore, C. neoformans treated with the small molecules led to accumulation of unspliced Prp8 precursor. The potency of these inhibitors is better than or comparable to the current frontline antifungal drugs. Mechanistic studies indicated that the small molecules inhibited Prp8 intein splicing by covalently binding to the Prp8 intein active-site residue Cys1, the nucleophile that initiates the protein splicing reaction.
Results
Development of Split-GFP–Based Prp8 Intein-Splicing Assay.
Previously, we developed a high throughput screening (HTS) assay based on split Renilla luciferase (RLuc-Prp8) (8). Although the RLuc-Prp8 assay is highly robust, it can potentially identify luciferase inhibitors. Although it is not surprising that compounds often inhibit multiple targets, some compounds may yield false positives by inhibiting luciferase instead of Prp8 intein splicing. One way to eliminate these false positives is to screen positive hits against luciferase itself. However, this method may potentially remove true inhibitors that display dual inhibition activities toward both Prp8 intein splicing and luciferase.
To better address this concern, we developed an intein-splicing assay with a split-GFP, using methods similar to those described previously (21, 25). We inserted the Prp8 intein between GFP residues G128 and D129 using a mega-PCR mutagenesis strategy (8, 21, 26). The GFP–Prp8 intein fusion protein (GFP–Prp8i) was expressed as inclusion bodies, refolded at pH 9.0 to prevent autosplicing, and purified using affinity and size-exclusion chromatography (8, 21).
The intein-splicing assay was initiated by diluting the purified, concentrated GFP–Prp8i into assay buffer at pH 8 in a 96-well plate. Reducing agents such as Tris (2-carboxyethyl) phosphine (TCEP) are required to reduce the nucleohilic cysteine (Cys1) and thereby trigger in vitro intein splicing (8). As expected, purified GFP–Prp8i did not generate fluorescence in the absence of TCEP (Fig. 1A). In contrast, addition of TCEP led to a significant increase in fluorescence, presumably because TCEP triggers Prp8 intein splicing to generate reconstituted full-length GFP (Fig. 1A) (buffer vs. TCEP). To rule out the possibility that TCEP may affect native GFP fluorescence, we measured WT GFP fluorescence with or without TCEP incubation. As shown in Fig. 1A, TCEP did not have any effect on the native GFP fluorescence.
Fig. 1.
Validation of the split-GFP–Prp8 intein-splicing assay. (A) The Prp8 intein-splicing assay based on split-GFP. GFP (200 nM) and GFP–Prp8i (200 nM) were used with DMF or cisplatin (40 µM) for fluorescence detection. n = 8. ***P < 0.001. (B) Dose–response fitting of inhibition of splicing of the GFP–Prp8i by cisplatin. GFP–Prp8i (200 nM) was used. Cisplatin was in twofold serial dilutions with concentrations ranging from 100 µM (30 µg/mL) to 0.78 µM (0.23 µg/mL). n = 3.
The Prp8 intein insertion site between G128 and D129 in GFP is not in a conserved region essential for GFP fluorophore formation. To eliminate the possibility that the observed fluorescence increase is due to unspliced GFP–Prp8i precursor refolding and maturation, we performed SDS/PAGE analysis of GFP–Prp8i in the presence and absence of TCEP (SI Appendix, Fig. S1A). The GFP–Prp8i precursor did not self-splice after overnight incubation in the absence of TCEP. Addition of TCEP triggered GFP–Prp8i splicing, resulting in an additional band near 25 KDa, which is consistent with the molecular mass of native full-length GFP. The GFP–Prp8i precursor did not become fluorescent regardless of TCEP addition (SI Appendix, Fig. S1 A and B). In contrast, the 25-KDa protein showed significant fluorescence upon TCEP addition, indicating that the fluorescence increase observed in the 96-well plate resulted from Prp8 intein splicing.
Application of Split-GFP Assay for Small-Molecule Screening.
To further validate the split-GFP assay, we used cisplatin as a control inhibitor (8). We first showed that addition of cisplatin did not affect native GFP fluorescence, (Fig. 1A). Notably, the fluorescence increase of the GFP–Prp8i fusion was significantly reduced by cisplatin (Fig. 1A). The assay had a Z′ score of 0.89, indicating that it is suitable for HTS (Z′ > 0.5) (27) (Fig. 1A). We next investigated dose-dependent inhibition of Prp8 intein splicing by cisplatin. Our data showed that cisplatin inhibited Prp8 intein splicing in a dose-dependent manner, with an IC50 of 6.7 µM (Fig. 1B). This IC50 value is comparable to that determined using the RLuc-Prp8 assay (2.5 µM) (8).
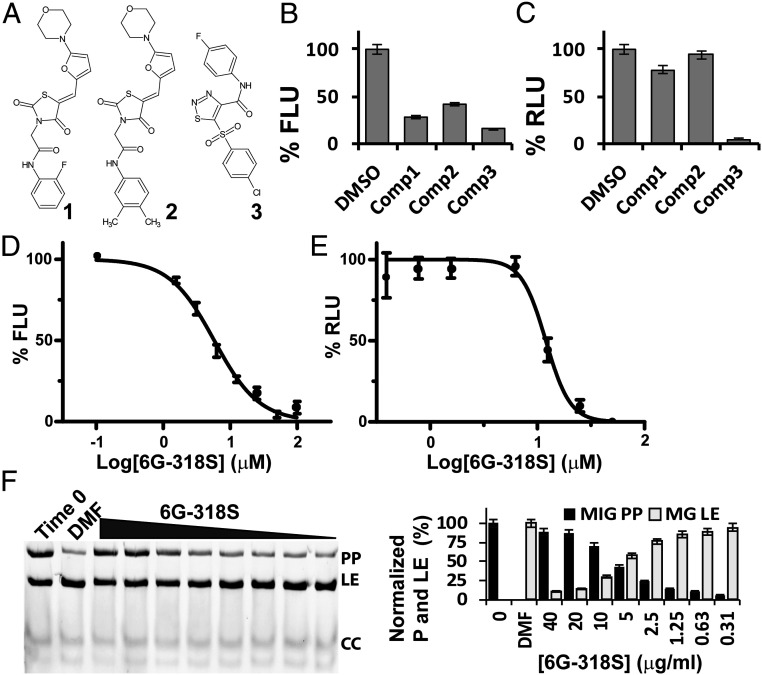
We next carried out a pilot screen of small-molecule inhibitors of Prp8 intein splicing. The compound library (Timtec) contained about 240 putative protease inhibitors (28) and 60 compounds from an in-house collection. At 20 µM, three compounds showed greater than 50% inhibition of the GFP–Prp8i signal (Fig. 2 A and B). The three compounds were further tested using the RLuc-Prp8 intein-splicing assay (8). Our results showed that only compound 3, 5-[(4-chlorophenyl)sulfonyl]-N-(4-fluorophenyl)-1,2,3-thiadiazole-4-carboxamide (commercial name 6G-318S) suppressed more than 50% of RLuc-Prp8 intein-splicing activity at 20 µM (Fig. 2C).
Fig. 2.
Pilot screening reveals a strong intein inhibitor. (A) Chemical structures of hit compounds. (B) Inhibition of split-GFP–Prp8 splicing by hit compounds at 20-µM concentration. (C) Inhibition of the split RLuc–Prp8 intein splicing by hit compounds at 20 µM concentration. (D and E) Dose-dependent inhibition of the split-GFP–Prp8i (200 nM) and RLuc–Prp8 (2 nM) intein splicing by 6G-318S, which was in twofold serial dilutions with concentrations ranging from 100 µM to 0.78 µM. n = 3. (F) Dose-dependent inhibition of MIG–Prp8 splicing by 6G-318S. (Left) SDS/PAGE analysis of each sample; (Right) relative percentage reduction of the MIG precursor upon treatment, compared to the starting material (T0). %P decrease is calculated as (%PT0 − %PSample)/(%PT0 − %PDMF) × 100, where %P indicates percent of the MIG precursor at time point 0 (PT0) and/or after ∼18 h of treatment with DMF (PDMF) or 6G-318S at different concentrations (PSample). n = 3.
Inhibition of Prp8 Intein Splicing by 6G-318S.
