Significance
Identification of transcription factors (TFs) is a starting point for the analysis of transcriptional regulatory systems of organisms. Here, we report the development of DeepTFactor, a deep learning-based tool that predicts TFs using protein sequences as inputs. We interpreted the reasoning process of DeepTFactor, confirming that DeepTFactor inherently learned DNA-binding domains of TFs. DeepTFactor predicted 332 TFs of E. coli K-12 MG1655, and three of them were experimentally validated by identifying genome-wide binding sites with ChIP-exo experiments. We provide DeepTFactor as a stand-alone program for researchers to analyze their own protein sequences of interest. It will serve as a useful tool for understanding the regulatory systems of organisms.
Keywords: ChIP-exo, deep learning, transcription factor, transcription regulation, y-ome
Abstract
A transcription factor (TF) is a sequence-specific DNA-binding protein that modulates the transcription of a set of particular genes, and thus regulates gene expression in the cell. TFs have commonly been predicted by analyzing sequence homology with the DNA-binding domains of TFs already characterized. Thus, TFs that do not show homologies with the reported ones are difficult to predict. Here we report the development of a deep learning-based tool, DeepTFactor, that predicts whether a protein in question is a TF. DeepTFactor uses a convolutional neural network to extract features of a protein. It showed high performance in predicting TFs of both eukaryotic and prokaryotic origins, resulting in F1 scores of 0.8154 and 0.8000, respectively. Analysis of the gradients of prediction score with respect to input suggested that DeepTFactor detects DNA-binding domains and other latent features for TF prediction. DeepTFactor predicted 332 candidate TFs in Escherichia coli K-12 MG1655. Among them, 84 candidate TFs belong to the y-ome, which is a collection of genes that lack experimental evidence of function. We experimentally validated the results of DeepTFactor prediction by further characterizing genome-wide binding sites of three predicted TFs, YqhC, YiaU, and YahB. Furthermore, we made available the list of 4,674,808 TFs predicted from 73,873,012 protein sequences in 48,346 genomes. DeepTFactor will serve as a useful tool for predicting TFs, which is necessary for understanding the regulatory systems of organisms of interest. We provide DeepTFactor as a stand-alone program, available at https://bitbucket.org/kaistsystemsbiology/deeptfactor.
A transcription factor (TF) is a sequence-specific DNA-binding protein that plays a major role in transcription initiation. TFs promote (or block) the RNA polymerase to regulate the rates of the transcription of a set of genes. Analyzing transcriptional regulation enables us to understand how an organism controls the expression of genes in response to genetic or environmental perturbations. Identification of TFs is a starting point for the analysis of transcriptional regulatory systems. TFs have been predicted by analyzing sequence homology with the DNA-binding domains of TFs which have already been characterized (1–3). Data-driven approaches, such as machine learning, have also been used to predict TFs (4, 5). Conventional machine learning models require a rigorous feature selection process that depends on the domain expertise, such as calculation of physicochemical properties of molecules and homology analysis of biological sequences (6). Meanwhile, deep learning inherently learns latent features from the rather raw representation of inputs for the specific task of solving biological problems of interest (7–9). We recently reported the development of DeepEC, which uses deep learning to identify enzyme commission (EC) numbers of enzymes with high accuracy at high speed (10). Although powerful, deep learning has been criticized for the difficulty with which its internal logic of the black box-like reasoning process can be understood. Several techniques have been devised to interpret how the deep learning model functions (11). Saliency methods, which calculate the gradients of prediction score with respect to input to visualize where the deep learning models are focused, can interpret deep learning models for visual understanding (11–16). Recently, saliency methods were also used to interpret deep learning models for biological problems, such as prediction of binding sites of RNA-binding proteins (17) and prediction of the potential energy function for protein conformation (18). Based on these advances in applying deep learning to biological problems, it was reasoned that deep learning could also be used to better classify TFs among proteins and predict presently unknown TFs.
In this study, we report the development of DeepTFactor, a deep learning-based tool for the prediction of TFs employing a convolutional neural network that has three subnetworks in parallel. DeepTFactor predicts TFs vs. non-TFs using protein sequences as inputs. We also interpreted the reasoning process of DeepTFactor using a saliency method. Using DeepTFactor, we predicted 332 TFs in the genome of Escherichia coli K-12 MG1655. Among them, three predicted TFs, one previously known TF and two previously unknown TFs belonging to y-ome, were experimentally validated. DeepTFactor outperformed previous TF prediction tools using homology analysis with known DNA-binding domains or using a conventional machine learning model. We also provide DeepTFactor as a stand-alone program and the results of a DeepTFactor analysis that predicted 4,674,808 TFs from 73,873,012 protein sequences in 48,346 genomes.
