Significance
Circadian rhythms are important in nearly all aspects of brain function. We previously identified diurnal rhythms in transcripts in human cortical regions that are impacted in aging and schizophrenia. Here, we identify diurnal rhythms in transcript expression across the human dorsal and ventral striatum in subjects without psychiatric or neurological disorders. We find some transcripts to be rhythmic across regions and some in only select regions. We also find distinct phase relationships across regions suggesting waves of transcript expression in specific cellular processes across the striatum. These studies establish a temporal map of rhythms across the human striatum and help in understanding the normal function of rhythms in these regions and how disruption could lead to pathology.
Keywords: human postmortem, circadian rhythms, striatum, gene expression
Abstract
The human striatum can be subdivided into the caudate, putamen, and nucleus accumbens (NAc). Each of these structures have some overlapping and some distinct functions related to motor control, cognitive processing, motivation, and reward. Previously, we used a “time-of-death” approach to identify diurnal rhythms in RNA transcripts in human cortical regions. Here, we identify molecular rhythms across the three striatal subregions collected from postmortem human brain tissue in subjects without psychiatric or neurological disorders. Core circadian clock genes are rhythmic across all three regions and show strong phase concordance across regions. However, the putamen contains a much larger number of significantly rhythmic transcripts than the other two regions. Moreover, there are many differences in pathways that are rhythmic across regions. Strikingly, the top rhythmic transcripts in NAc (but not the other regions) are predominantly small nucleolar RNAs and long noncoding RNAs, suggesting that a completely different mechanism might be used for the regulation of diurnal rhythms in translation and/or RNA processing in the NAc versus the other regions. Further, although the NAc and putamen are generally in phase with regard to timing of expression rhythms, the NAc and caudate, and caudate and putamen, have several clusters of discordant rhythmic transcripts, suggesting a temporal wave of specific cellular processes across the striatum. Taken together, these studies reveal distinct transcriptome rhythms across the human striatum and are an important step in helping to understand the normal function of diurnal rhythms in these regions and how disruption could lead to pathology.
Circadian rhythms are prominent in nearly all aspects of human physiology including brain function. During the daytime, many neurons are active to facilitate learning, alertness, movement, feeding, and other activities. At night, other neuronal processes take over to promote sleep, consolidate memories, and provide restorative actions. These rhythms in cellular function are primarily governed by a core molecular transcriptional/translational feedback loop, consisting of the CLOCK or NPAS2 proteins, which dimerize with ARNTL (also known as BMAL1), leading to the transcription of multiple genes, including the Period (PER) and Cryptochrome (CRY) genes (1). Over time, the PER and CRY proteins form a complex with other factors, enter the nucleus, and inhibit the activity of CLOCK/NPAS2 and ARNTL. Our lab and others have found that, in the human brain, there are many transcripts whose expression levels follow a circadian rhythm, presumably regulated by the core molecular clock (2–4). Most studies thus far have focused on cortical regions, and found that many of the same transcripts are rhythmic across prefrontal cortical regions with very similar phase (i.e., rhythms peak at roughly the same time of day) (2–4). Moreover, in cortical regions, the top molecular pathway represented by these rhythmic genes is typically “circadian rhythm signaling,” often followed by pathways involving second messenger signaling pathways (i.e., cAMP-mediated signaling, nNos signaling, and synaptic long-term depression) (4). A microarray study by Li et al. measured transcript rhythms across additional brain regions, including the amygdala, hippocampus, and nucleus accumbens (NAc), and found some similarities and differences across regions (2). For example, many of the core circadian clock genes were rhythmic in all regions. However, the clock-controlled gene DBP, for example, was rhythmic in all regions except for the amygdala, which also had the fewest rhythmic transcripts (2). They also found a large reduction in rhythmicity across brain regions in subjects with major depressive disorder (2). Studies from our group have found stark differences in rhythmic transcripts in cortical regions of subjects with schizophrenia compared with control subjects, as well as an impact of aging on rhythms, suggesting that dysregulation of these transcript rhythms may represent an important mechanism underlying psychiatric and neurological diseases (3, 4).
In humans, the striatum has been subdivided into three main regions that serve some overlapping and some distinct functions. In general, the ventral striatum (primarily NAc) is involved in reward and motivation, the caudate nucleus is associated with cognition and working memory, and the putamen is associated with motor control (5). Working together, these regions produce a sequential chain of events to produce goal-directed behavior, beginning with motivation for the goal, cognitive processing, and finally motor response (5). These striatal regions are composed of upward of 90% GABAergic medium spiny neurons (MSNs), which primarily project to the pallidal complex, substantia nigra, and ventral tegmental area. However, the axon collaterals from these MSNs can also terminate onto other striatal MSNs or interneurons, forming synaptic connections within and across striatal regions (5). The ventral striatum is unique in several ways. First, it contains islands of Calleja, particularly around the medial border of the NAc, which contain quiescent, immature cells, and are thought to function as an “endocrine striatopallidal system” (6). Second, the NAc receives projections from the amygdala and hippocampus, which are not present in dorsal striatal structures. Third, it has core and shell subregions with distinct roles in reward processing (5). The NAc has been strongly implicated in the regulation of goal-directed behaviors, reward value, and regulation of affective states, making it central to substance use and depressive disorders (7). While prefrontal cortical projections can be found in all three striatal regions, sensorimotor areas project primarily to the putamen (5). Indeed, activity in the lateral putamen is associated in imaging studies with repetitive and learned movements with little cognitive input (5). Thus, the putamen has been strongly implicated in movement disorders such as Parkinson’s disease (8). In contrast, the learning of sequential movements is associated with activity in more anterior striatal regions, including the caudate (9). The caudate also receives the majority of the projections from the dorsal prefrontal cortex and has been shown to be important in working memory tasks (10). Lesions to the caudate produce deficits in delayed response working memory tasks (11). The caudate has also been implicated in multiple psychiatric disorders. For example, schizophrenia is associated with early dysregulation of dopamine in the rostral caudate (12). Moreover, dysfunction of the caudate nucleus can lead to inefficient thalamic gating, resulting in hyperactivity in the orbitofrontal and anterior cingulate cortices, producing the symptoms of intrusive thoughts and anxiety associated with obsessive-compulsive disorder (13).
Studies of human striatal regions thus far have been limited by a small number of subjects, profiling of a single striatal region (or bulk striatal tissue), and profiling of all subjects regardless of time of death (TOD). One study performed a microarray analysis on the NAc of 26 subjects and looked at the impact of “loneliness” on gene expression (14). Another performed RNA sequencing (RNA-seq) in the dorsal striatum of 17 control subjects and 18 subjects with bipolar disorder (15). However, diurnal rhythms in transcript expression across multiple striatal regions have not been elucidated. Here, we used a TOD analysis to assess diurnal rhythms in transcript expression using RNA-seq data generated from NAc, caudate, and putamen from human subjects without psychiatric or neurological disorders (SI Appendix, Table S1 and Dataset S1). We find some similarities in rhythmic transcripts across regions, including phase concordance of many of the core molecular clock genes. We also find striking differences across regions. Notably, the putamen has a very large number of rhythmic transcripts compared to the caudate and the NAc. Furthermore, the top rhythmic transcripts in the NAc are primarily small nucleolar RNAs (snoRNAs) and long noncoding RNAs (lncRNAs), many of which are not rhythmic in the dorsal striatal regions. We further identify distinct phase relationships across regions suggesting “waves” of transcript expression in specific cellular processes across the striatum. Taken together, these studies establish a temporal map of transcriptome expression across the human striatum and identify potential mechanisms underlying rhythmic processes in specific regions.