Using a concentration series of 6G-318S, our results indicated that 6G-318S inhibited Prp8 intein splicing in a dose-dependent manner in both GFP–Prp8i and RLuc-Prp8 assays. The IC50 values determined by the two assays were 5.8 µM and 11.2 µM, respectively (Fig. 2 D and E). In addition, using in-gel GFP-fluorescence, we showed that 6G-318S inhibited GFP–Prp8i splicing in a dose-dependent manner (SI Appendix, Fig. S1B).
We used an additional fluorescence reporter system to investigate inhibition of Prp8 intein splicing by 6G-318S (8). This reporter system, MIG–Prp8, comprising a fusion protein of the Prp8 intein flanked by maltose binding protein (MBP) and GFP, allowed observation of functional Prp8 intein splicing in Escherichia coli lysate (8). Incubation of the MIG–Prp8 fusion protein precursor (PP) with TCEP triggered Prp8 intein splicing, leading to reduced PP and accumulation of ligated extein (LE) product MBP-GFP (MG) (Fig. 2F, time 0 vs. solvent dimethylformamide [DMF] control). C-terminal cleavage product (GFP) could also be visualized. Using the MIG–Prp8 intein construct, we showed in vitro inhibition of Prp8 intein splicing by 6G-318S, leading to accumulation of the MIG–Prp8 precursor PP and decrease of LE product in a dose-dependent manner (Fig. 2F). Quantification of the PP and LE bands indicated a perfectly inverse relationship (Fig. 2 F, Right). Overall, the data indicated that 6G-318S inhibits Prp8 intein splicing in vitro.
Derivatives of 6G-318S and Inhibition Selectivity.
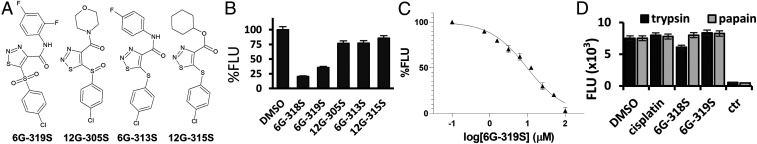
To explore 6G-318S analogs, we performed a substructure search and found four commercially available derivatives: namely 6G-319S, 12G-305S, 6G-313S, and 12G-315S (Fig. 3A). We performed the Prp8 intein-splicing inhibition assay using the GFP–Prp8i construct for these compounds at 20-µM concentration. Our results showed that none of the four compounds displayed better inhibition than 6G-318S, but that one derivative, 6G-319S, showed more than 50% inhibition of the Prp8 intein splicing (Fig. 3B). In dose–response inhibition of the Prp8 intein splicing using the GFP–Prp8i construct, 6G-319S registered an IC50 value of 9.7 µM (Fig. 3C), in the same range as 6G-318S (5.8 µM). Apparently, the aniline moiety and the phenylsulfone pharmacophore group are essential for these thiadiazole derivatives to maintain their inhibitory activities against Prp8 intein splicing. The other sulfide or sulfoxide analogs were found to be inactive.
Fig. 3.
Analogs of 6G-318S have lower inhibitory activity but are intein-specific. (A) Chemical structure of 6G-318S analogs. (B) Inhibition of the split-GFP–Prp8i splicing by the 6G-318S analogs at 20 µM. (C) Dose-dependent inhibition of the GFP–Prp8i intein splicing by 6G-319S, which was in twofold serial dilutions with concentrations ranging from 100 µM to 0.78 µM. n = 3. (D) No inhibition of the trypsin and papain protease activities by intein inhibitors. Compound concentration for cisplatin, 6G-318S, and 6G-319S was set at 400 µM. Control inhibitor AP-2HCl was at 3.5 µM. n = 3.
Intein self-splicing may resemble serine/cysteine protease activity in function by using nucleophilic residues to cleave peptide bonds. The identified inhibitors, 6G-318S and 6G-319S, were further evaluated for specificity using a counter-screening assay measuring inhibition of representative serine (trypsin) and cysteine (papain) proteases, as we described previously (29, 30). Cisplatin was also included. Using a FRET substrate Abz-RRRRSAG-nTyr developed previously (29, 30), we showed that at 400 µM, neither 6G-318S, 6G-319S, nor cisplatin significantly inhibited the protease activities of trypsin and papain (Fig. 3D). In contrast, control inhibitor antipain dihydrochloride (AP-2HCl) (31) at 3.5 µM completely abolished the protease activities (Fig. 3D). The results suggest that these compounds are specific to Prp8 intein splicing.
Inhibition of Intein Splicing and Growth of C. neoformans.
Using a broth microdilution assay (32), we evaluated the antifungal efficacy of 6G-318S and 6G-319S against the Prp8 intein-containing fungus C. neoformans H99 strain. We also used C. albicans, a fungus without the Prp8 intein, as a control. Our results demonstrated that the compounds 6G-318S and 6G-319S are potent inhibitors of C. neoformans (Table 1). The minimum inhibitory concentration (MIC) values (0.6 µg/mL and 1.3 µg/mL for 6G-318S and 6G-319S, respectively) (Table 1) were comparable to or better than those of the current first-line antifungal drugs, such as 5-fluorocytosine (5-FC), itraconazole (IZ), fluconazole (FZ), and amphotericin B (AmB) (33). In contrast, 6G-318S and 6G-319S did not inhibit growth of C. albicans (MIC >5 µg/mL) (Table 1). These results suggest that these compounds selectively inhibit the growth of Prp8 intein-containing fungi. We found the minimum fungicidal concentration (MFC) of 6G-318S to be 1.25 µg/mL for C. neoformans (SI Appendix, Fig. S2). Compounds exhibiting a MFC/MIC ratio <4 are considered fungicidal, and 6G-318S displays a ratio of 2, thus confirming that it is fungicidal (34).
Table 1.
Inhibition of pathogenic fungi by identified compounds using microdilution assay
| Compounds | MIC (µg/mL) | CC50 (µg/mL)/(µM) | TI | |
| C. neoformans | Candida albicans | A549 | Cne | |
| 6G-318S | 0.62 | 5.0 | 13.9 (35) | 22 |
| 6G-319S | 1.3 | 20.8 | 44.9 (108) | 35 |
TI, therapeutic index.
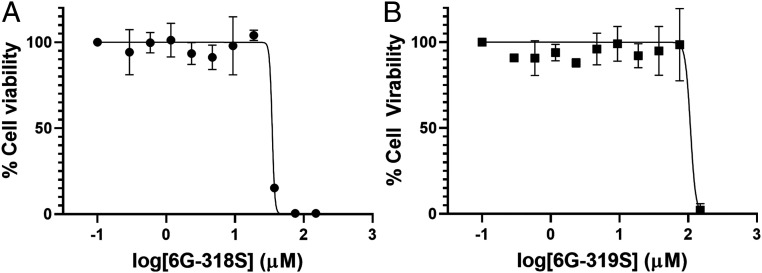
Compound 6G-319S differs from 6G-318S only by the addition of a fluoride to the phenyl ring (Figs. 2A and 3A). Both in vitro Prp8 intein splicing and in vitro fungal killing assays indicated that the introduction of a 2-F substituent on the aniline moiety (as with 6G-319S) led to a slight loss of potency (Figs. 2D and 3C and Table 1). However, this modification significantly improved its cytotoxicity profile (Fig. 4). As shown in Table 1, 6G-319S showed a 3.2-fold higher cytotoxicity value (CC50: Compound concentration required for 50% inhibition of cell viability) than 6G-318S. Values were 44.9 µg/mL (∼108 µM) for 6G-319 and 13.9 µg/mL (∼35 µM) for 6G-318S, indicating that 6G-319S is less toxic to human cells than 6G-318S. The results also demonstrated that these compounds have a good (>10) therapeutic index because the cytotoxicity CC50 values of compounds 6G-318S and 6G-319S are 22- and 35-fold, respectively, of their corresponding MIC values toward C. neoformans.
Fig. 4.
Improved cytotoxicity profile with 6G-319S. Cell viability assay with 6G-318S (A) and 6G-319S (B). A549 cells were incubated with various concentrations of the drugs and then assayed for viability at 48 h postincubation. Experimental data were fitted using a sigmoidal function. Cell viability with DMSO control was set as 100% viable. Medium alone with DMSO was set as 0% viability. n = 3.
