Significance
Uniquely human biology is the result of genetic differences between humans and other primates, but identifying the critical changes still poses a major challenge. We screened >32,000 human-specific substitutions in two classes of putative transcriptional enhancers implicated in human evolution for their effects on enhancer activity in human neural stem cells, a cell type fundamental for cortical development and expansion. We identify hundreds of substitutions that modify enhancer activity either alone or in combination with other variants. We find different patterns of substitution effects in different enhancer types. Our data support substitutions often interacting by modifying the same transcription factor binding sites. The functional substitutions we identify provide a rich set of candidate loci for studies of human-specific regulatory evolution.
Keywords: gene regulation, enhancer evolution, human accelerated regions, neurodevelopment, massively parallel enhancer assay
Abstract
Genetic changes that altered the function of gene regulatory elements have been implicated in the evolution of human traits such as the expansion of the cerebral cortex. However, identifying the particular changes that modified regulatory activity during human evolution remain challenging. Here we used massively parallel enhancer assays in neural stem cells to quantify the functional impact of >32,000 human-specific substitutions in >4,300 human accelerated regions (HARs) and human gain enhancers (HGEs), which include enhancers with novel activities in humans. We found that >30% of active HARs and HGEs exhibited differential activity between human and chimpanzee. We isolated the effects of human-specific substitutions from background genetic variation to identify the effects of genetic changes most relevant to human evolution. We found that substitutions interacted in both additive and nonadditive ways to modify enhancer function. Substitutions within HARs, which are highly constrained compared to HGEs, showed smaller effects on enhancer activity, suggesting that the impact of human-specific substitutions is buffered in enhancers with constrained ancestral functions. Our findings yield insight into how human-specific genetic changes altered enhancer function and provide a rich set of candidates for studies of regulatory evolution in humans.
Changes in developmental gene regulation are hypothesized to contribute to the evolution of novel human phenotypes (1–3). However, identifying specific genetic variants that altered regulatory activity in human development remains challenging. Previous studies using comparative genomics strategies have identified two classes of regulatory elements that may encode uniquely human functions. The first class are human accelerated regions (HARs), which are highly conserved across species but exhibit a significant excess of fixed sequence changes in humans (4–8). HARs are enriched near genes implicated in brain development, and several HARs have been shown to encode transcriptional enhancers with human-specific changes in activity (4, 6–10). The second class of elements are human gain enhancers (HGEs), which show increased levels of histone modifications associated with enhancer activity in developing human tissues compared with rhesus macaque and mouse (11, 12). Thousands of HGEs have been identified in the human embryonic cortex. Chromosome conformation studies demonstrate that HGEs and HARs target genes involved in neurogenesis, axon guidance, and synaptic transmission during human cortical development (13–16).
The exact genetic changes that drive novel human regulatory functions in HARs and HGEs, and the effect they have on enhancer activity both individually and in combination, remain to be fully determined. One approach used to discover functional single-base variants is the massively parallel reporter assay (MPRA) (17), in which a synthesized library of candidate regulatory elements is cloned in front of a reporter gene containing a random oligonucleotide barcode. High-throughput sequencing of the transcribed barcode collection is then used to quantify regulatory activity. A single MPRA experiment interrogates thousands of sequence changes at once, which provides the means to comprehensively analyze sequence variation within regulatory elements in a combinatorial manner. This technique has been used to measure the effect of saturating mutations in single enhancers and to identify single nucleotide variants that may alter enhancer activity (17–19).
A recent MPRA queried 714 HARs in human and chimpanzee neural progenitor cells (20). Forty-three percent of these HARs were found to act as enhancers in this assay, two-thirds of which showed differential activity between the human and chimpanzee versions. The study also dissected the sequence variation in seven of these HARs, showing that a combination of buffering and amplifying effects of single-base substitutions generated a conserved or modified regulatory output (20). However, previous studies have not distinguished between human-specific substitutions and other variation such as chimpanzee-specific substitutions or segregating variants, which are less likely to underlie the genetics of human-specific traits. Furthermore, HARs only capture a small fraction of human-specific substitutions in the genome that may alter regulatory function. The impact of human-specific substitutions on enhancer activity overall has not been investigated.
To gain insight into this question, we used MPRAs to identify the additive and nonadditive effects of over 32,000 human-specific substitutions in HARs and HGEs on enhancer activity. We chose to use H9-derived human neural stem cells (hNSCs), a model system for studying human neurogenesis (21) (Materials and Methods), as regulatory changes in neural precursors may have contributed to the expansion of the human cortex (1). We identified 11.7% of HARs and 33.9% of HGEs as active enhancers in human neurodevelopment, of which 27.5% and 34.6%, respectively, were differentially active between human and chimpanzee.
We then designed a second MPRA to quantify how 1,300 human-specific substitutions within differentially active HARs and HGEs act alone or in combination to alter enhancer activity. We found that pairs of interacting substitutions were often in close proximity, in line with a model where transcription factor binding is modified in concert by multiple substitutions. We identified a suite of transcription factors (TFs) implicated in enhancer activity in our assay, including cell cycle regulators. Our findings reveal mechanisms of enhancer evolution in humans and provide an entry point toward a functional understanding of the genetic changes underlying the evolution of the human neocortex.
Results
Identifying Changes in Enhancer Activity in HARs and HGEs.
We used a two-stage MPRA to screen the effects of human-specific substitutions within 1,363 HARs and 3,027 HGEs on enhancer activity (Fig. 1A and Dataset S1). To be included in our screen, we required substitutions to exhibit a derived sequence change in human compared to the orthologous position in chimpanzee, orangutan, rhesus macaque, and marmoset, which we required to exhibit the same, putatively ancestral, character state. We then used the Single Nucleotide Polymorphism Database (dbSNP) (build 144) to exclude human polymorphisms (Materials and Methods). This filtering scheme yielded 32,776 human-specific substitutions (termed “hSubs”) for our study.
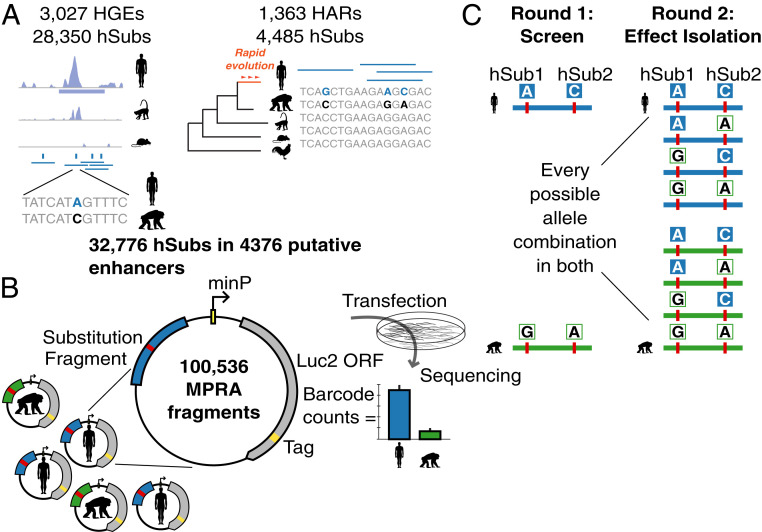
Fig. 1.
Experimental design. (A) We synthesized 137-bp MPRA fragments overlapping 32,776 hSubs in 3,027 HGEs and 1,363 HARs. (B) MPRA fragments (human in blue, chimpanzee in green) were cloned in front of a luc2 reporter gene and a random oligonucleotide barcode tag (in yellow). Sequencing and counting barcodes provide a quantitative measure of enhancer activity. (C) In stage 1 of the experiment, we screened 50,268 orthologous human–chimpanzee fragment pairs. In stage 2, the impact of genetic differences within all fragments exhibiting species-specific changes in activity was dissected by testing all possible combinations of hSubs and ancestral states in both the human and chimpanzee background reference sequences.
