Significance
Severe coronavirus (CoV) infections have been responsible for hundreds of deaths. Since the emergence of severely pathogenic CoVs, namely the severe acute respiratory syndrome (SARS) virus in 2003, the Middle East respiratory syndrome (MERS) virus in 2012, and the SARS-CoV-2 in 2019, no licensed vaccine or effective antiviral treatment has been developed. Bats are reservoir hosts for many human viruses, including SARS and MERS. The protein relationship between human betacoronaviruses and their bat-specific ancestors is still unknown. This work includes a detailed structure-function analysis of a bat viral macrodomain and provides key insights into features relevant in determining conserved protein functions. This information is vital to the design of macrodomain inhibitors and antiviral therapies.
Keywords: coronavirus, macrodomain, viral protein, crystal structure, ADP-ribose
Abstract
Macrodomains are proteins that recognize and hydrolyze ADP ribose (ADPR) modifications of intracellular proteins. Macrodomains are implicated in viral genome replication and interference with host cell immune responses. They are important to the infectious cycle of Coronaviridae and Togaviridae viruses. We describe crystal structures of the conserved macrodomain from the bat coronavirus (CoV) HKU4 in complex with ligands. The structures reveal a binding cavity that accommodates ADPR and analogs via local structural changes within the pocket. Using a radioactive assay, we present evidence of mono-ADPR (MAR) hydrolase activity. In silico analysis presents further evidence on recognition of the ADPR modification for hydrolysis. Mutational analysis of residues within the binding pocket resulted in diminished enzymatic activity and binding affinity. We conclude that the common structural features observed in the macrodomain in a bat CoV contribute to a conserved function that can be extended to other known macrodomains.
Coronaviruses (CoVs) are positive-sense RNA viruses that cause respiratory disease in humans. From 2002 to 2003, the severe acute respiratory syndrome (SARS) virus caused a pandemic claiming over 800 lives (1). A decade later, the Middle East respiratory syndrome (MERS) virus, isolated from an outbreak in Saudi Arabia, was found to cause respiratory and nephritic disease (2, 3). In late 2019, a new virus which is related to the SARS-like bat CoVs began to cause illness (4). The new virus, termed SARS-Cov-2, 2019-nCoV, or HCoV-19, has caused a pandemic spanning at least 160 countries, with over 8.3 million infected and over 440,000 fatalities (5, 6). Symptoms of COVID-19, a pneumonia-like condition, range from mild symptoms such as dry cough, sore throat, and fever to serious or fatal complications including organ failure, pulmonary edema, severe pneumonia, and acute respiratory distress (7). Mortality rates for CoV disease range from 35% for MERS, 10% for SARS, and up to 14.5% for COVID-19. The COVID-19 outbreak was declared a Public Health Emergency of International Concern on January 30, 2020.
These viruses belong to the beta genus of the CoV family and are further differentiated into lineages A through D. The SARS-CoV and SARS-CoV-2 belong to lineage B, and the MERS-CoV belongs to lineage C (4, 8, 9). The CoVs employ the host cell machinery to translate polyproteins, pp1a and pp1ab, that are cleaved by viral proteases into 16 nonstructural proteins (10). The largest of these, the nonstructural protein 3 (nsp3), contains a conserved macrodomain that is critical to CoV infection (11).
The macrodomain protein family shares a conserved structure, binds a variety of small and macromolecular ligands, and functions in cellular processes ranging from chromatin structure maintenance and dynamics to innate immune function, transcription, and apoptosis (12–20). Viral macrodomains exhibit a mixed α/β fold of 130 to 190 amino acids with a ligand-binding cleft most commonly used to bind mono-ADP ribose (ADPR) (MAR) and poly-ADPR (PAR) (21–23).
Viral macrodomains remove MAR and/or PAR modifications from acidic (D and E) residues by hydrolyzing the ester bond (i.e., deMARylation) (11, 24). The hepatitis E virus (HEV) macrodomain hydrolyzes PAR modifications from proteins efficiently in vitro through the association with another helicase, but similar dePARylation activities were not observed in other proteins (24). DeMARylation is conserved in macrodomains of all CoVs and other single-stranded RNA (ssRNA) viruses such as alphavirus, rubella virus, and HEV and are essential for infection (8, 11, 25–27). The SARS-CoV macrodomain’s deMARylation activity curtails host cell immune responses (28). So far, no cytosolic protein substrates have been identified for viral macrodomains.
Phylogenetic analysis describes the close relationship of the MERS-CoV to CoVs of the Pipistrellus and Neormicia bats, and the MERS-CoV shares a cellular surface receptor, glucose-regulated protein 78, with the bat-CoV HKU9 (29). Bat CoVs are considered to be the progenitors of the severely pathogenic human CoVs. Whole-genome analysis of SARS-CoV-2 shows significant similarity with known β-CoVs (30). We therefore aimed to determine which structural and functional features are conserved between bat and human CoVs. We based our work on the bat virus HKU4, a lineage C β-CoV which was isolated from the lesser bamboo bat, Tylonycteris pachypus, in 2007 (31).
Here, we report the X-ray crystal structures of the HKU4 macrodomain in complex with three small-molecule ligands: ADPR, a conserved ligand from the MacroD subfamily, and two analogs, ADP-glucose (ADPG) and NAD+. These compounds were chosen because ADPR is the product of protein deMARylation, NAD+ is an endogenous metabolite important for cellular homeostasis and a potential ligand for viral macrodomains, and ADPG is a structural analog of ADPR. The NAD+ and ADPG complexes provide structural and affinity differences important in the development of viral macrodomain inhibitors.
The HKU4 macrodomain shares the conserved macrodomain topology and belongs to the MacroD subfamily. We show that it is an enzyme capable of removing ADPR from proteins. We defined the active-site residues that are important for its ligand binding and enzyme activity. Furthermore, we employed in silico analysis to investigate modes of interaction with MARylated peptides.
Results
Crystallization and Structure.
