SUMMARY
Innate responses of myeloid cells defend against pathogenic bacteria via inducible effectors. Deoxyhypusine synthase (DHPS) catalyzes the transfer of the N-moiety of spermidine to the lysine-50 residue of eukaryotic translation initiation factor 5A (EIF5A) to form the amino acid hypusine. Hypusinated EIF5A (EIF5AHyp) transports specific mRNAs to ribosomes for translation. We show that DHPS is induced in macrophages by two gastrointestinal pathogens, Helicobacter pylori and Citrobacter rodentium, resulting in enhanced hypusination of EIF5A. EIF5AHyp was also increased in gastric macrophages from patients with H. pylori gastritis. Furthermore, we identify the bacteria-induced immune effectors regulated by hypusination. This set of proteins includes essential constituents of antimicrobial response and autophagy. Mice with myeloid cell-specific deletion of Dhps exhibit reduced EIF5AHyp in macrophages and increased bacterial burden and inflammation. Thus, regulation of translation through hypusination is a critical hallmark of the defense of eukaryotic hosts against pathogenic bacteria.
Graphical Abstract
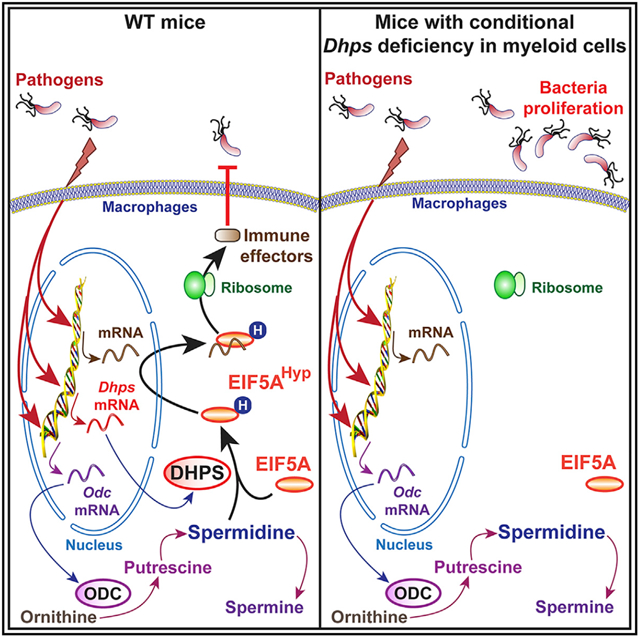
In Brief
Gobert et al. demonstrate that hypusination, a specific mechanism regulating translation, is induced in macrophages by bacteria. Hypusination is required for the translation of inducible antimicrobial effectors. Mice that specifically lack hypusination in macrophages are highly susceptible to Helicobacter pylori and Citrobacter rodentium, two pathogens of the gastrointestinal tract.
INTRODUCTION
The mammalian gastrointestinal tract is the home of the largest population of myeloid cells (Lee et al., 1985). These resident and infiltrating cells respond to inflammatory signals and to foreign antigens, thus exhibiting enhanced antimicrobial abilities that are needed for the clearance of pathogens (Hardbower et al., 2017; Niess et al., 2005; Wennerås et al., 2000). In this context, deciphering the molecular mechanisms that regulate the innate response of these immune cells is essential for a better understanding of the protective immunity against gastrointestinal pathogens.
The arginase-ornithine decarboxylase (ODC) metabolic pathway plays an essential role in the orchestration of the activation of macrophages (Latour et al., 2020). Arginase activity converts arginine into urea and ornithine, with the latter being metabolized by ODC into putrescine (Pegg, 2009). This first and shortest polyamine is then sequentially converted by spermidine synthase and spermine synthase into spermidine and spermine, respectively (Latour et al., 2020; Pegg, 2016). We have implicated putrescine in the pathophysiology of infection with Helicobacter pylori (Hardbower et al., 2017), a specific pathogen of the stomach responsible for diseases ranging from gastritis to gastric cancer (Peek et al., 2010). Using mice with myeloid-specific Odc knockout, we found that endogenous putrescine alters histone methylation/acetylation and dampens the transcription of the genes encoding proinflammatory markers (Hardbower et al., 2017). This results in the limitation of inflammation and the exacerbation of H. pylori burden (Hardbower et al., 2017). In addition, we have established that spermine reduces arginine uptake, which is needed for nitric oxide (NO) synthase 2 (NOS2) translation and NO-dependent killing of H. pylori, thus impairing immunity to the bacterium (Bussière et al., 2005; Chaturvedi et al., 2010). However, the importance of spermidine for the outcome of the infection remains unknown.