In the first stage of our screen, we sought to identify HARs and HGEs with potential changes in enhancer activity due to hSubs. We designed human–chimpanzee orthologous pairs of 137-bp sequence fragments centered on each substitution. Where two hSubs were in such close proximity that they would overlap the same fragment, we generated additional fragments centered on the midpoint between each pair of hSubs. This provided 100,536 fragments in total (Fig. 1A). To generate the MPRA library, we synthesized and cloned each fragment upstream of a minimal promoter driving the expression of a luc2 firefly luciferase open reading frame (ORF) tagged with a random oligonucleotide barcode (Fig. 1B and SI Appendix, Fig. S1). Each MPRA fragment was linked to 80 unique barcodes on average, yielding a library containing ∼8 million unique molecules. We included human and chimpanzee orthologous fragments in a single library to prevent batch effects that would confound potential species differences in activity.
We then carried out four replicate MPRAs. We transfected the library into hNSCs and collected both input plasmid DNA (pDNA) and total RNA (SI Appendix, Supplementary Text). Following complementary DNA (cDNA) synthesis, we used high-throughput sequencing to determine barcode counts in both the cDNA and pDNA fractions (SI Appendix, Supplementary Text). Barcode counts were summarized by fragment. We removed fragments with fewer than 12 barcodes as those showed high variance in barcode counts across replicates (SI Appendix, Fig. S2). A total of 78,487 fragments passed this threshold, hereafter called “measured fragments.” Measured fragments had on average 69.3 associated barcodes (SD = 62.4; 95% range, 13 to 233), and barcode counts showed high correlations between replicates (Spearman’s rank correlation, ρ = 0.87 to 0.89 for pDNA and ρ = 0.81 to 0.85 for cDNA, P < 1 × 10−300).
Fragment counts were highly correlated between the cDNA and pDNA fractions (ρ = 0.79 to 0.81, P < 1 × 10−300). This indicates that most fragments produced as much cDNA as predicted from their pDNA input, supporting that they did not exhibit enhancer activity in the screen. However, a subset of human and chimpanzee fragments do show increased levels of cDNA-derived barcode counts, suggesting they act as transcriptional enhancers (Fig. 2 A and B). To quantify enhancer activity for each fragment, we first calculated the ratio of cDNA counts over pDNA counts for each barcode. We then defined fragment activity as the log2 mean of activities of all barcodes assigned to that fragment. We found fragment activity to be robust between replicates (ρ = 0.73 to 0.78, P < 1 × 10−300). We then used one-tailed t tests to compare each fragment’s activity against the rest of the library to identify fragments showing significant levels of activity in the screen (Materials and Methods and SI Appendix, Fig. S3).
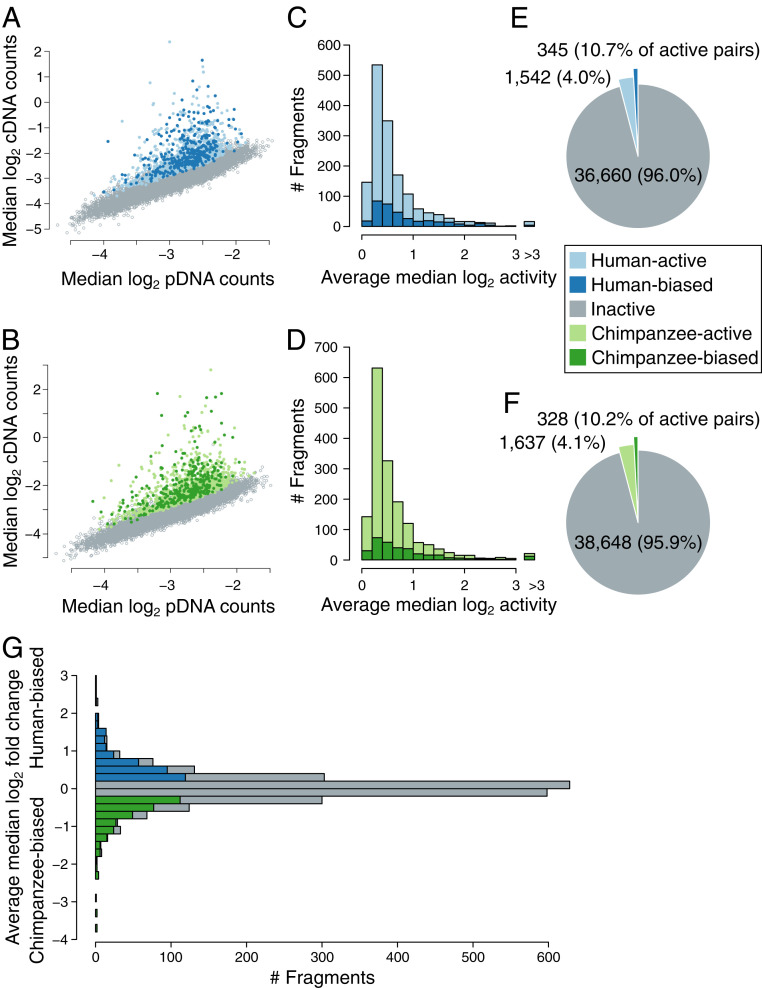
Fig. 2.
Quantifying enhancer activity using MPRA. (A and B) Comparing MPRA barcode counts in pDNA vs. cDNA fractions identifies fragments showing increased numbers of cDNA counts, indicative of enhancer activity. (A) Active human fragments are shown as light blue points and differentially active human fragments are shown in dark blue. (B) Active chimpanzee fragments are shown as light green points and differentially active chimpanzee fragments are shown in dark green. (C and D) The distribution of enhancer activity in all active and differentially active fragments in human (B) and chimpanzee (D). (E and F) The number of active and differentially active fragments detected from human (E) and chimpanzee (F). (G) The distribution of differences in activity between all active human and chimpanzee fragments. Human and chimpanzee fragments showing significant increases in activity are labeled in dark blue or dark green, respectively. Fragments showing nonsignificant activity differences are shown in gray.
We identified 3,202 fragments (4.1% of all measured fragments; SI Appendix, Table S1A) that showed significant enhancer activity (Fig. 2 C–F). Consistent with previous MPRA studies (17, 19, 22), most of these fragments showed modest activity; only 15.2% of active fragments showed an average activity higher than 2. Active fragments were more commonly found in regions with evidence of regulatory activity in hNSCs based on chromatin accessibility or histone H3 K27 acetylation (H3K27ac) peaks (SI Appendix, Table S2). Compared to HARs, HGEs contained a larger number of measured (36,656 vs. 7,884) and active fragments (2,161 [5.9%] vs. 349 [4.4%]).
We then sought to identify orthologous human and chimpanzee fragments that showed differential activity and that could be used to dissect the effects of human-specific substitutions in the second MPRA described below. We defined a set of 3,219 measured fragment pairs in which the human, chimpanzee, or both fragments were active (Materials and Methods). We applied two-tailed t tests in each replicate to identify significant differences in activity, required that fragment activity was biased in the same species direction in all replicates, and imposed a minimum average log2 difference threshold >0.2 (SI Appendix, Fig. S4). We identified 673 differentially active fragment pairs (23.5% of all active pairs; Fig. 2 E and F and SI Appendix, Table S1A). Most of these showed modest changes in activity between species (mean fold change = 1.58, 95% range, 1.17 to 3.76; Fig. 2G). However, 113 fragments (16.8%) showed a fold change larger than 2 and 15 fragments (2.2%) showed a fold change larger than 4 (Fig. 2G). We found that differentially active fragments were not more likely to be human biased than chimpanzee biased (P = 0.59, two-tailed binomial test).
