By systematic depletion of PCM proteins and centrioles in human cell lines, we reveal the cooperative role of the centriole and PCM scaffold proteins, pericentrin and CDK5RAP2, in the recruitment of CEP192 to spindle poles to promote bipolar spindle formation.
Abstract
The pericentriolar material (PCM) that accumulates around the centriole expands during mitosis and nucleates microtubules. Here, we show the cooperative roles of the centriole and PCM scaffold proteins, pericentrin and CDK5RAP2, in the recruitment of CEP192 to spindle poles during mitosis. Systematic depletion of PCM proteins revealed that CEP192, but not pericentrin and/or CDK5RAP2, was crucial for bipolar spindle assembly in HeLa, RPE1, and A549 cells with centrioles. Upon double depletion of pericentrin and CDK5RAP2, CEP192 that remained at centriole walls was sufficient for bipolar spindle formation. In contrast, through centriole removal, we found that pericentrin and CDK5RAP2 recruited CEP192 at the acentriolar spindle pole and facilitated bipolar spindle formation in mitotic cells with one centrosome. Furthermore, the perturbation of PLK1, a critical kinase for PCM assembly, efficiently suppressed bipolar spindle formation in mitotic cells with one centrosome. Overall, these data suggest that the centriole and PCM scaffold proteins cooperatively recruit CEP192 to spindle poles and facilitate bipolar spindle formation.
Introduction
Centrosomes nucleate and anchor microtubules, thereby facilitating efficient spindle formation and chromosome segregation during mitosis (Moritz et al., 1995; Kollman et al., 2011; Woodruff et al., 2017). The microtubule-organizing activity of centrosomes depends on the pericentriolar material (PCM) that surrounds one or two centrioles (Woodruff et al., 2014). Abnormalities in centrosome organization and function lead to chromosomal segregation errors; several mutations in centrosomal proteins have also been implicated in the development of diseases such as cancer (Nigg and Raff, 2009; Gönczy, 2015). In addition, PCM disorganization directly causes chromosome missegregation (Watanabe et al., 2019; Cosenza et al., 2017). Therefore, elucidating the function and organization of centrosome in mitosis will contribute to a better understanding of the mechanisms through which centrosomes dictate the spindle structure and support accurate chromosome segregation.
PCM contains a large number of proteins, such as the γ-tubulin ring complex (γ-TuRC), CDK5RAP2, CEP192, and pericentrin. During the G2/M transition, CEP192 recruits Aurora A and polo-like kinase 1 (PLK1) to centrosomes in a pericentrin-dependent manner; subsequently, CEP192 activates these kinases to promote microtubule nucleation and centrosome separation (Joukov et al., 2014). CEP192 also supports the organization of other PCM components for efficient bipolar spindle assembly (Gomez-Ferreria et al., 2007). PLK1 phosphorylates pericentrin to further recruit other PCM components to centrosomes, thereby increasing the microtubule nucleation activity of the centrosome during mitosis (Lee and Rhee, 2011). Microtubule nucleation activity depends on γ-TuRC (Zheng et al., 1995; Wieczorek et al., 2020; Liu et al., 2020; Consolati et al., 2020; Moritz et al., 1995; Kollman et al., 2011), the activity of which is up-regulated by the binding of CDK5RAP2 to γ-TuRC (Choi et al., 2010; Hanafusa et al., 2015). In addition to their functions in microtubule nucleation, previous studies have described pericentrin and CDK5RAP2 regulating spindle pole focusing and spindle orientation through the regulation of motor proteins or other spindle pole proteins (Lee and Rhee, 2010; Chavali et al., 2016; Tungadi et al., 2017; Chen et al., 2014).
During the G2/M phase, PCM expands around the pair of centrioles that form the structural core of the centrosome and increases its ability to nucleate microtubules. In Drosophila melanogaster and Caenorhabditis elegans, it has been reported that centrioles regulate the architecture and dynamics of PCM (Kirkham et al., 2003; Conduit et al., 2010; Erpf et al., 2019; Cabral et al., 2019; Sir et al., 2013; Alvarez-Rodrigo et al., 2019; Conduit et al., 2014). In addition, it has been shown that PCM disorganization causes precocious centriole disengagement during mitosis (Seo et al., 2015; Kim et al., 2015, 2019; Watanabe et al., 2019), which can result in impairment of spindle pole integrity (Watanabe et al., 2019). This cross-reactive interplay between centrioles and PCM complicates the analysis of the individual function of PCM at spindle poles independent of the involvement of centriolar machinery. The centriole-independent functions of PCM have been partially characterized in the acentriolar meiotic spindles of mouse oocytes. During meiotic spindle formation in mice, acentriolar microtubule-organizing centers are formed and merge into two equal spindle poles (Clift and Schuh, 2015; Schuh and Ellenberg, 2007). Conditional knockout of pericentrin induces spindle instability and severe meiotic errors that lead to pronounced female subfertility in mouse oocytes. These findings suggest that pericentrin assists in organizing functional spindle poles to achieve faithful chromosome segregation (Baumann et al., 2017). However, as the system of meiosis is particularly unique compared with that of mitosis, it is unclear whether acentrosomal spindle formation pathways can be directly compared between oocytes and somatic cells.
To evaluate the distinct functions of PCM in human somatic cells independently of centrioles, it is important to use an assay system that enables the analysis of mitotic spindles that lack centrioles. As centriole duplication requires PLK4 (Habedanck et al., 2005; Bettencourt-Dias et al., 2005), its specific inhibitor, centrinone, can be used to remove centrioles (Wong et al., 2015). Treatment with centrinone leads to progressive loss of centrioles and generates mitotic spindles with one or zero centrosomes. Using this strategy, we have previously shown the critical roles of nuclear mitotic apparatus protein (NuMA) in spindle bipolarization in early mitosis of cells without centrosomes (Chinen et al., 2020). Similarly, by using mitotic cells with one centrosome, Dudka et al. recently reported that centrosomes regulate the length of K-fibers and thereby alter their dynamics in a HURP-dependent manner (Dudka et al., 2019).