Synergy with Existing Drugs.
To test if the Prp8 intein-splicing inhibitor 6G-318S has synergistic effects with clinically used drugs, we first performed a drug combination study using the Sensititre YeastOne Y09 AST Kit (Thermofisher) that contains nine drugs: AmB, 5-FC, IZ, FZ, anidulafungin (AND), micafungin (MF), caspofungin (CAS), posaconazole (PZ), and voriconazole (VOR). We chose 6G-318S at 0.16 μg/mL, at which concentration C. neoformans is viable. As shown in SI Appendix, Fig. S3, addition of 6G-318S did not change the MICs for AND, MF, CAS, PZ, and FZ. In contrast, 6G-318S addition reduced MICs more than fourfold for AmB and twofold for 5-FC, IZ and VOR. It is noted that combination of AmB at the lowest available concentration in the test plate with 6G-318S was sufficient to suppress C. neoformans growth (SI Appendix, Fig. S3A). Therefore, the AmB MIC reduction may be higher than fourfold when combined with 6G-318S.
To further investigate a possible synergistic effect of 6G-318S with existing drugs, we carried out the checkerboard assay (35–37) to determine the fractional inhibitory concentration (FIC) (Table 2 and SI Appendix, Fig. S3C). In the FIC experiment, 6G-318G showed a synergistic effect (FIC 0 to 0.5) with AmB with a FIC index value of 0.37 (38). The combination of 6G-318G with 5-FC or IZ had an additive effect (FIC 0.5 to 1), with FIC indices of 0.63 and 1, respectively. The combination of 6G-318G and VOR had an indifferent effect (FIC 1 to 4), with a FIC index of 2.04.
Table 2.
Effect of 6G318S in combination with known antifungals
| Well position | Compounds | MIC-alone (µg/mL)* | MIC-comb (µg/mL)* | FIC index* | Effect |
| F6 | 6G-318S | 1.000 | 0.125 | 0.37 | Synergistic |
| AmB | 0.578 | 0.144 | |||
| D6 | 6G-318S | 1.000 | 0.500 | 0.63 | Additive |
| 5-FC | 8.100 | 2.025 | |||
| D6 | 6G-318S | 1.000 | 0.500 | 1.00 | Additive |
| IZ | 0.176 | 0.088 | |||
| G3 | 6G-318S | 1.000 | 0.063 | 2.04 | Indifferent |
| Vor | 0.044 | 0.087 |
MIC and FIC index were calculated by checkerboard assay, as illustrated in SI Appendix, Fig. S3. FIC index value of ≤0.5 is considered as synergy, >0.5 to 1 is additive, >1 to 4 is indifferent.
Inhibitors Bind Covalently to the Active Site of the Prp8 Intein.
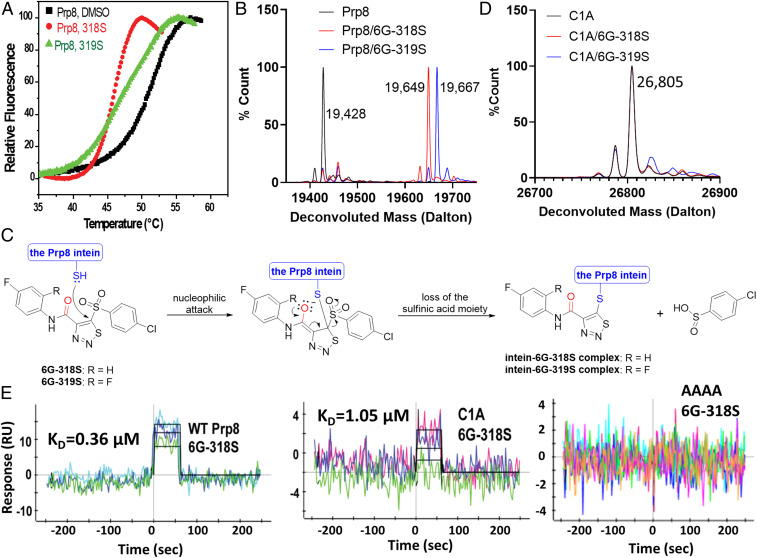
To investigate the mode of action, we first evaluated if the compounds bind to the Prp8 intein using a protein thermal shift assay (PTSA), as we described previously (26). Results of the PTSA confirmed that addition of the compounds induced large melting temperature (Tm) changes (Fig. 5A), indicating binding of the small-molecule inhibitor to the Prp8 intein. Interestingly, the binding of intein inhibitors decreased Tm of the Prp8 intein, indicating reduced protein stability. Covalent modification of active site residues by inhibitors could account for the reduced protein stability (see Fig. 5 B–D).
Fig. 5.
Direct binding of 6G-318S and 6G-319S to the Prp8 intein. (A) PTSA for binding of compounds to the Prp8 intein. ∆Tm was defined as Tm-drug − Tm-DMSO. (B) Deconvolution of MS spectra of the recombinant WT Prp8 intein (pXI version) and its complex with 6G-318S and 6G-319S. (C) Proposed mechanism of inhibition of the Prp8 intein splicing by compounds 6G-318S and 6G-319S. Chemical reaction as illustrated. (D) Deconvolution of MS spectra of the recombinant C1A Prp8 intein mutant (pXI version) and its complex with 6G-318S and 6G-319S. (E) Kinetic binding data. SPR sensograms are shown with different colors for the binding of 6G-318S at different concentrations to recombinant Cga Prp8 or mutant inteins (pXI version), which were coupled to a ProteOn GLH sensor chip. Global fitting of data to a 1:1 binding model is shown in black.
To further investigate the nature of inhibitor binding, we performed electrospray ionization–mass spectrometry (ESI-MS), using purified Cga Prp8 intein, as described previously (8). The Cga Prp8 intein only contains the intein residues without any flanking extein sequence, and is 88% identical to the Cne Prp8 intein. The Prp8 intein was incubated overnight with or without 6G-318S or 6G-319S and then subjected to ESI-MS analysis. As shown in Fig. 5B, deconvolution of the ESI-MS data revealed that untreated Prp8 intein had a molecular mass of 19,428 Da, exactly as predicted from its sequence. In contrast, the deconvoluted molecular masses for the Prp8 samples treated with 6G-318S and 6G-319S, respectively, shifted 221 Da and 239 Da, compared to that of untreated Prp8 intein, to 19,649 and 19,667, respectively. The mass differences were much smaller than the molecular masses for 6G-318S (396.98 Da) and 6G-319S (414.97 Da). These results suggest that the compounds react covalently with the Prp8 intein, possibly through the active site Cys1 thiol, and become truncated in the process.
The mass difference between the adducts of compounds 6G-318S and 6G-319S upon binding to the Prp8 intein is 18 Da, which is the exact mass of the extra fluoride atom for 6G-319S compared to 6G-318S. The results indicated that the two Prp8 intein inhibitors 6G-318S and 6G-319S followed the same chemistry to react with the Prp8 intein. Examination of the chemical structures of 6G-318S and 6G-319S suggested that the free thiol group of the catalytic Cys1 of the Prp8 intein likely attacks the carbon–carbon double bond of the thiadiazole moiety of the inhibitors, resulting in the cleavage of the bond between the phenylsulfonyl and thiadiazole ring (Fig. 5C). This would lead to a phenylcarbamoyl-thiadiazole adduct of 222.0 Da (6G-318S) or 240.0 Da (6G-319S) to form the intein–inhibitor complex, with the removal of the 4-chlorobenzenesulfinic acid fragment (176.6 Da), according to the mass spectrometry (MS) analysis. Covalent reaction of the thiadiazole ring of the adduct with the active site Cys1 led to the loss of one hydrogen, resulting in a final total mass addition of 221 Da for 6G-318S or of 239 Da for 6G-319S to the Prp8 intein, respectively.
