Significance
Mature cardiomyocytes are highly adapted to pump efficiently over billions of cycles. The mechanisms that regulate and coordinate the acquisition of the specialized features of mature cardiomyocytes in the postnatal period are not well understood. This knowledge gap hinders studies of disease pathogenesis and the maturation of stem cell–derived cardiomyocytes for cardiac regenerative medicine. Here, we studied the roles of sarcomeres, the contractile machinery of cardiomyocytes, in regulating cardiomyocyte maturation. Using an in vivo genetic mosaic approach to circumvent animal lethality and secondary consequences of cardiac dysfunction, we analyzed the contribution of sarcomeres to cardiomyocyte maturation in a physiological context. We identified an important role of sarcomeres in the regulation of signal transduction and gene transcription.
Keywords: cardiomyocyte maturation, actinin 2, MRTF, SRF, cardiomyopathy
Abstract
The paucity of knowledge about cardiomyocyte maturation is a major bottleneck in cardiac regenerative medicine. In development, cardiomyocyte maturation is characterized by orchestrated structural, transcriptional, and functional specializations that occur mainly at the perinatal stage. Sarcomeres are the key cytoskeletal structures that regulate the ultrastructural maturation of other organelles, but whether sarcomeres modulate the signal transduction pathways that are essential for cardiomyocyte maturation remains unclear. To address this question, here we generated mice with cardiomyocyte-specific, mosaic, and hypomorphic mutations of α-actinin-2 (Actn2) to study the cell-autonomous roles of sarcomeres in postnatal cardiomyocyte maturation. Actn2 mutation resulted in defective structural maturation of transverse-tubules and mitochondria. In addition, Actn2 mutation triggered transcriptional dysregulation, including abnormal expression of key sarcomeric and mitochondrial genes, and profound impairment of the normal progression of maturational gene expression. Mechanistically, the transcriptional changes in Actn2 mutant cardiomyocytes strongly correlated with those in cardiomyocytes deleted of serum response factor (SRF), a critical transcription factor that regulates cardiomyocyte maturation. Actn2 mutation increased the monomeric form of cardiac α-actin, which interacted with the SRF cofactor MRTFA and perturbed its nuclear localization. Overexpression of a dominant-negative MRTFA mutant was sufficient to recapitulate the morphological and transcriptional defects in Actn2 and Srf mutant cardiomyocytes. Together, these data indicate that Actn2-based sarcomere organization regulates structural and transcriptional maturation of cardiomyocytes through MRTF-SRF signaling.
Cardiomyocyte maturation is essential for the generation of robust cardiomyocytes that contract forcefully throughout postnatal life. The transcriptional, morphological, and functional hallmarks of mature cardiomyocytes have been well described (1, 2), but little is known about how cardiomyocytes acquire these features during development. This knowledge gap obscures the contribution of cardiomyocyte maturation to both cardiac disease pathogenesis and the loss of heart regenerative capacity. It also impairs efforts to engineer mature stem cell–derived cardiac tissues for cell therapy, in vitro disease modeling, and pharmacological testing.
The ultrastructural hallmarks of cardiomyocyte maturation include the expansion of sarcomeres, the specialized cytoskeletal structures that produce contractile force. In addition, the plasma membrane invaginates and forms transverse tubules (T-tubules), which facilitate action potential propagation and excitation–contraction coupling. Mitochondria grow massively in both number and size to enhance energy production capacity. The development of these intracellular structures is coupled with gene expression changes to establish the contractile, electrophysiological, and metabolic functions of adult cardiomyocytes (1, 2).
Mechanisms that orchestrate ultrastructural and transcriptional changes in cardiomyocyte maturation are beginning to emerge. Serum response factor (SRF) is a transcription factor that is essential for cardiomyocyte maturation (3). SRF directly activates key genes regulating sarcomere assembly, electrophysiology, and mitochondrial metabolism. This transcriptional regulation subsequently drives the proper morphogenesis of mature ultrastructural features of myofibrils, T-tubules, and mitochondria. Sarcomeres perform a central role of converting transcriptional information into ultrastructural changes, partly by providing scaffolds for the biogenesis of T-tubules and mitochondria. Although prior studies have suggested that sarcomeres also regulate signal transduction and thereby transcription (4, 5), whether this signaling role of sarcomeres contributes to cardiomyocyte maturation was not previously evaluated.
In noncardiomyocytes, the polymeric state of β-actin, a ubiquitous actin isoform, could regulate SRF transcription activity by modulating its cofactors myocardin-related transcription factor A and B (MRTFA/B; also known as MKL1/2). The monomeric form (G-actin) of β-actin interacts with MRTFs and retains them in the cytoplasm, inhibiting their transcriptional functions (6). The polymerization of β-actin into a filamentous state (F-actin), by contrast, decreases G-actin concentration, derepresses MRTFs, and activates SRF.
In cardiomyocytes, F-actin is a major component of the sarcomeric thin filaments and is built by the cardiac-specific isoform of α-actin (ACTC1). The assembly and organization of ACTC1 polymers into sarcomeres require α-actinin-2 (ACTN2), which directly interacts with ACTC1 filaments and organizes their alignment at Z-lines, which form the lateral boundaries of individual sarcomeres. Although MRTFA/B are known to be essential in postnatal cardiomyocytes (7), whether ACTC1 polymerization regulates MRTF-SRF signaling has not been determined.
Technical challenges have complicated studies to understand the physiological functions of sarcomere components: in vitro cell culture artificially perturbs sarcomere organization in cardiomyocytes, and organ-wide genetic manipulation of sarcomere genes often causes cardiac pathology or animal lethality. Consequently, although many sarcomere components, including ACTN2 and ACTC1, have been investigated by biochemical or cell culture methods, the in vivo functions of these molecules in development and homeostasis remain poorly explored.
To circumvent these issues, here we used adeno-associated virus (AAV)-mediated cardiac genetic mosaic analysis (8, 9) to study sarcomere functions in mice. This approach circumvents animal lethality and the secondary effects of heart dysfunction, enabling investigation of cell-autonomous functions of sarcomere genes within a physiological context. We hypothesized that ACTC1 polymerization regulates cardiomyocyte maturation through MRTF-SRF signaling. To test this hypothesis, we impaired ACTC1 polymerization by mutating ACTN2 and observing the effects on ACTC1 polymerization, MRTF-SRF signaling, and cardiomyocyte maturation.
Results
Cas9-Based Somatic Mutagenesis of Actn2 in Cardiomyocyte Maturation.
We previously established a Cas9/AAV9-based somatic mutagenesis (CASAAV) system (SI Appendix, Fig. S1A) to quickly determine the role of a given gene in cardiomyocyte maturation (8, 10). We produced two AAV vectors targeting Actn2. Each AAV expresses two independent guide RNAs (gRNAs) and Cre recombinase under the control of the cardiomyocyte-specific cardiac troponin T (cTnT) promoter (11, 12). These AAVs were injected into postnatal day one (P1) mice carrying a Cre-activated Cas9-2A-GFP (green fluorescent protein) allele (13) to trigger Cas9-based mutagenesis of Actn2. We verified successful depletion of ACTN2 protein in ∼60% of transduced (GFP+) cardiomyocytes at P30 by immunostaining (SI Appendix, Ta1B-C). GFP+ cardiomyocytes exhibited defective T-tubule formation and drastically reduced cell size (SI Appendix, Fig. S1 D–F), indicating cardiomyocyte maturation defects caused by ACTN2 depletion. These data encouraged us to mechanistically analyze the link between ACTN2 and cardiomyocyte maturation.
Generation of a Conditional Mutant Allele of Actn2.