Results
Rhythmic Transcript Expression in NAc, Caudate, and Putamen.
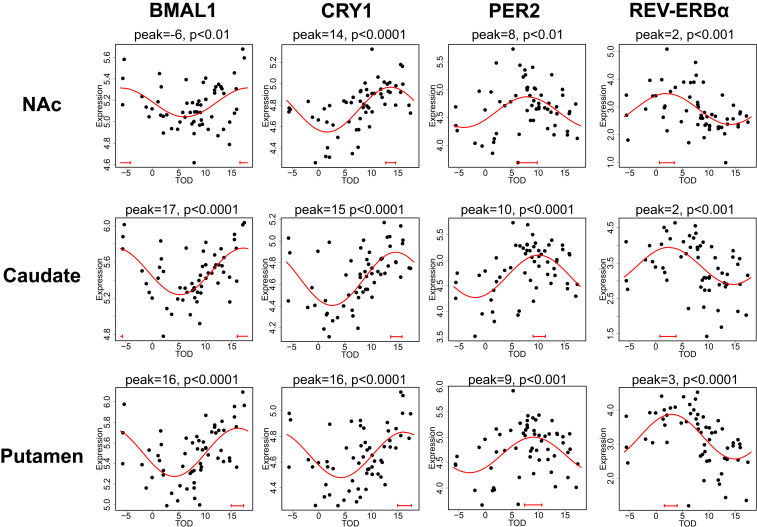
Nonlinear regression was used to detect circadian patterns of transcript expression based on individual subjects’ TOD. Sinusoidal curves were fitted to the expression data using the nonlinear least-squares method, and coefficient of determination (R2) was used as a measure of the goodness of fit. The empirical P value was estimated by comparing the observed R2 to the null distribution of R2 generated from 1,000 different TOD-permuted expression datasets. We detected many rhythmic transcripts in the striatum (Table 1 and Dataset S2), with more in the putamen [P < 0.01: n = 3,097 (20.8%)] compared to the caudate [P < 0.01: n = 1,053 (7.0%)] and NAc [P < 0.01: n = 1,344 (8.8%)]. This significance threshold (P < 0.01) captures many of the transcripts with higher amplitude and R2 values (SI Appendix, Fig. S1). Heat maps of expression levels ordered by TOD showed global rhythms in transcript expression across the three striatal regions (SI Appendix, Fig. S2). Many of the core circadian clock genes were identified as rhythmic in the NAc, caudate, and putamen. For example, the known circadian genes BMAL1, CRY1, PER2, and REV-ERBα all show strong expression rhythms in each region (Fig. 1). Furthermore, the peak time of expression for each transcript was consistent across regions, indicating phase concordance for these circadian genes. These peak times were also very similar to what our group has reported previously in human postmortem cortex (3, 4).
Table 1.
Comparison of the number of rhythmic transcripts in each striatal region at various significance thresholds
| NAc | Caudate | Putamen | |
| Total transcripts | 15,330 | 15,041 | 14,866 |
| No. of rhythmic transcripts (% of total) | |||
| q < 0.01 | 270 (1.8) | 9 (0.1) | 581 (3.9) |
| q < 0.05 | 664 (4.3) | 72 (0.5) | 3,212 (21.6) |
| P < 0.01 | 1,344 (8.8) | 1,053 (7.0) | 3,097 (20.8) |
| P < 0.05 | 3,057 (19.9) | 3,290 (21.9) | 6,153 (41.4) |
Fig. 1.
Circadian gene expression patterns of four canonical circadian clock genes in the NAc (Top), caudate (Middle), and putamen (Bottom). In the scatterplots, each dot represents a subject, with the x axis indicating TOD on a ZT scale (−6 to 18 h) and the y axis indicating level of transcript expression. The red line is the fitted sinusoidal curve. Confidence intervals (90%) of peak estimates are depicted by the red bar in each plot. All four clock genes show rhythms in each striatal region, with consistent peak times across regions. Peak time and P values are located above each scatterplot.
Characterization of the Top Rhythmic Transcripts in Each Region.
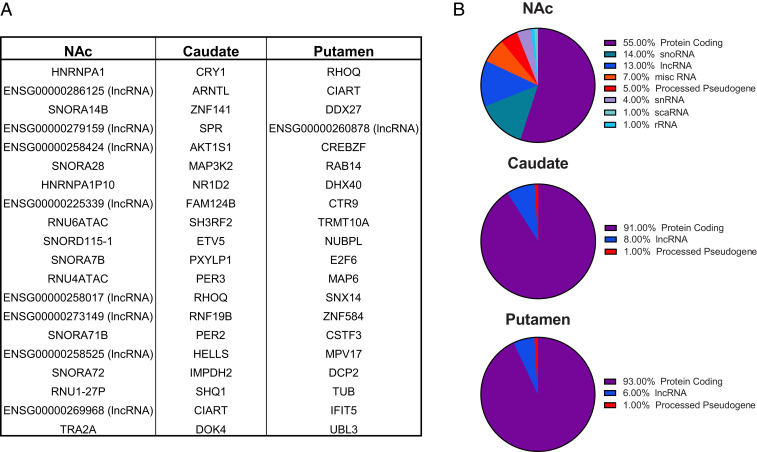
The top 20 transcripts with significant rhythms in expression are listed in Fig. 2A, with scatterplots of the top three rhythmic transcripts in each region depicted in SI Appendix, Fig. S3. Many of the top rhythmic transcripts in the caudate were core circadian clock genes, including CRY1, ARNTL, PER2, and PER3. CIART, which encodes the circadian protein CHRONO, was rhythmic in both the caudate and the putamen (Fig. 2A). Recently published studies from our group have identified CIART as a top rhythmic transcript in the human postmortem cortex (3, 4). Notably, many of the top rhythmic transcripts in the NAc were noncoding RNAs, including snoRNAs, small nuclear RNAs (snRNAs), and lncRNAs (Fig. 2A). Biotype charts (Fig. 2B) of the top 100 rhythmic transcripts showed that a majority of these top rhythmic transcripts in the caudate and putamen were protein-coding (91% and 93%, respectively), with a smaller number of lncRNAs (8% and 6%, respectively; Fig. 2B). In contrast, only 55% of the top rhythmic transcripts in the NAc were protein-coding transcripts (Fig. 2B). The rates of mapping to protein-coding transcripts, as well as the percentage of expressed protein-coding transcripts, appeared to be similar across regions (SI Appendix, Figs. S4A and S5B), suggesting that the lower percentage of rhythmic protein-coding transcripts in the NAc is not due to lower overall expression or mapping of these transcripts. Despite similar rates of mapping to expressed noncoding RNAs across regions (SI Appendix, Fig. S4A), many of the top rhythmic transcripts in the NAc were noncoding RNAs, with the majority being snoRNAs [14% of transcripts (Fig. 2B) and 26.5% of mapped reads (SI Appendix, Fig. S4B)], known for their role in guiding chemical modifications of other RNAs such as ribosomal and transfer RNAs (16). Overall, 76.7% (33 of 43) of the snoRNAs detected in the NAc were rhythmic, compared to 0% (0 of 14) in the caudate and 54.5% (6 of 11) in the putamen (SI Appendix, Fig. S6).
Fig. 2.