In this study, we show the cooperative roles of the centriole and PCM scaffold proteins in bipolar spindle formation in human cells. When PCM assembly was inhibited by depletion of the PCM scaffold proteins pericentrin and CDK5RAP2, we found that another PCM protein, CEP192, remained at the centriole wall, where it presumably promoted bipolar spindle formation. Furthermore, we induced the formation of mitotic spindles with only one centrosome by treating human cells with centrinone. We found that the one-centrosome cells formed a bipolar spindle that accumulated PCM components, including CEP192, at the acentriolar pole. In such cells, depletion of pericentrin or CDK5RAP2 compromised the formation of the acentriolar pole and significantly prolonged mitotic progression. In contrast, the artificial accumulation of PCM components at the acentriolar pole accelerated the mitotic progression in one-centrosome cells. These results demonstrate that the centriole and PCM scaffold proteins, pericentrin and CDK5RAP2, cooperatively assemble and retain CEP192 at the spindle poles and facilitate bipolar spindle formation.
Results
CEP192 at the centriolar wall is sufficient for organizing mitotic spindle poles
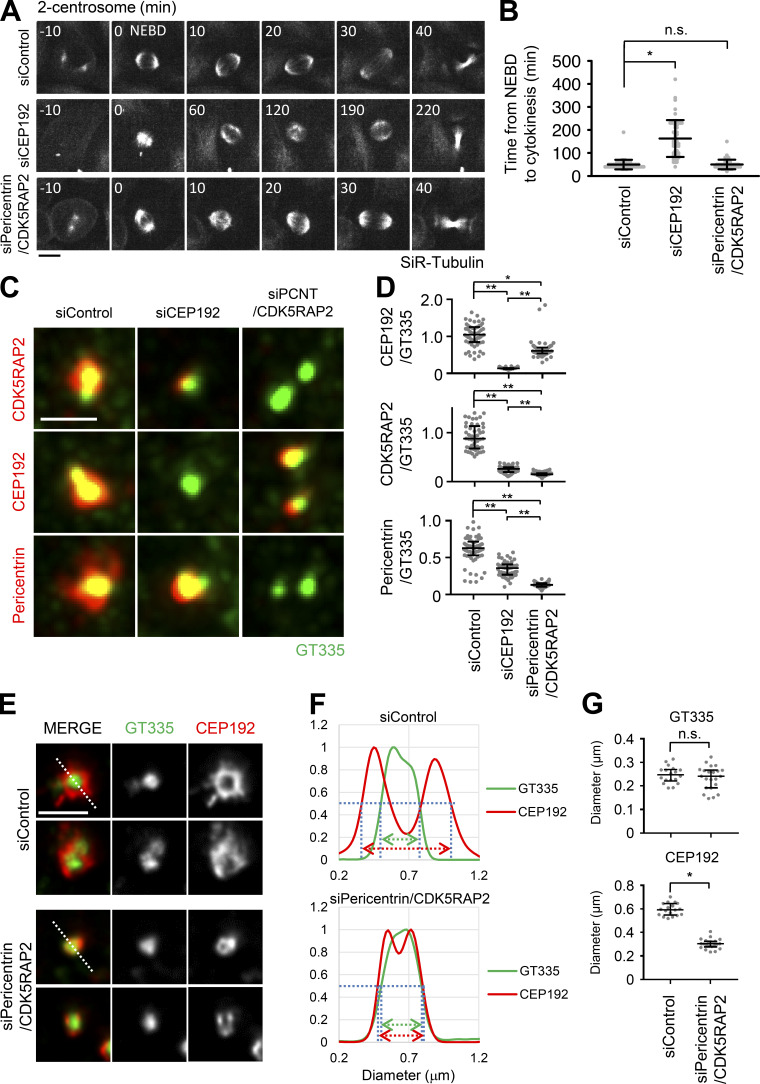
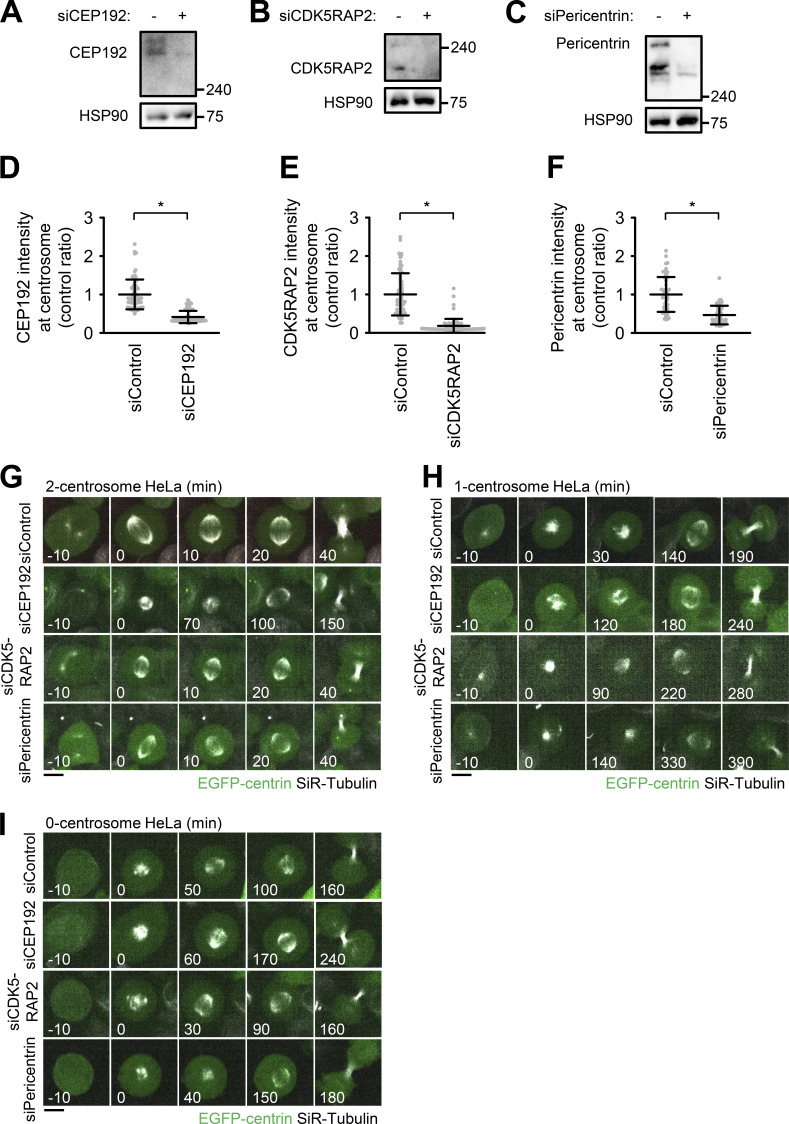
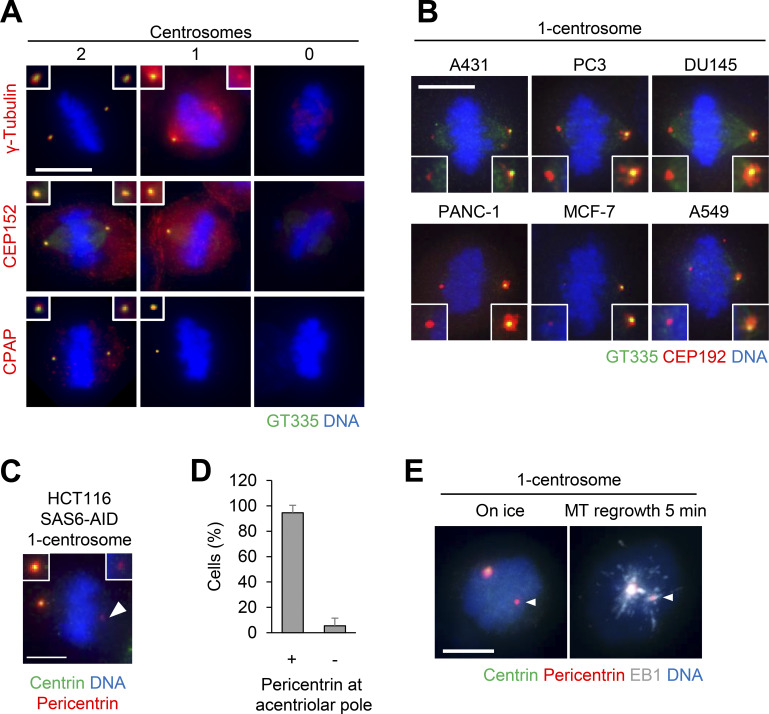
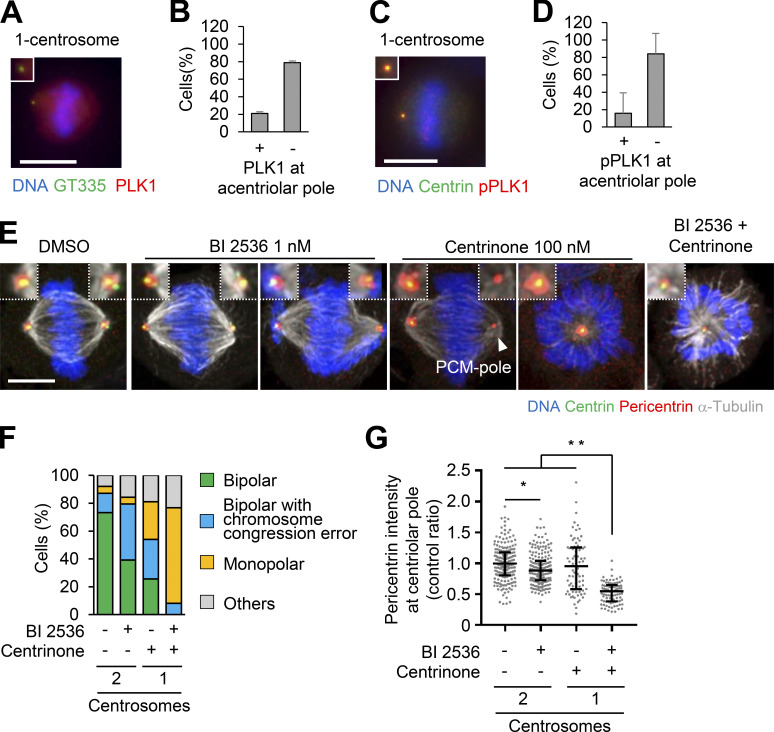
To understand the functions of PCM in bipolar spindle formation, we depleted the main components of PCM, such as CEP192, pericentrin, and CDK5RAP2, and observed bipolar spindle assembly in HeLa, A549, and RPE1 cells. As previously described, the depletion of CEP192 caused severe defects in bipolar spindle formation and prolonged mitotic duration (Fig. 1, A and B; Fig. S3; Videos 1 and 2). On the other hand, double depletion of pericentrin and CDK5RAP2 or their individual depletion had a limited effect on mitotic duration in HeLa cells (Fig. 1, A and B; Fig. 2 C; Fig. S1 G; Videos 1, 3, 15, 17, and 18) and bipolar spindle assembly in RPE1 and A549 cells (Fig. S3, A–D). These results suggest that CEP192, but not pericentrin or CDK5RAP2, is critical for mitotic progression. It has been suggested that pericentrin and CDK5RAP2 cooperatively recruit PCM components, including CEP192, at centrosomes (Kim and Rhee, 2014). Therefore, we observed the amount and localization of CEP192 at centrosomes upon depletion of pericentrin and CDK5RAP2. We found that a certain quantity of CEP192 remained at centrosomes in pericentrin/CDK5RAP2 double-depleted cells (Fig. 1, C and D). To further understand this mechanism, we used gated stimulated emission depletion (STED) microscopy to analyze the detailed localization pattern of CEP192 at centrosomes in pericentrin/CDK5RAP2 double-depleted cells (Fig. 1, E–G). Centrioles were marked by polyglutamylated centriolar microtubules. In control cells, CEP192 was detectable in the PCM clouds that surrounded mother centrioles (Fig. 1, E–G). In contrast, in pericentrin/CDK5RAP2 double-depleted cells, the reduced quantity of CEP192 was detectable only on centriolar walls. These results raise the possibility that CEP192 at the centriolar wall, rather than in the PCM cloud, is crucial for the microtubule-organizing center function of centrosomes.
Figure 1.
CEP192 at centriolar wall is sufficient for bipolar spindle formation. (A) Time-lapse observation of the structure of microtubules upon siRNA treatment against the indicated proteins. HeLa cells expressing EGFP-centrin1 and mCherry-NuMA were observed with a 40× objective. Gray represents SiR-tubulin. mCherry-NuMA and EGFP-centrin1 are not shown. Z-projections: 10 planes, 2.2 µm apart. Scale bar, 10 µm. Time zero corresponds to NEBD. (B) Mitotic duration, the time required from NEBD to cytokinesis, in A. Line and error bars represent the mean and SD (n ≥ 50 cells from two independent experiments). Kruskal–Wallis test was used to determine the significance of the difference. *, P < 0.05; n.s., not significant. (C) The localization of PCM proteins in mitotic spindles of the cells in which the indicated protein was depleted. Red and green represent PCM proteins (CDK5RAP2, CEP192, or pericentrin) and GT335, respectively. Z-projections of 10 sections, every 0.3 µm. Scale bar, 1 µm. (D) The signal intensity of PCM proteins on mitotic centrosomes of fixed HeLa cells was analyzed (n > 45 for each condition). Line and error bars represent median with interquartile range. Kruskal–Wallis test was used to determine the significance of the difference. *, P < 0.05; **, P < 0.0001. (E) STED images showing centriolar distribution of CEP192 in pericentrin/CDK5RAP2 double-depleted cells. HeLa cells were treated with control siRNA or pericentrin/CDK5RAP2 siRNA for 48 h and stained with the indicated antibodies. Scale bar, 1 µm. (F and G) Representative line intensity profiles (F) and measured diameters (G) of GT335 and CEP192. The line profiles were measured along the dotted lines in E. The profiles were fitted with double Gaussian curves, and the distances between the half-maximal intensity points at the far ends were measured as the diameters (schematically indicated with dotted lines and arrows in the profiles; fitted curves are not shown). Horizontal bars and error bars in the plots for the diameters represent median and interquartile range. n = 18 (for siControl) or 22 (for siPericentrin/CDK5RAP2) centrosomes; data from two independent experiments were pooled. Mann–Whitney U test was used to determine the significance of the difference. *, P < 0.0001; n.s., not significant.
Figure S3.
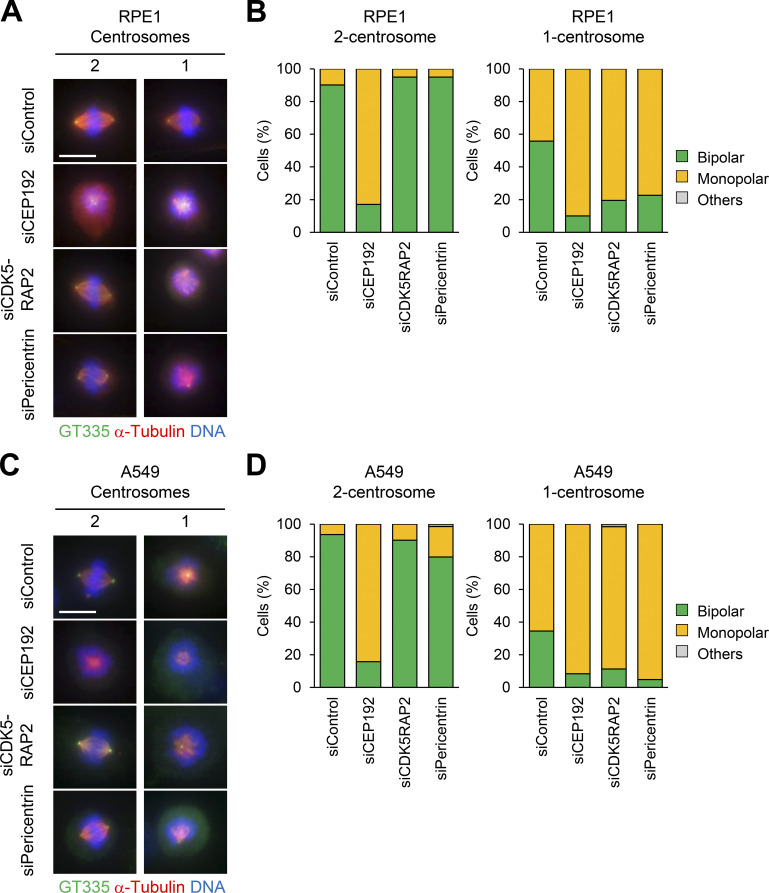
Pericentrin and CDK5RAP2 are crucial for bipolar spindle formation of one-centrosome cells. (A–D) Mitotic spindle structures in siPCM-treated RPE1 (A) and A549 (C) cells. DMSO-treated control mitotic spindles (two centrosomes) and centrinone-treated mitotic spindles (one centrosome). After siRNA treatment with or without 100 nM centrinone, mitotic spindle structures were observed. Green, red, and blue represent GT335, α-tubulin, and DNA, respectively. Z-projections: 31 planes, 0.5 µm apart. Scale bar, 10 µm. Frequency of mitotic spindle structures in RPE1 (B) and A549 (D) cells. Values are mean percentages from two independent experiments (n > 20 for each experiment).
Video 1.
Control HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with two centrosomes labeled with SiR-tubulin and observed with a 40× objective. Gray represents SiR-tubulin. mCherry-NuMA and EGFP-centrin1 are not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 1 A.
Video 2.
CEP192-depeleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with two centrosomes labeled with SiR-tubulin and observed with a 40× objective. Gray represents SiR-tubulin. mCherry-NuMA and EGFP-centrin1 are not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 1 A.
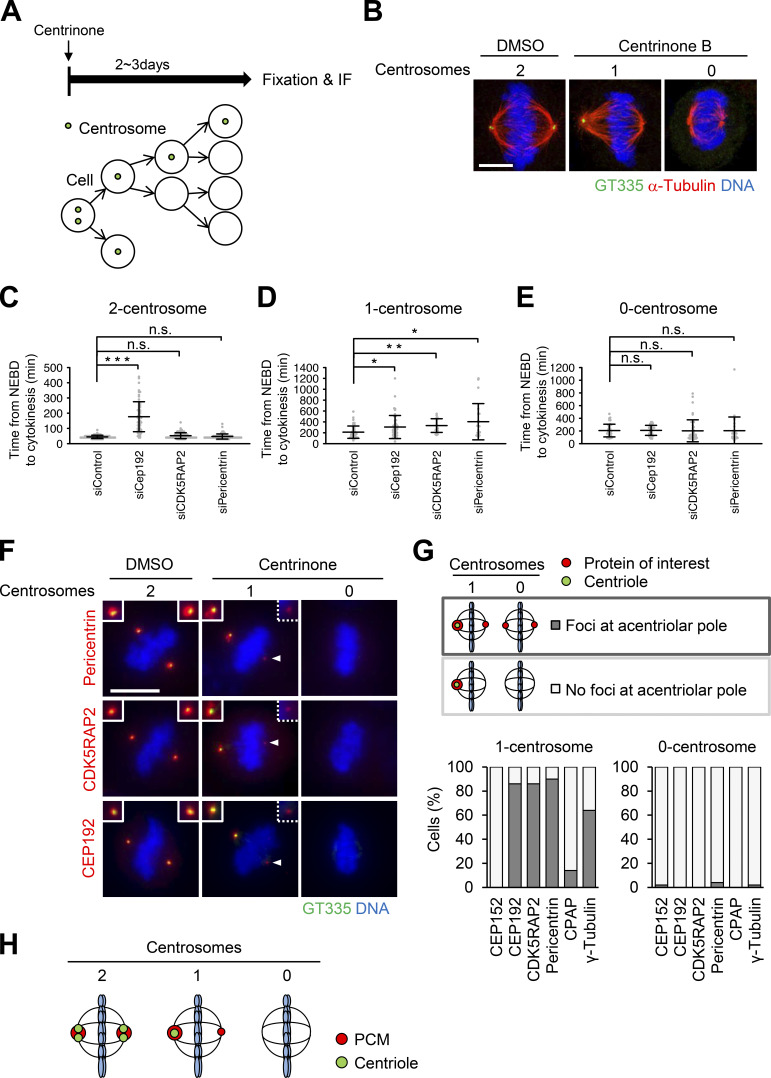
Figure 2.