Results
Construction of a Deep Neural Network for Transcription Factor Prediction.
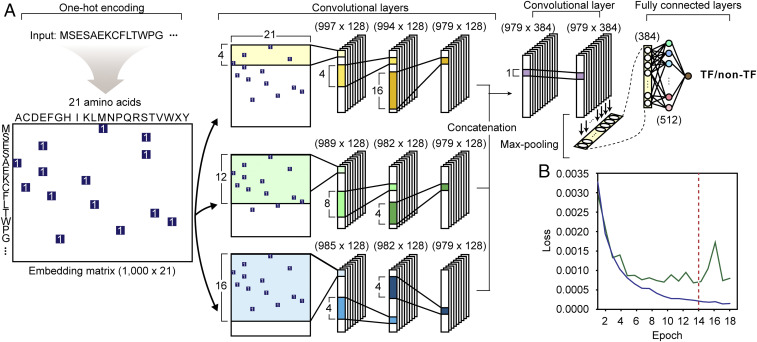
A deep neural network named DeepTFactor was constructed to predict TFs (Fig. 1A). DeepTFactor was designed to extract latent features using parallel subnetworks. The best performing network architecture was identified by testing 38 subnetworks that have different receptive fields of the last convolutional layer in the subnetworks (SI Appendix, Materials and Methods and Table S1). The constructed DeepTFactor has three subnetworks in parallel. Each subnetwork uses three convolutional layers having different sizes of filters to extract the latent features. The extracted features are processed by the subsequent max-pooling layer and fully connected layers to decide whether the input sequence is a TF. To train the neural network, protein sequences containing TF and non-TF sequences were retrieved from the Swiss-Prot dataset released April 2018 (19). The retrieved sequences were processed to be used for the inputs of the network (SI Appendix, Materials and Methods and Fig. S1). A dataset was constructed using 19,406 TF sequences and 58,218 (three times the number of TF sequences) non-TF sequences, which were randomly sampled from all (523,143) non-TF sequences. The dataset was split into the training dataset, validation dataset, and test dataset by the ratio of 8:1:1. In the splitting process, stratified sampling was performed to ensure that TF and non-TF sequences were evenly distributed into the datasets. Batch normalization, dropout, and early stopping were employed (Fig. 1B) to prevent the overfitting of the neural network (SI Appendix, Fig. S2). Batch size and learning rate were also optimized to obtain the best performing model (SI Appendix, Table S2). For the test dataset, DeepTFactor showed an accuracy, F1 score, and Matthews correlation coefficient (MCC) of 0.9773, 0.9541, and 0.9392, respectively (SI Appendix, Table S3).
Fig. 1.
Network architecture of DeepTFactor. An input protein sequence is embedded into a matrix by one-hot encoding. (A) A series of convolutional layers and fully connected layers extracts features from the embedded matrix, resulting in the prediction of whether or not the given protein sequence is a transcription factor. (B) The loss of DeepTFactor prediction along the training process. Blue and green lines indicate loss for the training dataset and validation dataset, respectively. The training process stopped if the validation loss does not decrease in five successive epochs. The red dashed line indicates the epoch where the early stopping occurred.
Comparison of the Prediction Performance of DeepTFactor with Other TF Prediction Tools.