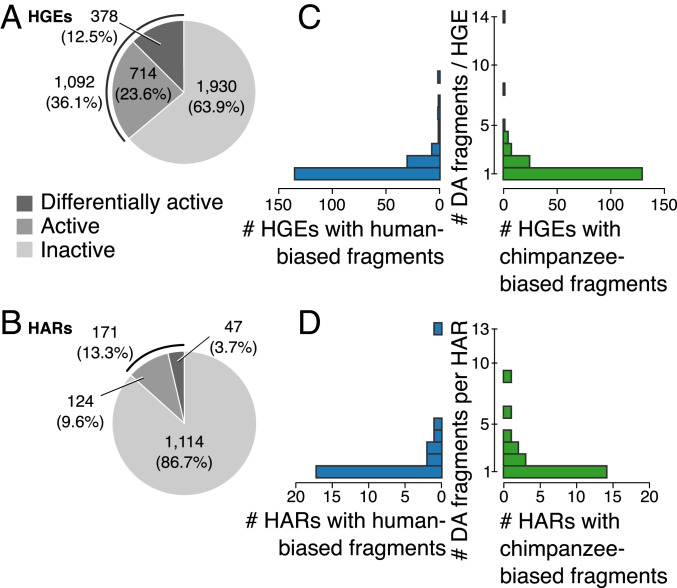
We then used these results to identify active and differentially active HARs and HGEs. We defined a HAR or HGE as active if it contained at least one active fragment and as differentially active if it contained at least one differentially active fragment. We found that 11.7% of HARs and 33.9% of HGEs were active and that 3.2% and 11.7%, respectively, were differentially active (Fig. 3 A and B). The number of human- and chimpanzee-biased HARs and HGEs are shown in Fig. 3 C and D. A substantial fraction of the putative enhancers we studied thus showed MPRA activity even though the overall number of fragments that showed activity was small, consistent with previous MPRA studies (19, 22). As was the case at the fragment level, there was no preference for enhancer activity to be biased in the direction of either species (Fig. 3 C and D). However, when we compared differentially active fragments with genomic regions that showed differential chromatin accessibility in neural progenitor cells derived from human and chimpanzee cortical organoids (23), we found agreement in the direction of species bias between the datasets. Five human-biased accessibility calls overlapped six differentially active MPRA fragments, five of which were human biased. Four chimpanzee-biased accessibility peaks overlapped four differentially active MPRA fragments, three of which were chimpanzee biased (Dataset S1). Overall, our MPRA captured changes in HAR and HGE activity in NSCs that potentially reflect species-specific regulatory differences in human corticogenesis.
Fig. 3.
Active and differentially active HARs and HGEs identified by MPRA. (A and B) The proportion of inactive, active, and differentially active HGEs (A) and HARs (B). (C and D) The distribution of the number of differentially active fragments per HGE (C) and HAR (D). Most species-biased HGEs and HARs contain only one differentially active MPRA fragment. There are an additional 18 HGEs that contain both human- and chimpanzee-biased fragments in different parts of their sequence (not included in C).
We compared our findings with those from a recent study (20) that screened 714 HARs (5, 6) in an MPRA using a lentiviral integration design in human and chimpanzee neural progenitor cells at two different time points after neural induction. While this study detected a larger number of active HARs, likely due to different criteria used for identifying significantly active fragments (Discussion), there was good agreement between the findings in both studies. Of the 76 HARs active in our study and tested in both studies, 51 (67%) were found active in both. Of the 17 HARs that contained differentially active fragment pairs in our study and were tested in both studies, 14 (82%) showed differential activity in both.
Isolating the Effects of Human-Specific Substitutions in HARs and HGEs.
We designed a second MPRA experiment to isolate the specific effects of each hSub in differentially active fragments. In addition, we included validation fragments (identical copies) for all fragments that showed evidence of activity in the first MPRA (including both fragments of a pair if only one was active. To maximize the number of potentially active sequences evaluated in the second MPRA, we also used relaxed thresholds to determine activity (Materials and Methods and SI Appendix, Fig. S3). After filtering out fragments with <12 barcodes, this second MPRA library contained a total of 22,860 measured fragments (93.1% of all designed). To increase the power of our screen, we included a larger number of barcodes per fragment compared to the first MPRA (mean = 179.5 barcodes per measured fragment; 95% range = 16 to 808). We carried out two replicate MPRAs in hNSCs and measured enhancer activity as described for the first MPRA.
The second MPRA replicated the results from the first MPRA well. Fragment activity was highly correlated between the two MPRAs (Pearson’s ρ range, 0.88 to 0.92, P < 1 × 10−300), similar to correlations of replicates within each MPRA (ρ, 0.91 [MPRA 1], 0.94 [MPRA 2], P < 1 × 10−300). In the second MPRA, we included a set of negative control fragments with comparable sequence content as experimental fragments from genomic regions that exhibit no evidence of enhancer activity. We then used one-tailed t tests to identify experimental fragments with significant activity compared to these negative controls (Materials and Methods and SI Appendix, Fig. S3). We chose this approach because the second library consisted solely of fragments with prior evidence of activity. This was in contrast to the first MPRA, in which most fragments were expected to be inactive and could thus be used as a null distribution for identifying significantly active fragments (SI Appendix, Fig. S5). Applying the same criteria for identifying significantly active fragments as in the first MPRA (Materials and Methods), we found that 69.7% of previously identified active fragments that were measured in both MPRAs replicated their activity as enhancers in the second MPRA (SI Appendix, Table S1B). Of the fragment pairs that were significantly differentially active in the first and measured again in the second MPRA, 89.2% were active, of which 66.9% were differentially active (two-tailed t test; SI Appendix, Table S1B).
Fragments that are differentially active between human and chimpanzee may contain both hSubs and additional background genetic variation, such as human polymorphisms and substitutions that did not meet our criteria for selection, that is, of less interest for understanding human regulatory evolution. To isolate the specific effects of individual hSubs from background variation, we generated fragments containing all possible combinations of the hSub and the corresponding chimpanzee allele on both the human and the chimpanzee background sequence for all differentially active fragment pairs (SI Appendix, Fig. S1B). We interrogated 1,366 hSubs using 14,429 combinatorial fragments. We then employed an ANOVA modeling scheme to identify hSub-specific effects on enhancer activity. We calculated hSub effect sizes as the fold change normalized by the SD. This approach distinguishes between additive effects, which sum up linearly, and interactive effects, where multiple changes modulate each other to produce an outcome different from the sum of effects. We identified 1) additive effects of hSubs; 2) additive background variation effects; 3) interactions between pairs of hSubs; and 4) interactions between hSubs and the background. We identified 401 hSubs that showed significant effects on fragment activity, either alone (additive; n = 315) or in combination with other hSubs or background variation (interactive; n = 120; note that a subset of individual hSubs can be scored as additive or interactive in two different fragments; Fig. 4).
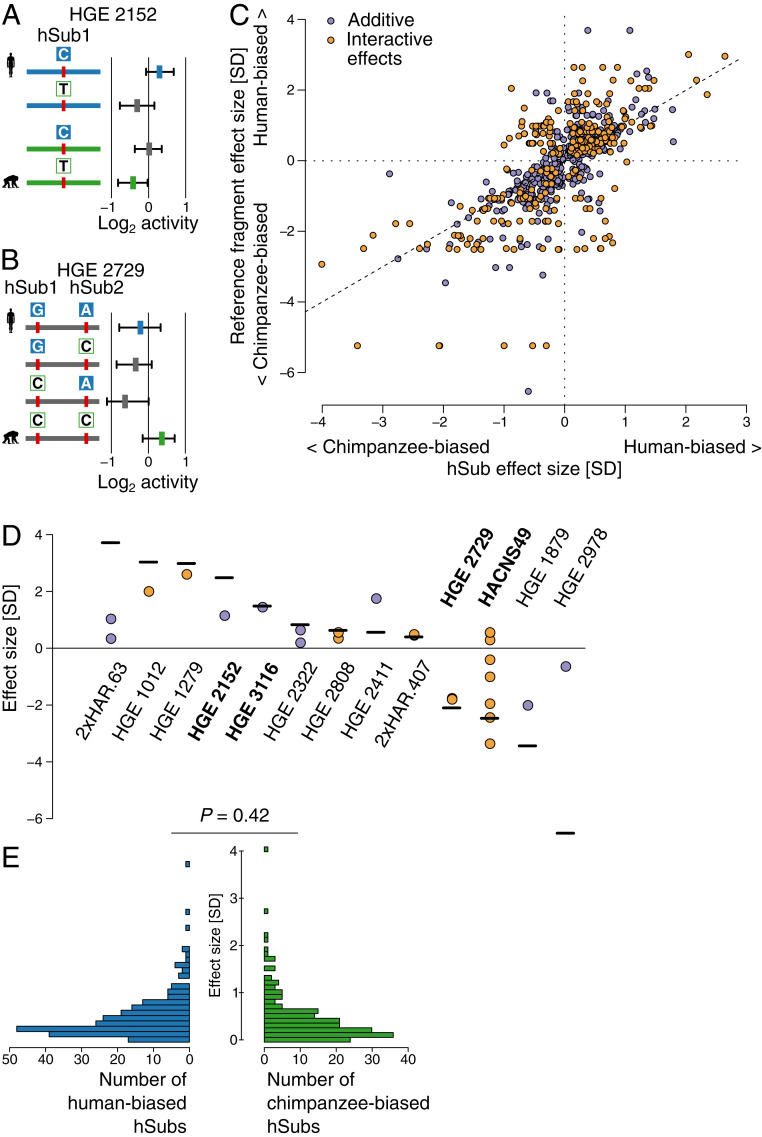
Fig. 4.