Characterization of the top rhythmic transcripts in the NAc, caudate, and putamen. (A) Lists of the top 20 rhythmic transcripts in the NAc, caudate, and putamen. Many of the top rhythmic transcripts in the caudate are core circadian clock genes. In the NAc, most of the top rhythmic transcripts are noncoding RNAs, including snoRNAs, snRNAs, and lncRNAs. Ensembl IDs are listed for the lncRNAs, as all these transcripts were not associated with Human Genome Organisation Gene Nomenclature Committee symbols. (B) Biotype charts of the top 100 rhythmic transcripts revealed that the majority of the top rhythmic transcripts in the caudate and putamen are protein-coding, with a small percentage of noncoding RNAs (i.e., lncRNAs). In the NAc, a larger percentage of the top rhythmic transcripts were noncoding RNAs, particularly snoRNAs (14%).
Given the enrichment of rhythmic snoRNAs in the NAc, we next performed a snoRNA target analysis on the 33 rhythmic snoRNAs in the NAc using an online interactive database tool called snoDB (Dataset S3) (17). Overall, we found that rhythmic snoRNAs in the NAc have a wide variety of targets, including mRNA, rRNA, snRNA, and other snoRNA targets. For example, one rhythmic snoRNA of interest, SNORD115-1, targets HTR2C mRNA (Dataset S3), which encodes for the serotonin 5-HT2C receptor (18). Together, given the targets, these highly rhythmic snoRNAs in the NAc could be regulating processes such as ribosomal biogenesis and mRNA splicing in a diurnal manner.
Pathway and Biological Process Enrichment for Rhythmic Transcripts in Each Region.
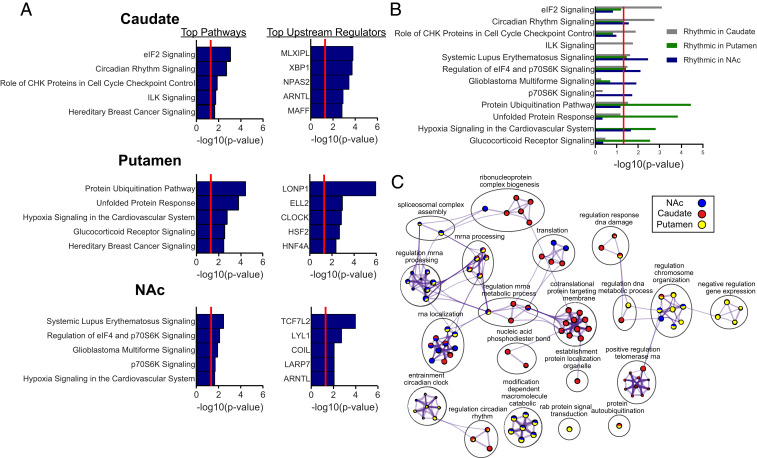
To gain additional insight into potential functional roles of rhythms in the striatum, we investigated pathways and biological processes enriched for rhythmic transcripts in each striatal region. As expected, “circadian rhythm signaling” was highly significant in the caudate, significantly enriched in the NAc, and trending toward significance in the putamen (P = 0.056; Fig. 3 A and B; Dataset S4 provides a complete list of pathways and upstream regulators for each region). Gene Ontology (GO) biological process enrichment for the top 1,000 rhythmic transcripts in each region revealed that the clusters “regulation of circadian rhythms” and “entrainment of the circadian clock” were also enriched in the caudate as well as the putamen (Fig. 3C; Dataset S5 provides a complete list of enriched processes within each cluster). Among the top predicted upstream regulators for rhythmic transcripts in the caudate were the core circadian clock genes NPAS2 and ARNTL, as well as XBP1, a transcription factor that was recently found to regulate 12-h rhythms in mouse liver (Fig. 3A) (19). The circadian protein CLOCK was also a predicted upstream regulator in the putamen, and ARNTL in the NAc (Fig. 3A). In addition to circadian-related pathways, biological processes, and upstream regulators, we also found evidence that rhythmic transcripts in striatal brain regions are involved in translation. For instance, the pathway “regulation of eIF4 and p70S6K signaling” was significant in all three striatal regions (Fig. 3 A and B). Rhythmic transcripts in the caudate seemed to be particularly enriched for translation, with the top pathway being “eIF2 signaling,” required for translation initiation, and strong enrichment for clusters related to translation (Fig. 3 A–C). Interestingly, the pathway “p70S6K signaling” was only significant in the NAc. This signaling pathway is involved in phosphorylation of the S6 ribosomal protein, which induces protein synthesis (20). Thus, rhythms in translation in the NAc might preferentially involve this mechanism. The cluster “mRNA processing,” which includes processes related to RNA splicing, was also highly enriched in all three regions. Taken together, all three regions showed strong rhythms in factors related to transcriptional regulation, mRNA processing, and translation.
Fig. 3.
Pathway and biological process enrichment for rhythmic transcripts in the NAc, caudate, and putamen. (A) Top five pathways and upstream regulators enriched for rhythmic transcripts in each striatal region (Dataset S4 provides a complete list of pathways and upstream regulators). A significance threshold of P < 0.01 (NAc, 1,344 transcripts; caudate, 1,053 transcripts; putamen, 3,097 transcripts) was used to determine rhythmicity for the transcript input list. The significance threshold for pathway and upstream regulator enrichment is P < 0.05 [−log10(P value) > 1.3 in plots]. (B) Pathway enrichment for all three regions shown on the same plot, revealing both distinct and overlapping pathway enrichment between regions. (C) GO biological process enrichment via Metascape for the top 1,000 rhythmic transcripts in each region. Meta-analysis was used to compare process enrichment across striatal regions, depicted in the Cytoscape network plots. Terms with P < 0.01, a minimum count of 3, and enrichment factor >1.5 were grouped into clusters based on their membership similarities. The most statistically significant term within a cluster was chosen to represent the cluster. The top 10 significant terms were chosen for visualization (Dataset S5 provides a complete list of all enriched processes within each cluster). The nodes are represented as pie charts, where the size of the pie is proportional to the total number of gene hits for that specific term. The pie charts are color-coded based on the identity of the gene list, where the size of the slice represents the percentage of transcripts enriched for each corresponding term. Similar terms (kappa score >0.3) are connected by edges.
We also found evidence of rhythmic transcripts in striatal brain regions related to immune signaling and cytokine production such as “systemic lupus erythematosus signaling” (Fig. 3 A and B). This is perhaps not surprising, as multiple studies have found strong circadian regulation of immune function in the brain and periphery in animal models (21). The top pathway represented by rhythmic transcripts in the putamen was “protein ubiquitination pathway,” which was also enriched to a lesser degree in the caudate (Fig. 3 A and B). Pathways related to cellular stress (e.g., “unfolded protein response” and “glucocorticoid receptor signaling”) were also highly enriched in the putamen, but not in the NAc or caudate, suggesting that, in the putamen specifically, there is a diurnal rhythm in protein folding and degradation. Predicted top upstream regulators represented by rhythmic transcripts included the mitochondrial matrix protease LONP1 (putamen), the glucose-dependent transcription factor MLXIPL (caudate), and the Wnt signaling transcription factor TCF7L2 (NAc; Fig. 3A).