We generated a floxed Actn2 allele (Actn2F), in which the genomic region containing exons 2, 3, and 4 was flanked by LoxP sequences (SI Appendix, Fig. S2A). Heterozygous (Actn2F/+) and homozygous (Actn2F/F) mice were fertile and exhibited normal heart function (SI Appendix, Fig. S2B). We expected that Cre-based recombination of this allele would remove exons 2 to 4 and result in joining exons 1 to 5, resulting in a frameshift mutation and reduction of Actn2 mRNA through nonsense-mediated decay. Treatment of Actn2F mice at P1 with AAV-Cre, an AAV vector expressing Cre driven by the cardiomyocyte-selective cTnT promoter (11), triggered these expected findings (see next section) as well as ectopic splicing between exon 1 and exon 7, which we denote as Actn2ΔE2-6 (SI Appendix, Fig. S2 C and D). This ectopic splicing preserved the open reading frame, and we confirmed the expression of this protein, at a much lower level than ACTN2 in control mice, by Western blot using an antibody that recognizes the C terminus of ACTN2 (SI Appendix, Fig. S2E).
ACTN2ΔE2-6 lacks amino acids 42 to 204 (SI Appendix, Fig. S2F), which cover the majority of the actin-binding domains of ACTN2. AAV-mediated expression of ACTN2ΔE2-6-GFP did not detectably alter cardiomyocyte morphological maturation or overall heart function (SI Appendix, Fig. S2 G–J). Thus, ACTN2ΔE2-6 is a hypomorphic mutant that does not exert a dominant-negative impact on cardiomyocyte maturation.
Mosaic Actn2 Mutation in the Heart.
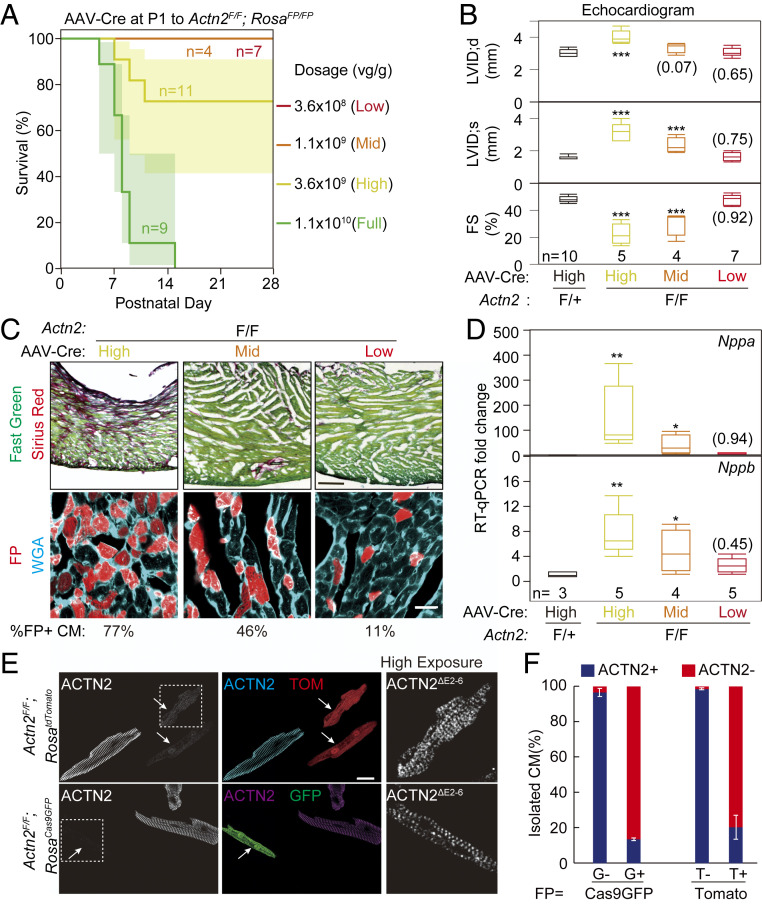
Actn2F alleles were combined with Cre-inducible fluorescent protein (FP) reporter alleles RosaCas9GFP (13) or RosatdTomato (14) so that FP reporters could be used as surrogate markers of AAV-Cre transduced cardiomyocytes. Full-dose (1.1 × 1010 viral genome per gram body weight [vg/g]) AAV-Cre treatment of P1 Actn2F/F pups resulted in death about 1 wk postinjection (Fig. 1A). By echocardiography, high- (3.6 × 109 vg/g) and mid-dose (1.1 × 109 vg/g) AAV-Cre triggered acute dilated cardiomyopathy, characterized by diminished left ventricular systolic function and dilatation (Fig. 1B). Picro sirius red and wheat germ agglutinin (WGA) staining revealed cardiac fibrosis in the high- and mid-dose AAV-Cre–treated hearts (Fig. 1C). Real-time qPCR detected the up-regulation of cardiac stress markers Nppa and Nppb in these groups with cardiac dysfunction (Fig. 1D). These data confirmed that ACTN2 is required to maintain normal heart function.
Fig. 1.
Characterization of mice with mosaic Actn2 mutation. (A) Survival curve of Actn2F/F;RosaFP/FP mice treated with low, mid, high, and full doses of AAV-Cre. (B) Effect of AAV-Cre dosage on heart function and chamber size at P30. Left ventricle (LV) function and size were assessed echocardiographically. FS, LV fractional shortening; LVID;s, LV internal diameter at end systole; LVID;d, LV internal diameter at end diastole. (C) Effect of AAV-Cre dosage on myocardial fibrosis. P30 heart sections were stained with sirius red/fast green (Top; scale bar, 200 µm) or wheat germ agglutinin (WGA, Bottom; scale bar, 20 µm). The fraction of FP+ cells is indicated below images. (D) Real-time qPCR (RTq-PCR) analysis of Nppa and Nppb in P30 heart ventricles. (E) Representative images of ACTN2 immunofluorescence in P30 FP+ and FP− cardiomyocytes that were isolated from the same heart. The arrows point to ACTN2 mutant cardiomyocytes. The boxed area is enlarged and overexposed to show the presence of mutant ACTN2 protein (scale bar, 20 µm). (F) Quantification of cardiomyocytes with decreased ACTN2 signal (ACTN2−) among FP+ and FP− cardiomyocytes. G, Cas9GFP; T, tdTomato. Error bar, SD. n = 3 hearts per group. Box plots are described in Materials and Methods. Two-tailed Student’s t test versus control: *P < 0.05, **P < 0.01, ***P < 0.001. Parentheses indicate nonsignificant P values.
Cardiac dysfunction triggers cascades of secondary effects that confound the study of cardiomyocyte maturation (8, 9). To circumvent this problem, we identified a low dose (3.6 × 108 vg/g) of AAV-Cre that recombined ∼11% of Actn2F/F cardiomyocytes without causing overall heart dysfunction, fibrosis, or activation of cardiac stress markers (Fig. 1 A–D). We treated Actn2F/F;RosaFP/FP mice with this dose of AAV-Cre at P1 and isolated cardiomyocytes at P30. Immunofluorescent staining of ACTN2 in these dissociated cardiomyocytes (Fig. 1E) showed a dramatic decrease of ACTN2 signal in 80 to 85% of FP+ cardiomyocytes (Fig. 1F), while over 95% FP− cardiomyocytes retained strong ACTN2 staining (Fig. 1F). Therefore, FPs reliably mark mutant cardiomyocytes in this system.
To characterize transcription in control and mutant cardiomyocytes, we purified FP+ cardiomyocytes from AAV-Cre–treated Actn2F/F and Actn2F/+ mice by fluorescence-activated cell sorting (FACS) (SI Appendix, Fig. S3A) and performed real-time qPCR and RNA sequencing (RNA-Seq) (SI Appendix, Fig. S3B). RNA-Seq analysis confirmed aberrant splicing between exons 1 and 7 and dramatically decreased expression of exons 2 to 6 in mutant cardiomyocytes (SI Appendix, Fig. S3 C and D). Exon 1 to exon 5 junctions were also observed, which likely triggered nonsense-mediated RNA decay and reduced expression of all other exons (SI Appendix, Fig. S3 C and D). Overall reduction of Actn2 mRNA was validated by real-time qPCR using a probe targeting exons 11 to 12 (SI Appendix, Fig. S3B). Consistent with these mRNA changes, the immunofluorescence signal of ACTN2ΔE2-6 protein in the mutant cardiomyocytes was very weak (Fig. 1E, right “High Exposure”) as compared to wild-type ACTN2 in control cardiomyocytes. The low level of the mutant ACTN2ΔE2-6 protein makes a dominant-negative effect unlikely. Thus, defects in Actn2 mutant cells should be interpreted as the consequence of effective ACTN2 depletion rather than the loss of just the actin-binding domain.