Cells with one centrosome can organize bipolar spindles in mitosis by forming a PCM-positive acentriolar spindle pole (PCM pole). (A) Schematic illustration of centrinone-induced removal of centriole. (B) DMSO-treated control mitotic spindles (two centrosomes) and centrinone B–treated centrosome-depleted spindles (one or zero centrosomes). Green, red, and blue represent GT335 (polyglutamylated centriole microtubules), α-tubulin, and DNA, respectively. Z-projections: 12 planes, each 0.13 µm apart. Scale bar, 5 µm. (C–E) Mitotic duration, the time required from NEBD to cytokinesis, in DMSO-treated two-centrosome (C), centrinone-treated one-centrosome (D) and zero-centrosome (E) cells in Fig. S1, G–I. Line and error bars represent the mean and SD (n ≥ 20 cells from two independent experiments). Kruskal–Wallis test was used to determine the significance of the difference. *, P < 0.05; **, P < 0.005; *** P < 0.0001; n.s., not significant. (F) Distribution of centrosomal factors in centriolar and acentriolar spindle poles. DMSO-treated control mitotic spindles (two centrosomes) and centrinone-treated mitotic spindles (one or zero centrosomes) in HeLa cells. Green, red, and blue represent GT335, the protein of interest (pericentrin, CDK5RAP2, or CEP192), and DNA, respectively. Z-projections: 21 sections, every 1 µm. Scale bar, 10 µm. Arrowheads indicate the PCM at the acentriolar spindle pole. (G) Quantification of pole patterns in F and Fig. S2 A. Values are presented as mean percentages from two independent experiments (n = 25 for each experiment). (H) Schematic illustration of PCM localization at spindle poles in two-, one-, or zero-centrosome cells.
Figure S1.
Depletion of PCM components in two-, one-, and zero-centrosome cells. (A–C) Western blot analysis of the efficiency of protein depletion of CEP192 (A), CDK5RAP2 (B), and pericentrin (C) after 48 h of siRNA transfection in HeLa cells. (D–F) Quantification of depleted centrosomal CEP192 (D), CDK5RAP2 (E), and pericentrin (F). Line and error bars represent the mean and SD. The Mann–Whitney U test (two tailed) was used to obtain a P value. *, P < 0.0001. (G–I) Time-lapse observation of the structure of microtubules upon siRNA treatment against the indicated proteins. DMSO-treated two-centrosome (G), centrinone-treated one-centrosome (H) and zero-centrosome (I) HeLa cells expressing EGFP-centrin1 and mCherry-NuMA were observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. Z-projections: 10 planes, 2.2 µm apart. Scale bar, 10 µm. Time zero corresponds to NEBD.
Video 3.
Pericentrin and CDK5RAP2-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with two centrosomes labeled with SiR-tubulin and observed with a 40× objective. Gray represents SiR-tubulin. mCherry-NuMA and EGFP-centrin1 are not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 1 A.
Video 15.
Control HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with two centrosomes labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 G.
Video 17.
CDK5RAP2-depeleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with two centrosomes labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 G.
Video 18.
Pericentrin-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with two centrosomes labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 G.
Cells with one centrosome form a bipolar spindle that accumulates PCM components at the acentriolar pole
To understand the functions of PCM independently of centrioles in human cells, we next induced the formation of mitotic spindles with one or zero centrosomes by treating HeLa cells with the PLK4 inhibitors centrinone or centrinone B (Fig. 2, A and B). Centrosomes were marked by polyglutamylated centriolar microtubules or centrin to determine their number. We depleted PCM components CEP192, pericentrin, and CDK5RAP2 in one- or zero-centrosome cells and observed their mitotic progression using live-cell imaging. As described above, the depletion of CEP192, but not pericentrin or CDK5RAP2, prolonged mitosis in cells with two centrosomes (Fig. 2 C; Fig. S1 A–G; Videos 15, 16, 17, and 18). On the other hand, interestingly, we found that depletion of pericentrin or CDK5RAP2, as well as CEP192, significantly prolonged mitotic duration in one-centrosome cells (Fig. 2 D; Fig. S1 H; Videos 19, 20, 21, and 22). In contrast, we found that depletion of pericentrin, CDK5RAP2, or CEP192 had a limited effect on mitotic progression in zero-centrosome cells (Fig. 2 E; Fig. S1 I; Videos 23, 24, 25, and 26). These results suggest that pericentrin and CDK5RAP2 are important for mitotic progression in one-centrosome cells, but not in two- or zero-centrosome cells.
Video 16.
CEP192-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with two centrosomes labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 G.
Video 19.
Control HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with one centrosome labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 H.
Video 20.
CEP192-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with one centrosome labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 H.
Video 21.
CDK5RAP2-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with one centrosome labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 H.
Video 22.
Pericentrin-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with one centrosome labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 H.
Video 23.
Control HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with zero-] centrosomes labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 I.
Video 24.
CEP192-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with zero centrosomes labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 I.
Video 25.
CDK5RAP2-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with zero centrosomes labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 I.
Video 26.
Pericentrin-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with zero centrosomes labeled with SiR-tubulin and observed with a 40× objective. Green and gray represent EGFP-centrin1 and SiR-tubulin, respectively. mCherry-NuMA is not shown. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S1 I.
We further analyzed the localization patterns of PCM proteins at spindle poles using immunofluorescence (IF) microscopy (Fig. 2, F and G; and Fig. S2, A–D). We found that the acentriolar spindle poles of one-centrosome cells incorporate a detectable amount of PCM components, such as pericentrin, CDK5RAP2, CEP192, and γ-tubulin (Fig. 2, F and G; and Fig. S2 A), but not CEP152 or CPAP (Figs. 2 G and S2 A). In this study, we termed the acentriolar spindle pole that contains PCM the “PCM pole.” In contrast, most spindle poles of zero-centrosome cells lacked PCM components, as previously described (Fig. 2, F and G; Fig. S2 A; Chinen et al., 2020). PCM components were consistently detectable at the acentriolar spindle poles in one-centrosome cells of various human cell lines (Fig. S2 B). Furthermore, the PCM pole was similarly observed in one-centrosome cells induced by SAS6 depletion using the auxin-inducible degron (AID) system (Fig. S2, C and D; Yoshiba et al., 2019), suggesting that this phenotype was not a specific result of PLK4 inhibition.
Figure S2.
Distribution of centrosomal factors in centriolar and acentriolar spindle poles. (A–D) Distribution of centrosomal factors in centriolar and acentriolar spindle poles. (A) DMSO-treated control mitotic spindles (two centrosomes) and centrinone-treated spindles (one or zero centrosomes) of HeLa cells. Green, red, and blue represent GT335, protein of interest (γ-tubulin, CEP152, or CPAP), and DNA, respectively. Z-projections: 21 planes, 1 µm apart. Scale bar, 10 µm. (B) PCM poles were observed in various cells. Green, red, and blue represent GT335, CEP192 and DNA, respectively. Z-projections: 40 planes, 0.3 µm apart. Scale bar, 10 µm. (C) PCM poles were observed in one-centrosome spindles induced by SAS6 depletion. Green, red and blue represent centrin, pericentrin, and DNA, respectively. Scale bar, 5 µm. Arrowheads indicate the PCM at the acentriolar spindle pole. (D) Quantification of pole patterns in C. Values are presented as mean percentages from three independent experiments (n = 20 for each experiment). Error bars represent SD. (E) Microtubule nucleation from the PCM pole. Following treatment with ice, microtubule nucleation (5 min at 25°C) was observed in one-centrosome cells. Gray, red, green, and blue in the merged image represent EB1, RFP (pericentrin), GFP (centrin), and DNA, respectively. Z-projections: 21 planes, 1 µm apart. Scale bar, 5 µm. MT, microtubule. Arrowheads indicate the PCM at the acentriolar spindle pole.
We next examined whether the PCM pole nucleates microtubules using a microtubule regrowth analysis. For this analysis, we immunostained the microtubule end binding protein 1 (EB1), which marks growing microtubule plus ends. When restarting the microtubule nucleation, the EB1 signals started developing around both centriolar and PCM poles, with PCM poles nucleating fewer microtubules (Fig. S2 E). Thus, PCM poles possess microtubule nucleation activity, although this activity appears slightly lower than that of centriolar poles. Collectively, these results suggest that one-centrosome cells assemble PCM at the acentriolar spindle pole, which harbors microtubule nucleation activity (Fig. 2 H).
The PCM pole is formed by either split of the PCM from the centriolar pole or accumulation of PCM
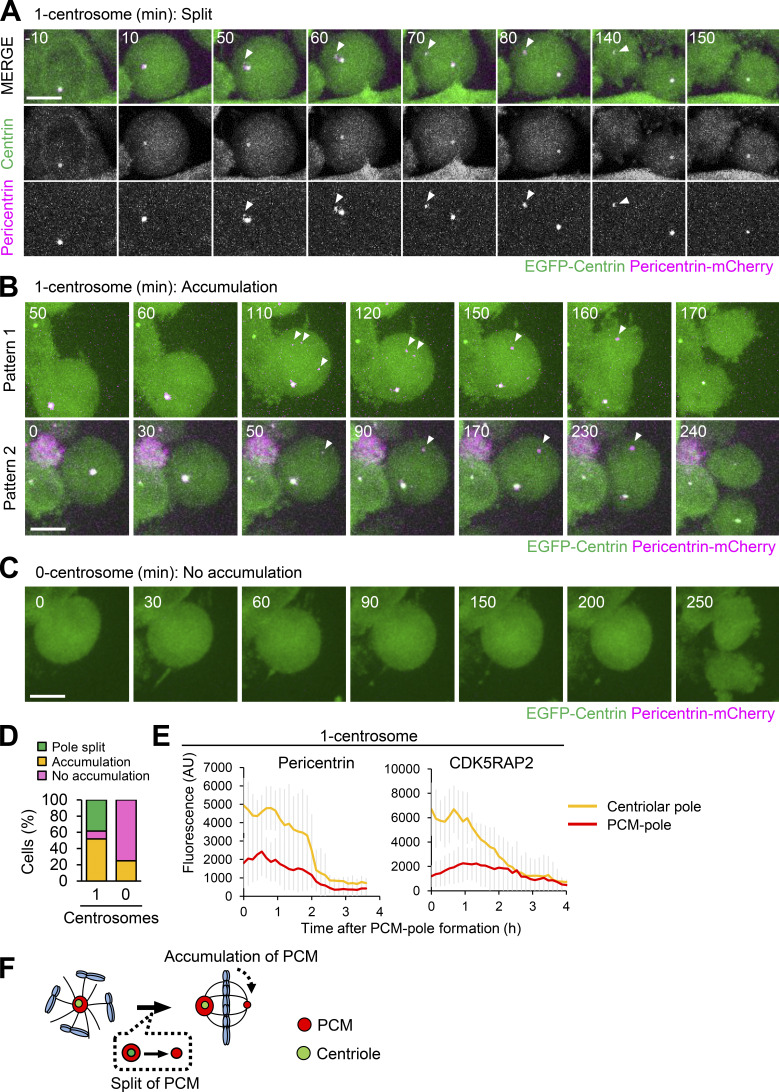
To understand the mechanism of PCM recruitment to the acentriolar pole in one-centrosome cells, we used time-lapse fluorescence microscopy to track the dynamics of endogenous pericentrin or CDK5RAP2 tagged with mCherry as markers of PCM. This strategy revealed that, at first, pericentrin accumulated at centriolar poles in early mitosis. Subsequently, one-centrosome cells formed pericentrin-positive PCM poles by either splitting of the PCM from the centriolar pole or de novo accumulation of PCM (38.5% and 51.9%, respectively; Fig. 3, A, B, and D; Videos 4, 5, and 6). These PCM poles disappeared after cytokinesis (Fig. 3, A, B, and E; Videos 4, 5, and 6), consistent with the observation that PCM proteins are disassembled after mitotic exit (Woodruff et al., 2014). On the other hand, a detectable amount of pericentrin did not accumulate at the acentriolar spindle poles in most zero-centrosome cells (Fig. 3, C and D; Video 7). Taken together, these observations suggest that one-centrosome cells initially accumulate PCM proteins around centrioles and subsequently generate the acentriolar pole by splitting and/or recruiting PCM components on the opposite side for bipolar spindle formation (Fig. 3 F).
Figure 3.
The PCM pole is formed by splitting PCM from the centriolar pole or by accumulation of PCM components. (A–D) HeLa cells expressing EGFP-centrin1 and pericentrin-mCherry were observed with a 60× objective. Magenta and green represent pericentrin-mCherry and EGFP-centrin1, respectively. Z-projections: 20 planes, 1.2 µm apart. Scale bars, 10 µm. Time zero corresponds to the beginning of mitotic cell rounding. (A) Splitting of the PCM components from the centriolar pole in one-centrosome cells. Arrowheads indicate the PCM at the acentriolar spindle pole. (B) PCM accumulation in one-centrosome cells. Arrowheads indicate the accumulation of PCM at acentriolar spindle poles. (C) Cell division in zero-centrosome cells without accumulation of PCM. (D) Quantification of patterns of PCM dynamics in A–C. Values are percentages of the total cells from 52 (for one-centrosome cells) or 24 (for zero-centrosome cells) cells from two independent experiments. (E) Averaged time courses of pericentrin-mCherry or CDK5RAP2-mCherry signals at the centriolar spindle pole and PCM pole of 10 cells. Time course data were aligned at PCM pole formation (0 h). Error bars represent SD; AU, arbitrary units. (F) Schematic illustration of PCM pole formation by splitting PCM from the centriolar spindle pole or by accumulation of PCM components.
Video 4.
Centrinone-treated HeLa cells expressing EGFP-centrin1 and pericentrin-mCherry with one centrosome observed with a 60× objective. Magenta and green represent pericentrin-mCherry and EGFP-centrin1, respectively. 20 × 1.2–μm z-stacks were acquired every 10 min. Playback is sped up 6,000× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 3 A.