All complexes formed well-defined crystals that adopted the triclinic space group P1, with diffraction limits of 1.35 Å resolution for the ADPR complex, 1.8 Å for the NAD+ complex, and 1.5 Å for the ADPG complex. The structure was determined by molecular replacement (Methods Summary). Data collection and refinement statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics
| Parameter | ADPR | NAD+ | ADPG |
| PDB code | 6MEA | 6MEB | 6MEN |
| Data collection | |||
| Space group | P1 | P1 | P1 |
| Cell dimensions | |||
| a, b, c, Å | 33.61, 41.64, 59.78 | 33.61, 41.56, 59.91 | 33.55, 41.69, 60.00 |
| α, β, γ, ° | 73.16, 88.29, 88.25 | 73.42, 88.56, 88.43 | 73.04, 88.36, 88.05 |
| Resolution, Å | 39.85–1.35 (1.37–1.35) | 33.59–1.80 (1.84–1.80) | 39.86–1.50 (1.53–1.50) |
| Unique reflections | 63,312 (2,865) | 28,245 (1,381) | 49,739 (2,476) |
| Completeness, % | 92.7 (86.2) | 98.1 (82.0) | 99.9 (100) |
| Multiplicity | 3.7 (3.4) | 3.6 (2.4) | 7.5 (7.4) |
| Rmerge, % | 12.1 (65.7) | 8.8 (16.6) | 12.9 (128.4) |
| Rpim, % | 7.4 (42.1) | 5.1 (11.8) | 5.1 (50.8) |
| CC (1/2) | 0.991 (0.662) | 0.992 (0.905) | 0.995 (0.721) |
| I/σ (I) | 9.8 (3.0) | 9.3 (4.3) | 9.1 (1.9) |
| Refinement | |||
| Resolution, Å | 33.59–1.35 (1.39–1.35) | 33.59–1.80 (1.85–1.80) | 29.21–1.50 (1.54–1.50) |
| Total reflections | 63,270 (4,552) | 28,243 (1,765) | 49,737 (3,695) |
| Completeness, % | 92.9 (89.8) | 98.1 (83.7) | 99.9 (100) |
| Rwork, % | 14.2 (27.9) | 19.0 (21.4) | 17.3 (25.9) |
| Rfree, % | 16.2 (29.4) | 23.2 (23.3) | 20.9 (32.3) |
| Wilson B, Å2 | 10.0 | 10.3 | 16.1 |
| Average B-factors, Å2 | |||
| Overall | 14.7 (2,884 atoms) | 16.6 (2,871 atoms) | 24.7 (2,739 atoms) |
| Protein | 12.7 (2,440 atoms) | 14.9 (2,417 atoms) | 23.1 (2,426 atoms) |
| Ligand | 9.7 (72 atoms) | 25.5 (88 atoms) | 44.3 (76 atoms) |
| Water | 29.0 (372 atoms) | 26.2 (366 atoms) | 33.9 (237 atoms) |
| rms deviations | |||
| rmsd bonds, Å | 0.007 | 0.010 | 0.014 |
| rmsd angles, ° | 1.40 | 1.50 | 1.65 |
| CC (Fo-Fc) | 0.97 | 0.94 | 0.97 |
| CC (Fo-Fc free) | 0.97 | 0.90 | 0.95 |
| Ramachandran | 100% favored (no outliers) | 99.1% favored (1 outlier) | 99.1% favored (no outlier) |
| Clash score | 1.79 | 5.80 | 4.00 |
| Molprobity score | 0.94 | 1.32 | 1.19 |
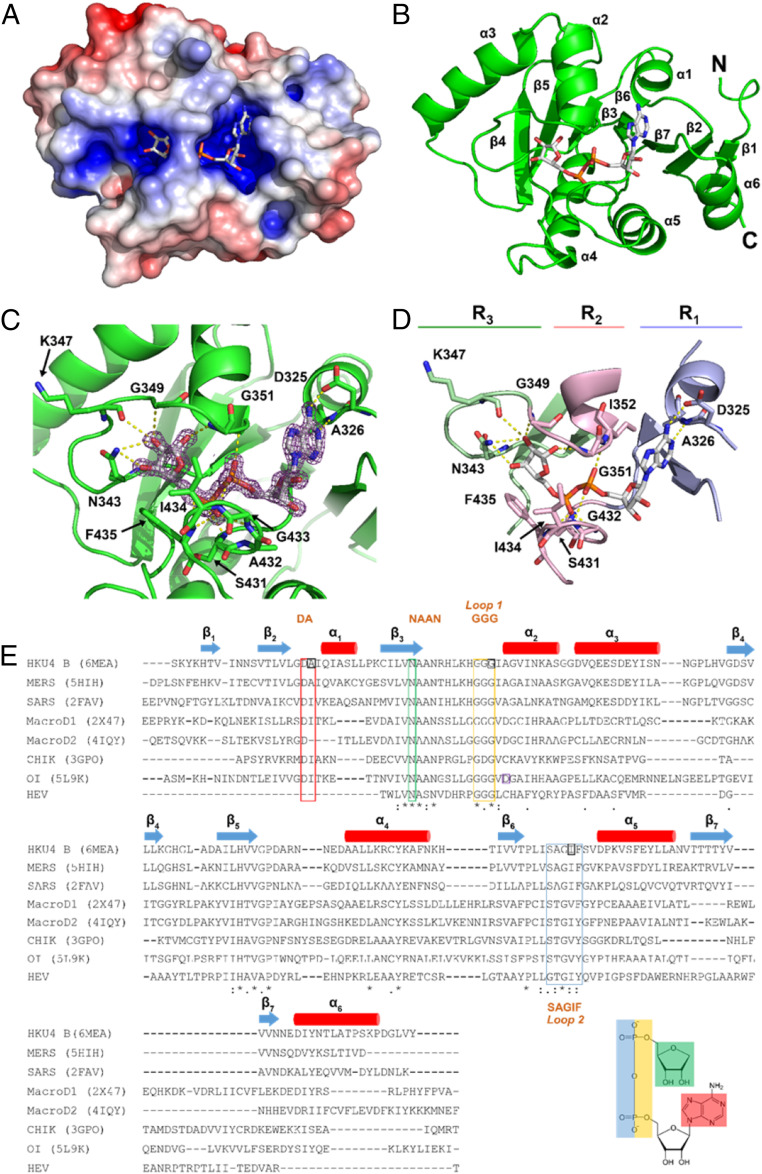
The protein exhibits a mixed α/β fold with seven β-strands enveloped by six α-helices. The fold is consistent with the MacroD subfamily (16). The α-helices surround the β-sheet with α1 to α3 on one side and α4 to α6 on the other. A positively charged cavity is formed by loops that connect β2 to α1, β3 to α2, and β6 to α5 (Fig. 1 A and B).
Fig. 1.
The crystal structure of HKU4 macrodomain in complex with ADPR. (A) Surface representation. (B) Ribbon representation with secondary structures and N and C termini labeled. (C) A close-up of the binding cavity with residues responsible for forming hydrogen bonds (yellow dashes). ADPR is displayed in stick format with an electron density omit map (purple) with a sigma value of 2.5. (D) Binding cleft with ADPR. The cleft is shaded in three regions: R1 (G324-S331, V426-L429, T454-N460), R2 (G349-V355, I430-V437), and R3 (V339-H348, L396-P401). Selected residues are shown in stick format and labeled. (E) Clustal Omega sequence alignment of viral macrodomains (21, 44, 47, 61, 77). The secondary structure of HKU4 macrodomain is displayed: β-strands (blue arrow) and α-helices (red cylinder). Conserved motifs of viral macrodomains are described by color: DA (red), NAAN (green), GGG (yellow), and SAGIF (blue). The residues selected for mutagenesis studies are shaded by orange squares. The residue hypothesized to serve as a catalyst for Oceanobacillus iheyensis macrodomain enzymatic activity is shaded by a purple square (61).
Macrodomain–ADPR Complex.
ADPR exhibits well-defined electron density with a map correlation coefficient of 0.94 (Figs. 1C and 2A). The binding pocket shape and charge complement the ligand’s ring system. The binding pocket has three distinct regions, which we designate as R1, R2, and R3 to orient the reader (Fig. 1D and SI Appendix, Fig. S1). The R1 region is formed by three loops: 1) the loop between β2 and the N terminus of α1, 2) the first four residues in the loop following β6, and 3) the loop between β7 and α6 (Fig. 1 D and E). In R1, the ADPR adenine forms two hydrogen bonds with a conserved aspartate (D325) and the backbone nitrogen of A326 (Fig. 1E and SI Appendix, Fig. S1).
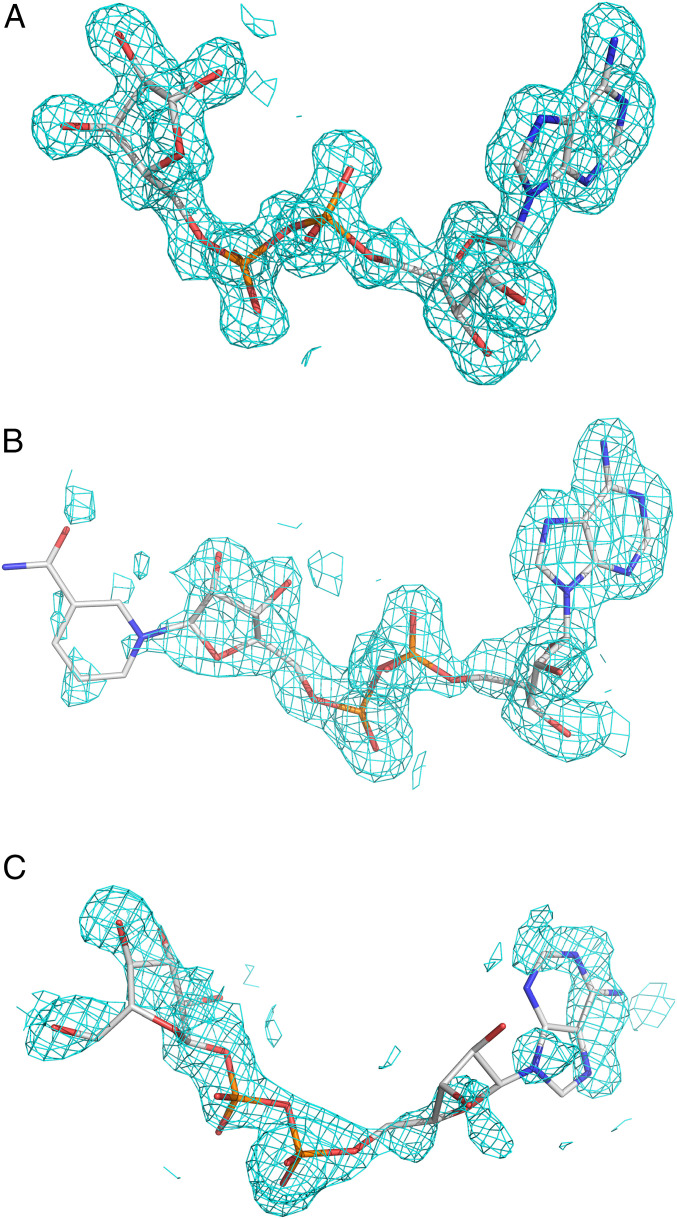
Fig. 2.