The performance of DeepTFactor was also compared with previously developed TF prediction tools, namely TFpredict (5) and P2TF (2). TFpredict classifies eukaryotic TF sequences from non-TF sequences using a support vector machine (SVM). TFpredict outperformed previously existing methods for the classification of TFs (5). P2TF, a database for analyzing prokaryotic TFs, provides a module to predict prokaryotic TFs using RPS-BLAST (2). Because TFpredict and P2TF were developed for eukaryotic and prokaryotic sequences, respectively, the performance of DeepTFactor, which predicts TFs of both eukaryotic and prokaryotic origins, was separately evaluated on each domain data. To ensure a fair performance comparison, protein sequences not used in the development of the three tools were retrieved from the Swiss-Prot dataset (released from April 2018 to April 2020). DeepTFactor outperformed TFpredict and P2TF in all categories except for prokaryotic sensitivity, which showed the same performance (Table 1). The performance of TFpredict was also compared by training the SVM on the same dataset that DeepTFactor was trained on (SI Appendix, Materials and Methods and Table S4). The trained TFpredict showed an accuracy, F1 score, and MCC of 0.9560, 0.8361, and 0.8058, respectively, for the test dataset. Since DeepTFactor showed higher performance over TFpredict, it can be inferred that the deep learning model performs better than the SVM-based model for the data previously unseen by the models. Deep learning also outperformed RPS-BLAST in predicting prokaryotic TFs.
Table 1.
Comparison of TF classification performance
| Domain | Tool | Accuracy | Specificity | Sensitivity | F1 score |
| Eukaryote* | DeepTFactor | 0.9801 | 0.9815 | 0.9521 | 0.8154 |
| TFpredict | 0.9287 | 0.9285 | 0.9341 | 0.5474 | |
| Prokaryote† | DeepTFactor | 0.9885 | 0.9900 | 0.9286 | 0.8000 |
| P2TF | 0.8686 | 0.8616 | 0.9286 | 0.2600 |
Performance comparison for eukaryotic protein sequences.
Performance comparison for prokaryotic protein sequences.
Interpretation of the Reasoning of DeepTFactor Using Integrated Gradients.
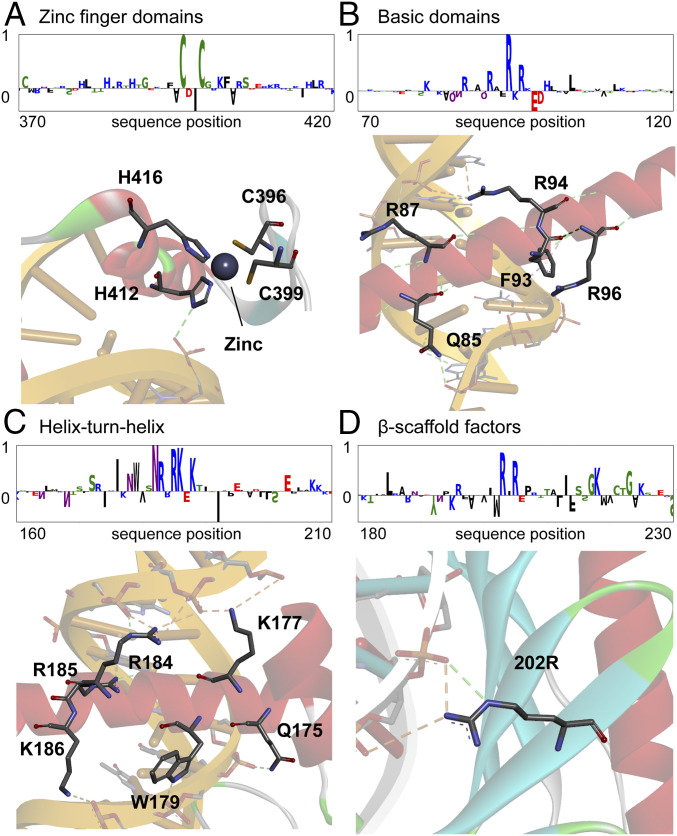
To understand how DeepTFactor predicts TFs, a saliency method of integrated gradients was employed to determine which parts of the input protein sequence DeepTFactor focuses on. Integrated gradients are the path integral of the gradients from the baseline to the input, where the baseline represents the absence of features in the input (15). Integrated gradients were used to trace important signals from an output of the deep neural network toward an input. When the gradients of the prediction score for Egr1, a TF of Mus musculus, were analyzed, DeepTFactor highlighted the zinc finger domain, a DNA-binding domain (Fig. 2A). Even though the information on the DNA-binding domain was not explicitly given during the training process, DeepTFactor inherently learned the DNA-binding domains of the TFs. The zinc finger domain as well as the other major DNA-binding domains of the TFs (i.e., basic domain, helix-turn-helix, and β-scaffold factors) were captured and learned by DeepTFactor (Fig. 2). Integrated gradients also detected multiple DNA-binding domains in a single TF (SI Appendix, Fig. S3). Thus, DeepTFactor understands the features of the TFs from the sequence data, rather than memorizing the labels of data. Since the integrated gradients mainly highlighted the DNA-binding domains, it was tested whether DeepTFactor was trained only to classify DNA-binding proteins, instead of trained to classify TFs. Using DNA-binding protein sequences including both TF sequences and non-TF sequences in the Swiss-Prot dataset (again employing the data not used for training DeepTFactor; released from April 2018 to April 2020), the performance of DeepTFactor on DNA-binding proteins was tested. The F1 score, MCC, and specificity obtained were 0.9095, 0.6801, and 0.6860, respectively (SI Appendix, Table S3). DeepTFactor showed reasonably high performance, differentiating TFs from non-TFs among the DNA-binding proteins.