Phase-set enrichment analysis (PSEA) (22) was also performed on the top 1,000 rhythmic transcripts in each striatal region to determine temporally coordinated expression of gene sets (SI Appendix, Figs. S7–S10 and Dataset S6). In the NAc, the majority of phase-enriched biological processes clustered at night around ZT19-20 (SI Appendix, Fig. S7 and Dataset S6). The processes with the most unimodal phase clustering were related to mRNA processing and splicing (SI Appendix, Figs. S7 and S10 and Dataset S6). In the caudate, there were many phase-enriched biological processes, all of which clustered between ZT21 and 24 prior to sunrise (SI Appendix, Fig. S8 and Dataset S6). The processes with the most unimodal phase clustering were related to translation and mitochondrial function (SI Appendix, Figs. S8 and S10 and Dataset S6). Finally, in the putamen, there was a subset of phase-enriched biological processes clustered around subjective dawn and subsets clustered at night (SI Appendix, Fig. S9 and Dataset S6). The processes with the most unimodal phase clustering around dawn were related to protein catabolism and cellular stress (SI Appendix, Figs. S9 and S10 and Dataset S6). The processes with the most unimodal phase clustering at night were related to mRNA processing and splicing, although the phase clustering was more widespread than in the NAc (SI Appendix, Figs. S9 and S10 and Dataset S6). Overall, these findings show significant phase clustering of specific cellular processes with different peak times across the striatum.
Rhythmic Overlap between Striatal Regions.
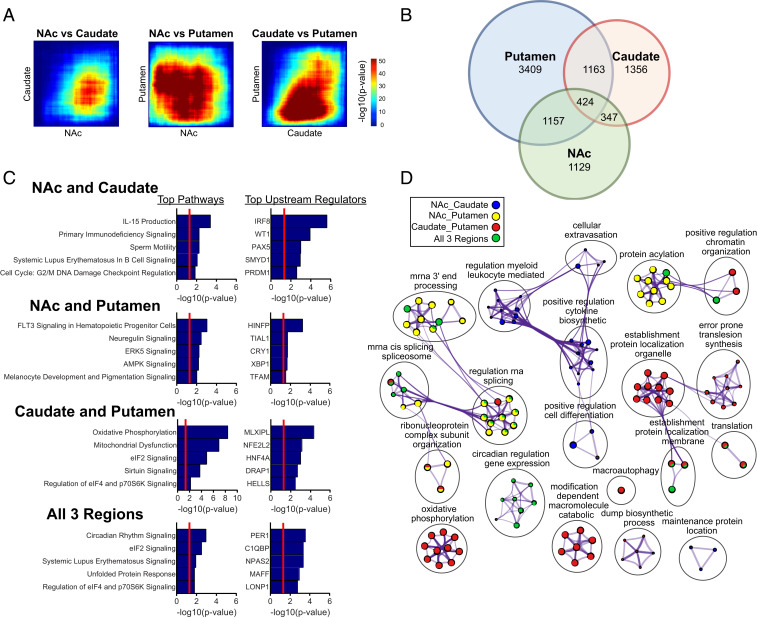
Next, we were interested in a more rigorous comparison of rhythms across striatal regions to determine rhythmic overlap and shared pathway and biological process enrichment. To determine rhythmic overlap, we employed rank–rank hypergeometric overlap (RRHO) analysis, which is a threshold-free approach that examines the degree of overlap of identified genes across two datasets. We found a high degree of overlap between the NAc and the putamen and the caudate and putamen (Fig. 4A). However, in contrast, there was a small degree of overlap between the NAc and caudate. Venn diagrams were also constructed for rhythmic transcripts at a significance threshold of P < 0.05 (Fig. 4B), showing that transcripts in common between only the NAc and caudate (347 transcripts) were fewer than between the NAc and putamen (1,157 transcripts) and caudate and putamen (1,163 transcripts). There were 424 transcripts in common between all three regions. Next, pathway and biological process enrichment was performed to determine the functional roles of these overlapped rhythmic transcripts (Fig. 4 C and D). Similar to our results obtained from analysis of individual regions, the top pathways represented by rhythmic transcripts in common between all three regions were enriched for pathways and processes related to circadian rhythms, translation, and RNA splicing (Fig. 4 C and D; Datasets S7 and S8 provide complete lists of pathways, upstream regulators, and processes for each region comparison). Among the top upstream regulators were the core circadian clock genes PER1 and NPAS2.
Fig. 4.
Overlap in rhythmic transcripts between striatal regions. (A) RRHO plot indicating a high degree of overlap in rhythmic transcripts between the NAc and putamen and the caudate and putamen. In contrast, there was a very small degree of overlap in rhythmic transcripts between the NAc and caudate. (B) Venn diagrams showing overlap of rhythmic transcripts at a significance threshold of P < 0.05. Consistent with the RRHO plots, transcripts in common between only the NAc and putamen (1,157 transcripts) and caudate and putamen (1,163 transcripts) show a higher degree of overlap than the NAc and caudate (347 transcripts). There were 424 transcripts in common between all three striatal regions. (C) Top five pathways and upstream regulators enriched for rhythmic transcripts in common between NAc and caudate, NAc and putamen, caudate and putamen, and all three regions (Dataset S7 provides a complete list of pathways and upstream regulators). Overlapping rhythmic transcripts were from the Venn diagram in B. (D) GO biological process enrichment via Metascape for the rhythmic transcripts in common between the NAc and caudate, NAc and putamen, caudate and putamen, and all three regions (Dataset S8 provides a complete list of all enriched processes within each cluster). Network plots were generated using the same parameters as in Fig. 3.
The top pathways and processes represented by rhythmic transcripts in common between only NAc and caudate related to immune function, including the “IL-15 production” pathway and “positive regulation of cytokine biosynthetic process” biological process cluster (Fig. 4 C and D). The top upstream regulator represented by transcripts rhythmic in both NAc and caudate was IRF8 (Fig. 4C), a transcription factor of the interferon regulatory factor family.
The top pathways represented by rhythmic transcripts in common between only NAc and putamen were related to growth factor signaling, including the “neuregulin signaling” and “ERK5 signaling” pathways (Fig. 4C). Among the top enriched biological processes for NAc and putamen were those related to RNA splicing and protein acetylation (Fig. 4D). The top upstream regulator represented by rhythmic transcripts in common between only NAc and putamen is HINFP, which encodes for histone H4 transcription factor.
The top pathways for rhythmic transcripts in common between only caudate and putamen were highly enriched for mitochondrial function, including “oxidative phosphorylation,” “mitochondrial dysfunction,” and “sirtuin signaling” pathways (Fig. 4C). Consistent with this finding, “oxidative phosphorylation” was among the top enriched clusters of biological processes (Fig. 4D), and one of the top enriched upstream regulators was NFE2L2, a transcription factor that plays a role in the response to oxidative stress (23).
Taken together, these results show a high degree of rhythmic overlap between the NAc and putamen and the caudate and putamen, with much lower overlap between the NAc and caudate. While pathways and processes related to circadian rhythms, translation, and mRNA processing were enriched for transcripts rhythmic in all three regions, rhythmic transcripts shared between the NAc and caudate were enriched for immune function, those between the NAc and putamen for energy metabolism and growth factor signaling, and those between the caudate and putamen for mitochondrial function.
Phase Relationships between Striatal Regions.