Dissecting additive and interactive effects of human-specific substitutions on enhancer activity. (A) An example locus showing additive effects of a single hSub and background variation. The boxplots at Right show median and quartiles of barcode distributions for each fragment. The blue box shows the activity of the human reference fragment, the green box the activity of the chimpanzee reference fragment, and gray boxes the activities of synthetic intermediates. Human hSub allele states are shown as filled blue boxes, and chimpanzee allele states are shown in green. The bar corresponding to each fragment is colored based on whether the human (blue) or chimpanzee (green) reference sequence includes additional background variation. Gray indicates that the human and chimpanzee references are identical other than the hSubs analyzed. (B) An example of an interaction effect between two hSubs. Colors correspond to those in A. See SI Appendix, Fig. S5 for additional examples. (C) All 315 additive and 120 interactive hSub effect sizes plotted against the reference allele effect size. Note that a subset of individual hSubs can be scored as additive or interactive in two different fragments. Each hSub is indicated by a colored circle. Positive values on either scale indicate human-biased activity, while negative values indicate chimpanzee-biased activity. All hSubs from the same reference fragment will have the same value on the y axis. Points along the diagonal indicate hSub effects that align with the reference fragment effect. (D) Comparison of reference fragment effect sizes (horizontal lines) with the hSubs within them (dots). The three most human- and chimpanzee-biased fragments and additional example loci are shown. Each locus is labeled by its HAR or HGE designation. The examples mentioned in the main text and those shown in A and B are labeled in bold text. In cases where hSub effects do not add up to the fragment effect size, background variation and statistical noise make up the remainder of the effect. SI Appendix, Fig. S7 shows all fragments. (E) Effect size distributions of human and chimpanzee hSubs. A Mann–Whitney U test indicates no difference in distribution.
There was no significant species bias in the direction or size of hSub effects (Mann–Whitney U = 13,137, P = 0.35 for additive effects; U = 1,705, P = 0.81 for interactive effects; Fig. 4E and Materials and Methods). For hSubs involved in two-way interactive effects with another hSub or with the background, we estimated the hSub effect size in both reference states (i.e., both the partner hSub or background variation states) and extracted the larger of the two effect sizes. This reference state is where interactive effects exert their main function, and most of the other reference effect sizes were negligibly small (65% of the smaller effect sizes were <0.2 SDs). Interactive effect sizes were significantly larger than additive effect sizes (additive: mean = 0.43 SDs; interactive: mean = 0.65 SDs; U = 13,666, P = 8.0 × 10−6). However, in both cases effect sizes were generally small (quartile range for additive effects, 0.16 to 0.55; for interactive effects, 0.26 to 0.75). Despite this, we did identify hSubs with large effects. Of the 315 hSubs with additive effects, 24 were >1 SD and two were >2 SDs (SI Appendix, Table S3). Of the 120 hSubs with two-way interactive effects, 16 effects were >1 SD and five were >2 SDs. The maximal effect sizes observed were associated with interactive effects. The most human-biased effect was 3.36 SDs and the most chimpanzee-biased effect 4.01 SDs. The largest additive effects were smaller at 2.26 (human biased) and 2.90 (chimpanzee biased) SDs.
An example of an hSub with an additive effect on enhancer activity is shown in Fig. 4A. This fragment contains one hSub and additional background sequence differences between the human and chimpanzee alleles. The T->C hSub has a major effect on fragment activity independent of the background sequence differences: the human-specific C allele is more active than the ancestral T allele. The background variation also contributes to differences in overall fragment activity, with the human allele being more active than the chimpanzee allele. These results illustrate the value of isolating the effects of bona fide human-specific substitutions, which are most relevant for understanding and characterizing regulatory changes that underlie uniquely human biology.
An example of interacting hSubs is shown in Fig. 4B. This fragment contains two hSubs and no background variation. The human (GA) and chimpanzee (CC) reference alleles are both more active than either of the synthetic intermediates (GC or CA). This shows how the activating effect of one hSub allele may depend on the allele state of another hSub. Only a specific combination of allele states leads to high activity, while other combinations lead to a reduced activity. The chimpanzee allele is the most active allele overall. Three additional examples are shown in SI Appendix, Fig. S6.
Isolating the effects of single hSubs reveals how multiple hSubs may combine to alter enhancer function (Fig. 4 C and D and SI Appendix, Fig. S7). hSub effects may largely align with the resulting fragment effect. In the simplest case, a single hSub in the absence of any background variation will cause the entire change in fragment activity (e.g., HGE 3116 in Fig. 4D). In more complex cases, individual hSub effects may point in the opposite direction of the difference in fragment activity, thereby buffering the overall fragment effect (e.g., HACNS49 in Fig. 4D). Such effects are then compensated for by other hSubs or by the background variation in the same fragment. Fragments that contained larger numbers of hSubs often showed more complex interactions between hSubs and the background, as can be seen by the large spread of hSub effects within some fragments (e.g., HACNS49 in Fig. 4D). Fragments containing more hSubs tended to have a greater overall effect size (Spearman’s rank correlation ρ = 0.12, P = 0.0080).
Effect Sizes of Human-Specific Substitutions Differ between HGEs and HARs.
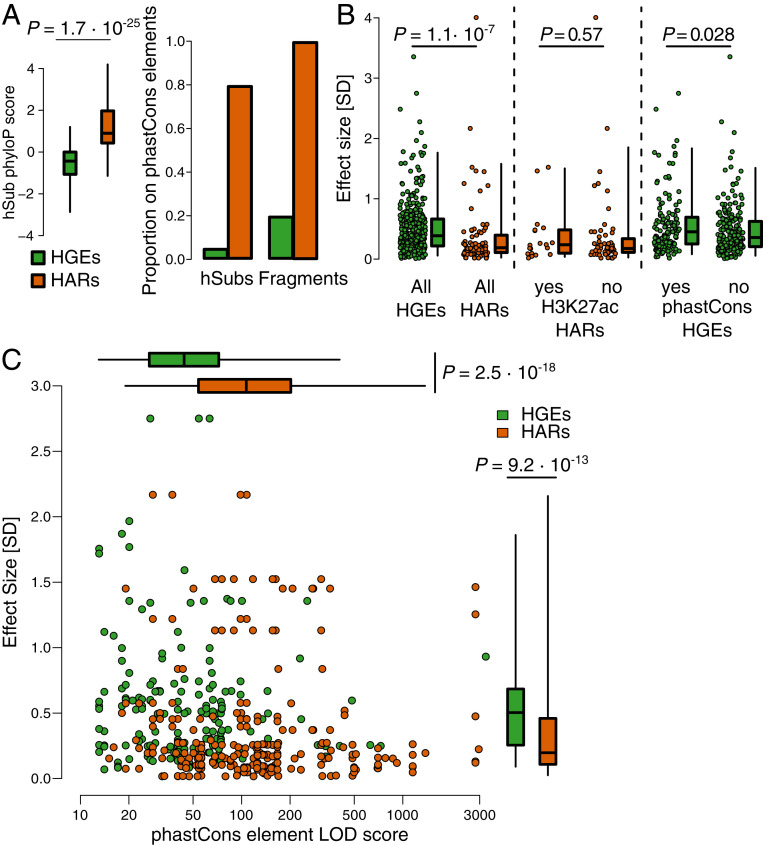
We found that hSubs in HGEs exhibited significantly larger overall effect sizes than hSubs in HARs (HGEs: mean = 0.51 SDs; HARs mean = 0.38 SDs; Mann–Whitney U = 19,211; P = 1.1 × 10−7; Fig. 5B). We considered several possible mechanisms that could account for this finding. First, a greater proportion of HGEs are active in our MPRA compared to HARs. HGEs were defined based on epigenetic signatures of enhancer activity in the developing human cortex at time points when substantial numbers of neural stem cells are present (12). In contrast, HARs were defined based on a significant excess of human-specific substitutions in otherwise deeply conserved regions, without reference to any potential biological function (5–8). HGEs may thus be more likely than HARs to show enhancer activity in an MPRA carried out in human neural stem cells. We therefore categorized HARs according to whether or not they showed evidence of endogenous activity based on H3K27ac marking in hNSCs (Materials and Methods). However, hSub effect sizes were not significantly different between HARs marked by H3K27ac and unmarked HARs (U = 611; P = 0.56; Fig. 5B).