Fig. 2.
Highlighted domains of the protein sequences by integrated gradients. Integrated gradients for the TF sequences highlighted DNA-binding domains. DeepTFactor detected major DNA-binding domains including (A) zinc finger domains (Protein Data Bank [PDB] ID code 1A1L), (B) basic domains (PDB ID code 1GD2), (C) helix-turn-helix (PDB ID code 1AKH), and (D) β-scaffold factors (PDB ID code 1CDW). The whole integrated gradient results are available in SI Appendix, Fig. S4. Sequence logos were generated using Logomaker (39).
Discovering the Uncharacterized Transcription Factors of the E. coli y-ome.
Although the E. coli K-12 MG1655 strain is the best studied model microorganism, 35% of the genes in its genome are still poorly annotated. A previous study compiled 1,600 genes of E. coli K-12 MG1655 as the y-ome, which is the set of genes with insufficient experimental evidence of specific functions for phenotypes (20). Thus, we aimed at predicting the TFs in the y-ome using DeepTFactor. Of the 4,248 protein-encoding genes, 332 genes were predicted to encode TFs, of which 200 were previously reported in RegulonDB (21). The remaining 132 genes for putative TFs contained 80 y-ome genes (SI Appendix, Fig. S5 and Dataset S1). Among the 80 y-ome genes, DeepTFactor was able to predict genes encoding TFs that have not been annotated with TF activities in the UniProt database (19). For example, YheO has been predicted to be a TF by DeepTFactor, even though it has no Gene Ontology annotation in the UniProt database. The TF activity of YheO was also confirmed in a recent study (22). For further validation of TFs predicted by DeepTFactor, three case studies were conducted. We selected three TFs for genome-wide chromatin immunoprecipitation with an exonuclease treatment (ChIP-exo) experiments as follows. As a positive control, we selected an already known TF (YqhC), which had not yet been fully characterized by a genome-wide experiment, so that we could further provide new insights on other potential regulations. We also selected previously uncharacterized TFs. Among the y-ome genes which have not been reported in RegulonDB, two predicted TFs (YahB and YiaU) were selected that have not been characterized yet. First, profile-based homology analysis was performed using hidden Markov models (23). It was confirmed that YqhC, YiaU, and YahB contained DNA-binding domains annotated by AraC-, LysR-, and LysR-type TFs, respectively (SI Appendix, Fig. S6 and Table S5). The DNA-binding motifs of the TFs were also identified using ScanProsite (24). The identified motifs coincided with the regions the integrated gradients highlighted (SI Appendix, Fig. S6).