Sigma-A-weighted difference maps (2Fo-Fc) for the ligands in the three crystal structures. Densities are contoured at 1.0 sigma level. (A) ADPR. (B) NAD+. (C) ADPG.
The second region (R2) is defined by two loops that stabilize the phosphate groups of ADPR by hydrogen bonding (Fig. 1D and SI Appendix, Fig. S1). Residues G349 to G351 (GGG motif) of loop 1 and S431 to F435 (SAGIF motif) of loop 2 are located in this region (Fig. 1E). The α-phosphate accepts hydrogen bonds from loops 1 and 2 via the backbone HN atoms of I352 and I434. The loops create a narrow groove around the diphosphate, forming a closed bridge whereby the loops close over the ligand via the I434 side chain. This complementarity results in the burial of 90% of ADPR’s surface area, rendering the ligand only 10% solvent accessible (Table 2).
Table 2.
Structural data of the HKU4 macrodomain complexes*
| Complex* | ADPR | NAD+ | ADPG |
| Interface residues | 29 | 29 | 28 |
| No. of hydrogen bonds | 11 | 8 | 9 |
| Average hydrogen bond length (Å) | 3.1 | 3.2 | 3.5 |
| Solvent-accessible surface area (%) | 10.1 | 21.4 | 14.8 |
Calculations for the structural and chemical properties of each complex interface were determined using PISA (78).
R3 comprises the loop region between β3 and α2. This region contains N340 to N343 (NAAN motif) that stabilize the distal ribose (Fig. 1 D and E and SI Appendix, Fig. S1). Collectively, the conserved residues among the MacroD-like macrodomains in humans, bats, archaea, and viruses, namely D325, N343, and G351, form a hydrogen bond network that orients ADPR into a conserved binding mode (Fig. 1E). The adenine and proximal ribose are accommodated in R1, the diphosphate in R2, and the distal ribose in R3.
Macrodomain–NAD+ Complex.
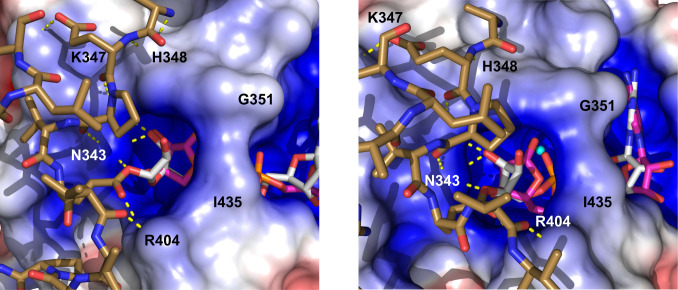
NAD+ has a map correlation coefficient of 0.88 (Fig. 2B). The electron density is well defined with the exception of the nicotinamide, which is moderately disordered (SI Appendix, Table S1). The groove accommodates the adenine and proximal ribose in R1, the diphosphate in R2, and the nicotinamide in R3 (Fig. 3A). The adenine is maintained in its conserved binding pocket, and the interactions of the diphosphate are similar to those of ADPR. However, the distal ribose forms fewer contacts with the binding site. Here, O3′’ interacts with A341 and A353 via a water molecule that is the likely nucleophile for deMARylation (see Discussion). The proposed catalytic water is stabilized by hydrogen bonding with the α-phosphate group and O3′’ of NAD+ (SI Appendix, Fig. S2B).
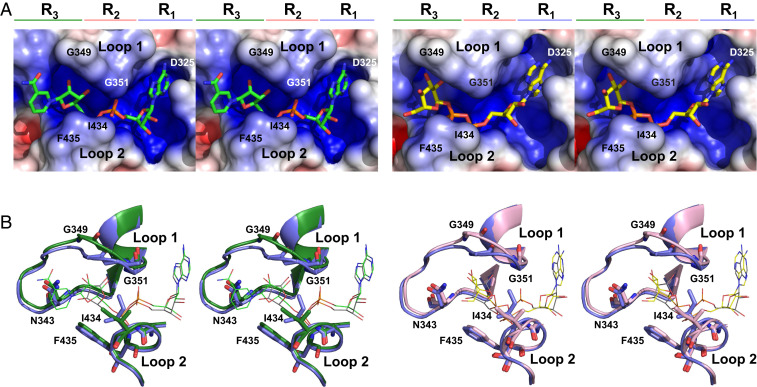
Fig. 3.
Binding cavity of macrodomain complexes. (A) Surface representation of macrodomain binding cavity with NAD+ (Left) and ADPG (Right). Select residues lining the binding cavity are labeled. (B) Wall-eye stereoview overlays of the binding cavity. On the left, the backbone and side chains of the protein are displayed as cartoons in complexes with NAD+ (green) and ADPR (blue). The ligands (NAD+, green; ADPR, gray) are shown as sticks. On the right, the backbone and side chains of the complexes with ADPR (blue) and ADPG (pink) are shown as cartoon and stick models. The ligands (ADPR, gray; ADPG, yellow) are displayed as sticks.
The nicotinamide fits on the outer edge of R3 and is located 3.9 to 5.0 Å away from the nearest residues, N343 and K347. There are no direct interactions between the nicotinamide ring and the protein. This is supported by the electron density observed for the nicotinamide ring, with weak electron density observed at the 1.0 sigma level (Fig. 2B), reflecting static disorder. The NAD+ binding pocket adopts an open conformation, with a 21% ligand solvent accessibility.
The NAD+ complex exhibits significant differences with respect to ADPR. The most dramatic difference is that the “closed” bridge over the binding site observed in the ADPR complex is replaced with an “open” site in the NAD+ complex, whereby the bridge is not formed and the ligand is much more solvent accessible (21% versus 10%).
Macrodomain–ADPG Complex.
In the ADPG complex, the ligand resides in the same regions of the binding pocket, but with an altered interaction profile (Figs. 2C and 3A and SI Appendix, Fig. S1). The binding pocket accommodates the glucopyranose in R3 in place of the distal ribose. The α-anomer of ADP-d-glucopyranose crystallized in complex with the protein and adopted an O,3B boat conformation. ADPG forms the lowest number of hydrogen bonds with the HKU4 macrodomain (Table 2 and SI Appendix, Fig. S1). The adenine is in an anti conformation with a χ = 115.8° torsion angle about the C1-N9 glycosidic bond. This places the adenine in a nonconserved orientation, forming a single hydrogen bond to the backbone amide HN of A326 in R1. An open conformation of the active site is observed (Fig. 3), with 14.2% solvent accessibility. ADPG displays a map correlation coefficient of 0.81 (Fig. 2C). The proposed catalytic water is also observed in this complex and is stabilized by hydrogen bonding with O4″ and O5″ of ADPG (SI Appendix, Fig. S2C).
The greatest structural difference between the open (NAD+, ADPG) and closed (ADPR) conformations is evident from movement of the loop residues. Relative to the ADPR complex, there is a change in conformation of both loops 1 and 2, with G349 to 350 changing backbone torsion angles (φ/ψ) by +18.4°/+115.4° and −82.3°/−169.6°, respectively. The Cα atom of G349 is displaced by 2.6 Å and that of G350 by 2.7 Å. In loop 2, the I434 side chain CD1 moves by 3.0 Å. I434’s χ1 dihedral angle rotates 120° from the gauche− rotamer (χ1 = −60°) to the gauche+ rotamer (χ1 = +60°) and extends further into the binding pocket, forming a steric bridge over the binding cleft, which we describe as the closed conformation (Fig. 3B). I434’s side chain rotation opens the binding site by ∼1.4 Å. The G350 to I434 Cα to Cα distance is 8.0 Å in the ADPR complex, while that in the NAD+ complex is 9.3 Å and in the ADPG complex is 9.4 Å. This increased distance reflects an accommodative opening of the binding site. Interactions characteristic to the open conformation include an interaction with K347 and contacts to the backbone rather than side chain of I352.
Ligand Binding Affinity.