Together, these data indicate that we have created a cardiac genetic mosaic model of Actn2 mutation. This system provides an unprecedented opportunity to perform loss-of-function analysis of the cell-autonomous roles of ACTN2-based sarcomere organization in vivo.
Impact of Actn2 on Morphological Maturation.
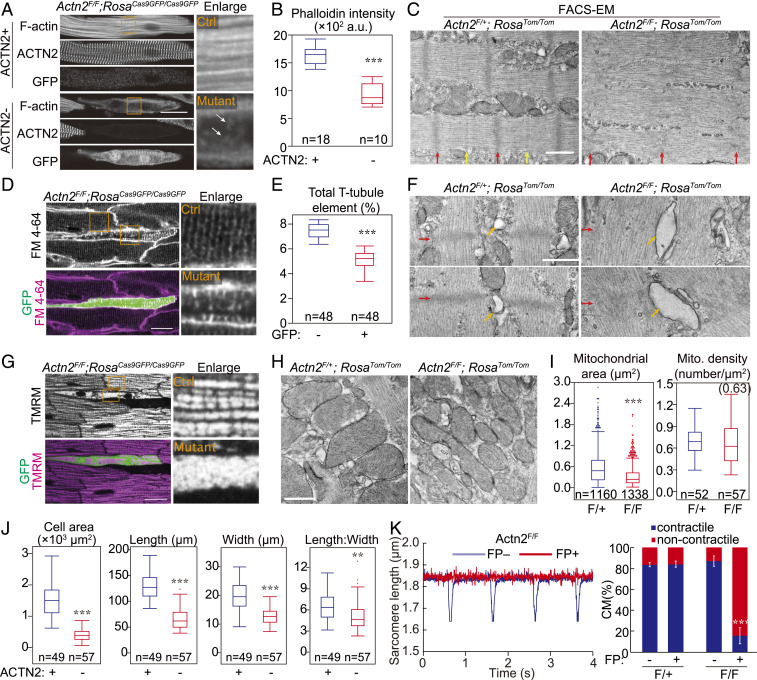
Because a major function of ACTN2 is to cross-link F-actin, we first examined the impact of Actn2 mutation on F-actin organization in cardiomyocytes. Compared to ACTN2+ cardiomyocytes, ACTN2− mutant cardiomyocytes exhibited drastically reduced F-actin as measured by phalloidin fluorescence intensity (Fig. 2 A and B). Residual F-actin was retained in the mutant cardiomyocytes at the cell periphery, while a small number of short F-actin fragments (Fig. 2A, arrows) could be observed in the cell interior, suggesting myofilament breakdown. To further investigate the impact of Actn2 mutation on sarcomere structures, we performed electron microscopy (EM) on cardiomyocytes that were purified by FACS (FACS–EM) (3, 15). Z-lines, the electron-dense boundaries of sarcomeres (Fig. 2C, red arrows), were dramatically diminished in the remnant myofibrils in mutant cardiomyocytes. Longitudinal sarcomere filaments were misaligned, and M-lines, an ultrastructural hallmark of mature sarcomeres (16), were not detected in mutant cardiomyocytes (Fig. 2C, yellow arrows).
Fig. 2.
The impact of Actn2 mutation on cardiomyocyte morphological maturation. Mosaic mutation of Actn2 was achieved by low-dose AAV-Cre treatment of Actn2F/F;RosaFP/FP mice. Control cells were either FP− or ACTN2+ cells from the same heart (A, D, G, J, and K) or FP+ cardiomyocytes from Actn2F/+;RosaFP/FP mice (C, F, and H). Mice were treated on P1 and analyzed at P30. In A, D, and G, orange boxed areas are enlarged to the right. (A) Phalloidin staining on control and Actn2 mutant cardiomyocytes. The arrows point to small F-actin fragments. (B) Quantification of fluorescence intensity of phalloidin staining. (C) EM analysis of sarcomere ultrastructure in FACS-sorted Tomato+ control and mutant cardiomyocytes. Red arrows, Z-lines. Yellow arrows, M-lines. (D) Confocal optical sections of hearts perfused with FM 4-64, a dye that labels the plasma membrane. The stained membranes within cardiomyocytes are T-tubules. (E) T-tubule quantification by AutoTT software. (F) EM analysis of T-tubules in FACS-sorted Tomato+ control and mutant cardiomyocytes. Red arrows, Z-lines. Orange arrows, T-tubules. (G) Confocal optical sections of hearts perfused with mitochondrial dye TMRM. Orange boxed areas are enlarged to the right. (H) EM analysis of mitochondria. (I) Quantification of mitochondrial area and density by EM. (J) Cardiomyocyte geometry, measured on dissociated cardiomyocytes. (K) Representative traces (Left) and quantification (Right) of contractile cardiomyocytes upon electrical stimulation. The error bar is SD. n = 3 hearts per group. (Scale bars, 20 µm for A, D, and G or 500 nM for C, F, and H.) Box plots are described in Materials and Methods. Two-tailed Student’s t test versus control: *P < 0.05, **P < 0.01, ***P < 0.001.
We next investigated the impact of Actn2 mutation on the maturation of other ultrastructures. T-tubules were imaged by confocal optical sectioning of hearts perfused with the plasma membrane dye FM 4-64 (15). FP+ mutant cardiomyocytes exhibited reduced T-tubule content as quantified by AutoTT software (17) (Fig. 2 D and E). Residual T-tubules in mutant cardiomyocytes appeared disorganized and showed greater signal intensity than control cardiomyocytes (Fig. 2D). FACS–EM analysis revealed dramatic dilatation of residual T-tubules in mutant cardiomyocytes (Fig. 2F, yellow arrows) adjacent to the remnant sarcomere Z-lines (Fig. 2F, red arrows).
During cardiomyocyte maturation, mitochondria greatly expand and adopt a highly organized pattern adjacent to sarcomeres. To study the effect of Actn2 mutation on structural maturation of mitochondria, we performed optical confocal sectioning of hearts perfused with the mitochondrial dye tetramethylrhodamine methyl ester (TMRM). TMRM signal intensity was retained in Actn2 mutants, indicating preserved mitochondrial membrane potential, but the spatial distribution of mitochondria was perturbed (Fig. 2G and SI Appendix, Fig. S4). FACS–EM analysis revealed a decreased mitochondrial cross-sectional area in Actn2 mutants, although mitochondrial density did not change (Fig. 2 H and I).
Enlarged size and elongated shape are other key morphological hallmarks of cardiomyocyte maturation. We measured the geometry of immunostained cardiomyocytes isolated from AAV-Cre–treated Actn2F/F;RosaFP/FP hearts. Mutant ACTN2− cardiomyocytes exhibited decreased cell area due to a reduction of both cell length and width (Fig. 3J). The length-to-width ratio also decreased, indicating that ACTN2 plays a more profound role in cardiomyocyte elongation than widening. We electrically stimulated Actn2 mutant and control cardiomyocytes and found that cardiomyocyte contraction was completely abolished in ∼80% of FP+ Actn2F/F cardiomyocytes (Fig. 2K), consistent with the frequency of FP+ cardiomyocytes with mutant ACTN2 (Fig. 1F).