To verify this hypothesis, we first evaluated if a Prp8 active site cysteine splicing-defective mutant (C1A) retained reactivity to the compounds. Using ESI-MS, we showed that the compounds failed to modify the C1A mutant (Fig. 5D), because no mass shifts were observed for the complexes, which is in contrast to those of the WT Prp8 intein (Fig. 5B). Moreover, liquid chromatography with tandem MS (LC-MS/MS) verified the predicted covalent modification of Cys1 of the Prp8 intein and a Cys-containing peptide by the 6G-318S fragment, both undergoing a mass shift of 221 Da, which is not affected by the choice of salt (SI Appendix, Figs. S4A and S5).
The Cga Prp8 intein used in the above experiments does not have extein residues flanking the intein due to the purification chemistry. Using an extein-containing Prp8 intein construct (8), we showed that a Prp8 intein with exteins was modified at the Cys1 position by 6G-318, in the same way as the Prp8 intein without exteins (SI Appendix, Fig. S4B).
To quantitate the molecular interaction between the splicing inhibitors and the Prp8 intein, we carried out a surface plasmon resonance (SPR) experiment to determine the binding affinity between 6G-318S and the Prp8 intein (Fig. 5 C and E). The results demonstrated that 6G-318S binds directly to the Prp8 intein with a binding affinity of 0.36 µM.
Previously, we showed that the Prp8 intein-splicing inhibitor cisplatin binds to the active site of the Prp8 intein (8). We also generated three inactive mutants involving active-site residues, C1A, H169A, and C1A/H62A/D95A/H169A (the AAAA mutant) (8). To further explore the mechanism of action, we evaluated the binding affinity of these mutants to 6G-318S. As shown (Fig. 5E and Table 3), mutation of the C-terminal active site residue H169 to alanine did not affect the binding affinity. In contrast, mutation of the N-terminal active site residue Cys1 to alanine reduced the binding affinity nearly threefold. Moreover, the AAAA mutant completely lost binding affinity to 6G-318S, indicating that the compound requires an additional intein feature for binding (Fig. 5E and Table 3). The SPR data were also consistent with the MS results showing that 6G-318S reacted with the free thiol group of the Prp8 intein.
Table 3.
Binding affinity between 6G-318S and the WT and mutant Prp8 inteins
| WT | C1A | H169A | AAAA | |
| KD (µM) | 0.36 | 1.05 | 0.23 | ND |
ND, Not detected. Values were determined by SPR (Fig. 5E).
Inhibition of Prp8 Intein Splicing in C. neoformans.
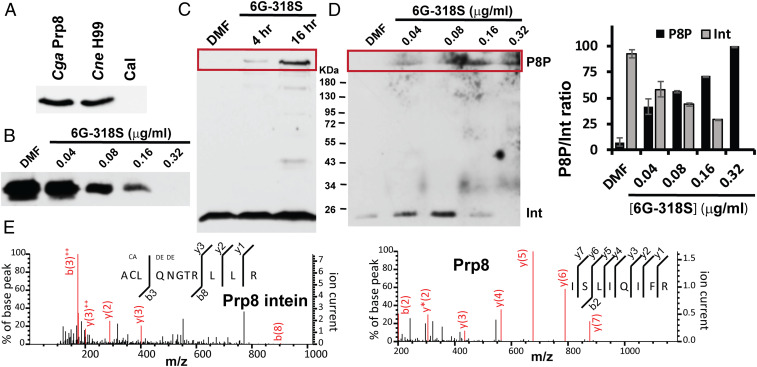
Using an anti-Prp8 intein antibody against the recombinant Prp8 intein, we performed Western blot analysis (Fig. 6). We used the recombinant Cga Prp8 intein comprising 170 amino acids as a positive control. We first confirmed that a protein of 20 KDa could be detected by immunoblot (Fig. 6A). The antibody serum could also detect intein from the cell lysate of C. neoformans H99. In contrast, no band could be detected for the cell lysate of inteinless C. albicans, implying that the antibody was specific for the Prp8 intein. Next, we performed Western blot analysis of cryptococcal samples with/without 6G-318S at different concentrations at 3 h posttreatment (Fig. 6B). We found that 6G-318S inhibited Prp8 splicing in vivo in C. neoformans, leading to a dose-dependent reduction of spliced Prp8 intein product (Fig. 6B). At 0.32 µg/mL, 6G-318S completely inhibited Prp8 splicing. The results were consistent with our fungal susceptibility studies, indicating that the MIC value for 6G-318S for the C. neoformans H99 strain was 0.62 µg/mL (Table 1).
Fig. 6.
6G-318S inhibits Prp8 intein splicing in vivo. (A) The antibody serum is specific to the Prp8 intein-containing fungi C. gattii and C. neoformans. Western blot analysis of cell lysates of C. gattii, C. neoformans, and C. albicans using a polyclonal anti-Prp8 intein serum. (B) Dose-dependent inhibition of Prp8 intein splicing in C. neoformans H99 by 6G-318S. (C) Time-course of inhibition of Prp8 intein splicing in C. neoformans H99 by 6G-318S. Western blot analysis of cell lysates of C. neoformans H99 treated with DMF or 6G-318S (0.16 μg/mL), using the polyclonal anti-Prp8 intein serum. (D) Dose-dependent accumulation of high molecular band, likely corresponding to Prp8 precursor in 6G-318S-treated C. neoformans H99 cells. (Left) Western blot analysis of cell lysates of C. neoformans H99 treated with DMF or 6G-318S at indicated concentrations, using the polyclonal anti-Prp8 intein serum; (Right) relative ratio between unspliced Prp8 protein precursor (P8P) and spliced intein (Int). n = 3. (E) MS/MS spectra obtained from the fragmentation of Prp8 precursor, including both Prp8 intein and extein peptides. Fragment ions corresponding to y- and b-ions were observed (red lines).
A light band corresponding to a protein with a size larger than 180 KDa was observed for the C. neoformans H99 cell lysate treated with 6G-318S for 4 h after Western blot with the Prp8 intein antibody serum (Fig. 6C). In contrast, this putative precursor band (∼292 KDa) was not observed for the DMF-treated control sample. Next, we performed Western blot analysis of the C. neoformans samples treated with different concentrations of 6G-318S or DMF control for 18 h. As shown in Fig. 6D, even after this long incubation period, samples treated with DMF did not show high molecular-weight protein using the anti-Prp8 intein serum in Western blot. In contrast, a dose-dependent increase of high molecular-weight protein was seen for samples treated with 6G-318S. Conversely, as the high molecular-weight protein accumulated, the spliced Prp8 intein was diminished. Overall, a clear inverse relationship was observed between the accumulation of the high molecular-weight protein and the decrease of the spliced Prp8 intein in vivo for samples treated with 6G-318S (Fig. 6 D, Right).
We suspect that the high molecular-weight protein represents unprocessed Prp8 protein precursor or branched intermediate (39), with a molecular mass of 291.9 KDa, which accumulates over time due to splicing inhibition by 6G-318S. To test this hypothesis, we excised the band and carried out MS proteomic analysis. Our MS results verified that the high molecular weight protein contained representative peptides from the Cne Prp8 precursor, including both the Prp8 intein and exteins (Fig. 6E). These results are consistent with our hypothesis that inhibitors preventing intein splicing lead to accumulation of unprocessed and nonfunctional precursor, ultimately resulting in fungal death.
Overexpression of the Prp8 Intein in C. neoformans Leads to Drug Resistance.
To further investigate the mechanism of inhibition in C. neoformans, we performed an overexpression experiment. We previously developed a system to overexpress WT and a nonsplicing AAAA Prp8 intein mutant in cryptococcal cells under the strong TEF1 promoter (8). The empty vector, WT, and the AAAA mutant plasmid were individually transformed into the C. neoformans H99 strain. We had shown that transformation of the empty vector, Prp8 WT intein and the AAAA mutant did not affect the growth of cryptococcal cells in the absence of an inhibitor (8). Using this system, we determined the MIC values of 6G-318S in these transformed cells. Our results showed that 6G-318S had identical MIC values with empty vector or AAAA mutant Prp8 intein-transformed H99 cells. In contrast, transformation of the WT Prp8 intein into H99 led to fourfold increase of MIC for 6G-318S, compared to H99 transformed with empty vector or the AAAA mutant. Importantly, our result was consistent with our hypothesis that the Prp8 intein is the intracellular target of 6G-318S.