Video 5.
Centrinone-treated HeLa cells expressing EGFP-centrin1 and pericentrin-mCherry with one centrosome observed with a 60× objective. Magenta and green represent pericentrin-mCherry and EGFP-centrin1, respectively. 20 × 1.2–μm z-stacks were acquired every 10 min. Playback is sped up 6,000× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 3 B, top panel (pattern 1).
Video 6.
Centrinone-treated HeLa cells expressing EGFP-centrin1 and pericentrin-mCherry with one centrosome observed with a 60× objective. Magenta and green represent pericentrin-mCherry and EGFP-centrin1, respectively. 20 × 1.2–μm z-stacks were acquired every 10 min. Playback is sped up 6,000× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 3 B, bottom panel (pattern 2).
Video 7.
Centrinone-treated HeLa cells expressing EGFP-centrin1 and pericentrin-mCherry with zero centrosome observed with a 60× objective. Magenta and green represent pericentrin-mCherry and EGFP-centrin1, respectively. 20 × 1.2–μm z-stacks were acquired every 10 min. Playback is sped up 6,000× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 3 C.
CDK5RAP2 and pericentrin are crucial for the bipolar spindle formation in one-centrosome cells
We next analyzed the specific role of PCM in cell division in one-centrosome cells. However, it is difficult to analyze the functions of some PCM pole components in this context. Among those appeared to localize at PCM poles (Fig. 2, F and G; and Fig. S2 A), for example, γ-tubulin also localizes along the whole spindle and regulates several pathways of microtubule nucleation in mitosis (Lecland and Lüders, 2014; Teixidó-Travesa et al., 2012). In addition, CEP192 is required for bipolar spindle formation in cells with two centrosomes (Zhu et al., 2008; Joukov et al., 2014). On the other hand, depletion of the PCM scaffold proteins CDK5RAP2 and pericentrin are known to have little effect on spindle formation in two- or zero-centrosome cells (Fig. 2, C–E; Fig. S1, G–I; Videos 15, 17, 18, 23, 25, and 26). Therefore, we selected CDK5RAP2 and pericentrin for further analysis of PCM poles in one-centrosome cells.
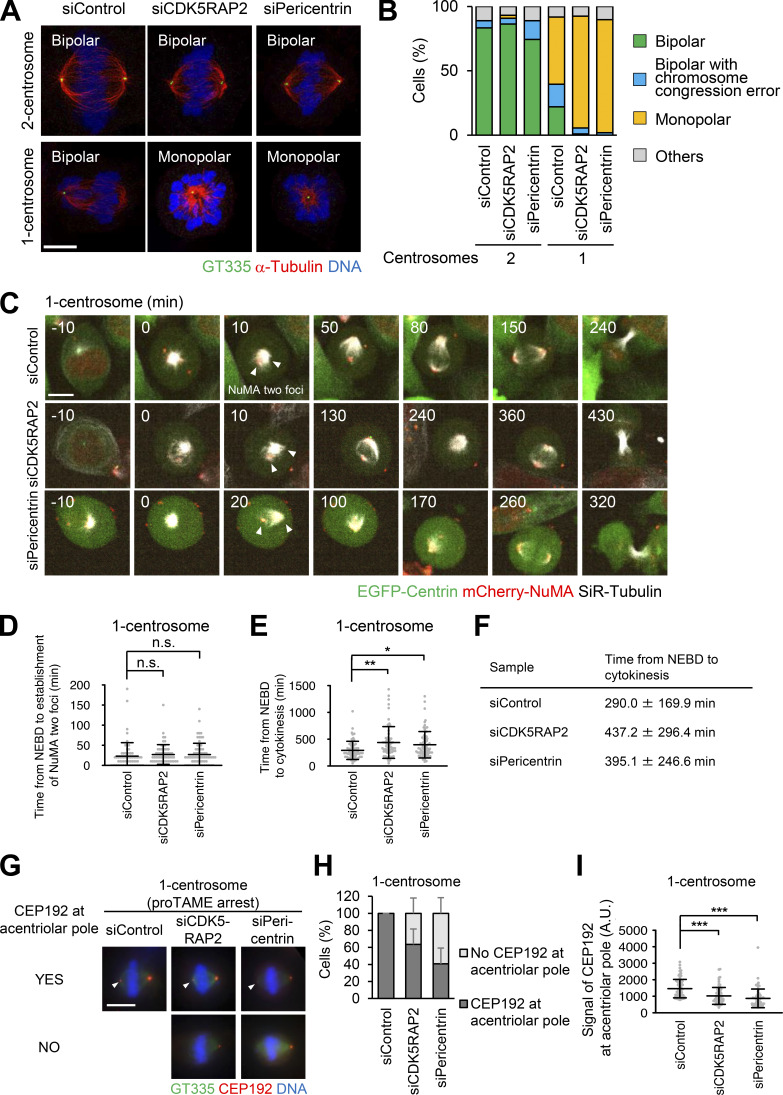
We found that depletion of CDK5RAP2 or pericentrin caused arrest of one-centrosome cells in mitosis with monopolar spindles; however, this effect was not observed in two-centrosome cells (Fig. 4, A and B). These results indicate that CDK5RAP2 and pericentrin play an important role in bipolar spindle formation specifically in one-centrosome cells, but not in two-centrosome cells. To further investigate this process, we tracked the dynamics of spindle poles in one-centrosome cells using time-lapse observation of NuMA tagged with mCherry. Upon depletion of CDK5RAP2 or pericentrin, the separation of two NuMA foci was normally detectable in early mitosis (Fig. 4, C and D; Videos 8, 9, and 10), while the time from nuclear envelope breakdown (NEBD) to cytokinesis was prolonged (Fig. 4, C, E, and F; Videos 8, 9, and 10). Next, by using proTAME (APC/C inhibitor) treatment, we arrested pericentrin or CDK5RAP2-depleted HeLa cells with one centrosome in metaphase to artificially induce bipolar spindle formation. Immunofluorescence analysis revealed that in these HeLa cells, the degree of CEP192 localization at the acentrosomal spindle poles was significantly reduced compared with that in control cells (Fig. 4, G–I). These results indicate that PCM scaffold proteins CDK5RAP2 and pericentrin are crucial for the recruitment of CEP192 at the acentrosomal spindle pole and bipolar spindle formation but are likely dispensable for the early step of spindle pole generation in one-centrosome cells.
Figure 4.
Pericentrin and CDK5RAP2 are crucial for spindle elongation and spindle bipolarization of one-centrosome cells. (A) Mitotic spindle structures upon siRNA treatment with or without 500 nM centrinone B. Green, red, and blue represent GT335, α-tubulin, and DNA, respectively. Z-projections: 5 planes, 0.3 µm apart. Scale bar, 5 µm. (B) Frequency of mitotic spindle structures after siRNA treatment against the indicated proteins in A. Values are presented as mean percentages. n > 86, data from two independent experiments were pooled. (C) Time-lapse observation of the structure of NuMA and microtubules upon siRNA treatment against the indicated proteins. Centrinone-treated one-centrosome HeLa cells expressing EGFP-centrin1 and mCherry-NuMA were observed with a 40× objective. Red, green, and gray represent mCherry-NuMA, EGFP-centrin1, and SiR-tubulin, respectively. Z-projections: 10 planes, 2.2 µm apart. Scale bar, 10 µm. Time zero corresponds to NEBD. Arrowheads indicate the separated two NuMA foci. (D) The time required for the initial establishment of two poles of NuMA in C. Line and error bars represent the mean and SD (n ≥ 60 cells from three independent experiments). Kruskal–Wallis test was used to determine the significance of the difference. n.s., not significantly different (P > 0.05). (E) Mitotic duration, the time required from NEBD to cytokinesis, in C. Line and error bars represent the mean and SD (n ≥ 60 cells from three independent experiments). Kruskal–Wallis test was used to determine the significance of the difference. *, P < 0.01; **, P < 0.001. (F) Table of the times from NEBD to cytokinesis in E. (G) Distribution of CEP192 in centriolar and acentriolar spindle poles in proTAME-treated one-centrosome cells. Green, red, and blue represent GT335, CEP192, and DNA, respectively. Z-projections: 21 sections, 0.5 µm apart. Scale bar, 10 µm. (H) Quantification of pole patterns in G. Values are presented as mean percentages from triplicates (n = 3, triplicates, n ≥ 20 cells for each assay). Error bars represent SD. (I) The signal intensity of CEP192 on acentrosomal spindle poles in G. Line and error bars represent the mean and SD (n = 3, triplicates, n ≥ 20 cells for each assay). Kruskal–Wallis test was used to determine the significance of the difference. ***, P < 0.0001. A.U., arbitrary units.
Video 8.
Control HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with one centrosome labeled with SiR-tubulin and observed with a 40× objective. Red, green, and gray represent mCherry-NuMA, EGFP-centrin1, and SiR-tubulin, respectively. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 4 C.
Video 9.
CDK5RAP2-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with one centrosome labeled with SiR-tubulin and observed with a 40× objective. Red, green, and gray represent mCherry-NuMA, EGFP-centrin1, and SiR-tubulin, respectively. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 4 C.
Video 10.
Pericentrin-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with one centrosome labeled with SiR-tubulin and observed with a 40× objective. Red, green, and gray represent mCherry-NuMA, EGFP-centrin1, and SiR-tubulin, respectively. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 4 C.
To verify whether pericentrin and CDK5RAP2 are important for bipolar spindle formation in other human cell lines with one centrosome, we observed the spindle structure of RPE1 and A549 cells upon depletion of pericentrin or CDK5RAP2. Through immunostaining, we found that in cells with one centrosome, depletion of pericentrin or CDK5RAP2 induced the formation of monopolar spindles (Fig. S3, A–D). These results further support the conclusion that the PCM proteins are required for bipolar spindle formation in one-centrosome cells.
Depletion of CEP57 promotes accumulation of PCM components at PCM poles and facilitates bipolar spindle formation in one-centrosome cells
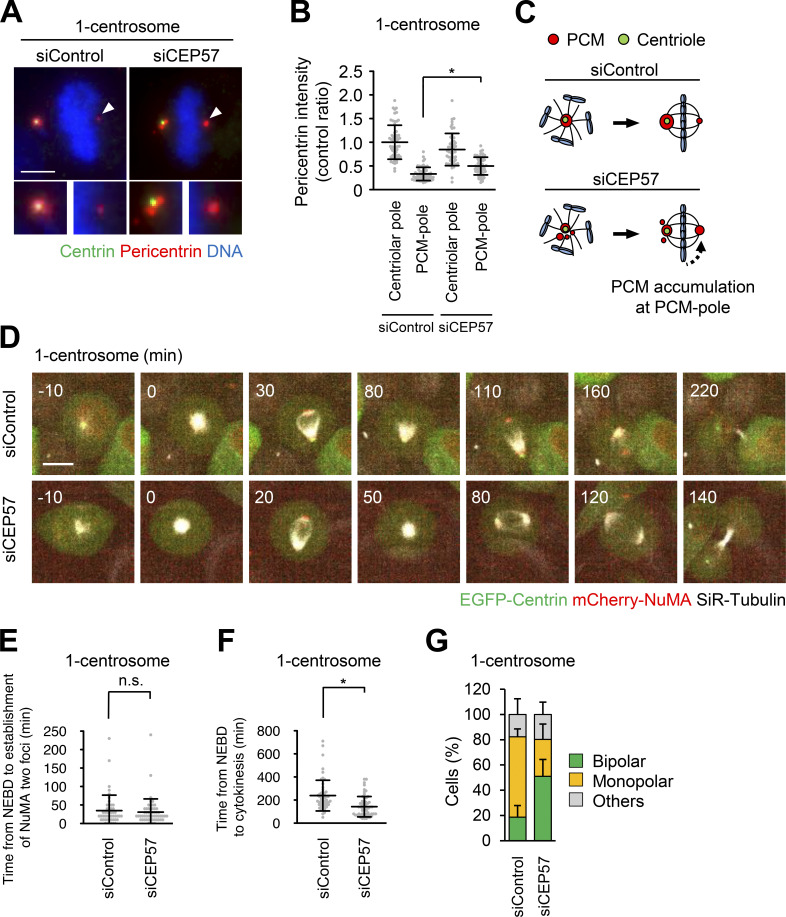
Next, we sought to further analyze the importance of PCM components at PCM poles for cell division in one-centrosome cells. Since siRNA-mediated depletion reduces the total expression level of CDK5RAP2 and pericentrin, it is difficult to analyze the function of the PCM components specifically at PCM poles (Fig. 4). Therefore, we used another approach, depleting CEP57, to indirectly manipulate the amount of PCM components at PCM poles. CEP57 provides a critical interface between the centriole and PCM, and depletion of CEP57 induces the fragmentation of PCM proteins in early mitosis of human cells (Watanabe et al., 2019). Given that 38.5% of one-centrosome cells assembled PCM poles by splitting PCM from the centrosome (Fig. 3, A and D; Video 4), we hypothesized that upon CEP57 depletion, the PCM that is dissociated from the centrosome could be incorporated into the acentriolar pole in one-centrosome cells. As expected, the amount of pericentrin at PCM poles was significantly increased, presumably due to the increased PCM fragmentation at centriolar poles after CEP57 depletion (Fig. 5, A–C). Subsequently, to analyze the effect of CEP57 depletion on the mitotic processes of one-centrosome cells, we performed time-lapse imaging of NuMA and microtubules. We found that depletion of CEP57 promoted bipolar spindle formation more efficiently than in control cells and thereby shortened the mitotic duration (Fig. 5, D–F; Videos 11 and 12). Under this condition, CEP57-depleted cells with one centrosome formed two separate NuMA foci, similar to siControl-treated one-centrosome cells, but established a bipolar spindle formation more efficiently (Fig. 5, E and G). Overall, these results suggest that accumulation of PCM components at PCM poles facilitates the bipolar spindle formation in one-centrosome cells.