Fig. 5.
Substitutions in HARs and HGEs differ in their effects on enhancer activity. (A) Both hSubs and fragments in HGEs (green) and HARs (orange) differ in their level of constraint. (B) hSubs in HGEs have significantly larger effect sizes than hSubs in HARs (Left). However, hSubs in HARs that show evidence of activity in hNSCs based on chromatin signatures do not have significantly larger effects than hSubs in inactive HARs (Middle). hSubs in fragments with evidence of constraint in HGEs show larger effects than hSubs in unconstrained fragments (Right). (C) HARs (orange) and HGEs (green) differ in effect size of their hSubs (shown on the y axis in the scatterplot and in the box plots on the Right) and in evolutionary conservation (measured as the LOD score of phastCons elements overlapping MPRA fragments; shown on the x axis in the scatterplot and in the box plots at the Top of the figure), but these two aspects are uncorrelated.
Second, HARs are substantially more conserved than HGEs, and are likely to encode regulatory functions that are still under some degree of constraint in humans. The effect of hSubs in HARs may be buffered due to this prior constraint. In contrast, HGEs include both constrained and unconstrained sequences, and hSubs in unconstrained regions may introduce novel enhancer activity without disrupting ancestral functions. To evaluate this hypothesis, we compared the distribution of constraint in HARs and HGEs using phastCons and phyloP (24, 25). As expected, sites with hSubs in HARs were more constrained than in HGEs (mean phyloPHGE = −0.57 vs. mean phyloPHAR = 1.20; U = 2,490; P = 6.2 × 10−25; Fig. 5A). Similarly, hSubs and MPRA fragments in HARs overlapped a higher proportion of constrained elements detected by phastCons than in HGEs (5.5% [hSubs] and 20.2% [fragments] for HGEs, 79.8% and 100% for HARs; Fig. 5A).
If constraint for prior function buffers the effect of hSubs on enhancer activity, we may expect that hSubs in constrained HGEs would show smaller effect sizes than those in unconstrained HGEs. However, we found the opposite to be true: hSubs in fragments that overlap phastCons elements showed a significantly larger effect size than those in fragments that did not overlap a phastCons element (U = 17,081, P = 0.028; Fig. 5B). While we detected an effect size difference between constrained and unconstrained HGEs, we did not find an overall correlation between hSub effect size and strength of constraint (Fig. 5C). In summary, while sequence constraint seems to play a role in determining hSub effect size, it affects hSub effect size differently in HGEs and HARs.
Human-Specific Substitutions Alter Predicted Transcription Factor Binding Sites in HARs and HGEs.
To identify TFs potentially driving human-specific enhancer activity in our MPRA, we mapped all vertebrate transcription factor binding site (TFBS) motifs from the JASPAR core database onto the human and chimpanzee genomes and tested for TFBS enrichment in MPRA fragments (SI Appendix, Supplementary Text). We identified 66 TFBS motifs that were enriched among all active fragments relative to all measured fragments (resampling test, PBH < 0.05; Dataset S2). However, with the notable exception of the cell cycle control factor TP53, we found no significant TFBS enrichments in differentially active, human- or chimpanzee-biased fragments (SI Appendix, Fig. S8 and Dataset S2). This suggests that there is a subset of transcription factors implicated in driving enhancer activity in our assay, and that there may not be a more specific subset driving species-biased activity. As not all predicted TFBSs may be bound by their corresponding factors in vivo, we repeated this analysis on TFBSs within regions of accessible chromatin in human neural stem cells and cortical organoids (assay for transposase-accessible chromatin using sequencing [ATAC-seq] data generated in this study and data from previous studies) (23, 26). Enriched motifs in accessible chromatin regions represented a subset of the motifs found to be enriched using all in silico predictions (Dataset S2). The smaller number of regions tested led to reduced statistical power for this analysis, such that only the test comparing active to all measured fragments returned significantly enriched TFBSs.
We next investigated if hSubs specifically altered TFBSs between human and chimpanzee enhancer sequences. We extracted TFBS predictions overlapping hSubs in active MPRA fragments and tested for overrepresentation of TFBS motifs predicted in the human or chimpanzee orthologs relative to the union of both sets. We found 4 TFBSs enriched among human sequences and 10 enriched among chimpanzee sequences (Dataset S2). Taken together, these analyses provide the basis to identify individual hSubs that putatively change TF binding in HARs and HGEs, with both additive and nonadditive effects.
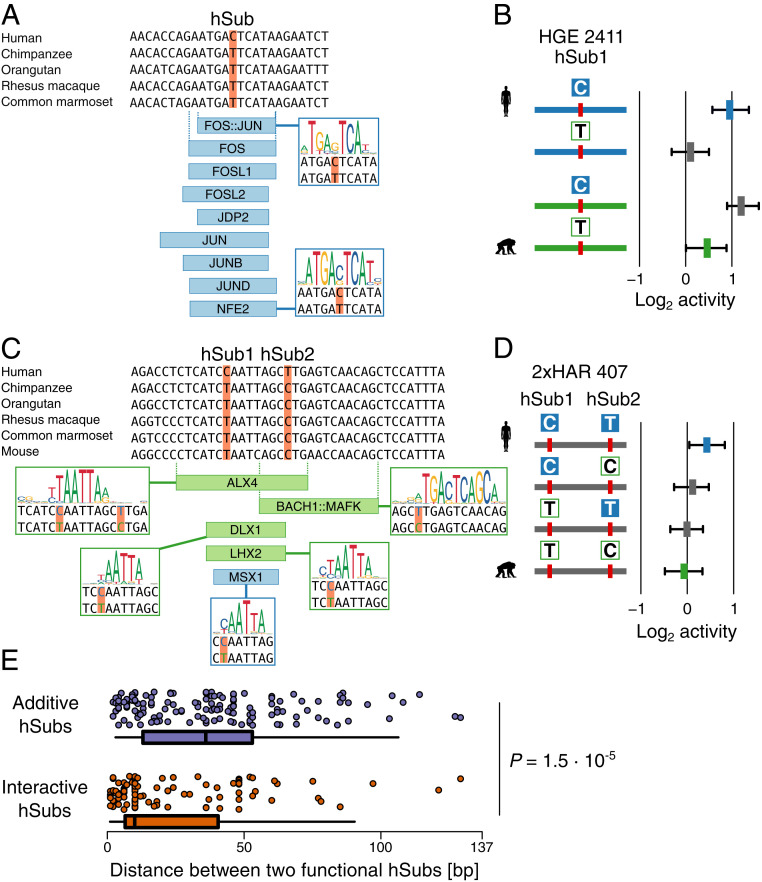
Three-quarters of the 401 hSubs with significant individual regulatory effects overlapped predicted TFBSs, including many of those enriched in active fragments. Of the hSubs that showed the largest effect sizes, one example is shown in Fig. 6A. In this case, an hSub with an additive effect (chr5: 108,791,729) generates a predicted TFBS for AP-1 TFs (FOS and JUN proteins) in the human sequence. Neither of those TFs has a predicted TFBS at the orthologous chimpanzee site. The hSub increases fragment activity by a factor of 1.98 (human-biased effect size = 1.8 SDs) in the human compared to the chimpanzee ortholog (Fig. 6B). AP-1 typically binds to promoter and enhancer sequences to activate target gene expression (27), providing a potential mechanism to explain the increased activity of this enhancer in our MPRA. AP-1 has also been suggested to control important cell cycle regulators such as cyclin D1 and TP53 (28). This hSub is located within a region of human-biased chromatin accessibility discovered in a comparative study of human and chimpanzee cortical organoids (23), supporting the possibility that this site contributes to altered regulatory activity in the human lineage.
Fig. 6.