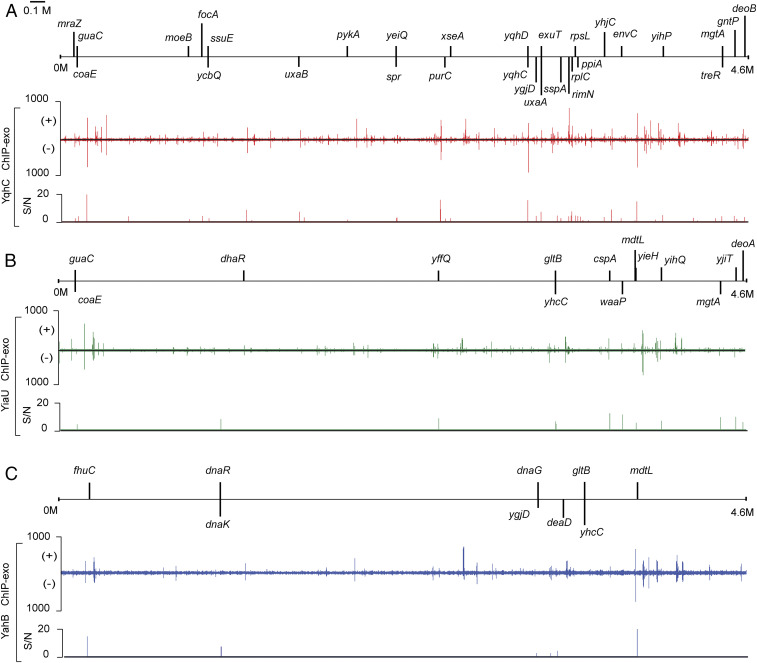
To identify genome-wide binding sites of the TFs, ChIP-exo experiments were performed (Fig. 3). Previously, Turner et al. (25) revealed that YqhC is a transcriptional activator that regulates the expression of the yqhD-dkgA operon in the presence of furfural. In this study, 25 binding peaks of YqhC were identified, including the yqhD promoter region (SI Appendix, Fig. S7 and Dataset S2). The divergent binding peak located between yqhC and yqhD confirmed that YqhC acts as a regulator of the yqhD-dkgA operon while autoregulating itself (25). To further identify the diverse role of YqhC, functional categories of clusters of orthologous groups (COGs) were analyzed for the 79 genes in 30 transcription units (TUs) that YqhC directly regulates (SI Appendix, Fig. S8). This set of target genes was enriched in a few COGs: translate, ribosomal structure, and biogenesis category as well as cell-cycle control, cell division, and chromosome partitioning (hypergeometric P value < 0.001). It was previously shown that YqhC regulates the expression of the yqhD-dkgA operon under the furfural stress condition (25). However, the ChIP-exo results suggested that 29 more TUs might be regulated by YqhC as can be seen from the 25 binding peaks (Fig. 3A). Thus, regulation of these 29 TUs is likely to occur under additional genetic and/or environmental perturbation conditions in addition to the furfural stress condition.
Fig. 3.
Genome-wide binding sites of YqhC, YiaU, and YahB. Overviews of binding profiles of (A) YqhC, (B) YiaU, and (C) YahB across the E. coli K-12 MG1655 genome. ChIP-exo experiments identified 25, 12, and 6 binding peaks for YqhC, YiaU, and YahB, respectively. (+) and (−) for the binding peak profiles indicate forward and reverse reads, respectively. S/N denotes the signal-to-noise ratio.
For the second case study, 12 binding peaks of YiaU were identified, including the promoter region of cspA (SI Appendix, Fig. S6), a major cold shock protein of E. coli (26). A binding peak in the intragenic region of the waaP encoding lipopolysaccharide core heptose (I) kinase was also detected (SI Appendix, Fig. S7). A recent study showed that YiaU binds to the promoter region of csgD, a regulator of biofilm formation (27). Considering that lipopolysaccharide participates in the early stage of biofilm formation (28, 29), we propose a potential role of YiaU for the regulation of biofilm formation in multiple stages.
For the third case study, the function of YahB was analyzed. This study experimentally verified previously unknown function of YahB in E. coli. The genome-wide binding of YahB revealed six binding peaks. For example, YahB binds to the promoter region of dhaR, a TF that controls the expression of the dhaKLM operon (SI Appendix, Fig. S7). The COG analysis of the 19 genes in 9 TUs directly regulated by YahB showed that YahB is involved in several processes including information storage/processing and metabolism/transport (SI Appendix, Fig. S8). All of the ChIP-exo results for the genome-wide binding sites of the TFs are available in Dataset S2.
Discussion
Characterizing unknown TFs with individual experiments such as ChIP-based experiments is tedious and not yet scalable (3, 22). Here, we report the development of DeepTFactor, a deep learning-based tool for the prediction of TFs. Although DeepTFactor was developed for protein sequences covering all domains of life, it outperformed the domain-specific TF prediction tools. Using DeepTFactor, we predicted 332 TFs in E. coli K-12 MG1655, further characterizing the genome-wide binding of three TFs (YqhC, YiaU, and YahB). We also analyzed the genome-wide binding sites for these three TFs.