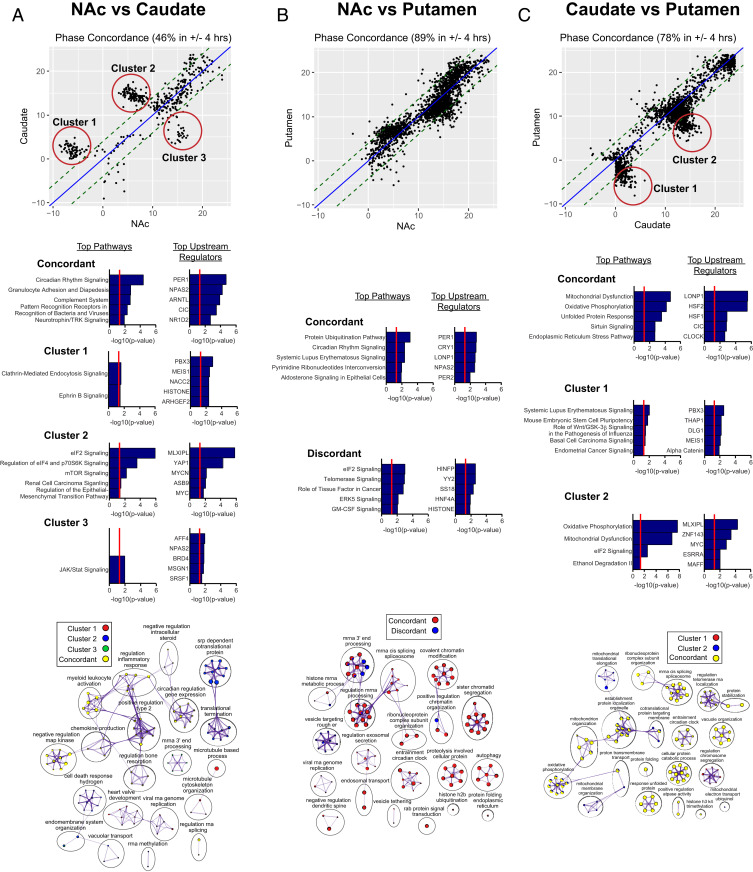
Next, we investigated phase relationships in rhythmic transcripts between striatal regions. For a given transcript, phases were plotted between the two regions, and transcripts were considered concordant if their phase differences fell within a window of ±4 h (SI Appendix, Fig. S11). Similar to the rhythmic overlap analysis, we found that there was low phase concordance between the NAc and caudate (46%; 215 concordant transcripts and 250 discordant transcripts; Fig. 5A). The top pathways and processes represented by concordant transcripts between the NAc and caudate were related to circadian rhythms (e.g., “circadian rhythm signaling” pathway and “circadian regulation of gene expression” biological process) and the immune system (e.g., the “complement system” pathway and “regulation of inflammatory response” process (Fig. 5A; Datasets S9 and S10 provide complete lists of pathways, upstream regulators, and processes for concordant and discordant transcripts). The top predicted upstream regulators were core circadian clock genes, including PER1, NPAS2, ARNTL, and NR1D2. Notably, the discordant transcripts appeared in three distinct clusters, which were analyzed separately for pathway and biological process enrichment (Fig. 5A). Peak time plots revealed similar findings, with distinct clusters of transcripts in the caudate related to translation and mitochondrial function that peak at different times (∼−12 to −5 and 0 to 10, respectively) relative to the NAc (SI Appendix, Fig. S12). The confidence intervals for peak estimates did not overlap for the majority of transcripts in each cluster, suggesting that this clustering is not due to poor phase estimation (SI Appendix, Table S2; Fig. S13 shows example scatterplots; peak estimate confidence intervals for all transcripts in each cluster are listed in Dataset S11). The three discordant clusters were enriched for different pathways and upstream regulators (Fig. 5A). Cluster 2 was unique in that these discordant transcripts were highly enriched for pathways and processes related to translation, including the “eIF2 signaling” and “regulation of eIF4 and p70S6K signaling” pathways and “translation termination” process cluster (Fig. 5A and Datasets S9 and S10).
Fig. 5.
Phase relationships between striatal regions. (A–C, Top) Phase concordance plots showing the phase relationship in rhythmic transcripts between the NAc and caudate (A), NAc and putamen (B), and caudate and putamen (C). For a given transcript, phases were plotted between the two regions, and transcripts were considered concordant if their phase differences fell within a window of ±4 h. (A, Top) There was low phase concordance between the NAc and caudate (46%; 215 concordant transcripts and 250 discordant transcripts). The discordant transcripts appeared in three distinct clusters. (B, Top) There was very high phase concordance between the NAc and putamen (89%; 1,715 concordant transcripts and 211 discordant transcripts). (C, Top) There was also high phase concordance between the caudate and putamen (78%; 1,482 concordant transcripts and 411 discordant transcripts). The discordant transcripts appeared in two distinct clusters. (A–C, Middle) Top five pathways and upstream regulators enriched for concordant and discordant transcripts, with discordant clusters analyzed separately (Datasets S9, S12, and S15 provide complete lists of pathways and upstream regulators for concordant and discordant transcripts in each region comparison). (A–C, Bottom) GO biological process enrichment via Metascape for concordant and discordant transcripts between regions (Datasets S10, S13, and S16 provide complete lists of enriched processes within clusters for each region comparison). Network plots were generated using the same parameters as in Figs. 3 and 4.
In comparing phase relationships between the NAc and putamen (Fig. 5B), we found very high phase concordance between the two regions (89%; 1,715 concordant transcripts and 211 discordant transcripts). Consistent with these findings, peak times appeared to be very similar between the NAc and putamen (SI Appendix, Fig. S12). The top pathways and processes represented by concordant transcripts between the NAc and putamen related to circadian rhythms, protein ubiquitination, and mRNA processing (including splicing; Fig. 5B; Datasets S12 and S13 provide complete lists of pathways, upstream regulators, and processes for concordant and discordant transcripts). The top upstream regulators were enriched for core circadian clock genes, including PER1, CRY1, NPAS2, and PER2. Clusters of discordant transcripts were not present in NAc and putamen comparison, so discordant transcripts were analyzed together for pathway and biological process enrichment analysis (SI Appendix, Table S2 and Dataset S14). The top pathways represented by discordant transcripts between the NAc and putamen were enriched for “eIF2 signaling” (Fig. 5B and Dataset S12). Biological enrichment analysis showed that the cluster of processes “vesicle targeting, rough ER to cis-Golgi” was enriched for discordant transcripts (Fig. 5B and Dataset S13).
In comparing phase relationships between the caudate and putamen (Fig. 5C), we found high phase concordance between the two regions (78%; 1,482 concordant transcripts and 411 discordant transcripts). The top pathways represented by concordant transcripts between the caudate and putamen were enriched for pathways related to mitochondrial function (e.g., “mitochondrial dysfunction,” oxidative phosphorylation,” and “sirtuin signaling”) and cellular stress (e.g., “unfolded protein response” and “endoplasmic reticulum stress pathway”; Fig. 5C; Dataset 15 provides a complete list of pathways and upstream regulators for concordant and discordant transcripts). Biological processes related to mitochondrial function, as well as circadian rhythms, cellular stress, and RNA spicing, were also enriched for concordant transcripts (Fig. 5C; Dataset S16 provides a complete list of enriched processes). Among the top enriched upstream regulators was CLOCK, the mitochondrial matrix protease LONP1, and the heat-shock transcription factors HSF1 and HSF2. The discordant transcripts appeared in two distinct clusters, which were enriched for different pathways and upstream regulators (Fig. 5C). Similar to the NAc and caudate comparisons, the confidence intervals for peak estimates did not overlap for the majority of transcripts in each cluster (SI Appendix, Table S2; Fig. S13 shows example scatterplots; peak estimate confidence intervals for all transcripts in each cluster are listed in Dataset S17). Similar to the concordant transcripts, cluster 2 was also enriched for pathways and processes related to mitochondrial function (Fig. 5C and Datasets S15 and S16). Unique to cluster 2 was enrichment for the clusters “inner mitochondrial membrane organization” and “mitochondrial translational elongation” (Fig. 5C and Dataset S16). Taken together, discordant clusters in the NAc and caudate and the caudate and putamen comparisons suggest a temporal wave of transcript expression in specific cellular processes across the striatum.