Changes in predicted transcription factor binding sites due to hSubs that alter enhancer activity. (A) Five-primate alignment over an hSub in HGE 2411. The alignment was derived from the 100-way Multiz alignment (GRCh37/hg19, University of Santa Cruz Genome Browser; http://genome.ucsc.edu) . Nine TFs are predicted to bind the human, but not the chimpanzee ortholog (TFBSs predicted to show increases in affinity are highlighted in blue). (B) MPRA activity of the hSub shown in A, as well as the activities of the chimpanzee ortholog and synthetic intermediates. The figure is labeled as in Fig. 4A. (C) Alignment of a pair of hSubs in 2xHAR 407 that overlaps a cluster of TFBSs (blue, stronger predicted binding in human; green, stronger predicted binding in chimpanzee). See SI Appendix, Fig. S9 for additional TFBSs in this locus that are not affected by the hSubs. (D) MPRA activity of the two hSubs shown in C and the corresponding chimpanzee ortholog and synthetic intermediates. (E) Distribution of physical distances between pairs of additive (Top, in blue) and interactive (Bottom, in orange) hSubs with regulatory effects. BH-corrected P values were calculated using a Mann–Whitney U test.
We next considered the impact of interactive hSubs on TFBS content. Substitutions that combine to alter enhancer activity could be due to multiple hSubs altering the same or physically adjacent TFBSs. Supporting this, interacting hSubs were significantly closer together than additive hSubs (Mann–Whitney U = 9362, P = 1.5 × 10−5; Fig. 6E). We found that 51.5% of interacting hSubs were clustered within 10 bp of each other, while additive hSubs did not show such clustering (19.7% of additive hSubs were within 10 bp). One example is shown in Fig. 6C, where two closely spaced hSubs overlap 14 predicted TFBSs and are in close proximity to an additional three. Four of the TFBSs (ALX4, BACH1::MAFK, DLX1, and LHX2) show increased predicted binding affinity for the chimpanzee sequence, one shows increased predicted affinity for MSX1 in human (Fig. 6C), while the rest show only marginal change in their predicted affinity for either allele (SI Appendix, Fig. S9). Both hSubs by themselves increase MPRA activity in human over chimpanzee (human-biased effect size of hSub1 = 0.22 SDs and of hSub2 = 0.20 SDs). Their combined effect on MPRA activity, however, is stronger than what would be expected from their individual effects, meaning their small individual effects combine to produce a stronger interactive effect (effect size = 0.58 SDs). BACH1::MAFK and DLX1 are known transcriptional repressors, suggesting the hSubs in this example may generate increased activity in human by disrupting recruitment of these factors (29–31). This pair of hSubs resides in a region of enriched H3K27ac in hNSCs, suggesting it acts as an enhancer in vivo (32).
Identifying Gene Targets of Differentially Active HARs and HGEs.
To identify genes potentially regulated by enhancers with human-specific gains in activity, we combined data from four different studies that used chromosome conformation capture in pluripotent stem cell-derived neural progenitor cells or primary human neocortical stem cells (Datasets S3 and S4) (13, 15, 33, 34). This dataset allowed us to infer regulatory interactions between differentially active enhancers and their putative target genes. We identified contacts between 111 differentially active enhancers harboring 95 hSubs with significant effects on activity and 195 genes expressed in hNSCs (Materials and Methods and Datasets S3 and S4). These genes were enriched for Gene Ontology categories related to cell differentiation and development (SI Appendix, Table S4).
Among the notable target genes was HES1, a transcription factor with a crucial role in controlling stem cell differentiation during neurogenesis as part of the Notch signaling pathway (35, 36). We found the HES1 gene to be in contact with the human-biased HGE 2152 in neural progenitor cells (33). Furthermore, HES1 TFBSs overlap hSubs in five different HGEs including HGE 2152 itself (Dataset S4). NOTCH1 is also contacted by the chimpanzee-biased HGE 3155 in developing human germinal zone tissue (15).
Discussion
Identifying genetic changes that altered molecular functions in human evolution is the essential first step toward understanding the origins of uniquely human traits. Here we used MPRAs to screen over 32,000 hSubs for their effects on the activity of putative transcriptional enhancers implicated in the evolution of the human cortex. We assayed 4,376 HARs and HGEs and identified members of each class that act as enhancers in our assay, as well as enhancers with differential activity between the human and chimpanzee orthologs.
HGEs were more frequently active than HARs. This is consistent with the fact that HGEs were identified based on epigenetic signatures of enhancer activity in the human, rhesus macaque, and mouse developing cortex, while HARs were originally identified using a comparative genomics approach without prior evidence of function (5–8, 12). However, the proportion of differentially active enhancers was similar in each class, indicating that HARs and HGEs both include a substantial proportion of enhancers with novel activity in humans. We also note that we may be underestimating the proportion of functional sequences in HGEs since we are focusing only on regions that include human-specific substitutions. In contrast, HARs were sampled more densely due to their deep conservation and high substitution density.
We then isolated the effects of 1,366 hSubs in differentially active fragments from each other and from background variation. This identified 401 hSubs with significant individual effects on enhancer activity. We found that most variants acted additively, while 30% showed interactions with other hSubs or with background variation, meaning that their effects were modulated by variants nearby. We also found pervasive additive and interactive background effects, indicating that segregating and chimpanzee-specific variants can have important consequences for enhancer activity differences between human and chimpanzee. The background effects we identified can obscure the effects of evolutionarily relevant hSubs, illustrating why it is important to distinguish the effects of segregating variants and fixed changes when studying human-specific biology.
We found that differentially active fragments overall, and the effects of hSubs specifically, were not biased toward increased enhancer activity in human, but instead showed an even amount of bias toward either species. In principle, substitutions within HARs could increase or decrease enhancer activity. However, HGEs were defined using functional evidence of increased enhancer activity during human corticogenesis. Although HGEs were ascertained based on comparisons to rhesus macaque and mouse, we expect HGEs to include human enhancers that show increased activity relative to chimpanzee. Moreover, our studies focused on hSubs, which are derived sequence changes in human compared to an ancestral primate state shared between chimpanzee and rhesus macaque. In this context, we may not expect many HGE fragments to exhibit increased activity in chimpanzee compared to human. There are several potential explanations for this finding. First, our assays test individual fragments within HGEs that are components of larger regulatory elements. In isolation, hSubs could increase or decrease activity, but in the context of the entire HGE may interact to increase activity overall. Our finding that hSubs and background variation interact in complex ways to alter activity at the fragment level provides support for this hypothesis. Second, HGEs were ascertained by comparing histone modification signatures in human and rhesus macaque developing cortex (12). The estimated divergence time of apes and old world monkeys is about three to four times greater than that of human and chimpanzee—∼28 vs. 8 million years (37). Only about one-quarter of HGEs may be expected to be human biased in comparison to chimpanzee, whereas the remaining three-quarters of HGEs may be expected to be shared between human and chimpanzee. Third, as HGEs were ascertained in primary samples from developing cortex, which include postmitotic neurons, the hNSCs we used may not fully reflect the cellular diversity of those tissues.
We found enriched TFBS motifs among MPRA fragments active in our study, potentially revealing TFs that substantially contribute to gene regulation in human corticogenesis. However, we could not identify a subset of TFs associated with species-specific enhancer activity, suggesting that changes in regulatory function for the enhancers we studied are driven by the same set of TFs as those that drive enhancer activity in general. We did identify TFBSs that showed changes due to hSubs and that were enriched in human versus chimpanzee active fragments. This supports that hSubs alter TFBS content in enhancers for specific transcription factors. Different TFBSs were uniquely enriched in human and chimpanzee fragments, implying that loss or gain of human enhancer activity due to hSubs involves changes in the recruitment of independent sets of TFs.
We also found that interacting hSubs were located significantly closer to each other than additive hSubs. This supports a model of TFBS evolution where variants interact by altering binding for the same TF or multiple TFs that bind in close proximity. For example, in the locus shown in Fig. 6 C and D, two close hSubs disrupt the predicted TFBSs of several transcriptional repressors, potentially underlying the higher activity of the human ortholog compared to chimpanzee. However, the small number of TFBS predictions overlapping multiple interacting hSubs precluded a systematic analysis.