Figure 5.
CEP57 depletion leads to an increase of PCM at the acentriolar pole and facilitates spindle bipolarization in one-centrosome cells. (A) Mitotic spindle pole structures of one-centrosome cells upon CEP57 depletion. Green, red, and blue represent centrin, pericentrin, and DNA, respectively. Z-projections: 20 planes, 0.5 µm apart. Scale bar, 5 µm. Arrowheads indicate the PCM at the acentriolar spindle pole. (B) The signal intensity of pericentrin on centrosomes or PCM poles in A. Line and error bars represent the mean and SD (n ≥ 50 cells from two independent experiments). Kruskal–Wallis test was used to determine the significance of the difference. *, P < 0.01. (C) Schematic illustration of CEP57-depletion-induced pericentrin accumulation at the PCM pole. (D) Time-lapse observation of NuMA structures and microtubules upon CEP57 depletion. Centrinone-treated one-centrosome HeLa cells expressing EGFP-centrin1 and mCherry-NuMA were observed with a 40× objective. Red, green, and gray represent mCherry-NuMA, EGFP-centrin1, and SiR-tubulin, respectively. Z-projections: 10 planes, 2.2 µm apart. Scale bar, 10 µm. Time zero corresponds to NEBD. (E) The time required for the initial establishment of two poles of NuMA in D. Line and error bars represent the mean and SD (n ≥ 50 cells from two independent experiments). The Mann–Whitney U test (two tailed) was used to obtain a P value. n.s., not significantly different (P > 0.05). (F) Mitotic duration, the time required from NEBD to cytokinesis, in D. Line and error bars represent the mean and SD (n ≥ 50 cells from two independent experiments). The Mann–Whitney U test (two tailed) was used to obtain a P value. *, P < 0.0001. (G) Frequency of mitotic spindle structures upon CEP57 depletion. Values are presented as mean percentages ± SD (n = 6, triplicates, two independent experiments, at least 29 spindles in each assay).
Video 11.
Control HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with one centrosome labeled with SiR-tubulin and observed with a 40× objective. Red, green, and gray represent mCherry-NuMA, EGFP-centrin1, and SiR-tubulin, respectively. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 5 D.
Video 12.
CEP57-depleted HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with one centrosome labeled with SiR-tubulin and observed with a 40× objective. Red, green, and gray represent mCherry-NuMA, EGFP-centrin1, and SiR-tubulin, respectively. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 5 D.
The activity of PLK1 is crucial for PCM pole assembly and bipolar spindle formation in one-centrosome cells
The accumulation of PCM components at centrosomes in mitosis is regulated by PLK1 activity (Haren et al., 2009; Lee and Rhee, 2011; Joukov et al., 2014). However, we found that PLK1 and phosphorylated PLK1 were not detected at most PCM poles in one-centrosome cells (Fig. 6, A–D). To determine if PLK1 was required for PCM pole assembly and subsequent bipolar spindle formation, we treated cells with a low dose of the PLK1 inhibitor BI 2536 (1 nM) and observed the amount of pericentrin at the centriolar pole and the spindle structure. Treatment of two-centrosome cells with a low dose of the PLK1 inhibitor caused chromosome congression errors and a slight reduction of pericentrin at centrosomes but did not affect bipolar spindle formation (Fig. 6, E–G). In contrast, in one-centrosome cells, PLK1 inhibition prevented PCM pole formation and led to the formation of monopolar spindles (Fig. 6, E and F). In addition, PLK1 inhibition greatly reduced the level of pericentrin at the centriolar pole compared with the level recorded in two-centrosome cells (Fig. 6 G). Together, these results suggest that PLK1 activity is crucial for PCM pole assembly and subsequent bipolar spindle formation in one-centrosome cells.
Figure 6.
PLK1 is crucial for PCM pole formation and bipolar spindle formation in one-centrosome cells. (A–D) PLK1 and phosphorylated PLK1 observed in one-centrosome cells. (A) Red, green, and blue represent PLK1, GT335, and DNA, respectively. Z-projections: 20 planes, 1 µm apart. Scale bar, 10 µm. (B) Frequency of localization of PLK1 in A. Values are presented as mean percentages from three independent experiments (n > 30 cells for each experiment). (C) Red, green, and blue represent phosphorylated PLK1, centrin, and DNA, respectively. Z-projections of 20 sections, every 1 µm. Scale bar, 5 µm. (D) Frequency of localization of phosphorylated PLK1 in C (n = 6, triplicates, two independent experiments, at least 20 cells in each assay). (E) Mitotic spindle structures upon PLK1 inhibition (1 nM of BI 2536) with or without 100 nM of centrinone. HeLa cells expressing EGFP-centrin1 and pericentrin-mCherry were observed with a 63× objective. Green, red, gray, and blue represent GFP (centrin1), RFP (pericentrin), α-tubulin, and DNA, respectively. Z-projections: 10 planes, 0.3 µm apart. Scale bar, 5 µm. (F) Frequency of mitotic spindle structures in E. Values are mean percentages from two independent experiments (n = 50 for each experiment). (G) The signal intensity of RFP (pericentrin) on GFP (centrin) of fixed mitotic HeLa cells expressing EGFP-centrin1 and pericentrin-mCherry (n > 45 for each condition). Line and error bars represent median with interquartile range. Kruskal–Wallis test was used to determine the significance of the difference. *, P < 0.05; **, P < 0.0001.
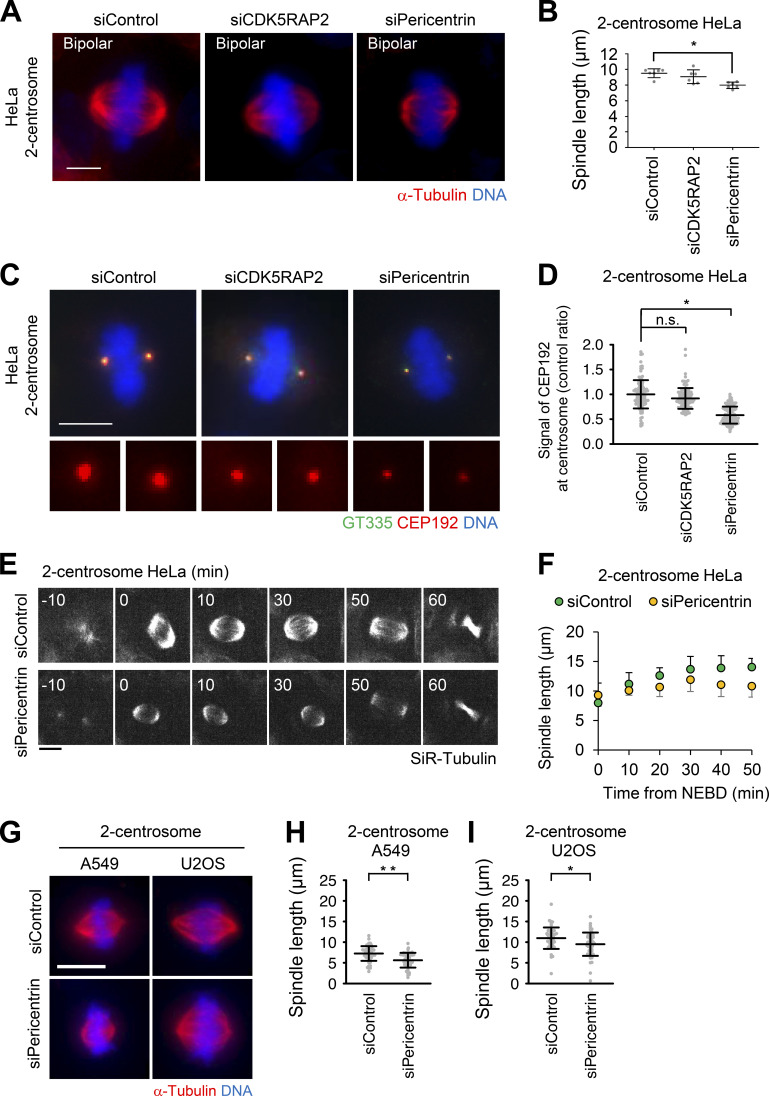
Pericentrin is crucial for bipolar spindle elongation in cells with two centrosomes
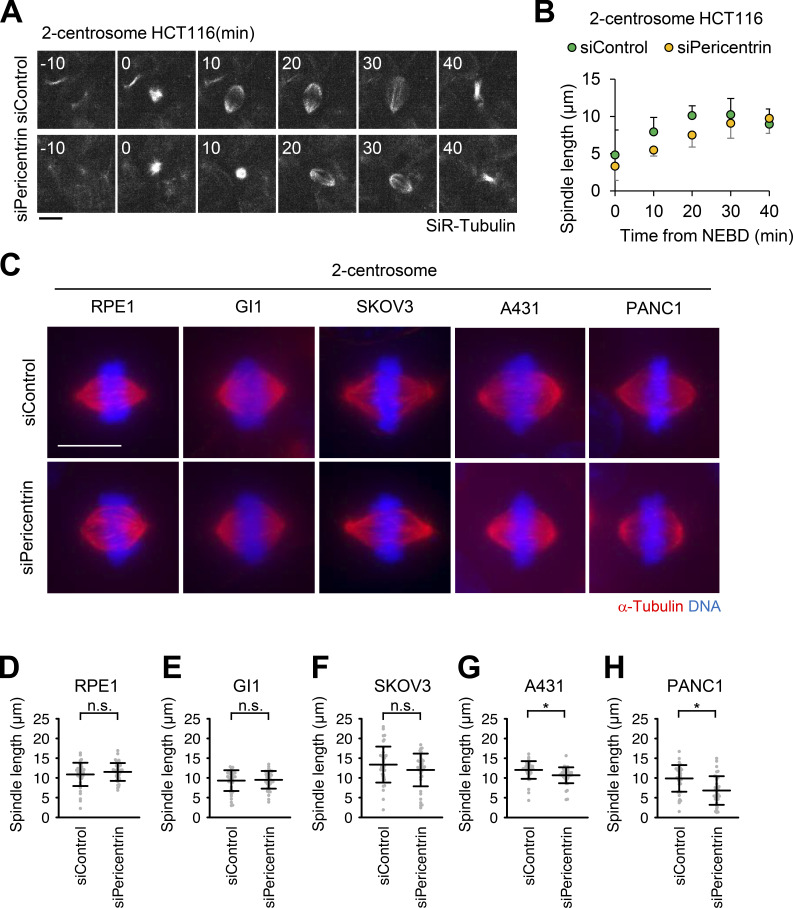
Although pericentrin and CDK5RAP2 are dispensable for efficient mitotic progression in cells with two centrosomes (Fig. 1, A and B; Fig. 2 C; and Fig. S1 G; Videos 1, 3, 15, 17, and 18), the detailed functions of these PCM components in bipolar spindle formation have not been carefully examined. We subsequently analyzed the spindle length upon depletion of pericentrin or CDK5RAP2 in HeLa cells. We found that depletion of pericentrin significantly reduced the spindle length compared with that of control cells, whereas depletion of CDK5RAP2 had a limited effect on the spindle length (Fig. 7, A and B). To further investigate this defect upon depletion of pericentrin, we performed live-cell imaging of mitotic spindle formation in HeLa and HCT116 cells. Depletion of pericentrin delayed the elongation of two spindle poles (Fig. 7, E and F; Fig. S4, A and B; Videos 13, 14, 27, and 28). These results suggest that pericentrin supports spindle elongation. In line with this result, IF analysis revealed that in pericentrin-depleted HeLa cells, the degree of CEP192 localization at the spindle poles was reduced; however, this was not observed in CDK5RAP2-depleted cells (Fig. 7, C and D). This observation implies that pericentrin more efficiently recruits CEP192 to centrosomes, thereby facilitating spindle elongation.
Figure 7.
Pericentrin is crucial for bipolar spindle elongation in cells with two centrosomes. (A) Mitotic spindle structures upon treatment with siRNA in cells with two centrosomes. Red and blue represent α-tubulin and DNA, respectively. Z-projections: 21 planes, 1 µm apart. Scale bar, 5 µm. (B) Quantification spindle length of HeLa cells (n = 6, triplicates, two independent experiments, at least 15 spindles in each assay). Line and error bars represent the mean and SD. Kruskal–Wallis test was used to determine the significance of the difference. *, P < 0.01. (C) CEP192 observed in two-centrosome cells. Green, red, and blue represent GT335, CEP192, and DNA, respectively. Z-projections: 20 planes, 0.5 µm apart. Scale bar, 10 µm. (D) The signal intensity of CEP192 on centrosomes in C. Line and error bars represent the mean and SD (n ≥ 50 cells from two independent experiments). Kruskal–Wallis test was used to determine the significance of the difference. *, P < 0.0001; n.s., not significantly different. (E) Time-lapse observation of the structure of microtubules upon depletion of pericentrin in two-centrosome HeLa cells were observed with a 40× objective. Gray represents SiR-tubulin. Z-projections: 10 planes, 2.2 µm apart. Scale bar, 10 µm. Time zero corresponds to mitotic onset. (F) Averaged time courses of the pole length at each time point in E. The length between two poles of spindle was measured from 40 cells from two independent experiments. Time course data were aligned at the time of the mitotic onset (0 min). Error bars represent SD. (G) Mitotic spindle structures of two-centrosome A549 and U2OS cells. Red and blue represent α-tubulin and DNA, respectively. Z-projections: 31 planes, 0.5 µm apart. Scale bar, 10 µm. The spindle length of A549 (H) and U2OS (I) cells upon depletion of pericentrin (n > 40 from two independent experiments). Line and error bars represent the mean and SD. The Mann–Whitney U test (two tailed) was used to obtain a P value. *, P < 0.005; **, P < 0.0001.