Spermidine has been shown to improve cardiac function, to be neuroprotective, and to dampen inflammation (Madeo et al., 2018), notably by stimulating autophagy and mitophagy (Eisenberg et al., 2009, 2016; Qi et al., 2016). Importantly, spermidine is required for the synthesis of the non-proteinogenic amino acid hypusine. The only known protein that contains hypusine is the eukaryotic translation initiation factor 5A (EIF5A; Park et al., 1981; Park and Wolff, 2018). Two isoforms of EIF5A exist: EIF5A1, referred to as eIF5A in this study, which is expressed universally; and eIF5A2, which is mainly present in testes, brain, and tumor cells (Jenkins et al., 2001; Nakanishi and Cleveland, 2016). Hypusine is formed by the action of two enzymes: deoxyhypusine synthase (DHPS), which transfers the N-terminal moiety of spermidine to the lysine-50 residue in EIF5A; and deoxyhypusine hydroxylase (DOHH) that completes hypusine biosynthesis by hydroxylation of the side chain (Figure 1A; Joe et al., 1995; Park et al., 2006). This post-translational modification of EIF5A, called hypusination, has been reported to be essential for two molecular events. On the one hand, EIF5AHyp binds specific mRNAs that contain a 5′-AAAUGU-3′ consensus sequence (Maier et al., 2010; Xu and Chen, 2001; Xu et al., 2004), and the EIF5AHyp/mRNA complex is then translocated to the cytoplasm by the nuclear transporter exportin-1 (Aksu et al., 2016), reaches ribosomes, and allows for translation (Park and Wolff, 2018). On the other hand, hypusinated EIF5A alleviates ribosome pausing during the translation of peptides enriched in diprolyl, diglycyl motifs and in basic and acidic amino acids (Gutierrez et al., 2013; Pelechano and Alepuz, 2017; Schuller et al., 2017). Hypusination is crucial for embryonic development, cell differentiation, and proliferation (Ganapathi et al., 2019; Nishimura et al., 2005; Park et al., 1997; Sievert et al., 2014). When small interfering RNA (siRNA) against EIF5A or mice with specific deletion of Dhps in pancreatic islet β cells were used, it was shown that hypusinated EIF5A leads to the development of glucose intolerance in inflammatory mouse models of diabetes, notably by supporting NOS2 translation (Levasseur et al., 2019; Maier et al., 2010). In addition, hypusination in macrophages supports the tricarboxylic acid (TCA) cycle and oxidative phosphorylation by promoting the translation of mitochondrial proteins and promotes alternative (M2) activation (Puleston et al., 2019). However, the role of hypusination in the innate response of macrophages during bacterial infection has not been determined. We hypothesized that protein translation by the DHPS/EIF5AHyp pathway is critical for the appropriate response of macrophages to invaders.
Figure 1. DHPS Expression and Activity in Macrophages.
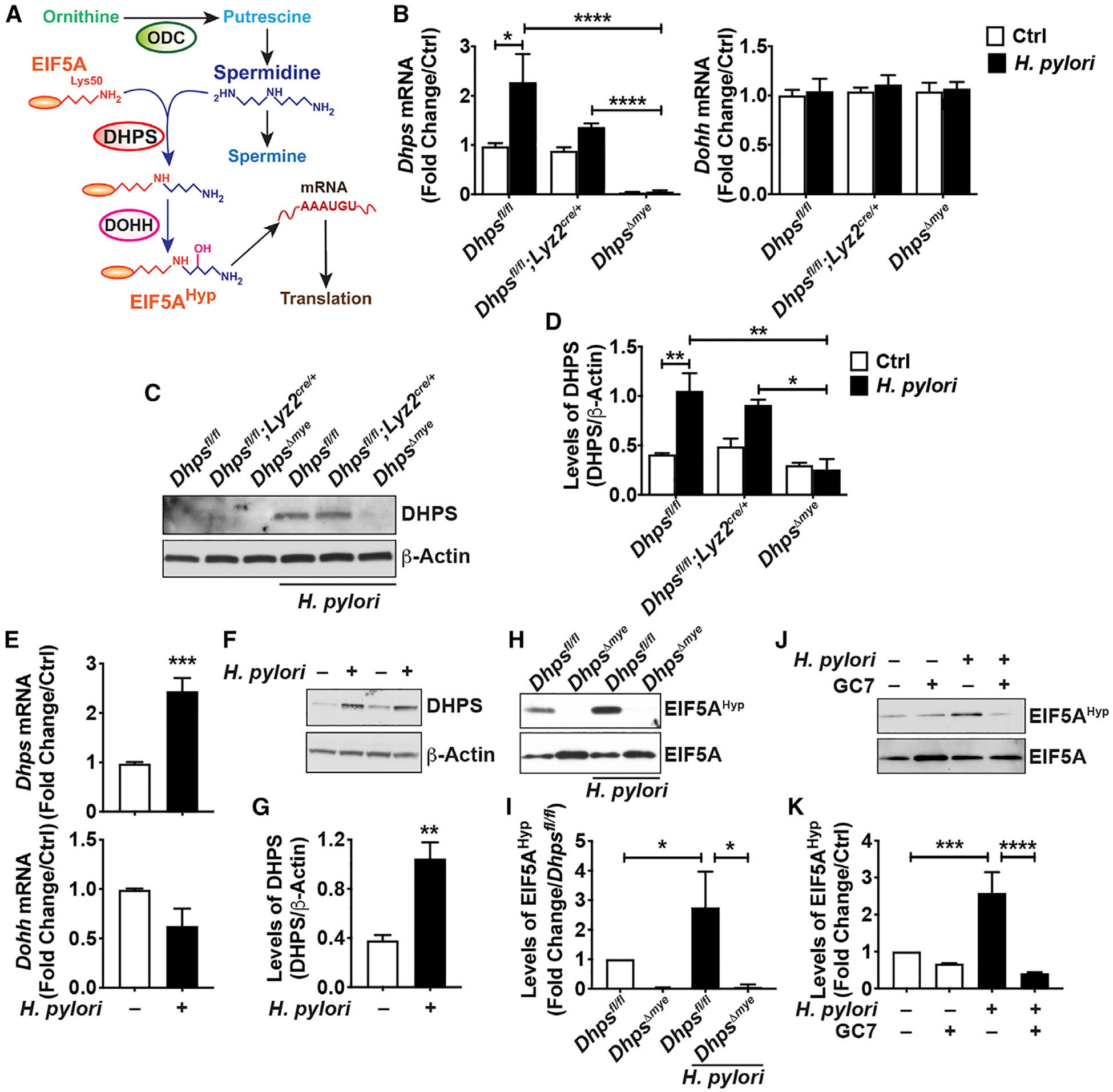