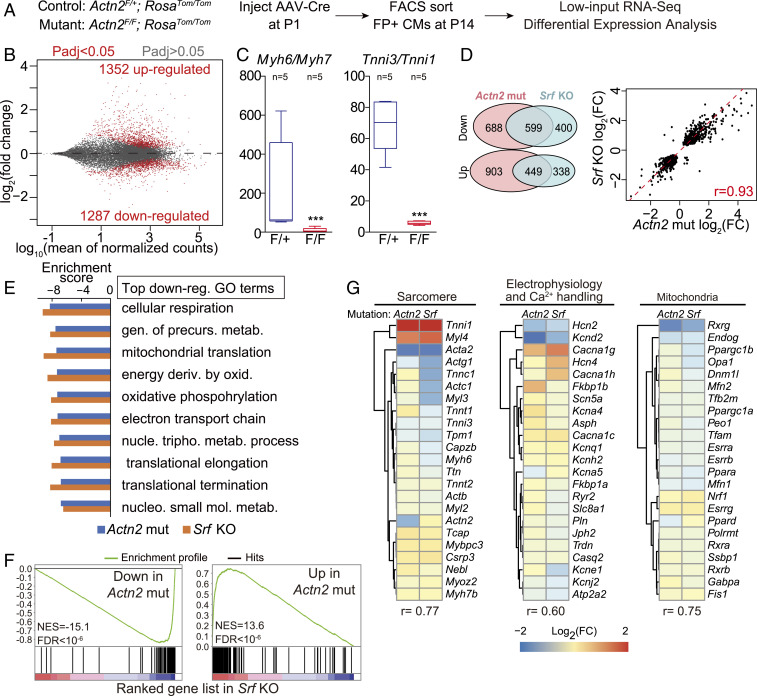
Fig. 3.
Cell-autonomous impact of Actn2 on transcription. (A) Experimental design to evaluate cell-autonomous regulation of the cardiomyocyte transcriptome downstream of Actn2. Low-dose AAV-Cre was administered to P1 mice. Transduced cardiomyocytes were isolated at P14, FACS purified, and analyzed by RNA-Seq. Control and mutant mice had the indicated genotypes. (B) M (log ratio)-versus-A (average) plot (MA plot) of RNA-Seq results. n = 5 per group. (C) Myh6/Myh7 and Tnni3/Tnni1 reads per kilobase million (RPKM) ratios in control versus mutant groups by RNA-Seq. (D) Differentially expressed genes in Actn2 mutation are compared to Srf KO. (Left) Overlap of significantly up- and down-regulated genes is depicted. (Right) Correlation analysis of the fold change of shared differentially expressed genes between Actn2 mutation and Srf KO is depicted. (E) Top GO terms are enriched among genes down-regulated in Actn2 mutation and Srf KO experiments. Negative scores mean down-regulation. (F) Enrichment of top up- and down-regulated genes in Actn2 mutation among genes with the most significant up- or down-regulation in Srf KO. The analysis was performed using GSEA. (G) Correlation of expression change of key sarcomere, electrophysiology/Ca2+ handling, and mitochondrial maturation genes between Actn2 mutation and Srf KO experiments. r, Pearson correlation coefficient.
Together, these data indicate that ACTN2 is critical for the establishment of specialized cardiomyocyte ultrastructure and morphology during maturation.
Impact of Actn2 on Transcription through MRTF-SRF Signaling.
Transcriptional regulation is essential to coordinate the diverse hallmarks of cardiomyocyte maturation. The wide spectrum of phenotypes in Actn2 mutant cardiomyocytes led us to hypothesize that ACTN2 regulates signal transduction and transcription beyond its canonical role as a structural protein. To test this hypothesis, we purified control and mutant cardiomyocytes by flow cytometry at P14 and performed RNA-Seq using an established protocol (3) (FACS–RNA-Seq, Fig. 2A and Dataset S1 and SI Appendix, Table S1). Five biological replicates for each group were clearly separated by principal component analysis (PCA, SI Appendix, Fig. S3E). Differential expression analysis identified 1,352 or 1,287 genes that were up- or down-regulated in mutant cardiomyocytes (Fig. 3B and Dataset S1). Sarcomeric gene isoform switching from Myh7 to Myh6 and from Tnni1 to Tnni3 are well-established hallmarks of transcriptional maturation in murine cardiomyocytes. RNA-Seq demonstrated a drastic decrease of Myh6/Myh7 and Tnni3/Tnni1 ratios in Actn2 mutants (Fig. 3C), which supported the role of Actn2 in transcriptional maturation.
To better interpret the defects in Actn2 mutants at the transcriptomic level, we purified wild-type P6 and P30 cardiomyocytes and performed RNA-Seq analysis to yield a reference dataset for the normal transcriptional changes between these two time points (SI Appendix, Fig. S5A and Table S1 and Dataset S1). P6 and P30 samples were well separated by PCA (SI Appendix, Fig. S5B). Over 3,000 genes were up- or down-regulated from P6 to P30 (SI Appendix, Fig. S5C and Dataset S1). Cell division and DNA replication were identified as the major biological processes that were down-regulated (SI Appendix, Fig. S5D), consistent with the exit of cardiomyocytes from the cell cycle during maturation. Tricarboxylic acid cycle and muscle contraction gene ontology (GO) terms were up-regulated (SI Appendix, Fig. S5D), agreeing with the increase of oxidative respiration and contractile functions during cardiomyocyte maturation. Gene set enrichment analysis (GSEA) (18) showed that the down-regulated genes in Actn2 mutants were greatly enriched among the genes up-regulated during maturation, while up-regulated genes in Actn2 mutants were enriched for genes down-regulated during maturation (SI Appendix, Fig. S5E). This result strongly indicates that Actn2 mutant cardiomyocytes are transcriptomically immature.
SRF is a key transcription factor regulating cardiomyocyte maturation (3). Therefore, we next compared Actn2 mutant and Srf knockout (KO) RNA-Seq data (3), which were produced at the same age using the same experimental and analytical protocols. Differentially expressed genes in these datasets were highly concordant: 60.0% of down-regulated and 57.1% of up-regulated genes in Srf KO cardiomyocytes were differentially expressed in the same direction in Actn2 mutant cardiomyocytes (Fig. 3D). Moreover, the relative changes in expression of these shared differentially expressed genes were highly correlated (Pearson’s r = 0.93) (Fig. 3D). GSEA (18) showed that the top 10 down-regulated GO terms, which were related to metabolism, mitochondria, and translation, were identical in the two experiments (Fig. 3E). The top 250 down- or up-regulated genes in Actn2 mutants were highly enriched among the most down- or up-regulated genes in Srf KO, respectively (Fig. 3F). We further performed correlation analysis using three panels of genes critical for mature cardiomyocyte sarcomeres, electrophysiology and Ca2+ handling, and mitochondria (Fig. 3G). Across these gene panels, there was excellent correlation (r > 0.6) between Actn2 mutant and Srf KO experiments. These data suggest that Actn2 and Srf function in the same pathway to regulate gene expression.
We previously mapped cis-regulatory elements that were bound by endogenous SRF protein in maturing murine hearts by biotin-mediated chromatin precipitation followed by sequencing (3). Based on these data, we found that ∼53% of down-regulated genes in Actn2 mutants were associated with SRF-bound promoters (within 1 kb of transcriptional start sites; SI Appendix, Fig. S6A). To functionally validate the impact of SRF on these promoters in vivo, we took the promoter of the actin filament binding protein tropomyosin 1 (Tpm1) as an example. The Tpm1 promoter is strongly bound by SRF in maturing P14 cardiomyocytes (SI Appendix, Fig. S6B). We designed an AAV reporter assay in which the Tpm1 promoter was positioned upstream of mCherry (SI Appendix, Fig. S6C). To make this reporter compatible with Cre-mediated mosaic gene inactivation, we inserted a loxP-stop-loxP cassette between the promoter and mCherry so that mCherry could be driven by the Tpm1 promoter only in Cre-expressing cells (SI Appendix, Fig. S6C). We first validated by fluorescence imaging that the mCherry expression was indeed dependent on codelivery of AAV-Cre (SI Appendix, Fig. S6D). Next, we coinjected AAV-Cre and the Tpm1-mCherry reporter into SrfF/+ and SrfF/F littermates. By both fluorescence imaging and real-time qPCR, SRF depletion significantly reduced mCherry expression (SI Appendix, Fig. S6 E and F). These data indicate that Tpm1 promoter activity is dependent on SRF, suggesting that Tpm1 regulation downstream of ACTN2 indeed depends on SRF.