Figure S4.
Pericentrin is critical for proper spindle elongation in two-centrosome cells. (A) Time-lapse observation of the structure of microtubules upon depletion of pericentrin. HCT116 cells were observed with a 40× objective. Gray represents SiR-tubulin. Z-projections: 10 planes, 2.2 µm apart. Scale bar, 10 µm. Time zero corresponds to mitotic onset. (B) Averaged time courses of the pole length at each time point in A. The length between two poles of spindle was measured from 40 cells from two independent experiments. Time course data were aligned at the time of the mitotic onset (0 min). Error bars represent SD. (C) Mitotic spindle structures of two-centrosome RPE1, GI1, SKOV3, A431, and PANC1 cells. Red and blue represent α-tubulin and DNA, respectively. Z-projections: 31 planes, 0.5 µm apart. Scale bar, 10 µm. (D–H) Quantification spindle length (n > 40 from two independent experiments) in C. Line and error bars represent the mean and SD. The Mann–Whitney U test (two tailed) was used to obtain a P value. *, P < 0.0005. n.s., not significantly different.
Video 13.
Control HeLa cells with two centrosomes labeled with SiR-tubulin and observed with a 40× objective. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 7 E.
Video 14.
Pericentrin-depleted HeLa cells with two centrosomes labeled with SiR-tubulin and observed with a 40× objective. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. 7 E.
Video 27.
Control HCT116 cells with two centrosomes labeled with SiR-tubulin and observed with a 40× objective. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S4 A.
Video 28.
Pericentrin-depleted HCT116 cells with two centrosomes labeled with SiR-tubulin and observed with a 40× objective. 10 × 2.2–μm z-stacks were acquired every 10 min. Playback is sped up 4,200× relative to real time. Image frames shown are maximum intensity projections at each time point. Related to Fig. S4 A.
Furthermore, we tested the effect of depletion of pericentrin on the spindle elongation of various cell types. The spindle length in pericentrin-depleted A549, U2OS, A431, and PANC1 cells was significantly shorter than that noted in control cells (Fig. 7, G–I; and Fig. S4, C, G, and H). On the other hand, in some cell types (RPE1, GI1, and SKOV3), the spindle length upon depletion of pericentrin was not altered compared with that observed in control cells (Fig. S4, C–F). These results suggest that pericentrin is required for efficient spindle elongation in certain cell lines with two centrosomes.
Discussion
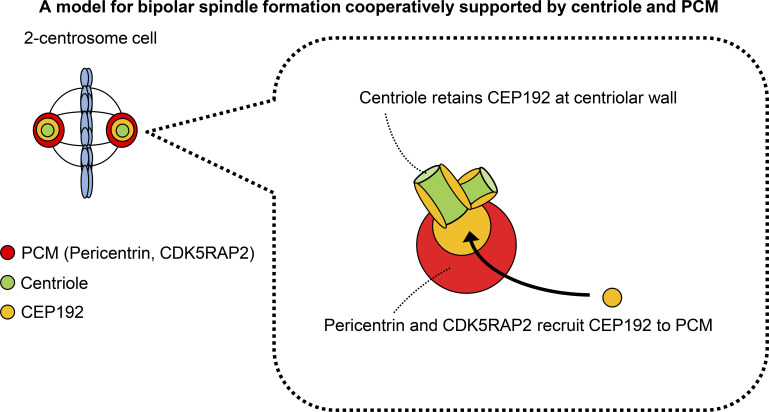
In this study, we show that the centriole and PCM scaffold proteins, pericentrin and CDK5RAP2, cooperate to recruit CEP192 at the spindle pole to facilitate bipolar spindle formation in several human cell lines. We found that, even in cells in which PCM assembly was suppressed, CEP192 remained at the centriole wall efficiently promoted bipolar spindle assembly (Fig. 1). Furthermore, cells with one centrosome formed a bipolar spindle with a PCM pole, which accumulates PCM proteins (including CEP192) at the opposite side of the centriolar spindle pole (Figs. 2 and 3). Consistently, the PCM pole assembly is critical for cell division in one-centrosome cells (Figs. 4 and 5). Overall, the findings in this study illustrate that the centriole and PCM scaffold proteins cooperatively promote bipolar spindle assembly through recruitment of CEP192 to the spindle pole (Fig. 8).
Figure 8.
The centriole and PCM cooperate to recruit CEP192 to the spindle pole to facilitate bipolar spindle formation. Schematic illustration of the assembly of CEP192 at the spindle pole by the centriole and PCM.
In interphase cells, CEP192 localizes at the centrioles and regulates the microtubule nucleation activity of centrosomes (O’Rourke et al., 2014). In G2/M phase, CEP192 is further recruited to PCM clouds by pericentrin (Joukov et al., 2014), promoting mitotic spindle formation. In pericentrin/CDK5RAP2 double-depleted cells, although the CEP192 localization was restricted on the centriolar wall, these cells efficiently completed mitosis (Fig. 1). These results suggest that a fraction of CEP192 remained at the centriolar wall is sufficient for its function in mitosis. A previous study suggested that CEP192 supports the sequential activation of PLK1 and Aurora kinase A (AURKA) at centrosomes (Joukov et al., 2014). Moreover, it has been shown that phosphorylated AURKA interacts with TPX2 and promotes spindle assembly (Joukov and De Nicolo, 2018). It is therefore possible that CEP192 at the centriole wall sufficiently activates the PLK1–AURKA pathway, thereby facilitating bipolar spindle formation.
We found that one-centrosome cells efficiently assembled PCM poles (Fig. 2, F and G; and Fig. S2, A–D). On the other hand, intriguingly, most zero-centrosome cells failed to assemble PCM proteins at the acentriolar poles in HeLa (Fig. 2, F and G), A549, DU145, HCT116, and PANC1 cells (Chinen et al., 2020). How does this difference occur? In one-centrosome cells, PLK1 was localized only at centriolar poles, but not at PCM poles (Fig. 6, A–D). However, the PLK1 kinase activity is somehow necessary for the assembly of PCM poles. It is possible that phosphorylation events at the centriole driven by the activity of PLK1 may provide a pool of PCM for the generation of the PCM pole. In contrast, zero-centrosome cells do not have the platform components (e.g., centrioles) for PCM assembly. Previous research indicated that in zero-centrosome cells, the activity of PLK1 in the cytoplasm was significantly increased (Takeda et al., 2020). However, in such cells, the PCM pole was not assembled at the spindle poles (Takeda et al., 2020). Together, these observations suggest that the centriole itself is important for PCM assembly, at least in the human cell lines that we experimentally tested.
The recent study has reported that acentrosomal spindle formation was promoted by the assembly of PCM proteins at spindle poles in zero-centrosome RPE1 and HeLa cells, but not in DLD1 and U2OS cells (Watanabe et al., 2020). The cause of these different properties in the PCM assembly between different cell types or HeLa cells from different laboratories has not been clarified yet (Chinen et al., 2020). One possible mechanism could be different expression levels of TRIM37, the ubiquitin ligase that targets CEP192, among cell types (Yeow et al., 2020, Meitinger et al., 2020). Another possibility could be the heterogeneity in genome-wide gene copy numbers, mRNAs, proteins, and protein turnover rates between HeLa cells in different laboratories (Liu et al., 2019). Further analysis of such cell-type–specific PCM assembly is required for an understanding of the mechanisms of mitotic spindle formation in human cells.
Knockdown experiments further revealed that CDK5RAP2 and pericentrin are crucial for cell division in one-centrosome cells. In addition, depletion of CEP57 augmented the assembly of PCM poles and facilitated mitotic progression in one-centrosome cells. These results indicate that the PCM proteins are required for PCM pole formation in one-centrosome cells and also raise the possibility that the balance of PCM quantities between two spindle poles may be a critical factor for proper mitotic progression in human cells. In line with this notion, it has been shown that in primary human malignancies, centrosome abnormalities such as centriole rosettes are frequently observed (Cosenza et al., 2017). These extra centrioles could lead to a greater accumulation of PCM proteins at the one centrosome, thereby increasing the nucleation of microtubules at this spindle pole and resulting in chromosome missegregation and aneuploidy. Our assay system may be useful for analyzing the balance of PCM quantities and the resulting microtubule nucleation between two spindle poles.
Previous clinical trials of PLK1 inhibitors have not been successful. Therefore, several studies have been performed to improve PLK1 inhibitor toxicity through combination with other inhibitors, such as α/β-tubulin inhibitors (Stehle et al., 2015; Weiß et al., 2016). Based on the vulnerabilities of one-centrosome cells described above, our study suggests the potential of dual inhibition of centriole duplication and PCM assembly as an attractive drug target for cancer therapies. The PLK1 inhibitor efficiently suppressed both PCM maturation and subsequent PCM pole formation in one-centrosome cells. The dual inhibition strategy, which inhibits both PLK1 and PLK4, might provide an alternative approach to targeting PLK1 in the development of anticancer drugs. In addition, recently, it was suggested that decreased centrosome numbers are associated with poorer response to chemotherapy and an increased invasive capacity of tumor cells in ovarian cancer (Morretton et al., 2019). Therefore, the strategy to suppress PCM assembly in centrosome-reduced cells may be an attractive method for targeting ovarian cancer cells that have a reduced number of centrosomes.
Materials and methods
Cell culture and transfection
HeLa and U2OS cells were obtained from the European Collection of Authenticated Cell Cultures. These cell lines were authenticated by short tandem repeat profiling at the European Collection of Authenticated Cell Cultures. hTERT RPE1 cells were obtained from the American Type Culture Collection (ATCC). HeLa cells stably expressing EGFP-centrin1 have been previously described (Tsuchiya et al., 2016). HeLa cells expressing mCherry-NuMA and EGFP–centrin1 have been previously described (Chinen et al., 2020). HeLa cells expressing pericentrin or CDK5RAP2 endogenously tagged with mCherry were generated using the CRISPR-Cas9 system, as previously described, with slight modifications (Natsume et al., 2016). gRNA oligos (pericentrin_gRNA_F: 5′-CACCGCTGTTTAATCATCGGGTGGC-3′ and pericentrin_gRNA_R: 5′-AAACGCCACCCGATGATTAAACAGC-3′, CDK5RAP2_gRNA_F: 5′-CACCGGGACTGCATGTTCCTGGAT-3′ and CDK5RAP2_gRNA_R: 5′-AAACATCCAGGAACATGCAGTCCC-3′) were hybridized and cloned into the BbsI site of pX330 (Addgene). To construct the donor plasmid for homology-directed repair, the homology arms of the pericentrin locus (chromosome 21: 47,864,730–47,865,813) or CDK5RAP2 locus (Chr9:120388937-120389538) were amplified (pBS2_pericentrin C-ter_InsF: 5′-GGTATCGATAAGCTTACCAGGTAATGCAAGTCCTCGCCG-3′ and pBS2_pericentrin C-ter_InsR: 5′-CGCTCTAGAACTAGTAGAATGCTCCGGGTTCCACTGA-3′, pBS2_CDK5RAP2 C-ter_InsF: 5′-CCCCCCCTCGAGGTCTGCTATTTTTACCAGTAAG-3′ and pBS2_ CDK5RAP2 C-ter_InsR: 5′-CTCTAGAACTAGTGGAAATCCAGGGGAAGACGTG-3′) from the genomic DNA of HeLa cells and cloned into pBluescript using the Infusion Cloning kit (Takara). A BamHI sequence with a silent mutation to prevent recutting was generated in the middle of the homology arm domain by mutagenesis PCR (pericentrin C-terminal silent BamHI_F: 5′-TACTTCAAAGAAATCTTGCCACCCGATGATTAAACAGGGATCCATAAAATGTCATGGCTCTTTCCTGCGA-3′, pericentrin C-terminal silent BamHI_R: 5′-GCCATGACATTTTATGGATCCCTGTTTAATCATCGGGTGGCAAGATTTCTTTGAAGTAGAATCTGCATATAAATAAAAATGAGG-3′, CDK5RAP2 C-ter silent BamHI_F: 5′-GGGGAGCTCATCCAGGAACATGCAGT-3′, CDK5RAP2 C-ter silent BamHI_R: 5′-CTGGATGAGCTCCCCCAGGCCTAAGC-3′). The mCherry cassette containing a hygromycin-resistant gene was introduced into the BamHI site in the middle of the homology arms. The plasmids were introduced into the HeLa cell line stably expressing EGFP-centrin1 (Tsuchiya et al., 2016) and isolated using the limited dilution method with hygromycin. A549, DU145, PC-3, PANC-1, GI-1, A431, and MCF-7 were obtained from the RIKEN BioResource Research Center. These cell lines were authenticated by short tandem repeat profiling at the RIKEN BioResource Research Center. HCT116 cells were obtained from the ATCC (CCL-247). HCT116 CMVOsTIR1 HsSAS6–AID cells have been previously described (Yoshiba et al., 2019). HCT116 cell lines were cultured in McCoy’s 5A medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. SKOV-3 was provided by Dr. Y. Nagumo (University of Tsukuba, Tsukuba, Japan). HeLa, U2OS, A549, GI-1, and A431 cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a 5% CO2 atmosphere. RPE1 cells was cultured in Ham’s F12/Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in a 5% CO2 atmosphere. DU145, PC-3, PANC-1, and SKOV-3 cells were cultured in RPMI1680 medium containing 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a 5% CO2 atmosphere. MCF-7 cells were cultured in MEM medium containing 10% fetal bovine serum, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in a 5% CO2 atmosphere. Transfection of siRNA constructs was conducted using Lipofectamine RNAiMAX (Life Technologies).