We employed isothermal titration calorimetry to measure the equilibrium dissociation constants of the ligands. The affinity (Kd) of ADPR is 14.1 µM (Fig. 4 and Table 3). No binding enthalpy was observed for ADPG or NAD+, suggesting weak or entropy-driven binding. Binding was confirmed qualitatively by nuclear magnetic resonance titrations, with estimated values in the millimolar (mM) range for ADPG and NAD+ (SI Appendix, Fig. S5).
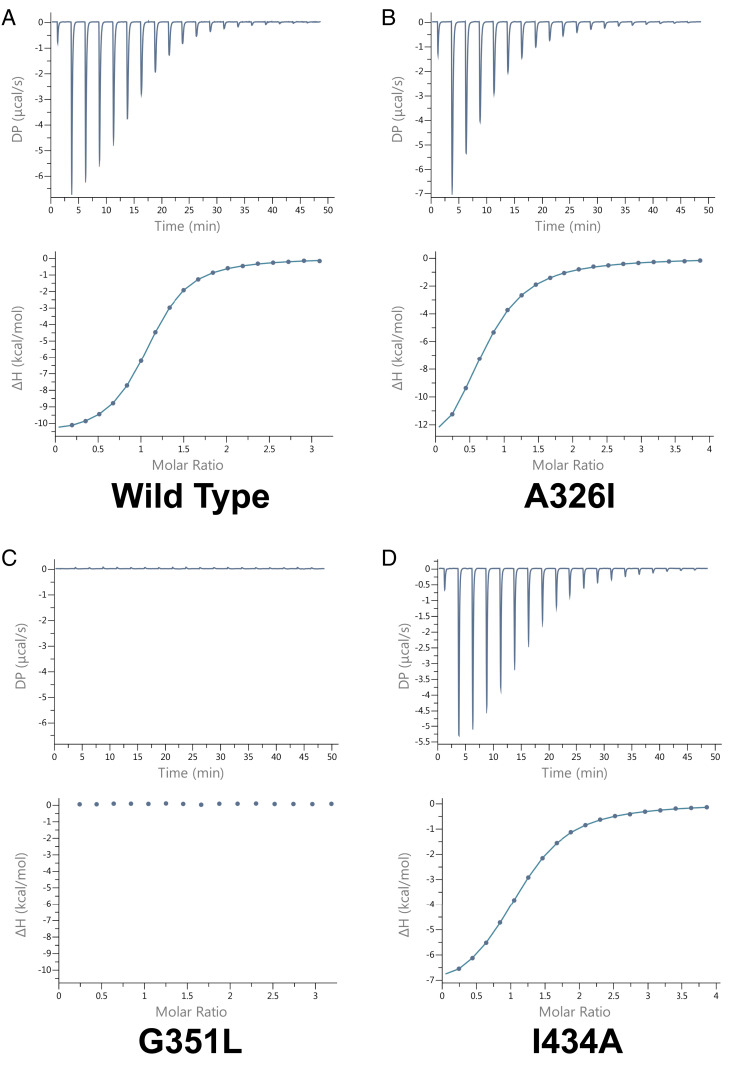
Fig. 4.
Isothermal titration calorimetry analysis of HKU4 macrodomain and ADPR. Titrations of (A) wild-type (average n sites = 1.06 ± 1.63 × 10−3, Kd = 1.41 × 10−5 ± 2.12 × 10−7, ΔH = −1.08 × 101 ± 3.03 × 10−2, offset = 1.06 × 10−1 ± 3.3 × 10−2 ΔG = −6.62, SEM = 0.06%), (B) A326I mutant (average n sites = 0.88 ± 9.40 × 10−3, Kd = 6.45 × 10−5 ± 1.22 × 10−6, ΔH = −1.35 × 101 ± 1.08 × 10−1, offset = 7.77 × 10−2 ± 6.03 × 10−2 ΔG = −5.72, SEM = 0.61%, P = 0.001), (C) G351L mutant, and (D) I434A mutant (average n sites = 1.11 ± 3.77 × 10−3, Kd = 4.1 × 10−5 ± 948 × 10−7, ΔH = −7.62 ± 5.13 × 10−2, offset = 1.53 × 10−2 ± 3.3 × 10−2 ΔG = −5.99, SEM = 4.1%, P = 0.02). Raw data (µcal/s) for each titration are displayed in each upper panel. Integration of the data are shown in each lower panel. All ligand binding assays were completed in triplicate. μcal/s, microcalories per second.
Table 3.
Thermodynamic data for ADPR binding in HKU4 nsp3 macrodomain
| Binding affinity (Kd), µM | Stoichiometry (N) | ΔH, kJ/mol | ΔG, kJ/mol | |
| WT | 14.1 ± 0.002 | 1.06 ± 0.002 | −10.8 ± 0.03 | −6.62 |
| A326I | 64.5 ± 0.001 | 0.879 ± 0.004 | −13.47 ± 0.001 | −5.72 |
| G351L | NDB | NDB | NDB | NDB |
| I434A | 41.0 ± 0.009 | 1.11 ± 0.004 | −7.62 ± 0.05 | −5.99 |
NDB, no detectable binding; WT, wild type.
To investigate residues that could play roles in binding, we created the mutations A326I, G351L, and I434A. By mutating A326 and G351 to bulkier residues, we interrogated whether adding steric interactions near the conserved D325 and in loop 1 would affect binding affinity. Conversely, we mutated I434 to a residue with a small side chain to probe the role of the steric bridge. All three mutants had a reduction in ADPR affinity: A326I bound with Kd 64.3 µM, I434A with Kd 41 µM, and no binding enthalpy was detected for the G351L mutant (Fig. 4 and Table 3).
A mutation of the conserved D325 was created but exhibited insoluble expression and could not be purified. Isothermal titration calorimetry assays on the G351L, I434A, and A426I mutant proteins binding to NAD+ or ADPG yielded uninterpretable results and could not be analyzed.
DeMARylating Enzyme.
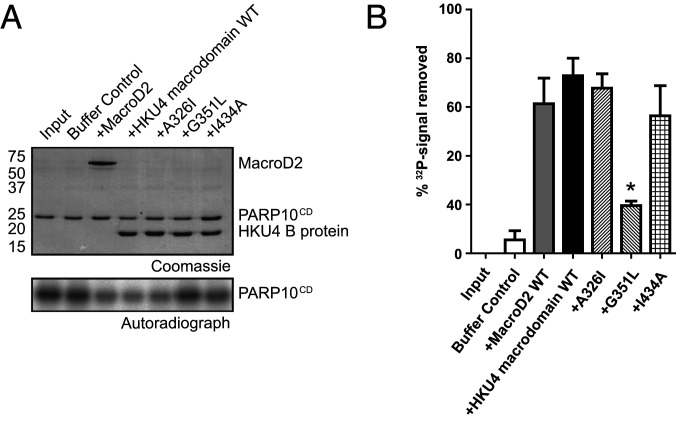
Catalytic activity was probed using a deMARylation assay which employs a model substrate, the catalytic domain of the autoMARylating PAR polymerase 10 (PARP10CD) (11, 32, 33). The work of McPherson et al. showed that PARP10CD is a substrate for MAR hydrolysis (11, 34, 35). PARP10 is induced upon CoV infection (34, 35) and has been used as a model substrate for alphaviral and coronaviral macrodomains (11, 24, 28). To prepare the MARylated substrate, 32P-labeled NAD+ was incubated with PARP10CD (11, 36). 32P-labeled PARP10CD was incubated with the HKU4 macrodomain. Buffer-only samples were used as negative controls, and MacroD2, a known deMARylating macrodomain, was used as a positive control. The HKU4 macrodomain removed 73% (SEM = 3.7%) of the 32P signal after a 1-h incubation, showing that it is an enzyme capable of deMARylating modified proteins (Fig. 5).
Fig. 5.
(A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and autoradiograph of deMARylation assay. (B) Quantification of deMARylation activity from enzymatic assay displaying results from wild-type mutants and controls. All enzyme assays were carried out in triplicate. *Statistically significant result.