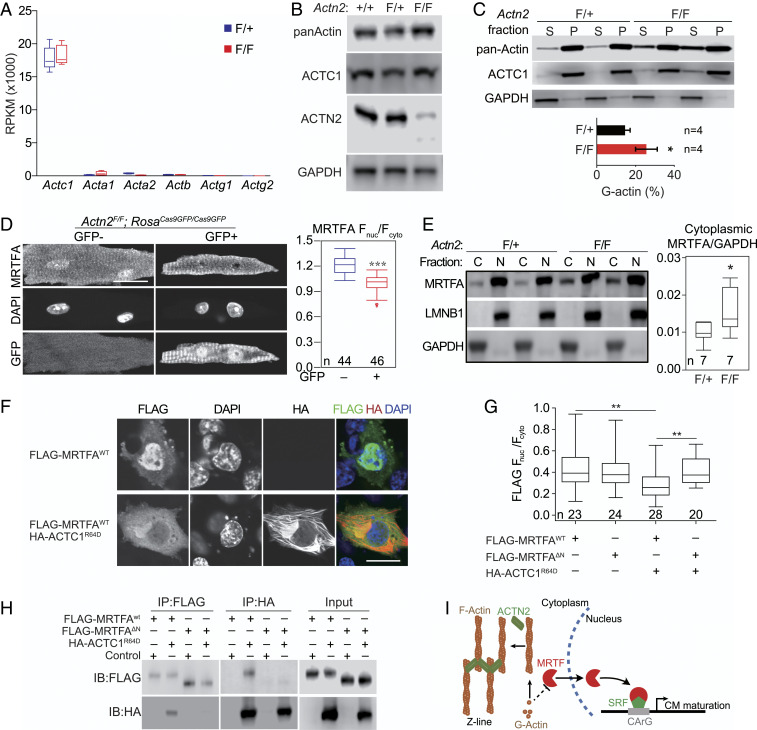
We hypothesized that Actn2 regulates gene transcription in cardiomyocyte maturation through the actin-MRTF-SRF signaling axis. To test this hypothesis, we first analyzed the cardiomyocyte RNA-Seq data to compare the transcript level of all six actin genes. A single isoform, Actc1, was responsible for ∼96.0% of total actin mRNA in cardiomyocytes (Fig. 4A). We next injected high-dose AAV-Cre into Actn2F/+ and Actn2F/F pups at P1 and collected heart tissue at P7 for biochemical actin analysis. This approach circumvented perturbation of the actin cytoskeleton during cardiomyocyte isolation and FACS purification. Although Actn2 mutation strongly reduced F-actin (Fig. 2 A and B), total actin (panActin) and ACTC1 protein levels were not changed (Fig. 4B). The decrease of F-actin and lack of change of total actin or the predominant actin isoform ACTC1 suggested that Actn2 mutation increased G-actin.
Fig. 4.
Impact of Actn2 mutation on actin polymerization and MRTF localization. (A) mRNA expression of all six actin isoforms in control and Actn2 mutant cardiomyocytes by RNA-Seq. (B) Western blot analysis of actin in P7 heart tissues treated with high-dose AAV-Cre at P1. (C) Western blot (Top) and densitometric quantification (Bottom) of F-actin:G-actin ratio by actin pelleting analysis. The error bar is SD. High-dose AAV-Cre was injected at P1, and the heart tissues were collected at P7. (D) MRTFA localization in cardiomyocytes isolated from hearts with mosaic Actn2 mutation. Mosaic dose AAV-Cre was given at P1, and cells were analyzed at P30. Cells were immunostained with an MRTFA antibody and imaged using a confocal microscope (scale bar, 20 µm). (Right) quantification of fluorescence intensity ratio between nuclear and cytoplasmic MRTFA signals. (E) Western blot analysis of MRTFA, LMNB1 (nuclear marker), and GAPDH (cytoplasmic marker) in nuclear (N) and cytoplasmic (C) fractions. High-dose AAV-Cre was injected at P1, and the heart tissues were collected at P7. (Right) Quantification of MRTFA Western blot signal in the cytoplasmic fraction, normalized to GAPDH. (F) Immunofluorescence images of NIH 3T3 cells expressing FLAG-MRTFA and HA-ACTC1R64D (scale bar, 20 µm). ACTC1R64D is an ACTC1 mutant that is expected to preferentially acquire the G-actin state. (G) Quantification of N and C MRTFA fluorescence intensity. MRTFA-ΔN is a mutant MRTFA in which an N-terminal fragment containing actin-binding motifs is deleted. (H) Reciprocal coimmunoprecipitation analysis of MRTFA and ACTC1 interaction in human embryonic kidney cells 293t (HEK293T) cells. The control is an empty plasmid that does not express FLAG- or HA-tagged proteins. IP, immunoprecipitation; IB, immunoblotting. (I) Model for ACTN2-MRTF-SRF sarcomere-to-nucleus signaling. ACTN2 promotes F-actin assembly into sarcomeres and decreases G-actin level, which allows MRTF to enter the nucleus and stimulate transcription through SRF. Box plots are described in Materials and Methods. Two-tailed Student’s t test versus control: *P < 0.05, ***P < 0.001.
To directly measure G-actin levels, we adopted an actin pelleting assay (19), in which ultracentrifugation pelleted F-actin from heart lysates while leaving G-actin in the supernatant. As a positive control, we treated heart lysates with Swinholide A, a small molecule that severs F-actin and triggers its depolymerization (20). This treatment significantly increased the amount of G-actin in the supernatant (SI Appendix, Fig. S7 A and B), validating this assay for analysis of actin polymeric states in heart tissue. The actin pelleting assay showed that most ACTC1 proteins were assembled into F-actin in control heart tissues (Fig. 4C and SI Appendix, Fig. S7C). Actn2 mutation significantly increased the fraction of ACTC1 G-actin by 1.7-fold (Fig. 4C).
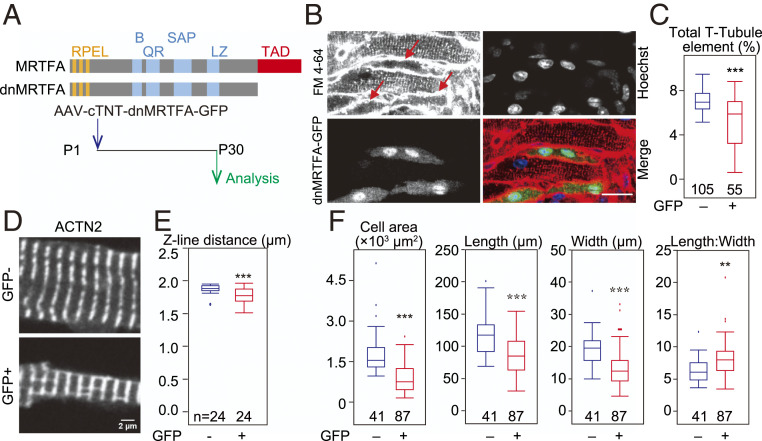
In other cell types, G-actin binds MRTF to reduce its nuclear localization. Therefore, we assessed the impact of Actn2 mutation on MRTF localization. Previous studies reported conflicting results about MRTF localization in cardiomyocytes (21, 22), likely due to its low protein levels in the heart and limited signal-to-noise ratio of available antibody staining. To circumvent these problems, we generated an AAV vector expressing MRTFA tagged with GFP. We transduced wild-type hearts with AAV-MRTFA-GFP, perfused the heart with Hoechst dye to label nuclei, and then imaged in situ by confocal optical sectioning. This method avoided potential artifacts arising from tissue processing, cardiomyocyte isolation, or nonspecific antibodies and clearly revealed predominant nuclear localization of MRTFA at P7, P14, and P30 (SI Appendix, Fig. S8A). We also stained wild-type dissociated cardiomyocytes with a widely used MRTFA antibody (23) and observed similar nuclear localization of endogenous MRTFA (SI Appendix, Fig. S8B). Antibody staining of dissociated cardiomyocytes transduced by AAV-MRTFA-GFP dramatically increased nuclear staining compared to nontransduced cardiomyocytes (SI Appendix, Fig. S8B), further authenticating this immunolocalization. Morphological analysis of MRTFA-GFP overexpressing cardiomyocytes demonstrated T-tubule disorganization and perturbed Z-line spacing (SI Appendix, Fig. S8 C–E), consistent with deleterious effects of both increased and decreased MRTF-SRF activity on cardiomyocyte maturation (3).