RNA interference
The following siRNAs were used: Silencer Select siRNA (Life Technologies) against CEP57 (s18692), CEP192 (s226819), CDK5RAP2 (s31429, s31430), pericentrin (s10136, s10138), and negative control #1 (4390843).
Chemicals
The following chemicals were used in this study: centrinone (PLK4 inhibitor, HY-18682; MedChem Express), Centrinone B (PLK4 inhibitor, gift from Dr. Andrew Shiau and Dr. Karen Oegema, University of California, San Diego, San Diego, CA), BI2536 (PLK1 inhibitor, A10134; AdooQ), proTAME (APC/C inhibitor, I-440; Boston Biochem), and SiR-tubulin (Microtubule probe, CY-SC002; SPIROCHROME).
Antibodies
The following primary antibodies were used in this study: rabbit polyclonal antibodies against CDK5RAP2 (IHC00063, immunofluorescence [IF] 1:500, Western blotting [WB] 1:1,000; Bethyl Laboratories), Cep192 (A302–324A, IF 1:1,000, WB 1:1,000; Bethyl Laboratories), Cep152 (A302–480A, IF 1:1,000; Bethyl Laboratories), CPAP/CENP-J (11517–1-AP, IF 1:100; Proteintech), α-tubulin (PM054, IF 1:300; MBL), γ-tubulin (T5192, IF 1:1,000; Sigma-Aldrich), pericentrin (ab4448, IF 1:500 or 1:1,000, WB 1:1,000; Abcam), PLK1 (A300-251A, IF 1:500; Bethyl Laboratories), and RFP (PM005, IF 1:1,000; MBL); and mouse mAbs against Centrin (04-1624, IF 1:1,000; Merck Milipore), EB1 (610534, IF 1:1,000, BD Biosciences), polyglutamylation modification (GT335, mAb; AG-20B-0020-C100, IF 1:1,000; AdipoGen), α-tubulin (T5168, IF 1:1,000; Sigma-Aldrich), HSP90 (610419, WB 1:5,000; BD Biosciences), and pT210-PLK1 (ab39068, IF 1:300; abcam). Fluorescein isothiocyanate–labeled anti-GFP was purchased from Abcam (ab6662, IF 1:250 or 1:500). The following secondary antibodies were used: Alexa Fluor 488 goat anti-mouse IgG (H+L) (A-11001, 1:500; Molecular Probes), Alexa Fluor 555 goat anti-rabbit IgG (H+L) (A-21428, 1:500; Molecular Probes), and Alexa Fluor 647 goat anti-mouse IgG (ab150115, 1:500; Abcam). Goat polyclonal antibodies-HRP against mouse IgG (W402B, WB 1:5,000–1:10,000; Promega), and goat polyclonal antibodies-HRP against rabbit IgG (W401B, WB 1:5,000–10,000; Promega).
Sample preparations for immunostaining
Cells were treated with 100 nM centrinone or 500 nM centrinone B for 1–3 d to induce acentrosomal cells (Fig. 2, B, F, and G; Fig. 6, A–D; and Fig. S2, A and B). To observe the microtubule nucleation from PCM poles (Fig. S2 E), first, HeLa cells expressing pericentrin-mCherry and EGFP-centrin1 were treated with 100 nM centrinone for 2 d. Then, cells were arrested in metaphase through treatment with 20 µM proTAME for 4 h and incubated on ice for 1 h to depolymerize microtubules. Subsequently, cells were incubated at 25°C for 5 min. For the Sas-6 depletion experiments using the AID system (Fig. S2, C and D), cells were incubated with 50 µM of indole-3-acetic acid for 2 d. To observe CEP192 localization at acentriolar poles (Fig. 4, G–I), cells were treated with 100 nM centrinone and siRNA for 2 d. Then, cells were arrested in metaphase through treatment with 20 µM proTAME for 6 h. To examine the effect of siRNA on acentriolar cells (Fig. 4, A and B; Fig. 5, A, B, and G; and Fig. S3), cells were treated with 100 nM centrinone or 500 nM centrinone B and siRNA for 2 d. For the chemical perturbation experiments (Fig. 6, E–G), cells were treated with 100 nM of centrinone and 1 nM BI 2536 for 2 d. In other experiments (Fig. 1, C–G; Fig. 7, A–D and G–I; Fig. S1, D–F; Fig. S4, C–H), cells were treated with siRNA for 2 d.
WB
For preparation of total cell lysates, cells were lysed in 1× SDS sample buffer. SDS–PAGE was performed using 6% or 10% polyacrylamide gels, followed by transfer on Immobilon-P membrane (Millipore Corporation). Blocking was performed in 2.5% skim milk in PBS containing 0.02% Tween (PBS-T) for 30 min at room temperature. The membrane was probed with the primary antibodies for 12–18 h at 4°C and washed with PBS-T three times. After that, the membrane was incubated with HRP-conjugated secondary antibodies for 1 h at room temperature and washed with PBS-T three times. The signals were detected with ECL Prime/Select reagents (GE Healthcare) or Chemi-Lumi One Ultra (Nacalai Tesque) via the ChemiDoc XRSþ system (Bio-Rad).
Sample preparations for live-cell imaging
For live-cell imaging, HeLa cells, HCT116 cells, HeLa cells expressing pericentrin-mCherry and EGFP-centrin1, HeLa cells expressing CDK5RAP2-mCherry and EGFP-centrin1, and HeLa cells expressing mCherry-NuMA and EGFP–centrin1 were cultured in 35-mm glass-bottom dishes (#627870; Greiner Bio-One) or 24-well SENSOPLATE (#662892; Greiner Bio-One) at 37°C in a 5% CO2 atmosphere.
To observe the dynamics of PCM (Fig. 3, A–E) in one-centrosome or zero-centrosome cells, cells were treated with 500 nM centrinone B. To test the effect of depletion of PCM proteins or CEP57 on one-centrosome cells (Fig. 4, C–F; and Fig. 5, D–F), cells were treated with siRNA with 100 nM centrinone for 2 d. To simultaneously observe one-centrosome cells and zero-centrosome cells (Fig. 2, C–E; and Fig. S1, G–I), after 1 d of treatment with 0.1% DMSO or 100 nM centrinone (to enrich zero-centrosome cells), HeLa cells expressing mCherry-NuMA and EGFP-centrin1 were treated with siRNA with 0.1% DMSO or 100 nM centrinone for 2 d. In other experiments (Fig. 1, A and B; Fig. 7, E and F; Fig. S4, A and B), cells were treated with siRNA for 2 d. Prior to imaging, cells were incubated with 50 nM SiR-tubulin for 3 h to visualize the microtubules.
Microscopy for IF analyses
For IF analyses, the cells cultured on coverslips (No. 1; Matsunami) were fixed using methanol at −20°C for 7 min and washed with PBS. The cells were permeabilized after fixation with PBS/0.05% TritonX-100 (PBSX) for 5 min and blocked in 1% BSA in PBSX for 30 min at room temperature. The cells were then incubated with primary antibodies for 7–24 h at 4°C, washed thrice with PBSX, and incubated with secondary antibodies and 0.2 µg/ml Hoechst 33258 (DOJINDO) for 45–60 min at room temperature. The cells were washed thrice with PBSX and mounted onto glass slides.
We counted the number of spindle patterns using a DeltaVision Personal DV-SoftWoRx system (Applied Precision) or an Axioplan2 fluorescence microscope (Carl Zeiss). Confocal microscopy images were captured by the Leica TCS SP8 system. For deconvolution for confocal microcopy images, Huygens essential software (Scientific Volume Imaging) was used.
STED images were taken using a Leica TCS SP8 STED 3X system with a Leica HCPL APO 100×/1.40 oil STED WHITE and a 660-nm laser line for depletion. Scan speed was set to 100 Hz with 5× line averaging in a 512 × 80 pixel format (pixel size, 15–20 nm). The Z interval was set to 180 nm. The STED images were processed by deconvolution using the Huygens Professional software (SVI).
Maximum intensity Z-projections of a representative picture for each condition were generated using the FIJI distribution of the ImageJ software. The number and step sizes of z-planes are described in the figure legends.
Microscopy for live imaging
A Confocal Scanner Box, Cell Voyager CV1000 (Yokogawa Electric) equipped with a 60× oil-immersion objective or CQ1 Benchtop High-Content Analysis System equipped with a 40× objective was used for live-cell imaging. Imaging was initiated 24–48 h after transfection, and images were acquired every 10 min for 24–48 h. Maximum intensity Z-projections of representative images for each condition were generated using the FIJI distribution of the ImageJ software. The number and step sizes of z-planes are described in the figure legends.
Statistical analysis
Statistical analyses were performed using the GraphPad Prism 7 software. P values were determined by nonparametric methods (Mann–Whitney U test for two independent samples or Kruskal–Wallis test for three or more independent samples). Details are described in the figure legends.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request.
Online supplemental material
Fig. S1 shows the efficiency of protein depletion and mitotic progression in HeLa cells upon depletion of centriole and PCM proteins. Fig. S2 shows the distribution of centrosomal factors in centriolar and acentriolar spindle poles. Fig. S3 shows the spindle structure of RPE1 and A549 cells upon depletion of centriole and PCM proteins. Fig. S4 shows the spindle length of various cells upon depletion of pericentrin. Video 1, Video 2, and Video 3 show the mitotic progression imaged using SiR-tubulin in HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with two-centrosome upon depletion of PCM proteins. Video 4, Video 5, Video 6, and Video 7 show the dynamics of endogenous pericentrin in HeLa cells expressing EGFP-centrin1 and pericentrin-mCherry with one or zero centrosomes. Video 8, Video 9, Video 10, Video 11, and Video 12 show the mitotic progression imaged using SiR-tubulin in HeLa cells expressing EGFP-centrin1 and mCherry-NuMA with one centrosome upon depletion of indicated proteins. Video 13, Video 14, Video 27, and Video 28 show the mitotic progression imaged using SiR-tubulin in HeLa cells (Video 13 and Video 14) or HCT116 cells (Video 27 and Video 28) with two-centrosome upon depletion of pericentrin. Video 15, Video 16, Video 17, Video 18, Video 19, Video 20, Video 21, Video 22, Video 23, Video 24, Video 25, and Video 26 show the mitotic progression imaged using SiR-tubulin in HeLa cells expressing EGFP-centrin1 and mCherry-NuMA upon depletion of centriole and PCM proteins.
Acknowledgments
We thank the Kitagawa laboratory members for fruitful discussions. We thank Dr. A. Shiau and Dr. K. Oegema at the Ludwig Institute for Cancer Research, University of California, San Diego, San Diego, CA for providing centrinone B. We are also thankful to Dr. Y. Nagumo for providing the SKOV-3 cells.
This work was supported by Japan Society for the Promotion of Science KAKENHI grants (24687026, 19H05651, 16H06168, 18K14705, and 17J02833); the Takeda Science Foundation; the Mochida Memorial Foundation for Medical and Pharmaceutical Research; and the Daiichi Sankyo Foundation of Life Science.
The authors declare no competing financial interests.
Author contributions: T. Chinen and D. Kitagawa designed the study; T. Chinen, K. Yamazaki, K. Fujii, K. Watanabe, Y. Takeda, and Y. Nozaki performed the experiments; and T. Chinen, K. Yamazaki, K. Hashimoto, S. Yamamoto, Y. Tsuchiya, and D. Kitagawa designed the experiments. T. Chinen, K. Yamazaki, Y. Takeda, and D. Takao analyzed the data; and T. Chinen, K. Yamazaki, and D. Kitagawa wrote the manuscript, which was reviewed by all authors.