Discussion
C. neoformans and C. gattii cause cryptococcal meningitis (CM) and pulmonary cryptococcosis, which are very difficult to treat. There are over 1 million annual cases of CM worldwide, with estimated deaths of 700,000 per year (9, 40–42). CM is seen most commonly in immunocompromised patients, such as those with HIV, severe combined immunodeficiency, or postorgan transplant status (43, 44). However, recent reports of infections caused by C. gattii in nonimmunocompromised people have raised concerns about the overall threat of the Cryptococcus species to public health (45–56).
Cryptococcal infection is usually treated by combination therapy, with administration time ranging from 12 to 14 wk to lifelong (57–62). Mainstay drugs for CM include AmB, 5-FC, triazoles (FZ, VOR, PZ, IZ), and CAS (in combination therapy only) (43, 57–59, 63, 64). These drugs kill fungi through various mechanisms, including interfering with membrane permeabilization (AmB) (65, 66), inhibition of 14α-demethylase (triazoles) (67), inhibition of DNA/RNA synthesis (5-FC) (68), and inhibition of (1→3)-β-d-glucan synthase (CAS) (69). However, CAS does not work for C. neoformans (70–73). Despite the availability of these antifungal drugs, mortality rates associated with cryptococcal infections often exceed 50%. In addition, these drugs are frequently associated with severe side effects, high toxicity, and many serious drug–drug interactions (74–76). Moreover, because of the lengthy treatment, drug resistance is a significant problem (16, 17).
The pathogenic cryptococcal fungi C. neoformans and C. gattii contain the Prp8 intein (77–80), which belong to the class 1 intein family (77, 79, 80). The Prp8 protein is essential and the most highly conserved protein of the spliceosome, which processes pre-mRNA into mRNA by RNA splicing. Previously, we found that cisplatin, an FDA-approved chemotherapeutic drug, could inhibit fungal Prp8 intein splicing in vitro and reduce the lung burden of C. neoformans in mice (8). We also determined the crystal structure of the Cne Prp8 intein in complex with cisplatin (7, 8), providing the structural basis for cisplatin inhibition of C. neoformans.
In the present study, we developed a split-GFP–based Prp8 intein-splicing assay suitable for high throughput screening. Using this assay, we performed a pilot screening of small molecules targeting Prp8 intein splicing and identified the candidate compounds 6G-318S and 6G-319S as potent inhibitors of Prp8 intein splicing. Both compounds appear specific to Prp8 intein splicing, and do not inhibit representative serine (trypsin) and cysteine (papain) proteases. We validated that these compounds bind covalently to the Prp8 intein active site residue Cys1, using MS, PTSA, mutagenesis experiments, and SPR techniques. Functional studies demonstrated that both compounds are fungicidal and potent inhibitors of the Prp8 intein-containing fungi C. neoformans and C. gattii, but not of C. albicans, which does not encode the Prp8 intein. Their MIC values are comparable to or better than the current frontline drugs for management of infections caused by C. neoformans and C. gattii. These Prp8 intein-splicing inhibitors also showed synergistic or additive effects with current frontline antifungals. More importantly, we showed that treatment of C. neoformans with 6G-318S led to accumulation of unspliced Prp8 protein precursor, supporting our hypothesis that splicing arrest leads to inhibition of the processing of Prp8 precursor protein. Finally, we demonstrated that overexpression of the Prp8 intein in C. neoformans but not an active-site C1A mutant led to resistance to 6G-318S treatment, reinforcing the contention that the compound acts by targeting the intein active site.
Microbial inteins are attractive drug targets (8, 19, 21). Because processed proteins carry out essential cellular functions in fungi, an intein-splicing inhibitor is a mechanistically novel antifungal agent. Intein-splicing inhibitors would have the following advantages over traditional antifungal drugs. 1) The Cryptococcus Prp8 inteins share high sequence identity (78–83%) (79). Particularly, the active site residues are absolutely conserved among the Prp8 inteins (79, 80). Therefore, it is likely that Prp8 intein-splicing inhibitors would be effective against all Cryptococcus fungal species. 2) Humans do not have inteins, suggesting that Prp8 intein inhibitors could be specific to the fungus. Our work demonstrates the potential of development of small-molecule inhibitors targeting Prp8 intein splicing and opens an avenue to develop a novel class of antifungals.
Materials and Methods
Compounds.
TCEP and cisplatin were from Sigma-Aldrich. Compounds 6G-318S, 6G-319S, 12G-305S, 6G-313S, and 12G-315S were from Key Organics.
Cloning, Expression, and Purification.
The split-GFP–Prp8i was constructed using a mega-PCR mutagenesis approach (26). Briefly, a PCR DNA fragment representing the Prp8 intein (in italics) flanked by GFP overlapping sequences (underlined) was amplified using the C. gattii genomic DNA as a template, with primers prp8_N_GFP_F: (CCTTGTTAATCGTATCGTGTTAAA-AGGTTGTCTGCAGAACGGTACCCG); and prp8_C_GFP_R: (CGAGAATGTTTC-CATCTTCTTTAAAATCTGAGTTGTGTAATACCAAATAGTCATGAC).
The overlapping PCR product was then used as a megaprimer for PCR mutagenesis with the split-GFP-RecA intein plasmid (21) as a template. After digestion with DpnI restriction enzyme, the PCR product was transformed into DH5α E. coli. Clones were verified by plasmid DNA sequencing. The expression, refolding, and purification of the GFP–Prp8i fusion protein were carried out as described previously (21). One exception is that prior to refolding, we treated the denatured and solubilized GFP–Prp8i inclusion body with 5,5′-dithiobis-(2-nitrobenzoic) acid (DTNB) to further prevent GFP–Prp8i self-splicing, by reacting with free cysteine at the intein active site.
The Cga MIG Prp8 and the Prp8 intein using pXI and pET28a vectors were constructed, expressed, and purified as described previously (8). The Flag-Prp8 intein and mutant constructs were cloned or synthesized into the pXL1-PTEF1 vector, as described previously (8) and transformed into the C. neoformans H99 strain, using a Bio-Rad electroporator.
Split-GFP–Prp8i Intein-Splicing Assay.
Using black- and clear-bottom 96-well plates, the DPNB-treated, refolded, and concentrated GFP–Prp8i fusion protein (0.5 µM) was incubated with compounds or DMF solvent control for 30 min at 25 °C in a 100 µL intein-splicing buffer (20 mM Bis-Tris propane, 0.5 M NaCl, 0.5 M arginine, 1 mM EDTA, and 0.05% CHAPS). Then 100 µL 2× TCEP was added to a final concentration of 20 µM. The reactions were incubated at 25 °C overnight. GFP–Prp8i splicing was monitored using a Bio-Tek FL800 microplate reader with excitation and emission wavelengths of 485 nm and 528 nm, respectively. Two readings were taken, one prior to addition of TCEP (I0) and one after incubation overnight with TCEP (If). Fluorescence/luminescence differences (∆Fluorescence or ∆Luminescence) were defined as If-I0 for each well. Relative GFP fluorescence-Prp8 (FLU) or RLuc-Prp8 luminescence (RLU, see below) was calculated by the following formula: % FLU (or % RLU) = (If-comp − I0-comp)/(If-DMF − I0-DMF) × 100. DMF control served as 100% splicing activity.
Split Luciferase Prp8 Intein-Splicing Assay.
The split RLuc–Prp8 fusion protein was carried out and monitored using a Veritas luminometer, as described previously (8).
Trypsin and Papain Protease Inhibition.
An Abz FRET substrate (Abz-RRRRSAG-nTyr) developed previously (29) was used as the substrate for inhibition of trypsin and papain proteases. In black- and clear-bottom 96-well plates, 1 µg trypsin (Sigma-Aldrich) was incubated with DMF or compounds at 400 µM at 25 °C for 30 min in a 70-µL reaction buffer containing 20 mM Tris, pH 8.0, 100 mM NaCl, 5% glycerol, and 0.05% CHAPS. For papain, 1 µg Papain (Sigma-Aldrich) was incubated with compounds at 400 µM at 25 °C for 30 min in a 70-µL reaction buffer composed of 1.1 mM EDTA, 0.067 mM β-mercaptoethanol, 5.5 mM cysteine. Substrate cleavage was monitored using a Bio-Tek FL800 microplate reader with excitation and emission wavelengths of 360 nm and 420 nm, respectively.