Next, we further analyzed the contributions of individual residues to the mechanism of deMARylation by the HKU4 macrodomain using site-specific mutants. Previously, we have shown that differences in enzymatic activity between wild-type and mutant proteins can be assessed by determining the extent of deMARylation at the 1-h time point (11, 37). Therefore, we chose the 1-h time point to further analyze individual residues contributing to the mechanism of deMARylation. To probe residue contributions, we used the mutant proteins described above: A326I, G351L, and I434A. The deMARylation levels for the mutants were lower than the wild type: with a 68% reduction of the 32P signal observed (SEM = 5.8%; P = 0.61) for A326I, 20% removed (SEM = 1.8%; P = 0.01) for G351L, and 56% removed (SEM = 12.4%; P = 0.33) for I434A (Fig. 5). Compared to wild type, A326I, G351L, and I434A mutants showed a 7%, 73%, and 23% reduction in activity. G351L maintained a low level of activity compared to the buffer control (P = 0.04), indicating that the mutation did not completely abolish its deMARylation capability. The statistically significant reduction of deMARylating activity observed with G351L indicates that loop 1 of the binding site is particularly important to enzymatic activity.
In Silico Docking.
Enzyme–substrate recognition was studied by in silico analysis. As a model peptide, an 18-residue sequence of human PARP1 (487 to 505; sequence 487AEPVEVVAPRGKSGAALS505) was selected. This peptide contains glutamate residues that are experimentally determined ADP ribosylation sites (38–40), which is consistent with the demonstrated specificity toward ADP ribosylated D and E residues observed for other viral macrodomains (11). Furthermore, the linker region is predicted to comprise a solvent-exposed flexible loop (39), which likely contains minimal secondary or tertiary structures that could bias docking. The peptide was ADP ribosylated with a single ADPR at one of two known sites (E488 or E491). The peptides were docked to the macrodomain and the 10 lowest-energy complexes were analyzed.
For the peptide MARylated at E491, the ADPR modification was positioned in the binding pocket (Fig. 6). The proposed catalytic water molecule was located near the β-phosphate of ADPR. There were consistent binding modes that were similar to the crystal structure. The peptide formed electrostatic interactions with the macrodomain surface. The predicted average affinity was −5.8 ± 0.23 kcal/mol, with an average RMSD of 6.04 ± 4.04 Å. The unmodified peptide side chains are predicted to form hydrogen bonds with protein residues in the β3 to α2, β5 to α4, and β6 to α5 regions. Because these interactions are not constrained, there is high variability. The multiple modes the modified peptide used to interact with the macrodomain substantially increased the RMSD values for the docking experiment compared to ADPR alone. Yet, ADPR adopted a similar binding mode, occupying the same site and forming identical hydrogen bonds as observed in the crystal complex. The adenine aligned similarly between the two complexes but with different O4′-C4-C5-O5′ torsion angles (−89.5° in the ADPR complex and 177.7° in the docked substrate) (SI Appendix, Fig. S6).
Fig. 6.
Lowest energy Autodock Vina docking pose of HKU4 B macrodomain and MARylated E491 PARP1 peptide. Peptide sequence: 487AEPVEVVAPRGKSGAALS505. View of ester bond linkage (Left) to E491 residue from MARylated PARP1 fragment. The residues that form hydrogen bonds to the fragment are shown by dashed lines (yellow) and are labeled. A side view of the binding cavity (Right) displays differences in distal ribose orientation and the position of the proposed catalytic water (cyan).
In contrast, the peptide MARylated at E488 showed multiple binding modes. These were inconsistent with the binding mode from the crystal structure, with the ADPR moiety located outside of the binding pocket. The modes had a weaker predicted affinity, −5.2 ± 0.2 kcal/mol, and an average RMSD of 13.9 ± 11.1 Å, indicating high structural variation.
In the E491 peptide–macrodomain complex, the R496 and K498 side chains of the peptide formed hydrogen bonds with H345 or N343 in the binding pocket (SI Appendix, Fig. S6). Compared to the E491 peptide, the E488 peptide made fewer interactions with the macrodomain. In this case, the positively charged side chains formed internal hydrogen bonds rather than interacting with the protein. Near the peptide N terminus, A487 and P489 formed hydrophobic interactions with the protein, which were not observed in the E488 peptide. This is likely to be due to the introduction of the bulky, charged ADP ribosyl moiety close to the N terminus. In addition, the unmodified E488 side chain formed backbone and side chain hydrogen bonds with the backbone and side chains of K347, H348, and G349. These interactions precluded any preferential docking of ADPR in the E488-modified peptide, suggesting that MARylation in this sequence context is not optimal for binding.
Discussion
Conserved and Unique Features of the Macrodomain Structure.
The HKU4 macrodomain structure shows high conservation of backbone and side chain orientations with respect to the group 2c virus MERS-CoV (Table 4). The structure is divergent compared to other viral proteins. A DALI search (41) showed high amino acid identity with the MERS macrodomain (61%) and low RMSD (0.6 Å), while the SARS macrodomain has lower identity (43%) and a higher RMSD (1.3 Å). The residues D325, N343, G351, and I434 are conserved among viral macrodomains such as HKU4, SARS, MERS, and HEV. Furthermore, ADPR-bound viral macrodomains maintain conserved positions of the ligand even in structures with a divergent key residue I434V as observed in CHIKV. In particular, the adenine binding previously observed in the CHIKV macrodomain shows conserved interactions. D325 is oriented in the same direction (toward the pocket) as observed in MERS-CoV and is divergent to that of SARS-CoV (21, 42, 43).
Table 4.
Structural analysis of HKU4 macrodomain complexed with ADPR with other viral macrodomain
| %ID | RMSD | %ID | RMSD | %ID | RMSD | |
| MERS-CoV (5HOL) | 61 | 0.6 | 62 | 0.5 | 61 | 0.6 |
| SARS-CoV (2FAV) | 43 | 1.3 | 44 | 1.3 | 43 | 1.3 |
| Feline-CoV (3EW5) | 32 | 1.8 | 32 | 1.9 | 32 | 1.7 |
| HCoV-229E (3EJG) | 31 | 2.0 | 29 | 2.0 | 29 | 2.0 |
| O. Iheyensis (5L9K) | 27 | 2.1 | 27 | 2.1 | 32 | 2.1 |
In contrast, A326 is unique to lineage C macrodomains. Other macrodomains possess bulkier residues adjacent to the conserved Asp in R1 and retain their ability to hydrolyze ADPR (Fig. 1E). In the MERS and HKU4 proteins, this residue is Ala, and this residue forms a hydrogen bond to the adenine N1 in both the ADPR and NAD+ complexes. A326 forms a hydrogen bond to the adenine N1 in both ADPR and NAD+ complexes. The mutation of this residue resulted in a slight decrease in binding affinity and slightly reduced enzymatic activity (the reduction in enzyme activity was not statistically significant) (Table 3 and Figs. 4 and 5). This residue therefore contributes to ligand binding and also may assist catalytic activity to a small degree in lineage C β-CoV.
Residues in loops 1 and 2 also contribute to binding and catalysis but are not unique to group 2c coronaviruses. G351 (loop 1) forms hydrogen bonds to the diphosphate in most macrodomains. The mutation of this residue to a bulky, hydrophobic amino acid (G351L) resulted in a significant decrease in ADPR affinity and deMARylation activity. We hypothesize that the introduction of a leucine sterically prevents ADPR from adopting the binding mode observed in the wild-type protein by changing the conformation of loop 1. This is indicated by the greatest decrease in binding affinity of all three mutants and a statistically significant decrease in enzymatic activity. This suggests the structural features of loop 1 are critical to function. In other studies of viral macrodomains, the mutation of the loop 1 residues also affected ADPR binding and catalysis (11).
I434 is a highly conserved residue in loop 2. The I434A mutant also showed a decrease in binding affinity and enzymatic activity. This is likely due to the increased solvent accessibility from an open binding site. Without a bulky aliphatic sidechain, loop 2 would be unable to form a steric bridge to loop 1, leaving the diphosphate binding region (R2) solvent exposed.
The low RMSD to the MERS macrodomain, the conserved identity and orientation of key residues (D325, N343, G351, and I434), and the formation of a closed bridge between loops 1 and 2 upon ADPR binding support the hypothesis that the HKU4 domain is a conserved viral macrodomain which is more structurally similar to its homolog of the same lineage (lineage C) than to a more distant relative (lineage B).
Open versus Closed Active Sites.
Although several macrodomains form a closed bridge over the binding cavity (e.g., FCoV, PARP15 macrodomain 2, Oryza sativa), only the β-CoVs form this bridge via an Ile in loop 2 (Table 5). For example, the nsp3 macrodomain from CHIKV adopts a closed conformation via a valine in loop 2, whereas the macrodomain 2 in PARP15 utilizes an arginine from loop 1. Based on currently available macrodomain structures, a closed loop involving a loop 2 residue is a conserved feature of viral macrodomains.