DeepTFactor not only outperforms other tools on predicting TFs from previously unseen sequence data but also predicts TFs in short inference time (0.2977 milliseconds per one sequence of interest). Due to the generalizability and scalability, DeepTFactor can predict TF sequences from newly sequenced data or vast amounts of less characterized sequences in databases in a high-throughput manner. To demonstrate such a use, we additionally predicted 4,674,808 TFs by analyzing 73,873,012 protein sequences in 48,346 genomes available from the National Center for Biotechnology Information (NCBI) Genome database, and provide https://zenodo.org/record/4264963 for further studies on the regulatory networks of the organisms for interested researchers. We also provide DeepTFactor as a stand-alone program for researchers to analyze their own sequences of interest: https://bitbucket.org/kaistsystemsbiology/deeptfactor.
Unraveling the black box of deep learning remains a challenge for deep learning-based applications in biology and biotechnology. To interpret the reasoning process of DeepTFactor, we applied integrated gradients, which detect the DNA-binding domains of the TF sequences together with other latent features yet unknown for the prediction of TFs. It was found that DeepTFactor considers the presence of the DNA-binding domain. However, the integrated gradients also highlighted some residues outside the DNA-binding domains (SI Appendix, Fig. S6). These highlighted residues might represent not yet characterized functional domains such as DNA-binding domains or might occur by chance due to imperfect network performance. Likewise, the applications of interpretable artificial intelligence (AI) are not limited to such detection. For example, understanding the reason why AI gives a wrong prediction can show engineers where to focus for further development of the deep learning model. If AI characterizes biological data well, it is also possible to learn the key features of the biological data that researchers fail to identify (16). To this end, various approaches to interpreting deep learning are actively being studied (30–35), including saliency methods such as integrated gradients (15), guided backpropagation (13), and Grad-CAM (16). Instead of analyzing such post hoc attentions of the trained neural network, interpreting attention-based neural networks that integrate trainable attention modules within the networks can also be used (36–38). With these developments, it is expected that interpretable AI will empower the use of deep learning for solving biological problems in this ever-increasing bio-big data era.
Materials and Methods
All the materials and methods conducted in this study are detailed in SI Appendix, Materials and Methods: preparation of the dataset for DeepTFactor development; construction and training of the deep neural network for TF prediction; construction and training of TFpredict; integrated gradient calculation; development environment; bacterial strains, media, and growth conditions; ChIP-exo experiment; and peak calling for the ChIP-exo dataset.
Supplementary Material
Acknowledgments
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021171118/-/DCSupplemental.
Data Availability.
Source code for DeepTFactor reported in this article is available at https://bitbucket.org/kaistsystemsbiology/deeptfactor. TF prediction results for NCBI genomes are available at https://zenodo.org/record/4264963. All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) https://www.ncbi.nlm.nih.gov/geo/ under accession no. GSE158683.
ChIP-exo data reported in this article have been deposited in the NCBI GEO (accession no. GSE158683). All study data are included in the article and supporting information.
References
- 1.Wilson D., Charoensawan V., Kummerfeld S. K., Teichmann S. A., DBD—Taxonomically broad transcription factor predictions: New content and functionality. Nucleic Acids Res. 36, D88–D92 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ortet P., De Luca G., Whitworth D. E., Barakat M., P2TF: A comprehensive resource for analysis of prokaryotic transcription factors. BMC Genomics 13, 628 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert S. A., et al. , The human transcription factors. Cell 172, 650–665 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Zheng G., et al. , The combination approach of SVM and ECOC for powerful identification and classification of transcription factor. BMC Bioinformatics 9, 282 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichner J., et al. , TFpredict and SABINE: Sequence-based prediction of structural and functional characteristics of transcription factors. PLoS One 8, e82238 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeCun Y., Bengio Y., Hinton G., Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Angermueller C., Pärnamaa T., Parts L., Stegle O., Deep learning for computational biology. Mol. Syst. Biol. 12, 878 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim G. B., Kim W. J., Kim H. U., Lee S. Y., Machine learning applications in systems metabolic engineering. Curr. Opin. Biotechnol. 64, 1–9 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Zou J., et al. , A primer on deep learning in genomics. Nat. Genet. 51, 12–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryu J. Y., Kim H. U., Lee S. Y., Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers. Proc. Natl. Acad. Sci. U.S.A. 116, 13996–14001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azodi C. B., Tang J., Shiu S.-H., Opening the black box: Interpretable machine learning for geneticists. Trends Genet. 36, 442–455 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Simonyan K., Vedaldi A., Zisserman A., Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv:1312.6034 (20 December 2013). [Google Scholar]