The MIG–Prp8 and Prp8–GFP In-Gel Splicing Assays.
The MIG–Prp8 and Prp8–GFP splicing assays were carried out as described previously (8). The samples were analyzed in SDS/PAGE without boiling. The gels were quantified using GFP fluorescence and Coomassie blue staining with the Bio-Rad Gel Doc EZ.
Cytotoxicity.
Cytotoxicity assays were carried out using human lung carcinoma cell line A549, as described previously (81). Briefly, the A549 cells (5 × 103) were incubated with/without compounds in a 96-well plate at 37 °C for 48 h. Cell viability was determined using the MTT assay (ATCC), as described previously (81, 82) (n = 3).
Western Blot.
Fresh colonies of C. neoformans were grown in yeast extract peptone dextrose (YPD) broth in an environmental shaker (250 rpm) overnight at 30 °C. The fungal cells (5 mL) were diluted in 125 mL YPD broth and grown for 1.5 h to a final cell density of 0.2 at OD530. The cells were distributed to test tubes (3 mL per tube). Then 3 mL of YPD medium containing DMF or twofold diluted compounds was added to the cells. The mixtures were incubated at 30 °C in a shaker at 250 RPM for up to 18 h. Samples were spun down and pellets were frozen for Western blot analysis.
Cryptococcal cells treated with DMF or compounds were washed twice with PBS buffer. Cells were spun down and supernatant was removed. For a 6-mL cell pellet, 30 µL of complete protease inhibitor mixture in PBS was added to the cells, followed by addition of 400 µL lysis buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 10% glycerol) and 500-µL glass beads. The fungal cells were lysed by rigorous beating for 2 min three times. The cell lysate was spun down. Supernatant was mixed with SDS/PAGE loading buffer (1:1). The mixtures were boiled at 95 °C for 10 min, followed by centrifugation at 15,000 rpm for 10 min. Samples were analyzed using 12% SDS/PAGE. The separated samples were blotted to a PVDF membrane and subjected to Wester blot analysis using an antibody against the Prp8 intein (7).
Mass Spectrometry.
The ESI-MS experiment was carried out as described previously (8). Briefly, the Cga Prp8 intein in pXI and pET28a backgrounds to yield intein only or intein flanked by extein sequence (30 µM) was incubated at 25 °C with compounds (300 µM) in a binding buffer (50 mM sodium phosphate, 100 mM NaCl, pH 7.0) for ∼18 h. MS of the incubated samples was carried out on an LTQ Orbitrap Velos instrument (Thermo Scientific) in the positive ion mode.
LC-MS/MS was carried out as described previously (29). Briefly, the protein band of interest on SDS/PAGE gel was manually excised, processed, and treated with 25 ng of sequencing grade modified trypsin (Sigma-Aldrich), as described previously (29). The digested peptides were extracted, separated, desalted, and subjected to LC-MS/MS analysis with a Jupiter C18 column (3 μm, 100 μm ID × 150 mm; Phenomenex), using a QSTAR XL Hybrid LC/MS/MS System, as described previously (29).
MS data acquisition was performed using Analyst QS 1.1 software (ABSciex) in positive ion mode for information-dependent acquisition analysis. The MASCOT 2.5 from Matrix Science was used to query fungal protein and contaminant data subsets using the following parameters: Peptide mass tolerance, 0.3 Da; MS/MS ion mass tolerance, 0.3 Da; and allowing up to two missed cleavages. Several variable modifications were applied, including methionine oxidation and cysteine carbamidomethylation. Only significant scores for the peptides defined by the Mascot probability analysis (http://www.matrixscience.com/help/scoring_help.html#PBM) greater than “identity” with 95% confidence were considered for the peptide identification.
PTSA.
The PTSA was conducted using an Applied Biosystem 7500 Fast Real-Time PCR System (ThermoFisher Scientific) from 25 to 80 °C, with methods similar to those described previously (29). Briefly, the Prp8 intein at a final concentration of 2.5 µM in 1× PBS was mixed with each compound to attain a 4.8-µM final concentration in 1.6% DMF for 30 min at 25 °C. PTSA was carried out in the MicroAmp Fast Optical 96-Well Reaction Plate (ThermoFisher Scientific). Thermal denaturation was monitored using SYPRO Orange (Life Technologies) according to the manufacturer’s manual. The Tm was calculated, using a Derivative model using the Protein Thermal Shift Software v1.0 (ThermoFisher Scientific). Compounds were considered to be binders when ∆Tm > 0.5 °C.
Surface Plasmon Resonance.
SPR was used to determine the affinity and kinetic analyses of the interactions between each drug and the Prp8 intein at 25 °C using a ProteOn XPR36 SPR instrument (Bio-Rad). The Prp8 intein and mutants were immobilized onto a ProteOn GLH sensor chip (2,500 to 7,000 RU) (Bio-Rad). A threefold dilution series of compounds was injected as the analytes. A blank surface blocked by ethanolamine was used as the control surface. The experiment was carried out at a flow rate of 100 µL/min using a PBSTD buffer containing 1× PBS, 0.005% surfactant P20, and 5% DMSO. Association (ka) and dissociation (kd) rates, as well as the dissociation constant (KD), were obtained by global fitting of the SPR data from multiple concentrations to a simple 1:1 Langmuir binding model, using the ProteOn Manager software suite (Bio-Rad).
Fungal Susceptibility Test.
The fungal susceptibility test was carried out with a compound concentration series as described previously (8). The broth microdilution method was used to determine the compound MIC values against C. neoformans. In brief, serial twofold dilutions of the 6G-318SS (20 to 0.03 µg/mL) were made in 100 µL RPMI-1640 medium with MOPS in 96-well U-bottom plates. The C. neoformans H99 strain (100 µL of 0.5 × 105/mL) in log phase was added to each well. After incubating the cells at 30 °C for 48 h, the cell culture was visualized to determine MIC. The MIC break-point was defined as no visible fungal growth by naked eye or by determining the 90% growth inhibition using absorbance at 630 nm, compared with no inhibitor control in a 96-well plate.
To determine MFC, the cultured cells in selected wells were first suspended by gentle pipetting. Then 50 μL of the cell culture suspension from each well was plated onto the Sabouraud dextrose agar (SDA) plates and further incubated for 48 h. MFC is defined as the minimum fungicidal concentration where there is no growth after plating on the SDA plates. The ratio of MFC/MIC ≤ 4 is considered as fungicidal.
For resistance studies, C. neoformans H99 strain transformed with an empty pXL1-PTEF1 vector, the Prp8-pXL1-PTEF1 (WT Prp8) construct, or the AAAA mutant were used, as described previously (8). MICs for selected compounds were determined for these transformed cells as described above.
Sensititre YeastOne Y09 AST Assay.
Fungal susceptibility to a combination of compounds was carried out using the Sensititre YeastOne Y09 AST assay kit (ThermoFisher), according to the manufacturer’s manual. Briefly, fungal C. neoformans cells in exponential growth phase were diluted to a final OD530 of 0.1. Then 20-µL diluted cells were further diluted into 10 mL YPD as a stock. Prior to cell addition, a Sensititre YeastOne Y09 AST assay plate was prepared with 6G-318S (100 µL) at 2× final concentration (0.16 µg/mL) or DMF control in YPD. The stock C. neoformans (100 µL, ∼200 cells) were dispensed into the compound-ready 96-well plate. The fungal cells (treated or with DMF) were incubated at 30 °C for 48 h. The cell cultures were visualized by nacked eye. The MIC break points were determined according to manufacturer’s instructions.
Checkerboard Assay.