Next we treated Actn2F/F;RosaCas9GFP/Cas9GFP mice with low-dose AAV-Cre at P1 and immunostained cardiomyocytes isolated from P30 hearts with the MRTFA antibody. The control and mutant cardiomyocytes were isolated from the same heart, stained in the same dish, and imaged in parallel using the same confocal parameters. The control and mutant cardiomyocytes were distinguished by GFP signal, and MRTFA nuclear and cytoplasmic signals were quantified by fluorescence intensity. Quantification of the nuclear-to-cytoplasmic MRTFA signal ratio showed that it was significantly lower in Actn2 mutant cardiomyocytes (Fig. 4D).
To validate this result using an orthogonal method that circumvented potential confounding effects of cardiomyocyte isolation, we performed subcellular fractionation of Actn2F/F;RosaCas9GFP/Cas9GFP and Actn2F/+;RosaCas9GFP/Cas9GFP ventricles that were treated with high-dose AAV-Cre at P1 and collected at P7. Nuclear and cytoplasmic markers LMNB1 and GAPDH were found in the expected subcellular fractions (Fig. 4E), confirming clean separation of these compartments. MRTFA was enriched in the nuclear fraction, and quantification showed that cytoplasmic MRTFA was greater in Actn2 mutant hearts as compared to controls (Fig. 4E). This observed change in cytoplasmic MRTFA likely under-represents the actual change because of the incomplete depletion of ACTN2 and the diluting effect of MRTFA in nonmyocytes and nontransduced cardiomyocytes in the tissues.
We further tested whether ACTC1 interacts with MRTFA and regulates in nuclear localization. An arginine-to-aspartate mutation at the 62nd residue of β-actin (ACTBR62D) has been widely used to mimic the G-actin state of β-actin (24). This residue, corresponding to the 64th residue of ACTC1, and neighboring amino acids are 100% conserved in ACTC1. We expressed MRTFA in NIH 3T3 cells and observed nuclear MRTFA localization. However, coexpression of MRTFA with ACTC1R64D was sufficient to reduce MRTFA nuclear localization (Fig. 4F). In contrast, ACTC1R64D was not able to perturb the nuclear localization of MRTFAΔN, a mutant lacking MRTFA’s canonical actin-binding domains (Fig. 4G). The interaction between MRTFA actin-binding domains and ACTC1R64D was further validated by reciprocal coprecipitation analysis (Fig. 4H).
We wondered if perturbation of other genes involved in sarcomere assembly would similarly disrupt ACTC1-MRTF-SRF signaling and impair cardiomyocyte maturation. Therefore, we used CASAAV to achieve mosaic inactivation of the actin filament binding protein TPM1. Cardiomyocytes with TPM1 depletion exhibited defective T-tubule maturation, F-actin/ACTN2 organization, and MRTFA nuclear localization (SI Appendix, Fig. S9 A–G). Thus, TPM1 and likely other genes required for sarcomere assembly also regulate the ACTC1-MRTF-SRF axis.
Together, these data indicate that, in maturing cardiomyocytes, ACTN2 promotes ACTC1 polymerization and assembly into sarcomeres, which enhances nuclear localization of MRTF and MRTF-SRF transcriptional activity (Fig. 4I). High Actn2 expression (SI Appendix, Fig. S9H), low G-actin levels (SI Appendix, Fig. S7C), and nuclear MRTF localization (SI Appendix, Fig. S8A) were observed throughout cardiomyocyte maturation. Collectively, these findings suggest that ACTC1-MRTF-SRF signaling is continuously active to promote cardiomyocyte maturation in vivo.
Perturbation of Cardiomyocyte Maturation by MRTF Inhibition.
To directly test the role of MRTFs in cardiomyocyte maturation, we used low-dose AAV to express a dominant-negative mutant of MRTFA (dnMRTFA) (25). This MRTFA variant lacks the transactivation domain (TAD, Fig. 5A) and competes with endogenous MRTFA and MRTFB for interaction with SRF. As a result, this approach inhibits both MRTF isoforms. dnMRTFA-GFP was found mainly in the cell nucleus (Fig. 5B), confirming that the low G-actin level in wild-type cardiomyocytes favors nuclear localization of MRTFA as well as dnMRTFA-GFP. We observed severe T-tubule defects in dnMRTFA-GFP–expressing cardiomyocytes (Fig. 5 B and C). These cells also exhibited perturbed ACTN2 organization, characterized by the presence of longitudinal ACTN2 staining (Fig. 5D) and decreased distance between Z-lines (Fig. 5E). Maturational cardiomyocyte hypertrophy was also perturbed by dnMRTFA (Fig. 5F). We further confirmed these phenotypes by mosaic inactivation of both Mrtfa and Mrtfb. Mrtfa−/−; MrtfbF/F; Rosa26Cas9GFP were treated with a mosaic dose of AAV-Cre. The Mrtfa/Mrtfb double mutant cardiomyocytes exhibited similar T-tubule and Z-line spacing phenotypes (SI Appendix, Fig. S10 A–E). These morphological phenotypes caused by MRTF inhibition were almost identical to observations in Srf KO cardiomyocytes (3), indicating a key role of MRTFs in the activation of SRF activity, which is essential for the organization of ACTN2 and for proper cardiomyocyte maturation.
Fig. 5.
Perturbation of morphological maturation by dnMRTFA. (A) Design of dnMRTFA-GFP overexpression experiments. RPEL, actin-binding domain. (B) Basic region. QR, glutamine-rich region; SAP, SAP domain; LZ, leucine zipper domain. (B) In situ confocal optical sectioning of hearts with mosaic dnMRTFA-GFP expression. Hearts were perfused with Hoeschst dye and FM4-64 (scale bar, 20 µm). The arrows point to treated cells with defective T-tubule organization. (C) AutoTT quantification of T-tubule content of dnMRTFA-GFP expressing and nonexpressing cardiomyocytes. (D) Immunofluorescent staining of ACTN2 in isolated cardiomyocytes. (E) Quantification of Z-line distance. (F) Size and aspect ratio of dnMRTFA-GFP and control cardiomyocytes that were isolated from hearts with mosaic dnMRTFA expression. Box plots are described in Materials and Methods. Two-tailed Student’s t test versus control: *P < 0.05, **P < 0.01, ***P < 0.001.