Our study also suggests that HARs and HGEs encode regulatory changes with distinct evolutionary histories and potential biological effects. Notably, the effect sizes of hSubs in HGEs were on average larger than those in HARs. We had expected that hSubs in HARs might show larger effect sizes as they may be the result of positive selection for novel functions. Differences in the degree of sequence constraint between HARs and HGEs may explain our findings. HARs are highly constrained sequences, and remaining constraints on their ancestral functions may mitigate the effects of human-specific sequence changes within them. In contrast, HGEs are more weakly constrained, and many HGEs arose within placental mammals (11). Such young enhancers have been shown to have weakly conserved regulatory activity and often exhibit changes in their activity across species (11, 38, 39). The larger effect size that we observed for hSubs in HGEs may reflect their increased evolutionary and functional plasticity compared to HARs. We also found that hSubs in constrained regions of HGEs had modestly larger effects than hSubs in unconstrained regions. We hypothesize that hSubs in constrained regions within HGEs may be altering ancestral regulatory functions, which may modify the activity of the preexisting element. In contrast, hSubs in unconstrained regions may be giving rise to novel, but weak, enhancer activity, without a prior regulatory function to provide a “boost” for their effects.
Our study design entails several caveats that merit discussion. Although we are comparing the effects of human–chimpanzee sequence differences, we performed our MPRAs in human neural progenitor cells only. It is possible these sequence changes may exhibit different effects in chimpanzee cells due to differences in trans factors. However, a recent comparative MPRA performed in both human and chimpanzee cells found very few differences attributable to differences in the cellular environment (20). Furthermore, the episomal nature of our assay may, in some instances, lead to differences in the observed activity in comparison to results obtained from an MPRA design using chromosomal integration (40), or even integration in the native chromatin context. The composition of the MPRA plasmid might also have effects on the observed MPRA activity. It has been proposed that promoter choice (41) and the presence of additional sequence motifs influencing transcription initiation and pausing (42) can affect MPRA activity. Enhancer fragments that are inactive in our assay might be active if their native target promoter was used.
We found hundreds of hSubs with individual effects on regulatory activity, many of which were modest relative to our strongest observed effects. This is consistent with the hypothesis that uniquely human cortical features are, in part, polygenic traits that result from many genetic changes of small effect (43). Mammalian cortical development is a prime example of a polygenic trait, given the large numbers of genes involved which interact in complex gene regulatory networks (1). However, it is important to note that the impact of an hSub on enhancer activity in an MPRA may not reflect its impact in the native genomic context during cortical development. The hSubs we characterized here may have substantial biological effects despite their modest effects on enhancer activity in our MPRA. Further studies, including genetic models where the effects of hSubs can be determined in vivo, will be required to address this question.
We identified 424 HARs and HGEs with human-specific changes in enhancer activity in human neural stem cells, as well as individual sequence changes that contribute to those regulatory innovations. These findings now enable detailed experimental analyses of candidate loci underlying the evolution of the human cortex, including in humanized cellular models and humanized mice. Comprehensive studies of the HARs and HGEs we have uncovered here, both individually and in combination, will provide novel and fundamental insights into uniquely human features of the brain.
Materials and Methods
Target Selection and Initial MPRA Library Design.
We selected genomic regions with potential human-specific regulatory activity by taking all human accelerated conserved noncoding sequences (8) and human accelerated regions (version 2) (5) (throughout the text collectively referred to as HARs) and all human gain enhancers, that is, regions that show markedly higher chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) H3K27ac or H3K4me2 signal in human compared to mouse and rhesus macaque during corticogenesis (12) (Dataset S1). Within these regions in human genome version GRCh37/hg19, we collected all human–chimpanzee substitutions that were fixed for the chimpanzee state in a primate alignment and likely to be fixed or nearly fixed in human populations (not present as a SNP marked as “common” in dbSNP build 144, excluding indels). This resulted in a list of 32,776 human-specific substitutions, or hSubs, that were queried in the MPRA. Detailed descriptions of our MPRA library design, construction, and experimental protocols are provided in SI Appendix, Supplementary Text and Extended Methods.
Data Analysis of the First MPRA.
After summarization, pDNA and cDNA counts were normalized by library size and log2 transformed. Very small data values showed a Poisson-like distribution, which is why we excluded barcodes with an average pDNA barcode count across replicates below −5.25 (SI Appendix, Fig. S13). We then normalized each cDNA value by its associated pDNA value to calculate each barcode’s “activity.” We summarized barcodes by fragment by calculating the median of all barcode values associated with a fragment in the pDNA fraction and the cDNA fraction, as well as the median of the cDNA/pDNA ratio (i.e., the fragment activity as defined above) for each fragment in each replicate. We also summarized replicates from the same cell batch further in two groups of replicates (“lineages”). Subsampling numbers of barcodes per fragment showed that correlations between replicates stabilized if a fragment had 12 or more barcodes (SI Appendix, Fig. S2), which we used as a cutoff for downstream analyses. Variance between replicates in pDNA counts, cDNA counts, or activity and in barcode or fragment counts was calculated using Spearman’s rank correlation.
To test for fragment activity, we determined the point of maximal density (µ) of the log2 activity density distribution in each replicate. This was found to be the most conservative distribution summary (SI Appendix, Fig. S14) and also accounted for an apparent artifact in the activity distribution of one of the replicates. We then tested the distribution of each fragment’s log2 activity barcodes against µ using a one-tailed t test to identify active fragments per replicate (SI Appendix, Fig. S3). This approach is appropriate, because pDNA values are effectively log-normally distributed (SI Appendix, Fig. S15). More extreme than log-normal values in the cDNA fraction identified by the t test are called significantly active. We are not able to identify fragments with potentially repressive activity, as there were few fragments in the cDNA fraction with substantially fewer barcode counts than in the pDNA fraction, likely because the baseline activity of the minimal promoter was already low (SI Appendix, Figs. S5 and S15). We applied Benjamini–Hochberg (BH) multiple testing correction and accepted a fragment as active if it had a PBH < 0.05 in at least two replicates. We further required that all replicates had a cDNA count larger than the pDNA count. These criteria resulted in very few active fragments and we developed more permissive criteria to increase the number of fragments for designing the second MPRA and for determining the set of active fragment pairs for differential activity testing. Note, however, that all reported statistics are based on the stringent set of criteria described above. For the permissive criteria, we relaxed the PBH-value cutoff to 0.1 and applied a one-tailed Mann–Whitney U test to the two lineages of replicates to account for potential nonnormal data distributions (SI Appendix, Fig. S3 and Table S1).
For differential activity testing, we included all orthologous fragment pairs that were measured in both species and active (according to the permissive criteria) in at least one of the species. We tested for differential activity by applying two-tailed t tests of activity of the human allele vs. that of the chimpanzee allele in each replicate. A fragment pair was accepted as differentially active if it had a PBH < 0.05 in at least two replicates. We further required that every replicate was biased in the direction of the same species and that the average log2 difference over all replicates was >0.2. For designing the second MPRA we again used more permissive criteria: We relaxed the PBH cutoff to 0.1 and applied a Mann–Whitney U test to the two lineages of replicates to account for nonnormal data distributions. The resulting fragments were selected for dissecting the effects of linked variants in the second MPRA. All of our scripts are publicly available (Data Availability).
During the course of our study, several methods for the analysis of MPRA data were published (44, 45). We compared the performance of these methods with ours and found our approach to be more conservative, but otherwise similar with regards to the results obtained (SI Appendix, Supplementary Text).
Design and Experimental Procedures for the Second MPRA.
Based on the results of the first MPRA, we designed a second MPRA library with two major components. First, we included all active fragments for replication (2,704 orthologous fragment pairs). Second, for all differentially active fragments, we designed artificial fragments that included all possible combinations of hSub states on both human and chimpanzee background sequences (14,963 fragments). In 972 loci (i.e., differentially active fragment pairs from the first MPRA), this library covered 1,366 hSubs. This library also contained additional negative controls. Detailed descriptions of design, experimental procedures, and data preparation are described in SI Appendix, Supplementary Text.
Data Analysis for the Second MPRA.