- 13.Springenberg J. T., Dosovitskiy A., Brox T., Riedmiller M., Striving for simplicity: The all convolutional net. arXiv:1412.6806 (13 April 2015). [Google Scholar]
- 14.Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A., “Learning deep features for discriminative localization” in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE Computer Society, Los Alamitos, CA, 2016), pp. 2921–2929. [Google Scholar]
- 15.Sundararajan M., Taly A., Yan Q., Axiomatic attribution for deep networks. arXiv:1703.01365 (13 June 2017). [Google Scholar]
- 16.Selvaraju R. R., et al. , Grad-CAM: Visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 128, 336–359 (2016). [Google Scholar]
- 17.Ghanbari M., Ohler U., Deep neural networks for interpreting RNA-binding protein target preferences. Genome Res. 30, 214–226 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du Y., Meier J., Ma J., Fergus R., Rives A., Energy-based models for atomic-resolution protein conformations. arXiv:2004.13167 (27 April 2020). [Google Scholar]
- 19.UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghatak S., King Z. A., Sastry A., Palsson B. O., The y-ome defines the 35% of Escherichia coli genes that lack experimental evidence of function. Nucleic Acids Res. 47, 2446–2454 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos-Zavaleta A., et al. , RegulonDB v 10.5: Tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 47, D212–D220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y., et al. , Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655. Nucleic Acids Res. 46, 10682–10696 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gough J., Karplus K., Hughey R., Chothia C., Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313, 903–919 (2001). [DOI] [PubMed] [Google Scholar]
- 24.de Castro E., et al. , ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner P. C., et al. , YqhC regulates transcription of the adjacent Escherichia coli genes yqhD and dkgA that are involved in furfural tolerance. J. Ind. Microbiol. Biotechnol. 38, 431–439 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Jiang W., Hou Y., Inouye M., CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272, 196–202 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Ogasawara H., et al. , Novel regulators of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli K-12. Microbiology (Reading) 166, 880–890 (2020). [DOI] [PubMed] [Google Scholar]
- 28.O’Toole G., Kaplan H. B., Kolter R., Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Genevaux P., Bauda P., DuBow M. S., Oudega B., Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 172, 1–8 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Ma J., et al. , Using deep learning to model the hierarchical structure and function of a cell. Nat. Methods 15, 290–298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adebayo J., et al. , Sanity checks for saliency maps. arXiv:1810.03292 (28 October 2018). [Google Scholar]
- 32.Strodthoff N., Wagner P., Wenzel M., Samek W., UDSMProt: Universal deep sequence models for protein classification. Bioinformatics 36, 2401–2409 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shrikumar A., Greenside P., Kundaje A., Learning important features through propagating activation differences. arXiv:1704.02685 (10 April 2017). [Google Scholar]
- 34.Kavvas E. S., Yang L., Monk J. M., Heckmann D., Palsson B. O., A biochemically-interpretable machine learning classifier for microbial GWAS. Nat. Commun. 11, 2580 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svensson V., Gayoso A., Yosef N., Pachter L., Interpretable factor models of single-cell RNA-seq via variational autoencoders. Bioinformatics 36, 3418–3421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jetley S., Lord N. A., Lee N., Torr P. H. S., Learn to pay attention. arXiv:1804.02391 (6 April 2018). [Google Scholar]
- 37.Chen L., et al. , TransformerCPI: Improving compound-protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments. Bioinformatics 36, 4406–4414 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Valeri J. A., et al. , Sequence-to-function deep learning frameworks for engineered riboregulators. Nat. Commun. 11, 5058 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tareen A., Kinney J. B., Logomaker: Beautiful sequence logos in Python. Bioinformatics 36, 2272–2274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source code for DeepTFactor reported in this article is available at https://bitbucket.org/kaistsystemsbiology/deeptfactor. TF prediction results for NCBI genomes are available at https://zenodo.org/record/4264963. All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) https://www.ncbi.nlm.nih.gov/geo/ under accession no. GSE158683.
ChIP-exo data reported in this article have been deposited in the NCBI GEO (accession no. GSE158683). All study data are included in the article and supporting information.