Table 5.
Significant structural and functional features of viral macrodomains
| Macrodomain (PDB ID)* | R1 | R2 | Hydrolase | ADPR affinity (KD), µM |
| π–π interactions (adenine) | Bridge | |||
| HKU4 (6MEA) | x (Asn) | Closed (loop 2 Ile) | ✔ | 14 |
| SARS (2FAV) | x (Asn) | Closed (loop 2 Ile) | ✔ | 24 (21) |
| MERS (5DUS) | x (Asn) | Closed (loop 2 Ile) | — | 2.95 (42) |
| CHIKV (3GPO) | x (Arg) | Closed (loop 2 Val) | ✔ | 5 (44) |
| FCoV (3EW5) | ✔ (Tyr) | Closed | — | 400 (79) |
| IBV (3EWP) | x | Closed | X | — (80) |
| VEEV (3GQO) | x | — | ✔ | 3.9 (44) |
| HCoV-229E (3EWR) | ✔ | Closed | ✔ | 28.9 (80) |
| MHV† | x (Asn) | Open | ✔ | — |
| HEV† | x (Leu) | Open | ✔ | >50 (21) |
| SFV† | x (Arg) | Open | ✔ | — |
| SINV† | ✔ (Tyr) | Closed | ✔ | — |
| FIPV† | ✔ (Tyr) | Closed | ✔ | — |
| ONNV† | x (Arg) | Closed | ✔ | — |
| MacroD2 (4IQY) | ✔ | Open | ✔ | 0.15 (47) |
| PARP-15 MD2 (3V2B) | ✔ | Closed (loop 1 Arg) | — | — (81) |
| PARP-14 MD3 (4ABK) | ✔ | Open | — | 1.9 (81) |
| PARG (5A7R) | ✔ | Closed | ✔ | — (82) |
| Oryza sativa (5LW0) | x | Closed | — | — |
The closed conformation is observed in all β-CoV and other viral macrodomains. For example, CHIKV and HCoV-229E adopt closed conformations with ligand solvent accessibility values of 11.8% and 7.9%, respectively (44, 45). In contrast, most macrodomains with open loop conformations also exhibit much lower ADPR binding affinity; for example, HEV and SFV macrodomains have not been shown to bind ADPR or bind weakly (46). An exception to this trend includes human MacroD2, which employs Y190, from loop 2, to form a hydrogen bond with the distal ribose, resulting in a higher affinity despite the open conformation (47). In the HKU4 structures, ligand accessibility values were 10% (closed) in the ADPR complex, which is significantly lower than the 21% accessibility value in the NAD+ complex (open). The closed structure may be employed by β-CoV macrodomains because they lack residues to form π–π interactions with the adenine moiety (Table 5). This feature may increase ligand affinity and assist in enzyme activity by optimizing the ADPR orientation. Ligand solvent accessibility is important in inhibitor rational design (48, 49). Lower solvent accessibility of the ligand often correlates with the strength of binding and number of inhibitor–protein interactions (50). The correlations between buried surface area and inhibitor potency have been shown (51–53).
In contrast, the HKU4 macrodomain adopts an open conformation similar to HEV and MacroD2 when other ligands (NAD+ or ADPG) are bound. This indicates a different conformation in which ligands may enter or exit the active site. An interaction with K347 and contacts to the backbone rather than side chain of I352 are found in the two open complexes. Specific stabilization of this conformation could be employed to select an inactive conformation of the protein and produce macrodomain inhibitors. Binding of the carbazole inhibitor GeA-69 to the allosteric site of PARP14 macrodomain 2 was accompanied by a loop movement, showing that stabilization of specific loop conformations can be employed in macrodomain inhibitor design (54).
Recently, conformational plasticity of the VEEV macrodomain loops 1 and 2 was observed (55). A transition between the apo and ADPR-bound forms of this protein occurred on the microsecond to millisecond timescale, and several residues in the loop underwent chemical shift perturbations upon ADPR binding. Malet et al. observed both open and closed loop conformations (44). Our data further support that conformational changes in these loops are related to binding and catalysis.
NAD+ Binding.
NAD+ is accommodated with a set of changes in the interaction profile. This is likely because the distal ribose in NAD+ positions its C1′ away from the cavity to accommodate nicotinamide via a rotation along the C4′ to C5′ bond. Compared to ADPR, there is a loss of two hydrogen bonds to the distal ribose. The RMSD between corresponding ligand positions in ADPR and NAD+ is 2.73 Å. In addition, fewer water molecules are present in the binding site to provide water-mediated interactions. These considerations correspond to the observed weaker binding affinity estimated for NAD+ in the mM range.
ADPG Binding.
Of the three ligands in this study, ADPG forms the lowest number of hydrogen bonds. The adenine does not interact with D325 and instead forms hydrogen bonds with the backbone of A326. This likely results from the change in conformation of the adenine moiety (rotation of 20° about the glycosidic bond). Unlike the other two ligands, the β-phosphate of ADPG shows no direct interactions. Accommodating the larger pyranose ring in this ligand appears to disrupt the conserved adenine binding mode (44).
All ligands display interactions with the SAGIF motif where S431 and F435 act as hydrogen bond donors, suggesting these are key parts of the pharmacophore. While S431’s side chain oxygen forms a hydrogen bond with the main chain oxygen of Pro401, S431’s main chain amide nitrogen forms a hydrogen bond with O1B of the ADPR β-phosphate. A similar interaction, S431 NH-β-phosphate, was observed in the MERS-CoV-ADPR complex (42, 56). This suggests that the backbone nitrogen atoms of these residues form a key part of an ADPR pharmacophore, which also includes G349, N343, and I434. It is intriguing to observe that the binding pocket optimized to accommodate a pentose sugar can be occupied by a hexose, glucopyranose; however, only two hydroxyls of Glc (O2 and O3) form hydrogen bonds to the pocket.
The glucose adopts an O,3B conformation, which is ∼4 to 6 kcal/mol higher in energy than the 4C1 conformation (57, 58). Though this conformation is likely stabilized by van der Waals interactions (A342 and F435) and hydrogen bonding (K347 and N343), this probably contributes to the low-affinity binding of ADPG.
DeMARylating Enzyme.
Viral macrodomains were previously shown to act as mono-ADP ribosylhydrolases, and this deMARylation contributes to the antiviral innate immune response (17, 59). In contrast to the PARG enzymes, viral macrodomains so far exhibit primarily deMARylation activity and limited dePARylation activity in vitro (11, 24, 60). The most notable exception is the macrodomain in HEV, which exhibits robust dePARylation activity upon interactions with a viral-encoded helicase (24). Therefore, we focused on investigating the potential deMARylation activity of the HKU4 macrodomain.
Stabilized by D325 and A326 in R1, the backbone of residues in loop 1 and loop 2, and N343 in R3, a modified MARylated peptide is optimally positioned for enzymatic cleavage. We hypothesize that the water molecule that is positioned near the α-phosphate in all complexes is set up for a nucleophilic attack on the C1″ of the distal ribose (SI Appendix, Figs. S2 and S3). The ester bond to the ADPR modification is hydrolyzed by the water molecule, and the product is released. In an alternative mechanism proposed for bacterial macrodomains, enzymatic deacetylation occurs via a nucleophilic water molecule that is deprotonated by a catalytic aspartate (61). Since viral macrodomains do not have acidic residues positioned in helix 2, this mechanism is not likely.
Compared to the MacroD2 control, the wild-type HKU4 protein has a lower binding affinity for ADPR (MacroD2, 0.15 μM; HKU4, 14 μM) but displays a 19% higher hydrolase activity. These data are consistent with studies of the CHIKV (alphavirus) macrodomain, which also indicated that binding and hydrolysis are partially separable with contributions from different residues (11, 37, 59, 62); for example, the mutation of residue Y114 in CHIKV led to reduced hydrolysis but increased ADPR binding. In HKU4, this residue is F435; the mutation of the adjacent I434 showed slight effects on both binding and hydrolysis. In HKU4, mutation of G351L led to undetectable binding but residual hydrolase activity. Additional interactions with a MARylated peptide, such as those shown in the modeling experiments (Fig. 6), could contribute to catalysis.