To determine the impact on potency of 6G-318S in combination with known antifungals, a checkerboard assay was carried by the microbroth dilution method in a 96-well plate as per Clinical and Laboratory Standards Institute (CLSI)-M60. A combination of 6G-318S (0.063-4.0 µg/mL) with AmB (0.005 to 4.62 µg/mL) or 5-FC (0.032 to 32.4 µg/mL) or IZ (0.003 to 2.820 µg/mL) or VOR (0.0003 to 0.349 μg/mL) were used in 96-well flat bottom plates for absorbance reading and round-bottom tissue-culture plates for visual examination. Compounds were initially dissolved in DMSO and diluted in RPMI-1640-MOPS media four times the targeted final concentration and 50 µL was added per well. The second compound was similarly diluted and 50 µL was added per well, to a final volume of 100 µL per well. C. neoformans H99 inoculum (100 µL) prepared from freshly grown cultures and diluted to an absorbance of 0.1 at 530 nm in water and further diluted (1:500) in RPMI-1640-MOPS was added to each well. To determine the fungicidal concentration of a compound, the wells with no visible growth were serially diluted, plated on SDA, and incubated at 30 °C for 48 to 72 h for the recovery of colony-forming units. Additionally, the cell density of treated and untreated wells was also estimated by reading absorbance at 630 nm using an EL808 reader (Bio-Tek). Percentage of inhibition compared to that of the no-drug control well was calculated. MIC is defined as the minimum concentration of drugs required to inhibit 90% of the growth. FIC index value is calculated as the ratio between MIC of compounds in combination and the sum of MIC of individual drug alone. The FIC index value is then used to categorize the interaction of the two compounds tested. An FIC index value of ≤0.5 is considered as synergy, 0.5 to 1 is additive, >1 to 4 is indifferent, and >4 is antagonistic (38).
Statistical Analysis.
All experiments were performed in triplicate unless specified otherwise. GFP fluorescence in gel was quantified using the Bio-Rad Gel Doc EZ system and Image Lab software #1709690 (Bio-Rad). One-way ANOVA and student t tests were used to carry out statistical analyses with the Prism software.
Supplementary Material
Acknowledgments
We thank the core facilities at the Wadsworth Center, including the Biochemistry Core for surface plasmon resonance analysis, the Applied Genomic Technologies Core for DNA sequencing, and the Tissue Culture Core for media preparation; Xiaojiang Li for help in the preparation of seed culture of various fungi; Drs. Xiaorong Lin and Youbao Zhao at the University of Georgia for the pXL1-PTEF1 plasmid; and Drs. Brian Callahan and Timothy S. Owen at Binghamton University, Dr. Elaheh Movahed at the Wadsworth Center, Dr. Kim McClive-Reed at Health Research, Inc. for helpful suggestions and discussions, and Matt Stanger for handling the manuscript. This work was supported by NIH Grants AI140726 and AI141178 (to H.L.), and GM44844 (to M.B.). J. Zhou is also supported by the John D. Stobo, M.D. Distinguished Chair Endowment Fund.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008815118/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Mills K. V., Johnson M. A., Perler F. B., Protein splicing: How inteins escape from precursor proteins. J. Biol. Chem. 289, 14498–14505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranko A. S., Wlodawer A., Iwaï H., Nature’s recipe for splitting inteins. Protein Eng. Des. Sel. 27, 263–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eryilmaz E., Shah N. H., Muir T. W., Cowburn D., Structural and dynamical features of inteins and implications on protein splicing. J. Biol. Chem. 289, 14506–14511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novikova O., Topilina N., Belfort M., Enigmatic distribution, evolution, and function of inteins. J. Biol. Chem. 289, 14490–14497 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novikova O., et al. , Intein clustering suggests functional importance in different domains of life. Mol. Biol. Evol. 33, 783–799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CDC , Basic TB Facts, https://www.cdc.gov/tb/topic/basics/default.htm. Accessed 20 March 2016.
- 7.Green C. M., et al. , Spliceosomal Prp8 intein at the crossroads of protein and RNA splicing. PLoS Biol. 17, e3000104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z., et al. , Cisplatin protects mice from challenge of Cryptococcus neoformans by targeting the Prp8 intein. Emerg. Microbes Infect. 8, 895–908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perfect J. R., Fungal diagnosis: How do we do it and can we do better? Curr. Med. Res. Opin. 29 (suppl. 4), 3–11 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Brown G. D., et al. , Hidden killers: Human fungal infections. Sci. Transl. Med. 4, 165rv113 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Gullo A., Invasive fungal infections: The challenge continues. Drugs 69 (suppl. 1), 65–73 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Tuite N. L., Lacey K., Overview of invasive fungal infections. Methods Mol. Biol. 968, 1–23 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Gandhi N. R., et al. , Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368, 1575–1580 (2006). [DOI] [PubMed] [Google Scholar]
- 14.WHO , Management of MDR-TB: A Field Guide: A Companion Document to Guidelines for Programmatic Management of Drug-Resistant Tuberculosis: Integrated Management of Adolescent and Adult Illness (IMAI) (World Health Organization, 2009). [PubMed] [Google Scholar]
- 15.Nathan C., Taming tuberculosis: A challenge for science and society. Cell Host Microbe 5, 220–224 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Pfaller M. A., Antifungal drug resistance: Mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 125(suppl. 1)S3–S13 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Ghannoum M. A., Rice L. B., Antifungal agents: Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12, 501–517 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard S. J., et al. , Frequency and evolution of Azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15, 1068–1076 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulus H., Protein splicing inhibitors as a new class of antimycobacterial agents. Drugs Future 32, 973–984 (2007). [Google Scholar]
- 20.Zhang L., Zheng Y., Callahan B., Belfort M., Liu Y., Cisplatin inhibits protein splicing, suggesting inteins as therapeutic targets in mycobacteria. J. Biol. Chem. 286, 1277–1282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan H., et al. , Exploring intein inhibition by platinum compounds as an antimicrobial strategy. J. Biol. Chem. 291, 22661–22670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manohar S., Leung N., Cisplatin nephrotoxicity: A review of the literature. J. Nephrol. 31, 15–25 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Barabas K., Milner R., Lurie D., Adin C., Cisplatin: A review of toxicities and therapeutic applications. Vet. Comp. Oncol. 6, 1–18 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Paken J., Govender C. D., Pillay M., Sewram V., Cisplatin-associated ototoxicity: A review for the health professional. J. Toxicol. 2016, 1809394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gangopadhyay J. P., Jiang S. Q., Paulus H., An in vitro screening system for protein splicing inhibitors based on green fluorescent protein as an indicator. Anal. Chem. 75, 2456–2462 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Brecher M., et al. , A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PLoS Pathog. 13, e1006411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iversen P. W., et al. , “HTS assay validation” in Assay Guidance Manual, Markossian S., Eds. (Eli Lilly and Company and the National Center for Advancing Translational Sciences, 2004). [PubMed] [Google Scholar]
- 28.Owen T. S., et al. , Förster resonance energy transfer-based cholesterolysis assay identifies a novel hedgehog inhibitor. Anal. Biochem. 488, 1–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z., et al. , Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res. 27, 1046–1064 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z., et al. , Erythrosin B is a potent and broad-spectrum orthosteric inhibitor of the flavivirus NS2B-NS3 protease. Antiviral Res. 150, 217–225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suda H., Aoyagi T., Hamada M., Takeuchi T., Umezawa H., Antipain, a new protease inhibitor isolated from actinomycetes. J. Antibiot. (Tokyo) 25, 263–266 (1972). [DOI] [PubMed] [Google Scholar]
- 32.Rex J. H., et al. , Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts (Cold Spring Harbor Laboratory, ed. 3, 2008). [Google Scholar]
- 33.Archibald L. K., et al. , Antifungal susceptibilities of Cryptococcus neoformans. Emerg. Infect. Dis. 10, 143–145 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hazen K. C., Fungicidal versus fungistatic activity of terbinafine and itraconazole: An in vitro comparison. J. Am. Acad. Dermatol. 38, S37–S41 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Fothergill A. W., “Antifungal susceptibility testing: Clinical laboratory and standard institute (CLSI) methods” in Interactions of Yeasts, Moulds and Antifungal Agents: How to Detect resistance, Hall G. S., Ed. (Human Press, New York, 2012), pp. 65–75. [Google Scholar]