Discussion
In this manuscript, we utilized a TOD approach to identify transcripts with 24-h rhythms in the human NAc, caudate, and putamen taken from the same subjects. Our findings reveal multiple transcripts in each region of the human striatum that display a significant 24-h rhythm in expression. Some of these transcripts are common among all three areas. These include many of the core circadian clock genes. In addition, these core circadian clock genes are generally in phase across all regions, with peaks and troughs at the same time of day. These peak times are also similar to what our group has reported previously in human postmortem cortex (3, 4). In addition to core clock genes, many of the transcripts that are rhythmic in all three brain regions are involved in mRNA processing, particularly RNA splicing, and appear to be in phase across regions. Many posttranscriptional processes, including RNA splicing, are regulated in a diurnal manner, and in turn contribute to circadian gene expression (24, 25). Indeed, the upstream regulators of transcripts that are phase-concordant and those that are rhythmic across all three regions are largely core clock genes, suggesting that these rhythms are controlled by the molecular clock via rhythms in transcription.
There are also some striking differences in rhythmic transcripts in the striatum. First, there are many more significantly rhythmic transcripts in the putamen compared to the caudate and the NAc. The putamen is primarily involved in motor control, including preparation and execution, as well as motor learning (5). The putamen is also involved in some forms of category learning and implicit learning (5). Humans have strong diurnal rhythms in nearly every physiological process including motor activity and motor learning (26, 27), which may be driven to some extent by molecular rhythms in the putamen. In contrast, the caudate has many fewer rhythmic transcripts, with the core circadian genes at the top of the list. While the caudate is also involved in motor control, it plays an important role in several cognitive processes, including procedural learning, decision making, associative learning, and inhibitory control of actions (5). Activity in the caudate is increased with spatial and motor memory tasks, and it is thought to integrate spatial information with motor behaviors (5). The caudate is also involved in the storing and processing of memories (5). It is not clear why the caudate would have fewer rhythmic transcripts compared to the putamen. It is possible that it is due in part to its function or need for temporal flexibility. It is also possible that there are different phases of rhythms across cell types in the caudate (neuronal vs. glial, for example), which would flatten rhythms in whole tissue homogenate. Cell type-specific analysis will need to be done in future studies to determine if this is a possibility.
The NAc has a very different complement of rhythmic transcripts, with many of the top rhythmic transcripts being noncoding RNAs, including snoRNAs and lncRNAs. In general, there are more of these transcripts detected in the NAc than in the dorsal striatal regions, and those that are expressed in dorsal striatum are generally not rhythmic or have lower amplitude rhythms. The snoRNAs are small RNA molecules that facilitate posttranscriptional modifications of other RNAs (primarily ribosomal RNAs, transfer RNAs, or other noncoding RNAs) and may also be involved in RNA splicing (28). In general, there are two subclasses of snoRNAs, the antisense C/D box and the H/ACA box RNAs, based on their sequence (29). Our analysis found that the majority of the rhythmic snoRNAs in the NAc fall into the H/ACA box category. They have a common secondary structure termed a hairpin–hinge–hairpin–tail structure (29). H/ACA box snoRNAs are often involved in the process of pseudouridylation of residues in rRNA, in which uridine is converted to a pseudouridine isoform that allows the uridine to form an additional hydrogen bond outside of its bond with adenine, resulting in a more complex rRNA tertiary structure that gives it increased function within the ribosomal complex (30). In addition to targeting rRNAs, many of the rhythmic snoRNAs target other snoRNAs for modifications that likely alter their function (30). snoRNA sequences are generally found within introns of host genes, and, interestingly, we find that many of the host genes of rhythmic snoRNAs are involved in mitochondrial function. One interesting rhythmic snoRNA in the NAc is SNORD115-1, which targets the mRNA for the HTR2C serotonin receptor. Studies have found that this particular snoRNA is involved in RNA editing, resulting in an exon-skipping event, as well as other mechanisms governing alternative splicing of the HTR2C mRNA (18, 31). It is also interesting that deletion of the chromosomal region that contains SNORD115-1 results in Prader–Willi syndrome, a developmental disorder associated with many symptoms, including learning difficulties, weight gain, behavioral problems, sleep problems, and poor muscle tone (32). The extent to which other snoRNAs interact with their targets is unclear. The fact that these snoRNAs are highly rhythmic only in the NAc is striking and suggests that this particular region of the brain may use this mechanism to control rhythms in rRNA function (and ultimately protein synthesis) and perhaps alternative splicing of HTR2C and other target genes at particular times of day. Interestingly, many snoRNAs were recently found to have 24-h rhythms in the mouse liver (33). Moreover, a study in cell culture found that BMAL1 associates with NOP58, a protein that binds SNORD118 to regulate pre-rRNA (34). The authors find that BMAL1 regulates NOP58/SNORD118 nucleolar levels, leading to rhythms in pre-rRNA processing. They also show that BMAL1 ablation decreases the size and number of nucleoli per nucleus and hypothesize that BMAL1 plays an important role in nucleolar function (34). Future studies in animal models are needed to understand the functional role of rhythmic snoRNAs and other noncoding RNAs within the NAc.
When we examined the rhythmic transcripts and the pathways and biological processes that were enriched in each region, we found enrichment for pathways and processes related to circadian rhythms, translation, and mRNA processing in all regions. There were also some differences in pathway and biological process enrichment, and the times at which they peak, across the striatum. The eIF2 signaling pathway (translational initiation) is particularly enriched in the caudate, and translation-related biological processes in this region appear to be phase-clustered between ZT21 and 24 prior to sunrise. The protein ubiquitination pathway and unfolded protein response is largely specific to the putamen, and processes related to cellular stress in this region are phase-clustered around subjective dawn. Biological processes related to mRNA processing and splicing are also phase-clustered at night in the putamen, as well as NAc. Inflammation-related pathways are most highly represented in the NAc. It is also interesting that the NAc is the only region that has an enrichment for p70S6K signaling, a pathway involved in phosphorylation of the S6 ribosomal protein. These protein-coding genes in this pathway may work in concert with, or be regulated by, the noncoding RNAs discussed above to regulate translation in the NAc via a mechanism that does not occur in the dorsal striatal regions. These analyses point toward common and distinct processes in each region in which rhythmic regulation is most important, perhaps for control of neuronal activity in that region, or for the response to prolonged or abnormal activity, which could lead to problems such as neuroinflammation in susceptible regions. Interestingly, when examining transcripts that are rhythmic in both caudate and putamen (but not NAc), the top enriched pathways and biological processes are involved in mitochondrial function. This is in contrast to our prior work in the human dorsolateral prefrontal cortex, which found that, in healthy control subjects, these transcripts were generally not rhythmic, but subjects with schizophrenia actually gained a rhythm in these transcripts (4). This suggests that rhythms in mitochondrial function may be differentially regulated across brain regions, and, if these rhythms are synchronized in a region that normally does not have rhythms, it could lead to pathological effects. Future studies will be needed to determine if subjects with schizophrenia have stronger or weaker rhythms in mitochondrial-related genes in the dorsal striatum.