Similar to the first MPRA, fragments were defined as active at a PBH < 0.05 in a one-tailed t test in both replicates. We tested the activity of experimental fragments against the distribution of negative controls (see above). This was necessary because the second library consisted solely of fragments with prior evidence of activity. While this would mean that more transcript is present in the sample, sequencing to a degree comparable between libraries leads to lower sequencing depth relative to the same fragments in the first library or to the pDNA library (SI Appendix, Fig. S5). This is in contrast to the first MPRA, in which most fragments were expected to be inactive and their distribution could thus be used as a null distribution for identifying active fragments. For the same reason we did not require the cDNA value to be larger than the pDNA value. Testing for differential activity between species, we used two-tailed t tests between orthologous sequences and accepted a fragment as differentially active with PBH < 0.05 in both replicates. All replicates of significant fragments agreed in the direction of species bias and showed an average log2 fold change >0.2. We compared the results of the second round of MPRA with the results from the first round by 1) forming Pearson’s product-moment correlation between the activities (that is, log2 [pDNA/cDNA]) of each replicate and by 2) comparing the numbers of fragments or fragment pairs found to be significantly active, or differentially active, in either round.
To identify hSub-specific enhancer effects, we applied an ANOVA test per locus (i.e., differentially active fragment from the first MPRA) with each hSub, the background variation, and their interactions as factors and the fragment activity as the response variable. Note that locus complexity, and thus model complexity, varied across fragments, from one hSub and no background variation up to seven hSubs with background. For each significant factor, we extracted its mean effect and its effect size in SDs according to ref. 46. For two-way interactions, we calculated effect size relative to the background state in which the factor showed its larger effect (i.e., of the two alternative states of the interacting hSub or background sequence). Significant hSubs were then annotated using comparative genomic data including hNSC H3K27ac ChIP-seq (32), ATAC-seq (this study and refs. 23, 26), and hNPC Hi-C (13, 15, 33, 34) data. ATAC-seq data were generated in triplicate according to ref. 47 and sequenced on an Illumina HiSeq 2500 (2 × 100 bp). Reads were mapped using Bowtie2 (option -X 2000) and open chromatin regions were called using MACS2 (options -B–nomodel–shift -25–extsize 50). Overlap with hNSC H3K27ac ChIP-seq is calculated using peak calls from ref. 32.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institute of General Medical Sciences (GM094780) and funds from the Kavli Institute for Neuroscience at Yale University to J.P.N., a Research Fellowship (UE 194/1-1) from the Deutsche Forschungsgemeinschaft (DFG) to S.U., an NSF Graduate Research Fellowship (DGE-1122492) to A.A.K., and an Autism Speaks Dennis Weatherstone Predoctoral Fellowship to E.G.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007049118/-/DCSupplemental.
Data Availability.
MPRA and ATAC-seq data have been deposited under Gene Expression Omnibus (GEO) accession GSE140983. Additional data used in this study are deposited under GEO accession GSE57369 (hNSC RNA-seq and H3K27ac histone ChIP-seq). The code used to analyze the data is deposited at GitHub: https://github.com/NoonanLab/Uebbing_Gockley_et_al_MPRA.
References
- 1.Geschwind D. H., Rakic P., Cortical evolution: Judge the brain by its cover. Neuron 80, 633–647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King M.-C., Wilson A. C., Evolution at two levels in humans and chimpanzees. Science 188, 107–116 (1975). [DOI] [PubMed] [Google Scholar]
- 3.Reilly S. K., Noonan J. P., Evolution of gene regulation in humans. Annu. Rev. Genomics Hum. Genet. 17, 45–67 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Capra J. A., Erwin G. D., McKinsey G., Rubenstein J. L. R., Pollard K. S., Many human accelerated regions are developmental enhancers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20130025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindblad-Toh K. et al.; Broad Institute Sequencing Platform and Whole Genome Assembly Team; Baylor College of Medicine Human Genome Sequencing Center Sequencing Team; Genome Institute at Washington University , A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard K. S., et al. , An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443, 167–172 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Pollard K. S., et al. , Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2, e168 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prabhakar S., Noonan J. P., Pääbo S., Rubin E. M., Accelerated evolution of conserved noncoding sequences in humans. Science 314, 786 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Haygood R., Babbitt C. C., Fedrigo O., Wray G. A., Contrasts between adaptive coding and noncoding changes during human evolution. Proc. Natl. Acad. Sci. U.S.A. 107, 7853–7857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prabhakar S., et al. , Human-specific gain of function in a developmental enhancer. Science 321, 1346–1350 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotney J., et al. , The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell 154, 185–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reilly S. K., et al. , Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347, 1155–1159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajarajan P., et al. , Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science 362, eaat4311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Torre-Ubieta L., et al. , The dynamic landscape of open chromatin during human cortical neurogenesis. Cell 172, 289–304.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Won H., et al. , Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Won H., Huang J., Opland C. K., Hartl C. L., Geschwind D. H., Human evolved regulatory elements modulate genes involved in cortical expansion and neurodevelopmental disease susceptibility. Nat. Commun. 10, 2396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melnikov A., et al. , Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 30, 271–277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Arensbergen J., et al. , High-throughput identification of human SNPs affecting regulatory element activity. Nat. Genet. 51, 1160–1169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulirsch J. C., et al. , Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell 165, 1530–1545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu H., et al. , Massively parallel dissection of human accelerated regions in human and chimpanzee neural progenitors. 10.1101/256313 (29 January 2018). [DOI]
- 21.Gage F. H., Mammalian neural stem cells. Science 287, 1433–1438 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Tewhey R., et al. , Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell 165, 1519–1529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanton S., et al. , Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Pollard K. S., Hubisz M. J., Rosenbloom K. R., Siepel A., Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siepel A., et al. , Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vierstra J., et al. , Global reference mapping of human transcription factor footprints. Nature 583, 729–736 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu R., et al. , The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell 54, 541–552 (1988). [DOI] [PubMed] [Google Scholar]
- 28.Shaulian E., Karin M., AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4, E131–E136 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Chiba S., et al. , Homeoprotein DLX-1 interacts with Smad4 and blocks a signaling pathway from activin A in hematopoietic cells. Proc. Natl. Acad. Sci. U.S.A. 100, 15577–15582 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitamuro T., et al. , Bach1 functions as a hypoxia-inducible repressor for the heme oxygenase-1 gene in human cells. J. Biol. Chem. 278, 9125–9133 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Sun J., et al. , Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 21, 5216–5224 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cotney J., et al. , The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 6, 6404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung I., et al. , A compendium of promoter-centered long-range chromatin interactions in the human genome. Nat. Genet. 51, 1442–1449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song M., et al. , Cell-type-specific 3D epigenomes in the developing human cortex. Nature 587, 644–649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imayoshi I., Sakamoto M., Yamaguchi M., Mori K., Kageyama R., Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J. Neurosci. 30, 3489–3498 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizutani K., Yoon K., Dang L., Tokunaga A., Gaiano N., Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449, 351–355 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Schrago C. G., Voloch C. M., The precision of the hominid timescale estimated by relaxed clock methods. J. Evol. Biol. 26, 746–755 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Berthelot C., Villar D., Horvath J. E., Odom D. T., Flicek P., Complexity and conservation of regulatory landscapes underlie evolutionary resilience of mammalian gene expression. Nat. Ecol. Evol. 2, 152–163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villar D., et al. , Enhancer evolution across 20 mammalian species. Cell 160, 554–566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue F., et al. , A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Res. 27, 38–52 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muerdter F., et al. , Resolving systematic errors in widely used enhancer activity assays in human cells. Nat. Methods 15, 141–149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tippens N. D., et al. , Transcription imparts architecture, function and logic to enhancer units. Nat. Genet. 52, 1067–1075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyle E. A., Li Y. I., Pritchard J. K., An expanded view of complex traits: From polygenic to omnigenic. Cell 169, 1177–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashuach T., et al. , MPRAnalyze: Statistical framework for massively parallel reporter assays. Genome Biol. 20, 183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myint L., Avramopoulos D. G., Goff L. A., Hansen K. D., Linear models enable powerful differential activity analysis in massively parallel reporter assays. BMC Genomics 20, 209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen J., Statistical Power Analysis for the Behavioral Sciences (Lawrence Erlbaum Associates, ed. 2, 1988). [Google Scholar]
- 47.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MPRA and ATAC-seq data have been deposited under Gene Expression Omnibus (GEO) accession GSE140983. Additional data used in this study are deposited under GEO accession GSE57369 (hNSC RNA-seq and H3K27ac histone ChIP-seq). The code used to analyze the data is deposited at GitHub: https://github.com/NoonanLab/Uebbing_Gockley_et_al_MPRA.