- 36.Hai T. P., et al. , The combination of tamoxifen with amphotericin B, but not with fluconazole, has synergistic activity against the majority of clinical isolates of Cryptococcus neoformans. Mycoses 62, 818–825 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis R. E., Diekema D. J., Messer S. A., Pfaller M. A., Klepser M. E., Comparison of Etest, chequerboard dilution and time-kill studies for the detection of synergy or antagonism between antifungal agents tested against Candida species. J. Antimicrob. Chemother. 49, 345–351 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Doern C. D., When does 2 plus 2 equal 5? A review of antimicrobial synergy testing. J. Clin. Microbiol. 52, 4124–4128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z., et al. , Structure of the branched intermediate in protein splicing. Proc. Natl. Acad. Sci. U.S.A. 111, 8422–8427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perfect J. R., Bicanic T., Cryptococcosis diagnosis and treatment: What do we know now. Fungal Genet. Biol. 78, 49–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y., et al. , The Cryptococcus neoformans transcriptome at the site of human meningitis. MBio 5, e01087–e13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhai B., Wu C., Wang L., Sachs M. S., Lin X., The antidepressant sertraline provides a promising therapeutic option for neurotropic cryptococcal infections. Antimicrob. Agents Chemother. 56, 3758–3766 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brouwer A. E., et al. , Combination antifungal therapies for HIV-associated cryptococcal meningitis: A randomised trial. Lancet 363, 1764–1767 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Husain S., Wagener M. M., Singh N., Cryptococcus neoformans infection in organ transplant recipients: Variables influencing clinical characteristics and outcome. Emerg. Infect. Dis. 7, 375–381 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacDougall L., et al. , Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the pacific northwest, USA. Emerg. Infect. Dis. 13, 42–50 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fyfe M., et al. , Cryptococcus gattii infections on Vancouver Island, British Columbia, Canada: Emergence of a tropical fungus in a temperate environment. Can. Commun. Dis. Rep. 34, 1–12 (2008). [PubMed] [Google Scholar]
- 47.Kronstad J. W., et al. , Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat. Rev. Microbiol. 9, 193–203 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartlett K. H., et al. , A decade of experience: Cryptococcus gattii in British Columbia. Mycopathologia 173, 311–319 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Bartlett K. H., Kidd S. E., Kronstad J. W., The emergence of Cryptococcus gattii in British Columbia and the Pacific Northwest. Curr. Infect. Dis. Rep. 10, 58–65 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Upton A., et al. , First contemporary case of human infection with Cryptococcus gattii in Puget Sound: Evidence for spread of the Vancouver Island outbreak. J. Clin. Microbiol. 45, 3086–3088 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Byrnes E. J., 3rd, et al. , A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathog. 7, e1002205 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Byrnes E. J. 3rd, Bartlett K. H., Perfect J. R., Heitman J., Cryptococcus gattii: An emerging fungal pathogen infecting humans and animals. Microbes Infect. 13, 895–907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Byrnes E. J., 3rd, et al. , Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 6, e1000850 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Byrnes E. J., Heitman J., Cryptococcus gattii outbreak expands into the Northwestern United States with fatal consequences. F1000 Biol. Rep. 1, 62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datta K. et al.; Cryptococcus gattii Working Group of the Pacific Northwest , Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg. Infect. Dis. 15, 1185–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Byrnes E. J., 3rd, et al. , First reported case of Cryptococcus gattii in the southeastern USA: Implications for travel-associated acquisition of an emerging pathogen. PLoS One 4, e5851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perfect J. R., et al. , Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 50, 291–322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boulware D. R. et al.; COAT Trial Team , Timing of antiretroviral therapy after diagnosis of cryptococcal meningitis. N. Engl. J. Med. 370, 2487–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rajasingham R., Rolfes M. A., Birkenkamp K. E., Meya D. B., Boulware D. R., Cryptococcal meningitis treatment strategies in resource-limited settings: A cost-effectiveness analysis. PLoS Med. 9, e1001316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perea S., Patterson T. F., Antifungal resistance in pathogenic fungi. Clin. Infect. Dis. 35, 1073–1080 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Zuger A., Louie E., Holzman R. S., Simberkoff M. S., Rahal J. J., Cryptococcal disease in patients with the acquired immunodeficiency syndrome. Diagnostic features and outcome of treatment. Ann. Intern. Med. 104, 234–240 (1986). [DOI] [PubMed] [Google Scholar]
- 62.Boelaert J. R., Goddeeris K. H., Vanopdenbosch L. J., Casselman J. W., Relapsing meningitis caused by persistent cryptococcal antigens and immune reconstitution after the initiation of highly active antiretroviral therapy. AIDS 18, 1223–1224 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Cornely O. A. et al.; AmBiLoad Trial Study Group , Liposomal amphotericin B as initial therapy for invasive mold infection: A randomized trial comparing a high-loading dose regimen with standard dosing (AmBiLoad trial). Clin. Infect. Dis. 44, 1289–1297 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Schiller D. S., Fung H. B., Posaconazole: An extended-spectrum triazole antifungal agent. Clin. Ther. 29, 1862–1886 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Baginski M., Czub J., Amphotericin B and its new derivatives—Mode of action. Curr. Drug Metab. 10, 459–469 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Gray K. C., et al. , Amphotericin primarily kills yeast by simply binding ergosterol. Proc. Natl. Acad. Sci. U.S.A. 109, 2234–2239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hitchcock C. A., Dickinson K., Brown S. B., Evans E. G., Adams D. J., Interaction of azole antifungal antibiotics with cytochrome P-450-dependent 14 alpha-sterol demethylase purified from Candida albicans. Biochem. J. 266, 475–480 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Polak A., Scholer H. J., Mode of action of 5-fluorocytosine and mechanisms of resistance. Chemotherapy 21, 113–130 (1975). [DOI] [PubMed] [Google Scholar]
- 69.Deresinski S. C., Stevens D. A., Caspofungin. Clin. Infect. Dis. 36, 1445–1457 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Chen S. C., Slavin M. A., Sorrell T. C., Echinocandin antifungal drugs in fungal infections: A comparison. Drugs 71, 11–41 (2011). [DOI] [PubMed] [Google Scholar]
- 71.Kartsonis N. A., Nielsen J., Douglas C. M., Caspofungin: The first in a new class of antifungal agents. Drug Resist. Updat. 6, 197–218 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Maligie M. A., Selitrennikoff C. P., Cryptococcus neoformans resistance to echinocandins: (1,3)beta-glucan synthase activity is sensitive to echinocandins. Antimicrob. Agents Chemother. 49, 2851–2856 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dupont B., Pialoux G., Amphotericin versus fluconazole in cryptococcal meningitis. N. Engl. J. Med. 326, 1568, author reply 1568–1569 (1992). [PubMed] [Google Scholar]
- 74.Gubbins P. O., McConnell S. A., Penzak S. R., Drug Interactions in Infectious Diseases (Humana Press, Totowa, NJ, 2001). [Google Scholar]
- 75.Moen M. D., Lyseng-Williamson K. A., Scott L. J., Liposomal amphotericin B: A review of its use as empirical therapy in febrile neutropenia and in the treatment of invasive fungal infections. Drugs 69, 361–392 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Kauffman C. A., Fungal infections. Proc. Am. Thorac. Soc. 3, 35–40 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Butler M. I., Goodwin T. J., Poulter R. T., A nuclear-encoded intein in the fungal pathogen Cryptococcus neoformans. Yeast 18, 1365–1370 (2001). [DOI] [PubMed] [Google Scholar]
- 78.Butler M. I., Poulter R. T., The PRP8 inteins in Cryptococcus are a source of phylogenetic and epidemiological information. Fungal Genet. Biol. 42, 452–463 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Liu X. Q., Yang J., Prp8 intein in fungal pathogens: Target for potential antifungal drugs. FEBS Lett. 572, 46–50 (2004). [DOI] [PubMed] [Google Scholar]
- 80.Pearl E. J., Tyndall J. D., Poulter R. T., Wilbanks S. M., Sequence requirements for splicing by the Cne PRP8 intein. FEBS Lett. 581, 3000–3004 (2007). [DOI] [PubMed] [Google Scholar]
- 81.Chen H., et al. , Selective inhibition of the West Nile virus methyltransferase by nucleoside analogs. Antiviral Res. 97, 232–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brecher M., et al. , Identification and Characterization of novel broad-spectrum inhibitors of the flavivirus methyltransferase. ACS Infect. Dis. 1, 340–349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.