We decided to employ RRHO to further explore the potential overlap in rhythmic transcripts across brain regions. Surprisingly, we find that the NAc and putamen, and the caudate and putamen, have the largest overlap in rhythmic transcripts, with much lower overlap between the NAc and caudate. This is evident in the phase relationships as well. The phase concordance between the NAc and putamen is very high (89%), meaning that these regions have many of the same rhythmic transcripts and they generally cycle in phase with one another. Transcripts in phase with one another are enriched for circadian rhythm signaling and RNA splicing, while transcripts out of phase are enriched for eIF2 signaling and the secretory pathway. The phase concordance between the caudate and putamen is a bit lower at 78%, while the concordance between the NAc and caudate is slightly less than half. When looking at the concordance plots, we identified distinct clusters of transcripts that are out of phase with one another. There are two distinct discordant clusters when examining the caudate and putamen phase relationships. Pathway analyses of these clusters reveal that cluster 1 is primarily enriched for factors related to cell growth and plasticity, while cluster 2 is primarily enriched for genes related to mitochondrial function. When examining the NAc versus caudate phase relationships, we identified three distinct discordant clusters enriched for different pathways and processes. While cluster 1 is enriched for clathrin-mediated endocytosis signaling and cluster 3 for JAK/Stat signaling, cluster 2 is highly enriched for pathways and processes related to translation, as well as other signaling pathways. It is unclear why these transcripts would be out of phase, but it suggests a possible “wave” of function for these processes across time of day in the striatum. Indeed, when we look at peak times in common rhythmic transcripts across regions, we find that transcripts involved primarily in translation and mitochondrial function generally peak at different times in the caudate compared to other regions. Future studies are needed to understand how modulation of time of day in these signaling pathways and protein syntheses alters brain activity and function.
One limitation of our study is the relatively small sample size. Due to the limited sample size and the nature of the study design, the pathway and process enrichment analyses in our study are somewhat exploratory and use P value cutoffs as statistical thresholds for determining rhythmicity, similar to what we have reported previously (4). Another limitation is the imbalance between the sexes (majority male), with limited statistical power to reliably measure potential sex differences in rhythmicity. This difference is driven by the fact that our brain bank contains many more samples from male subjects than female subjects. Future studies will focus on utilizing larger sample sizes in male and female subjects to specifically test whether any of the rhythmicity patterns are sex-specific.
Taken together, these studies define a temporal map of diurnal transcript rhythm architecture across three striatal regions in the human brain. The fact that this tissue was all processed from the same individuals gives us confidence in the findings. These studies are important in helping us understand the normal function of these regions and how disruption to these rhythms could lead to pathology. It also gives us an opportunity to look for processes that could be protective against insult and perhaps lead to novel therapeutics. These results are also informative regarding the appropriate timing of medications that impact striatal regions, as they show that it could be more or less effective to hit particular targets at specific times of day (35). We are particularly intrigued by the high level of rhythmicity in noncoding RNAs specifically within the NAc. It will be interesting to follow up on these results to determine the importance and function of rhythms in these transcripts in this region, which is so important for reward, motivation, and salience. Future studies will also help determine what drives these rhythms in particular regions, including inputs from specific brain areas, role of the core pacemaker in the suprachiasmatic nucleus, the molecular mechanisms of transcript regulation, and coordination between rhythms in transcription and translation.
Materials and Methods
Human Postmortem Brain Samples.
NAc, caudate, and putamen tissue samples were obtained through the University of Pittsburgh Brain Tissue Donation Program and the NIH NeuroBioBank. The absence of lifetime psychiatric disorders was determined by an independent committee of experienced clinicians using information from clinical records, toxicology results, and standardized psychological autopsy (36). Subjects were included based on three criteria: 1) known TOD within a 4-h window and meeting the criteria of rapid death, 2) age less than 65 y, and 3) postmortem interval less than 30 h. A total of 60 subjects met these criteria, and summary subject and tissue characteristics are described in SI Appendix, Table S1 (information for each subject listed in Dataset S1). One subject (ID no. 13250) was excluded from all analyses due to overall low expression levels and low correlation with the other subjects (n = 59 for all analyses). Further details are provided in SI Appendix, SI Methods.
TOD Analysis in the Zeitgeber Time Scale.
To analyze rhythmic gene expression, the TOD for each subject was normalized to a zeitgeber time (ZT) scale. TOD was first converted to coordinated universal time (UTC) by adjusting time zone and daylight savings time. The sunrise time was calculated by adjusting UTC with longitude, latitude, and elevation of death place. Each subject’s TOD was set as ZT = t hours after previous sunrise (if t < 18) or before next sunrise (if t ≥ −6).
RNA-Seq and Data Preprocessing.
Total RNA was extracted from the tissue samples using a combination of TRIzol (Invitrogen) and RNeasy Lipid Tissue Mini Kit (Qiagen). RNA quantity and quality were assessed using fluorometry (Qubit RNA Broad Range Assay Kit and Fluorometer; Invitrogen) and chromatography (Bioanalyzer and RNA 6000 Nano Kit; Agilent), respectively. The NAc and dorsal striatum samples had average RNA integrity number values of 8.0 and 7.7, respectively. Libraries were prepped for RNA-seq using the TruSeq Stranded Total RNA Sample Preparation Kit (Illumina). Paired-end dual-indexed sequencing (75 bp) was performed using the NextSeq 500 platform (Illumina). Raw and processed RNA-seq data were deposited into the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under accession no. GSE160521. Further details on data preprocessing are provided in SI Appendix, SI Methods.
Rhythmicity Analyses.
Nonlinear regression was used to detect circadian patterns of gene expression based on individual TOD as described previously (3, 4) (SI Appendix, SI Methods). Heat maps for transcripts exhibiting circadian rhythms were generated for each brain region (n = 59; P < 0.01; SI Appendix, Fig. S2). Expression levels were Z-transformed for each transcript, and the transcripts were ordered by the time at which they peak. Each column represents a subject, and the subjects were ordered by their TOD. Scatter plots were also generated for individual transcript rhythms. Each dot represents a subject, with the x axis indicating TOD on a ZT scale and the y axis indicating level of transcript expression. The red line is the fitted sinusoidal curve. Scatter plots were generated for the top three rhythmic transcripts in each region (SI Appendix, Fig. S3) and core circadian clock genes (Fig. 1).
Phase concordance plots were also generated to investigate differences in phase between NAc and caudate, NAc and putamen, and caudate and putamen. Adaptively weighted (AW) Fisher’s method was used to combine multiple P values across the regions (37), which provides a meta-analyzed P value for each transcript with increased statistical power and generates weight indicators to reflect the consistency of circadian signals in the two regions [i.e., (1,1) showing rhythmic in both regions, (1,0) and (0,1) reflecting rhythmicity in only one of them]. Meta-analyzed P values were corrected by Benjamini–Hochberg procedure. Transcripts with meta-analyzed q value <0.05 and AW Fisher weight (1,1) were used for phase concordance plots. For a given transcript, phases were plotted between the two regions. Transcripts were considered concordant if their phase differences fell within a window of ±4 h. This time window was justified by a univariate Gaussian mixture model on the signed phase difference, whose decision boundaries between components are around −4 and +4 h (SI Appendix, Fig. S11). The model was fitted to the phase difference of the two brain region comparisons with distinct discordant clusters (NAc and caudate, caudate and putamen). Details on the Gaussian mixture model and peak time analysis are provided in SI Appendix, SI Methods.
Pathway Enrichment and Upstream Regulator Analysis.
Ingenuity Pathway Analysis software (Qiagen) was used to identify molecular pathways and potential upstream regulators enriched in identified transcript lists. Further details are provided in SI Appendix, SI Methods.
Biological Process Enrichment.
Biological process enrichment was performed using the Web-based portal Metascape (38) (performed June 2020) and visualized using Cytoscape (39). Process enrichment was accomplished using GO biological processes as the ontology source. Within Metascape, the multigene list meta-analysis option was used to compare process enrichment across multiple regions (38). Further details are provided in SI Appendix, SI Methods.