References
- Alvarez-Rodrigo, I., Steinacker T.L., Saurya S., Conduit P.T., Baumbach J., Novak Z.A., Aydogan M.G., Wainman A., and Raff J.W.. 2019. Evidence that a positive feedback loop drives centrosome maturation in fly embryos. eLife. 8:e50130 10.7554/eLife.50130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, C., Wang X., Yang L., and Viveiros M.M.. 2017. Error-prone meiotic division and subfertility in mice with oocyte-conditional knockdown of pericentrin. J. Cell Sci. 130:1251–1262. 10.1242/jcs.196188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., and Glover D.M.. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207. 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Cabral, G., Laos T., Dumont J., and Dammermann A.. 2019. Differential Requirements for Centrioles in Mitotic Centrosome Growth and Maintenance. Dev. Cell. 50:355–366.e6. 10.1016/j.devcel.2019.06.004 [DOI] [PubMed] [Google Scholar]
- Chavali, P.L., Chandrasekaran G., Barr A.R., Tátrai P., Taylor C., Papachristou E.K., Woods C.G., Chavali S., and Gergely F.. 2016. A CEP215-HSET complex links centrosomes with spindle poles and drives centrosome clustering in cancer. Nat. Commun. 7:11005 10.1038/ncomms11005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.-T., Hehnly H., Yu Q., Farkas D., Zheng G., Redick S.D., Hung H.-F., Samtani R., Jurczyk A., Akbarian S., et al. . 2014. A unique set of centrosome proteins requires pericentrin for spindle-pole localization and spindle orientation. Curr. Biol. 24:2327–2334. 10.1016/j.cub.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen, T., Yamamoto S., Takeda Y., Watanabe K., Kuroki K., Hashimoto K., Takao D., and Kitagawa D.. 2020. NuMA assemblies organize microtubule asters to establish spindle bipolarity in acentrosomal human cells. EMBO J. 39:e102378 10.15252/embj.2019102378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.K., Liu P., Sze S.K., Dai C., and Qi R.Z.. 2010. CDK5RAP2 stimulates microtubule nucleation by the γ-tubulin ring complex. J. Cell Biol. 191:1089–1095. 10.1083/jcb.201007030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift, D., and Schuh M.. 2015. A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat. Commun. 6:7217 10.1038/ncomms8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit, P.T., Brunk K., Dobbelaere J., Dix C.I., Lucas E.P., and Raff J.W.. 2010. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 20:2178–2186. 10.1016/j.cub.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Conduit, P.T., Richens J.H., Wainman A., Holder J., Vicente C.C., Pratt M.B., Dix C.I., Novak Z.A., Dobbie I.M., Schermelleh L., et al. . 2014. A molecular mechanism of mitotic centrosome assembly in Drosophila. eLife. 3 e03399 10.7554/eLife.03399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consolati, T., Locke J., Roostalu J., Chen Z.A., Gannon J., Asthana J., Lim W.M., Martino F., Cvetkovic M.A., Rappsilber J., et al. . 2020. Microtubule Nucleation Properties of Single Human γTuRCs Explained by Their Cryo-EM Structure. Dev. Cell. 53:603–617.e8. 10.1016/j.devcel.2020.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosenza, M.R., Cazzola A., Rossberg A., Schieber N.L., Konotop G., Bausch E., Slynko A., Holland-Letz T., Raab M.S., Dubash T., et al. . 2017. Asymmetric Centriole Numbers at Spindle Poles Cause Chromosome Missegregation in Cancer. Cell Rep. 20:1906–1920. 10.1016/j.celrep.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Dudka, D., Castrogiovanni C., Liaudet N., Vassal H., and Meraldi P.. 2019. Spindle-Length-Dependent HURP Localization Allows Centrosomes to Control Kinetochore-Fiber Plus-End Dynamics. Curr. Biol. 29:3563–3578.e6. 10.1016/j.cub.2019.08.061 [DOI] [PubMed] [Google Scholar]
- Erpf, A.C., Stenzel L., Memar N., Antoniolli M., Osepashvili M., Schnabel R., Conradt B., and Mikeladze-Dvali T.. 2019. PCMD-1 Organizes Centrosome Matrix Assembly in C. elegans. Curr. Biol. 29:1324–1336.e6. 10.1016/j.cub.2019.03.029 [DOI] [PubMed] [Google Scholar]
- Gomez-Ferreria, M.A., Rath U., Buster D.W., Chanda S.K., Caldwell J.S., Rines D.R., and Sharp D.J.. 2007. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 17:1960–1966. 10.1016/j.cub.2007.10.019 [DOI] [PubMed] [Google Scholar]
- Gönczy, P. 2015. Centrosomes and cancer: revisiting a long-standing relationship. Nat. Rev. Cancer. 15:639–652. 10.1038/nrc3995 [DOI] [PubMed] [Google Scholar]
- Habedanck, R., Stierhof Y.D., Wilkinson C.J., and Nigg E.A.. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146. 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Hanafusa, H., Kedashiro S., Tezuka M., Funatsu M., Usami S., Toyoshima F., and Matsumoto K.. 2015. PLK1-dependent activation of LRRK1 regulates spindle orientation by phosphorylating CDK5RAP2. Nat. Cell Biol. 17:1024–1035. 10.1038/ncb3204 [DOI] [PubMed] [Google Scholar]
- Haren, L., Stearns T., and Lüders J.. 2009. Plk1-dependent recruitment of γ-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One. 4:e5976 10.1371/journal.pone.0005976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov, V., and De Nicolo A.. 2018. Aurora-PLK1 cascades as key signaling modules in the regulation of mitosis. Sci. Signal. 11:eaar4195 10.1126/scisignal.aar4195 [DOI] [PubMed] [Google Scholar]
- Joukov, V., Walter J.C., and De Nicolo A.. 2014. The Cep192-organized aurora A-Plk1 cascade is essential for centrosome cycle and bipolar spindle assembly. Mol. Cell. 55:578–591. 10.1016/j.molcel.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., and Rhee K.. 2014. Importance of the CEP215-pericentrin interaction for centrosome maturation during mitosis. PLoS One. 9:e87016 10.1371/journal.pone.0087016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Lee K., and Rhee K.. 2015. PLK1 regulation of PCNT cleavage ensures fidelity of centriole separation during mitotic exit. Nat. Commun. 6:10076 10.1038/ncomms10076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kim J., and Rhee K.. 2019. PCNT is critical for the association and conversion of centrioles to centrosomes during mitosis. J. Cell Sci. 132:jcs225789 10.1242/jcs.225789 [DOI] [PubMed] [Google Scholar]
- Kirkham, M., Müller-Reichert T., Oegema K., Grill S., and Hyman A.A.. 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 112:575–587. 10.1016/S0092-8674(03)00117-X [DOI] [PubMed] [Google Scholar]
- Kollman, J.M., Merdes A., Mourey L., and Agard D.A.. 2011. Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12:709–721. 10.1038/nrm3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecland, N., and Lüders J.. 2014. The dynamics of microtubule minus ends in the human mitotic spindle. Nat. Cell Biol. 16:770–778. 10.1038/ncb2996 [DOI] [PubMed] [Google Scholar]
- Lee, S., and Rhee K.. 2010. CEP215 is involved in the dynein-dependent accumulation of pericentriolar matrix proteins for spindle pole formation. Cell Cycle. 9:775–784. 10.4161/cc.9.4.10667 [DOI] [PubMed] [Google Scholar]
- Lee, K., and Rhee K.. 2011. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol. 195:1093–1101. 10.1083/jcb.201106093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Mi Y., Mueller T., Kreibich S., Williams E.G., Van Drogen A., Borel C., Frank M., Germain P.L., Bludau I., et al. . 2019. Multi-omic measurements of heterogeneity in HeLa cells across laboratories. Nat. Biotechnol. 37:314–322. 10.1038/s41587-019-0037-y [DOI] [PubMed] [Google Scholar]
- Liu, P., Zupa E., Neuner A., Böhler A., Loerke J., Flemming D., Ruppert T., Rudack T., Peter C., Spahn C., et al. . 2020. Insights into the assembly and activation of the microtubule nucleator γ-TuRC. Nature. 578:467–471. 10.1038/s41586-019-1896-6 [DOI] [PubMed] [Google Scholar]
- Meitinger, F., Ohta M., Lee K.Y., Watanabe S., Davis R.L., Anzola J.V., Kabeche R., Jenkins D.A., Shiau A.K., Desai A., and Oegema K.. 2020. TRIM37 controls cancer-specific vulnerability to PLK4 inhibition. Nature. 585:440–446. 10.1038/s41586-020-2710-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, M., Braunfeld M.B., Sedat J.W., Alberts B., and Agard D.A.. 1995. Microtubule nucleation by γ-tubulin-containing rings in the centrosome. Nature. 378:638–640. 10.1038/378638a0 [DOI] [PubMed] [Google Scholar]
- Morretton, J.-P., Herbette A., Cosson C., Mboup B., Latouche A., Gestraud P., Popova T., Stern M.-H., Nemati F., Decaudin D., et al. . 2019. Centrosome amplification favours survival and impairs ovarian cancer progression. bioRxiv. 623983 10.1101/623983 [DOI] [Google Scholar]
- Natsume, T., Kiyomitsu T., Saga Y., and Kanemaki M.T.. 2016. Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep. 15:210–218. 10.1016/j.celrep.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Nigg, E.A., and Raff J.W.. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- O’Rourke, B.P., Gomez-Ferreria M.A., Berk R.H., Hackl A.M.U., Nicholas M.P., O’Rourke S.C., Pelletier L., and Sharp D.J.. 2014. Cep192 controls the balance of centrosome and non-centrosomal microtubules during interphase. PLoS One. 9:e101001 10.1371/journal.pone.0101001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh, M., and Ellenberg J.. 2007. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 130:484–498. 10.1016/j.cell.2007.06.025 [DOI] [PubMed] [Google Scholar]
- Seo, M.Y., Jang W., and Rhee K.. 2015. Integrity of the Pericentriolar Material Is Essential for Maintaining Centriole Association during M Phase. PLoS One. 10:e0138905 10.1371/journal.pone.0138905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir, J.H., Pütz M., Daly O., Morrison C.G., Dunning M., Kilmartin J.V., and Gergely F.. 2013. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J. Cell Biol. 203:747–756. 10.1083/jcb.201309038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle, A., Hugle M., and Fulda S.. 2015. Eribulin synergizes with Polo-like kinase 1 inhibitors to induce apoptosis in rhabdomyosarcoma. Cancer Lett. 365:37–46. 10.1016/j.canlet.2015.04.011 [DOI] [PubMed] [Google Scholar]
- Takeda, Y., Yamazaki K., Hashimoto K., Watanabe K., Chinen T., and Kitagawa D.. 2020. The centriole protein CEP76 negatively regulates PLK1 activity in the cytoplasm for proper mitotic progression. J. Cell Sci. 133(19):jcs241281. [DOI] [PubMed] [Google Scholar]
- Teixidó-Travesa, N., Roig J., and Lüders J.. 2012. The where, when and how of microtubule nucleation - one ring to rule them all. J. Cell Sci. 125:4445–4456. 10.1242/jcs.106971 [DOI] [PubMed] [Google Scholar]
- Tsuchiya, Y., Yoshiba S., Gupta A., Watanabe K., and Kitagawa D.. 2016. Cep295 is a conserved scaffold protein required for generation of a bona fide mother centriole. Nat. Commun. 7:12567 10.1038/ncomms12567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tungadi, E.A., Ito A., Kiyomitsu T., and Goshima G.. 2017. Human microcephaly ASPM protein is a spindle pole-focusing factor that functions redundantly with CDK5RAP2. J. Cell Sci. 130:3676–3684. 10.1242/jcs.203703 [DOI] [PubMed] [Google Scholar]
- Watanabe, K., Takao D., Ito K.K., Takahashi M., and Kitagawa D.. 2019. The Cep57-pericentrin module organizes PCM expansion and centriole engagement. Nat. Commun. 10:931 10.1038/s41467-019-08862-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, S., Meitinger F., Shiau A.K., Oegema K., and Desai A.. 2020. Centriole-independent mitotic spindle assembly relies on the PCNT-CDK5RAP2 pericentriolar matrix. J. Cell Biol. 219:e202006010 10.1083/jcb.202006010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiß, L.M., Hugle M., Romero S., and Fulda S.. 2016. Synergistic induction of apoptosis by a polo-like kinase 1 inhibitor and microtubule-interfering drugs in Ewing sarcoma cells. Int. J. Cancer. 138:497–506. 10.1002/ijc.29725 [DOI] [PubMed] [Google Scholar]
- Wieczorek, M., Urnavicius L., Ti S.-C., Molloy K.R., Chait B.T., and Kapoor T.M.. 2020. Asymmetric Molecular Architecture of the Human γ-Tubulin Ring Complex. Cell. 180:165–175.e16. 10.1016/j.cell.2019.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. . 2015. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 348:1155–1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff, J.B., Wueseke O., and Hyman A.A.. 2014. Pericentriolar material structure and dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130459 10.1098/rstb.2013.0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff, J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A., and Hyman A.A.. 2017. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell. 169:1066–1077.e10. 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]
- Yeow, Z.Y., Lambrus B.G., Marlow R., Zhan K.H., Durin M.A., Evans L.T., Scott P.M., Phan T., Park E., Ruiz L.A., et al. . 2020. Targeting TRIM37-driven centrosome dysfunction in 17q23-amplified breast cancer. Nature. 585:447–452. 10.1038/s41586-020-2690-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba, S., Tsuchiya Y., Ohta M., Gupta A., Shiratsuchi G., Nozaki Y., Ashikawa T., Fujiwara T., Natsume T., Kanemaki M.T., and Kitagawa D.. 2019. HsSAS-6-dependent cartwheel assembly ensures stabilization of centriole intermediates. J. Cell Sci. 132:jcs217521 10.1242/jcs.217521 [DOI] [PubMed] [Google Scholar]
- Zheng, Y., Wong M.L., Alberts B., and Mitchison T.. 1995. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 378:578–583. 10.1038/378578a0 [DOI] [PubMed] [Google Scholar]
- Zhu, F., Lawo S., Bird A., Pinchev D., Ralph A., Richter C., Müller-Reichert T., Kittler R., Hyman A.A., and Pelletier L.. 2008. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr. Biol. 18:136–141. 10.1016/j.cub.2007.12.055 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this study are available from the corresponding authors upon request.