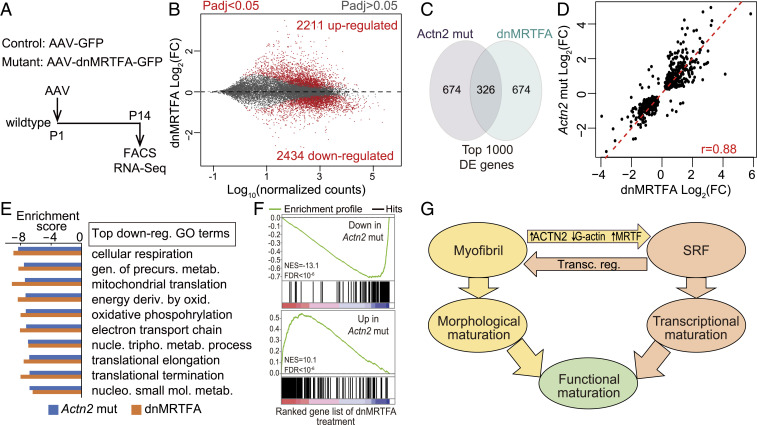
We next performed FACS–RNA-Seq to characterize the impact of low-dose AAV-dnMRTFA-GFP on transcription using AAV-GFP–treated cardiomyocytes as control (Fig. 6A and SI Appendix, Table S1 and Dataset S1). Five biological replicates of control versus treated cardiomyocytes were well separated by PCA (SI Appendix, Fig. S10F and Table S1). Statistical analysis revealed 2,434 down-regulated genes and 2,211 up-regulated genes (Fig. 6B and Dataset S1). A total of 33% of genes differentially expressed in dnMRTFA treatment and Actn2 mutation overlapped (Fig. 6C), and the relative expression change of these shared differentially expressed genes were highly correlated (r = 0.88; Fig. 6D). The down-regulated genes were enriched for the same top-ranked GO terms, as assessed by GSEA (Fig. 6E). This correlation was further validated by GSEA analysis of dnMRTFA data using custom gene sets containing the top up- and down-regulated genes in Actn2 mutants (Fig. 6F). These data demonstrate similar impacts of Actn2 mutation and MRTF inhibition on transcription in cardiomyocyte maturation and support the hypothesis that they function in the same signaling pathway.
Fig. 6.
Transcriptomic perturbation by dnMRTFA. (A) Design of RNA-Seq analysis of dnMRTFA treatment. Mice were treated with low-dose AAV-dnMRTFA-GFP or AAV-GFP. GFP+ cardiomyocytes were purified by FACS and analyzed by RNA-Seq. (B) MA plot of gene expression in dnMRTFA-GFP compared to GFP cardiomyocytes. n = 5 per group. (C) Venn diagram comparing genes differentially expressed in Actn2 mutation or dnMRTFA expression. (D) Correlation analysis of genes differentially expressed genes in both Actn2 mutation and dnMRTFA expression experiments. r, Pearson correlation coefficient. (E) The 10 GO terms most enriched among genes down-regulated in dnMRTFA-treated or Actn2 mutant cardiomyocytes were the same. (F) The top 250 up- or down-regulated genes in Actn2 mutants were highly enriched among the up- or down-regulated genes, respectively, in dnMRTFA-expressing cardiomyocytes. The analysis was performed using GSEA. (G) Model of mutual potentiation between sarcomere assembly and MRTF-SRF signaling to coordinate cardiomyocyte maturation.
Discussion
Sarcomere Z-lines function as signal transduction hubs to modulate cardiac disease pathogenesis (5), but whether sarcomeres regulate signaling during heart development was unclear. Manipulation of sarcomere components in animals often impacts overall heart function and consequently results in secondary effects that complicate the analysis of sarcomere functions. In this study, we combined AAV-mediated genetic mosaic analysis (9) with a newly generated floxed allele of Actn2 to efficiently disassemble sarcomeres in cardiomyocytes in vivo while maintaining overall normal cardiac function. This approach revealed a cell-autonomous role for ACTN2 in the ultrastructural maturation of cardiomyocytes, which was coupled with a profound impact on gene transcription.
We identified at least one molecular mechanism that contributes to this sarcomeric regulation of transcription: sarcomere assembly activates MRTF-SRF signaling by promoting ACTC1 polymerization. Although the importance of actin-MRTF-SRF signaling is established in noncardiomyocytes, it was unknown if this mechanism was active in cardiomyocytes. The actin cytoskeleton of cardiomyocytes is highly specialized, and the major cardiac actin isoform, ACTC1, was not previously connected to MRTFs. Here, we show that sarcomeres use actin-MRTF-SRF signaling to modulate SRF activity during cardiomyocyte maturation. Together with the knowledge that SRF signaling also activates sarcomere assembly (3), these two signaling axes potentially build a positive feedback loop to ensure robust and coordinated maturation of both cardiomyocyte ultrastructure and transcription (Fig. 6G).
Actn2 mutation resulted in additional transcriptional dysregulation that did not overlap with Srf KO or dnMRTF treatments (Figs. 3D and 6C), suggesting the regulation of other molecular pathways. For example, Actn2 could potentially regulate CSRP3 (also known as MLP), which was reported to shuttle between Z-lines and nucleus (26) to maintain normal cardiac function (27). Alternatively, sarcomere disassembly could impair cell contractility and induce transcriptional changes through other mechanotransduction pathways. Future studies are required to further dissect the mechanisms that govern ACTN2-dependent transcriptional regulation.
Cardiac pathological remodeling is characterized by changes that overlap with cardiomyocyte maturation defects in several aspects. One representative change is the reactivation of a fetal-like transcription program. By performing mosaic analysis, we excluded the interference of noncell autonomous pathological changes on our study of cardiomyocyte maturation. However, we recognize that genetic mosaics cannot exclude cell-autonomous activation of pathological stress responses. Identification of such stress responses and their impact on cardiomyocyte maturation would be critical to more precisely define the mechanisms that promote or perturb cardiomyocyte maturation.
Human sarcomere gene mutations, including ACTN2 mutations, are well-known causes of inherited cardiomyopathies, such as dilated cardiomyopathy and hypertrophic cardiomyopathy. While conventional mechanistic studies of these diseases have focused on sarcomere contractile function or calcium sensitivity, the discovery of ACTN2-ACTC1-MRTF-SRF signaling suggests altered signal transduction could contribute to disease pathogenesis caused by a subset of sarcomere mutations. This hypothesized mechanism also predicts that disruption of the coordinated cardiomyocyte maturation program could contribute to cardiac disease pathogenesis. Consistent with this idea, a recent study demonstrated cardiomyocyte maturation defects and congenital heart malformation as a consequence of disease-causing mutations in MRTFB, MYH7 (a major component of mature sarcomere thick filaments in humans), and an additional genetic modifier (28).
Cardiomyocytes that are derived from nonmyocytes or stem cells are widely used in disease modeling, pharmacological tests, tissue engineering, and cell therapy trials. These cells exhibit immature phenotypes in vitro, imposing a major bottleneck on cardiac regenerative medicine (29). Many studies have used ACTN2 staining patterns as a key hallmark of in vitro cardiomyocyte maturation. Here, we provide an insight that improved ACTN2 organization mechanistically contributes to cardiomyocyte maturation and is not merely a quantifiable end point. We propose that sarcomere maturation is a key prerequisite to overall cardiomyocyte maturation.
Materials and Methods
Please see SI Appendix, Supplemental Materials and Methods for additional information.
Mice.
All animal strains and procedures were approved by the Institutional Animal Care and Use Committee of Boston Children’s Hospital. Actn2F mice were generated by homologous recombination in embryonic stem cells. RosaCas9GFP/Cas9GFP (13) and RosaTomato/Tomato (14) were imported from the Jackson Laboratory. Mrtfa−/− (30) and MrtfbF/F (31) mice were reported previously.
AAV.
AAV9 was prepared as previously described (3, 8). AAV titer was quantified by real-time PCR using a fragment of the cardiac troponin T promoter DNA to make a standard curve. PCR primers for AAV quantification were 5′-TCGGGATAAAAGCAGTCTGG-3′ and 5′-CCCAAGCTATTGTGT-GGCCT-3′. AAV was injected into P1 pups subcutaneously. The P1 pups were anesthetized in an isoflurane chamber before injection, and 3.6 108 vg/g was used in all mosaic analyses.