PSEA.
PSEA was used to identify biologically related gene sets showing coordinated expression (22). Further details are provided in SI Appendix, SI Methods.
RRHO Analysis.
RRHO is a threshold-free approach that identifies the overlap between two lists of transcripts ranked by their −log10(P value) (40, 41). This approach avoids an arbitrary threshold in conventional Venn diagram approaches. RRHO was used to identify overlap between significantly rhythmic transcripts between the NAc and caudate, NAc and putamen, and caudate and putamen.
Supplementary Material
Acknowledgments
We thank Ryan Logan, John Enwright III, and William MacDonald for useful discussions. This project was funded by a Brain and Behavior Foundation Independent Investigator Award (to C.A.M.) and NIH Grants MH111601 (to C.A.M. and G.C.T.) and MH120907 (to K.D.K.). Brain tissue for this study was provided by the University of Pittsburgh Brain Tissue Donation Program and the NIH NeuroBioBank.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016150118/-/DCSupplemental.
Data Availability.
Raw and processed RNA-seq data reported in this paper were deposited into the NCBI GEO database (accession no. GSE160521). The code used in these analyses and instructions is available at GitHub (https://github.com/weiiizong/Circadian-analysis). All other data are included in the main text and SI Appendix.
References
- 1.Takahashi J. S., Molecular neurobiology and genetics of circadian rhythms in mammals. Annu. Rev. Neurosci. 18, 531–553 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Li J. Z., et al. , Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc. Natl. Acad. Sci. U.S.A. 110, 9950–9955 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen C. Y., et al. , Effects of aging on circadian patterns of gene expression in the human prefrontal cortex. Proc. Natl. Acad. Sci. U.S.A. 113, 206–211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seney M. L., et al. , Diurnal rhythms in gene expression in the prefrontal cortex in schizophrenia. Nat. Commun. 10, 3355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haber S. N., Corticostriatal circuitry. Dialogues Clin. Neurosci. 18, 7–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer G., Gonzalez-Hernandez T., Carrillo-Padilla F., Ferres-Torres R., Aggregations of granule cells in the basal forebrain (islands of Calleja): Golgi and cytoarchitectonic study in different mammals, including man. J. Comp. Neurol. 284, 405–428 (1989). [DOI] [PubMed] [Google Scholar]
- 7.Cooper S., Robison A. J., Mazei-Robison M. S., Reward circuitry in addiction. Neurotherapeutics 14, 687–697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickson D. W., Neuropathology of Parkinson disease. Parkinsonism Relat. Disord. 46 (suppl. 1), S30–S33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boecker H., et al. , Role of the human rostral supplementary motor area and the basal ganglia in motor sequence control: Investigations with H2 15O PET. J. Neurophysiol. 79, 1070–1080 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Elliott R., Dolan R. J., Differential neural responses during performance of matching and nonmatching to sample tasks at two delay intervals. J. Neurosci. 19, 5066–5073 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partiot A., et al. , Delayed response tasks in basal ganglia lesions in man: Further evidence for a striato-frontal cooperation in behavioural adaptation. Neuropsychologia 34, 709–721 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Weinstein J. J., et al. , Pathway-specific dopamine abnormalities in schizophrenia. Biol. Psychiatry 81, 31–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Casale A., et al. , Functional neuroimaging in obsessive-compulsive disorder. Neuropsychobiology 64, 61–85 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Canli T., et al. , Differential transcriptome expression in human nucleus accumbens as a function of loneliness. Mol. Psychiatry 22, 1069–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pacifico R., Davis R. L., Transcriptome sequencing implicates dorsal striatum-specific gene network, immune response and energy metabolism pathways in bipolar disorder. Mol. Psychiatry 22, 441–449 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Dupuis-Sandoval F., Poirier M., Scott M. S., The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip. Rev. RNA 6, 381–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouchard-Bourelle P., et al. , snoDB: An interactive database of human snoRNA sequences, abundance and interactions. Nucleic Acids Res. 48, D220–D225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kishore S., Stamm S., The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311, 230–232 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Pan Y., et al. , 12-h clock regulation of genetic information flow by XBP1s. PLoS Biol. 18, e3000580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung J., Kuo C. J., Crabtree G. R., Blenis J., Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 69, 1227–1236 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Haspel J. A., et al. , Perfect timing: Circadian rhythms, sleep, and immunity–An NIH workshop summary. JCI Insight 5, 131487 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang R., Podtelezhnikov A. A., Hogenesch J. B., Anafi R. C., Discovering biology in periodic data through phase set enrichment analysis (PSEA). J. Biol. Rhythms 31, 244–257 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Ryoo I. G., Kwak M. K., Regulatory crosstalk between the oxidative stress-related transcription factor Nfe2l2/Nrf2 and mitochondria. Toxicol. Appl. Pharmacol. 359, 24–33 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Benegiamo G., Brown S. A., Panda S., RNA dynamics in the control of circadian rhythm. Adv. Exp. Med. Biol. 907, 107–122 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Torres M., Becquet D., Franc J. L., François-Bellan A. M., Circadian processes in the RNA life cycle. Wiley Interdiscip. Rev. RNA 9, e1467 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Horowski R., Benes H., Fuxe K., Striatal plasticity and motor learning-importance of circadian rhythms, sleep stages and dreaming.. Parkinsonism Relat. Disord. 10, 315–317 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Roenneberg T., et al. , A marker for the end of adolescence. Curr. Biol. 14, R1038–R1039 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Hombach S., Kretz M., Non-coding RNAs: Classification, biology and functioning. Adv. Exp. Med. Biol. 937, 3–17 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Massenet S., Bertrand E., Verheggen C., Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 14, 680–692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMahon M., Contreras A., Ruggero D., Small RNAs with big implications: New insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip. Rev. RNA 6, 173–189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bratkovič T., Modic M., Camargo Ortega G., Drukker M., Rogelj B., Neuronal differentiation induces SNORD115 expression and is accompanied by post-transcriptional changes of serotonin receptor 2c mRNA. Sci. Rep. 8, 5101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angulo M. A., Butler M. G., Cataletto M. E., Prader-willi syndrome: A review of clinical, genetic, and endocrine findings. J. Endocrinol. Invest. 38, 1249–1263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aitken S., Semple C. A., The circadian dynamics of small nucleolar RNA in the mouse liver. J. R. Soc. Interface 14, 20170034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cervantes M., et al. , BMAL1 associates with NOP58 in the nucleolus and contributes to pre-rRNA processing. iScience 23, 101151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruben M. D., et al. , A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine. Sci. Transl. Med. 10, eaat8806 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glausier J. R., Kelly M. A., Salem S., Chen K., Lewis D. A., Proxy measures of premortem cognitive aptitude in postmortem subjects with schizophrenia. Psychol. Med. 50, 507–514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seney M. L., et al. , Opposite molecular signatures of depression in men and women. Biol. Psychiatry 84, 18–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y., et al. , Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plaisier S. B., Taschereau R., Wong J. A., Graeber T. G., Rank-rank hypergeometric overlap: Identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38, e169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cahill K. M., Huo Z., Tseng G. C., Logan R. W., Seney M. L., Improved identification of concordant and discordant gene expression signatures using an updated rank-rank hypergeometric overlap approach. Sci. Rep. 8, 9588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed RNA-seq data reported in this paper were deposited into the NCBI GEO database (accession no. GSE160521). The code used in these analyses and instructions is available at GitHub (https://github.com/weiiizong/Circadian-analysis). All other data are included in the main text and SI Appendix.