While all three mutations affected ligand binding, statistically significant reductions in hydrolase activity were not observed until mutations were made in the R2 region (G351L). Enzymatic activity decreased by 73% for the G351L mutation, which suggests that imposing a bulky residue into the pocket has the greatest effect on catalysis. Since all solved β-CoV complexes present a steric bridge across the binding cleft, the I434A mutation was made and showed a lesser decrease in activity. This indicates that activity is also related to the presence of a bulky hydrophobic residue at this position, which may influence the shape of the binding pocket and the orientation and solvent accessibility of the ligand.
Hydrophobic interactions between a ligand and binding pocket have often been exploited to design tight-binding inhibitors. For example, interactions between an Ile side chain of the inhibitor and the HIV protease binding pocket contribute about 3 kcal/mol to the free energy of binding (63). Hydrophobic inhibitor–protein interactions are important in the recognition of arylsulfonamides by carbonic anhydrase and of peptides by human dipeptidyl peptidase IV (48, 64). Furthermore, lipidation of CoV fusion inhibitors increased their inhibitory potency, and hydrophobic interactions were predicted to contribute to the binding of SARS-CoV-2 Mpro inhibitors (65, 66). Therefore, interactions with I434 and other loop 2 residues (e.g., L429, A432, and F435) could be exploited to design macrodomain inhibitors containing lipid moieties, aryl, or alkyl groups.
NAD+ does not form the same interaction with loop 1 and does not orient its ribosidic bond near the catalytic water (SI Appendix, Fig. S2). As a result, the HKU4 macrodomain does not hydrolyze NAD+ to ADPR.
Interaction with Substrates.
Molecular modeling has been used extensively to predict conformations, interactions, and dynamics of protein–ligand complexes (67). For example, modeling of the mono-ADP ribosylhydrolase ARH3 revealed residues that participate in the binding and demodification of ADPR (68). To investigate peptide–macrodomain binding, we conducted in silico docking and found that MARylated peptides can be accommodated in the binding pocket of the macrodomain. A model 18-residue peptide sequence derived from the BRCT-WGR linker region of PARP1, in which ADP ribosylation sites have been extensively explored, was chosen for this study. Each peptide was modified at only one site, namely E488 or E491, which are well-characterized ADP ribosylation sites in PARP1 (38, 39, 69). This left the remaining 17 amino acids unconstrained by secondary or tertiary structure and free to interact with the protein. Based on the lower variation in binding modes, greater predicted binding energy, higher number of intermolecular interactions, and positioning of ADPR in the pocket, it is reasonable to suggest that the E491-modified sequence provides interactions that are more similar to a physiological substrate.
While it is known that viral macrodomains possess the ability to bind and deMARylate proteins, three-dimensional structural elucidations of such complexes have not been determined. This model aids in that understanding by predicting several modes that would allow for the macrodomain to hydrolyze ADPR. The peptide-binding pocket could be specifically targeted to disrupt coronavirus activity for developing potential antiviral treatments. The preference for the peptide MARylated at E491 versus E488 suggests ways in which substrates might be preferentially recognized and thus provide specificity. Cellular PARPs 12 and 14, or their substrates, are possible physiological targets of this macrodomain (70). This may include components of replication–transcription complexes or stress granules (SGs). For example, MARylation of G3BP1, a major SG component, was lost upon overexpression of CHIKV nsp3 (71). Other potential substrates are viral nonstructural proteins, such as the CHIKV nsp2 viral protease, which is MARylated by PARP10 and activated by deMARylation (72).
Conclusions
This study provides a detailed structural analysis of a bat lineage C β-CoV macrodomain. The HKU4 macrodomain shares structural and functional features with other viral macrodomains, such as a conserved ADPR binding pocket and hydrolase activity. We show that loop 1 interactions, particularly G351, are key for binding adenine-containing ligands. The binding cavity adopts a closed conformation with ADPR, that is not observed with its analogs (NAD+ and ADPG), by forming a steric bridge between loops 1 and 2. Although the affinity for ADPR is lower than for other macrodomains, the HKU4 macrodomain is shown to be an efficient deMARylating hydrolase. The binding site is able to accommodate two analogs that provide starting points for inhibitor design and insights into key interactions.
It is evident that the β-CoVs share common features that can be exploited in the development of antiviral treatments. While interactions with A326 appear to be unique to lineage C β-CoVs, there are conserved structural features of bat macrodomains such as a closed steric bridge and functional features such as deMARylating modified substrates. If structural and functional features are retained in the β-CoV genus as they cross the species barrier, then it is reasonable to suggest that the features described in the paper may be consistent with CoVs that emerge from bats in the future.
Methods Summary
Protein Expression and Purification of HKU4 Macrodomain.
The HKU4 macrodomain and mutants were expressed in the Escherichia coli BL21(DE3) pLysS strain using the pET15b-TEV NESG (DNASU, clone EvNO00336943) vector (44). Protein purification employed nickel affinity chromatography followed by dialysis against 4M urea, tag cleavage with tobacco etch virus protease, a second affinity chromatography step, and size-exclusion chromatography. Crystals were grown using 20 to 25% polyethylene glycol (PEG) 3350 in Hepes buffer at pH 7.0 or 7.5. Diffraction datasets were collected either on our home source or at the Advanced Photon Source on the Southeast Regional Collaborative Access Team 22ID beam line. For data processing, we used a combination of HKL3000, XDS, Xia2, and Mosflm (73–76). DeMARylation assays were carried out with the method of McPherson et al. (11, 36). Detailed methods for structure refinement and functional studies are described in the SI Appendix. Coordinates and structure factors have been deposited in the Protein Databank.
Supplementary Material
Acknowledgments
The work conducted in this study was supported by the NIH Grant R35GM119456, University of Alabama at Birmingham Faculty Startup Funding, and the University of Alabama at Birmingham Department of Chemistry. C.C.S.D. and N.S. are partially funded through R01DE029007. This work is based on research conducted at the Southeast Regional Collaborative Access Team 22-ID beamline, which is funded by the National Institute of General Medical Sciences from the NIH (P41 GM103403). The deMARylation assay was developed through NIH R01GM104135 (A.K.L.L.) and T32CA009110 (R.L.M.). This research used resources of the Advanced Photon Source. Use of the Advanced Photon Source is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract number W-31-109-Eng-38. Portions of this work were excerpted from the following PhD thesis: Hammond, R., “Macrodomain mystery: investigating the structure-function link in novel Tylonycteris HKU4 and Rousettus HKU9 coronavirus proteins,” PhD thesis, University of Alabama at Birmingham, Birmingham, AL. Ann Arbor: ProQuest Dissertations and Theses, 2018 (87).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004500118/-/DCSupplemental.
Data Availability.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank [ID codes 6MEA (ADPR complex), 6MEB (NAD+ complex), and 6MEN (ADPG complex)].
References
- 1.Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M. R., Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 78, 9977–9986 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Groot R. J., et al. , Middle East respiratory syndrome coronavirus (MERS-CoV): Announcement of the coronavirus study group. J. Virol. 87, 7790–7792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E., van Doremalen N., Falzarano D., Munster V. J., SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 14, 523–534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu A., et al. , Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27, 325–328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Center C. R., Mortality analyses (2020). https://coronavirus.jhu.edu/data/mortality. Accessed 10 July 2020.
- 6.WHO , Coronavirus disease (COVID-2019) situation report 150 (2020). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed 18 June 2020.