CASAAV.
The protocols to perform CASAAV experiments were previously described (8, 10). The gRNA sequences in this study are as follows: Actn2, 5′-GATGGTCCAGATCATACCCA-3′, 5′-TCGCCCACAGAAATAGTCGA-3′, 5′-GAC-ATAGTTGTACTGCACGC-3′, and 5′-CCGCGACCTGCTCCTCGACC-3′; Tpm1, 5′-TCTGAACAGACGCATCCAGC-3′ and 5′-AGGCTGCAGATGAGAGTGAG-3′.
In Situ Heart Optical Confocal Sectioning.
In situ cardiac imaging was performed as previously described (15). Hearts were perfused with FM 4-64 (Invitrogen, 13320), TMRM (Life Technologies), or Hoechst 33342 (Invitrogen) dyes as indicated. The heart was then positioned on a glass-bottom dish and immediately imaged using an Olympus FV3000RS inverted confocal microscope.
FACS–EM.
Isolated cardiomyocytes in suspension were fixed with 4% paraformaldehyde for 30 min at room temperature. The fixed cells were sorted on a BD AriaII SORP cell sorter with a 100-µm nozzle. After FACS, the cells were fixed again in 2% formaldehyde and 2.5% glutaraldehyde in 0.1 M of Sodium Cacodylate buffer, pH 7.4, overnight at 4 °C. The cell pellets were next processed for routine transmission EM protocol.
RNA-Seq and Data Analysis.
Total RNA from FACS-sorted cardiomyocytes was reverse transcribed, and full-length complementary DNA was specifically amplified for eight PCR cycles using SMART-Seq v4 Ultra Low Input RNA Kit (Clontech). DESeq2 (32) was used to perform statistical analysis of differential gene expression, with differentially expressed genes defined by adjusted P < 0.05. An adjusted P value of 0.05 was used as the cutoff to identify differentially regulated genes.
Statistical Analysis.
Statistical analysis and plotting were performed using JMP software (SAS Institute). Statistical tests are indicated in each figure legend. The numbers in parentheses in figures indicate nonsignificant P values. The bar plots show mean ± SD. In the box plots, horizontal lines indicate the median and 25th and 75th quantiles; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; and dots represent possible outliers.
Supplementary Material
Acknowledgments
We appreciate the technical support by Dana-Farber Flow Cytometry Core, Harvard Medical School (HMS) Electron Microscopy Core, HMS Biopolymers Facility, and Boston Children’s Hospital (BCH) Animal Facility. We thank Dr. Guido Posern for sharing the MRTFA antibody. This project was supported by NIH (R01HL146634 and UM1HL098166 to W.T.P., R01AR044345 to A.H.B, and K99HL148309-01A1 to Y.G.), the American Heart Association (postdoctoral fellowship 18POST33960037 to Y.G.), and charitable support from the Boston Children’s Hospital Department of Cardiology. Mouse genotyping and phenotyping was supported by the resources of the Intellectual and Developmental Disabilities Research Center (IDDRC) Molecular Genetics Core funded by U54HD090255 from the National Institute of Child Health and Human Development (NICHD) of NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008861118/-/DCSupplemental.
Data Availability.
RNA-Seq data generated for this study have been deposited in the Gene Expression Omnibus database under the accession code GSE136096. Actn2F mice, produced in Dr. Alan Beggs’s laboratory, can be obtained through a material transfer agreement.
References
- 1.Guo Y., Pu W. T., Cardiomyocyte maturation: New phase in development. Circ. Res. 126, 1086–1106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karbassi E., et al. , Cardiomyocyte maturation: advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 17, 341–359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Y., et al. , Hierarchical and stage-specific regulation of murine cardiomyocyte maturation by serum response factor. Nat. Commun. 9, 3837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pyle W. G., Solaro R. J., At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ. Res. 94, 296–305 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Frank D., Frey N., Cardiac Z-disc signaling network. J. Biol. Chem. 286, 9897–9904 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vartiainen M. K., Guettler S., Larijani B., Treisman R., Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316, 1749–1752 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Mokalled M. H., et al. , Myocardin-related transcription factors are required for cardiac development and function. Dev. Biol. 406, 109–116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Y., et al. , Analysis of cardiac myocyte maturation using CASAAV, a platform for rapid dissection of cardiac myocyte gene function in vivo. Circ. Res. 120, 1874–1888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y., Pu W. T., Genetic mosaics for greater precision in cardiovascular research. Circ. Res. 123, 27–29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.VanDusen N. J., Guo Y., Gu W., Pu W. T., CASAAV: A CRISPR‐based platform for rapid dissection of gene function in vivo. Curr. Protoc. Mol. Biol. 120, 31.11.1–31.11.14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werfel S., et al. , Rapid and highly efficient inducible cardiac gene knockout in adult mice using AAV-mediated expression of Cre recombinase. Cardiovasc. Res. 104, 15–23 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Prendiville T. W., et al. , Novel roles of GATA4/6 in the postnatal heart identified through temporally controlled, cardiomyocyte-specific gene inactivation by adeno-associated virus delivery of Cre recombinase. PLoS One 10, e0128105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platt R. J., et al. , CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madisen L., et al. , A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B., Zhang C., Guo A., Song L.-S., In situ single photon confocal imaging of cardiomyocyte T-tubule system from Langendorff-perfused hearts. Front. Physiol. 6, 134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fürst D. O., Obermann W. M., van der Ven P. F., Structure and assembly of the sarcomeric M band. Rev. Physiol. Biochem. Pharmacol. 138, 163–202 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Guo A., Song L.-S., AutoTT: automated detection and analysis of T-tubule architecture in cardiomyocytes. Biophys. J. 106, 2729–2736 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A., et al. , Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White J. R., Naccache P. H., Sha’afi R. I., Stimulation by chemotactic factor of actin association with the cytoskeleton in rabbit neutrophils. Effects of calcium and cytochalasin B. J. Biol. Chem. 258, 14041–14047 (1983). [PubMed] [Google Scholar]
- 20.Bubb M. R., Spector I., Bershadsky A. D., Korn E. D., Swinholide A is a microfilament disrupting marine toxin that stabilizes actin dimers and severs actin filaments. J. Biol. Chem. 270, 3463–3466 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Ho C. Y., Jaalouk D. E., Vartiainen M. K., Lammerding J., Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature 497, 507–511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trembley M. A., et al. , Mechanosensitive gene regulation by myocardin-related transcription factors is required for cardiomyocyte integrity in load-induced ventricular hypertrophy. Circulation 138, 1864–1878 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Descot A., et al. , Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Mol. Cell 35, 291–304 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Posern G., Sotiropoulos A., Treisman R., Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell 13, 4167–4178 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cen B., et al. , Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 23, 6597–6608 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boateng S. Y., et al. , Cardiac dysfunction and heart failure are associated with abnormalities in the subcellular distribution and amounts of oligomeric muscle LIM protein. Am. J. Physiol. Heart Circ. Physiol. 292, H259–H269 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Arber S., et al. , MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell 88, 393–403 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Gifford C. A., et al. , Oligogenic inheritance of a human heart disease involving a genetic modifier. Science 364, 865–870 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X., Pabon L., Murry C. E., Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 114, 511–523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S., Chang S., Qi X., Richardson J. A., Olson E. N., Requirement of a myocardin-related transcription factor for development of mammary myoepithelial cells. Mol. Cell. Biol. 26, 5797–5808 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mokalled M. H., Johnson A., Kim Y., Oh J., Olson E. N., Myocardin-related transcription factors regulate the Cdk5/Pctaire1 kinase cascade to control neurite outgrowth, neuronal migration and brain development. Development 137, 2365–2374 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data generated for this study have been deposited in the Gene Expression Omnibus database under the accession code GSE136096. Actn2F mice, produced in Dr. Alan Beggs’s laboratory, can be obtained through a material transfer agreement.