- 7.Sohrabi C., et al. , World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 76, 71–76 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snijder E. J., et al. , Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331, 991–1004 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Boheemen S., et al. , Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio 3, e00473-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Báez-Santos Y. M., St John S. E., Mesecar A. D., The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Res. 115, 21–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McPherson R. L., et al. , ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc. Natl. Acad. Sci. U.S.A. 114, 1666–1671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahel D., et al. , Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feijs K. L., Forst A. H., Verheugd P., Lüscher B., Macrodomain-containing proteins: Regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat. Rev. Mol. Cell Biol. 14, 443–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han W., Li X., Fu X., The macro domain protein family: Structure, functions, and their potential therapeutic implications. Mutat. Res. 727, 86–103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buschbeck M., et al. , The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol. 16, 1074–1079 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Rack J. G., Perina D., Ahel I., Macrodomains: Structure, function, evolution, and catalytic activities. Annu. Rev. Biochem. 85, 431–454 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Fehr A. R., Jankevicius G., Ahel I., Perlman S., Viral macrodomains: Unique mediators of viral replication and pathogenesis. Trends Microbiol. 26, 598–610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung A. K. L., McPherson R. L., Griffin D. E., Macrodomain ADP-ribosylhydrolase and the pathogenesis of infectious diseases. PLoS Pathog. 14, e1006864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H.-J., et al. , The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180, 567–582 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pehrson J. R., Fried V. A., MacroH2A, a core histone containing a large nonhistone region. Science 257, 1398–1400 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Egloff M. P., et al. , Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 80, 8493–8502 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karras G. I., et al. , The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen M. D., Buckle A. M., Cordell S. C., Löwe J., Bycroft M., The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330, 503–511 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Li C., et al. , Viral macro domains reverse protein ADP-ribosylation. J. Virol. 90, 8478–8486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parvez M. K., The hepatitis E virus ORF1 ‘X-domain’ residues form a putative macrodomain protein/Appr-1″-pase catalytic-site, critical for viral RNA replication. Gene 566, 47–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorbalenya A. E., Koonin E. V., Lai M. M. C., Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, α- and coronaviruses. FEBS Lett. 288, 201–205 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kusov Y., Tan J., Alvarez E., Enjuanes L., Hilgenfeld R., A G-quadruplex-binding macrodomain within the “SARS-unique domain” is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology 484, 313–322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fehr A. R., et al. , The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. MBio 7, e01721-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ithete N. L., et al. , Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg. Infect. Dis. 19, 1697–1699 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou P., et al. , A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo P. C., et al. , Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 81, 1574–1585 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleine H., et al. , Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell 32, 57–69 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Yu M., et al. , PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene 24, 1982–1993 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Heer C. D., et al. , Coronavirus and PARP expression dysregulate the NAD Metabolome: a potentially actionable component of innate immunity. bioRxiv: 10.1101/2020.04.17.047480 (6 October 2020). [DOI] [PMC free article] [PubMed]
- 35.Atasheva S., Frolova E. I., Frolov I., Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication. J. Virol. 88, 2116–2130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palazzo L., et al. , ENPP1 processes protein ADP-ribosylation in vitro. FEBS J. 283, 3371–3388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abraham R., et al. , Both ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain affect neurovirulence in mice. MBio 11, e03253-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daniels C. M., Ong S. E., Leung A. K., The promise of proteomics for the study of ADP-ribosylation. Mol. Cell 58, 911–924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gagné J. P., et al. , Quantitative site-specific ADP-ribosylation profiling of DNA-dependent PARPs. DNA Repair (Amst.) 30, 68–79 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Chapman J. D., Gagné J. P., Poirier G. G., Goodlett D. R., Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 12, 1868–1880 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Holm L., Rosenström P., Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, W545-9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho C. C., Lin M. H., Chuang C. Y., Hsu C. H., Macro domain from Middle East respiratory syndrome coronavirus (MERS-CoV) is an efficient ADP-ribose binding module: Crystal structure and biochemical studies. J. Biol. Chem. 291, 4894–4902 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michalska K. et al., Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes. IUCrJ 7, 814–824 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malet H., et al. , The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J. Virol. 83, 6534–6545 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piotrowski Y., et al. , Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property. Protein Sci. 18, 6–16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neuvonen M., Ahola T., Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 385, 212–225 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jankevicius G., et al. , A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 20, 508–514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Araki M., et al. , Hydrophobic interactions at subsite S1′ of human dipeptidyl peptidase IV contribute significantly to the inhibitory effect of tripeptides. Heliyon 6, e04227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao Y., Li L., Improved protein-ligand binding affinity prediction by using a curvature-dependent surface-area model. Bioinformatics 30, 1674–1680 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Gromiha M., Ahmad S., Role of solvent accessibility in structure based drug design. Curr. Computeraided Drug Des. 1, 223–235 (2005). [Google Scholar]
- 51.Homan K. T., Tesmer J. J., Molecular basis for small molecule inhibition of G protein-coupled receptor kinases. ACS Chem. Biol. 10, 246–256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Breitenlechner C., et al. , Protein kinase A in complex with rho-kinase inhibitors Y-27632, Fasudil, and H-1152P: Structural basis of selectivity. Structure 11, 1595–1607 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Engh R. A., Bossemeyer D., Structural aspects of protein kinase control-role of conformational flexibility. Pharmacol. Ther. 93, 99–111 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Moustakim M., et al. , Discovery of a novel allosteric inhibitor scaffold for polyadenosine-diphosphate-ribose polymerase 14 (PARP14) macrodomain 2. Bioorg. Med. Chem. 26, 2965–2972 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makrynitsa G. I., et al. , Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose. J. Struct. Biol. 206, 119–127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lei J., Kusov Y., Hilgenfeld R., Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 149, 58–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Panczyk K., Plazinski W., Pyranose ring puckering in aldopentoses, ketohexoses and deoxyaldohexoses. A molecular dynamics study. Carbohydr. Res. 455, 62–70 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Plazinski W., Lonardi A., Hünenberger P. H., Revision of the GROMOS 56A6(CARBO) force field: Improving the description of ring-conformational equilibria in hexopyranose-based carbohydrates chains. J. Comput. Chem. 37, 354–365 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Alhammad Y. M. O., Fehr A. R., The viral macrodomain counters host antiviral ADP-ribosylation. Viruses 12, 384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eckei L., et al. , The conserved macrodomains of the non-structural proteins of Chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci. Rep. 7, 41746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zapata-Pérez R., et al. , Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis. Open Biol. 7, 160327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abraham R., et al. , ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc. Natl. Acad. Sci. U.S.A. 115, E10457–E10466 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nauchitel V., Villaverde M. C., Sussman F., Solvent accessibility as a predictive tool for the free energy of inhibitor binding to the HIV-1 protease. Protein Sci. 4, 1356–1364 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Snyder P. W., et al. , Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase. Proc. Natl. Acad. Sci. U.S.A. 108, 17889–17894 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia S., et al. , Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 30, 343–355 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Islam R., et al. , A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn., 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Forli S., et al. , Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 11, 905–919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abplanalp J., et al. , Proteomic analyses identify ARH3 as a serine mono-ADP-ribosylhydrolase. Nat. Commun. 8, 2055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tao Z., Gao P., Liu H. W., Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: Analysis and implications. J. Am. Chem. Soc. 131, 14258–14260 (2009). [DOI] [PubMed] [Google Scholar]
- 70.Grunewald M. E., et al. , The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog. 15, e1007756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jayabalan A. K., Griffin D. E., Leung A. K. L., Alphavirus nsP3 ADP-ribosylhydrolase activity disrupts stress granule formation. bioRxiv: 10.1101/629881 (20 June 2019). [DOI] [PMC free article] [PubMed]
- 72.Krieg S., et al. , Mono-ADP-ribosylation by ARTD10 restricts Chikungunya virus replication by interfering with the proteolytic activity of nsP2. bioRxiv: 10.1101/2020.01.07.896977 (8 January 2020). [DOI]
- 73.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M., HKL-3000: The integration of data reduction and structure solution–From diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winter G., xia2: An expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 (2009). [Google Scholar]
- 76.Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G., iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen D., et al. , Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J. Biol. Chem. 286, 13261–13271 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Wojdyla J. A., et al. , Structure of the X (ADRP) domain of nsp3 from feline coronavirus. Acta Crystallogr. D Biol. Crystallogr. 65, 1292–1300 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu Y., et al. , Crystal structures of two coronavirus ADP-ribose-1′'-monophosphatases and their complexes with ADP-ribose: A systematic structural analysis of the viral ADRP domain. J. Virol. 83, 1083–1092 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Forst A. H., et al. , Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure 21, 462–475 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Lambrecht M. J., et al. , Synthesis of dimeric ADP-ribose and its structure with human poly(ADP-ribose) glycohydrolase. J. Am. Chem. Soc. 137, 3558–3564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang J., et al. , The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roy A., Kucukural A., Zhang Y., I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y., I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.The UniProt Consortium , UniProt: The universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hammond R., "Macrodomain Mystery: Investigating the Structure-function Link in Novel Tylonycteris HKU4 and Rousettus HKU9 Coronavirus Proteins," PhD thesis, University of Alabama at Birmingham, Birmingham, AL. Ann Arbor: ProQuest Dissertations and Theses, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank [ID codes 6MEA (ADPR complex), 6MEB (NAD+ complex), and 6MEN (ADPG complex)].