(A) Schematic representation of the synthesis and role of EIF5AHyp.
(B) mRNA expression of Dhps and Dohh in BMmacs from Dhpsfl/fl, Dhpsfl/fl;Lyz2cre/+, and DhpsΔmye mice stimulated or not with H. pylori (Hp) PMSS1 for 6 h; n = 3 mice per genotype.
(C and D) DHPS protein levels were assessed by western blot in the same cells infected or not for 24 h (C); representative data of macrophages isolated from three different mice per genotype. The densitometric analysis of DHPS was performed from the three animals/group (D).
(E) Dhps and Dohh mRNA levels in RAW 264.7 cells infected or not with Hp for 6 h; n = 7 independent experiments.
(F and G) Representative immunoblot for DHPS using protein lysates from RAW 264.7 cells ± Hp (F); densitometry from five independent western blots is shown in (G).
(H–K) Levels of EIF5AHyp and EIF5A in Dhpsfl/fl- and DhpsΔmye-derived BMmacs (H and I) and in RAW 264.7 macrophages (J and K) infected or not with Hp for 24 h. The densitometry was determined from three immunoblots for RAW 264.7 cells and BMmacs. For all the panels, each bar represents the mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
See also Figures S1–S3.
In this report, we show that the enzyme DHPS is induced by H. pylori in macrophages. This results in increased hypusination of EIF5A in vitro and in mouse and human tissues with H. pylori gastritis. Using mice with specific deletion of Dhps in myeloid cells, and macrophages treated with the DHPS inhibitor N1-guanyl-1,7-diaminoheptane (GC7), we identified a series of inducible proteins regulated by hypusination in infected macrophages. These effectors are notably involved in the antimicrobial response and autophagy. Deletion of DHPS in myeloid cells resulted in higher survival of phagocytized bacteria and increased colonization in vivo by the gastric pathogen H. pylori and by the colonic pathogen Citrobacter rodentium, the rodent equivalent of the human pathogen enteropathogenic Escherichia coli. Thus, in addition to the classical transcriptional response of macrophages, we emphasize herein that the regulation of translation through hypusination is a critical hallmark of the defense of the mammalian host against pathogenic bacteria.
RESULTS
Pathogenic Bacteria Stimulate DHPS Induction
We used the Lyz2-Cre driver to generate mice with specific deletion of Dhps in the myeloid cell lineage, including macrophages, dendritic cells, and granulocytes. Based on the conceptual framework of the role of hypusination in innate immunity, we first assessed the macrophage response to H. pylori infection. In response to this pathogen, bone-marrow-derived macrophages (BMmacs) from Dhpsfl/fl mice exhibited a significant increase in Dhps mRNA expression, whereas the Dohh level was not affected by the infection (Figure 1B). The expression of Dhps in control or infected macrophages from Dhpsfl/fl;Lyz2cre/cre mice (hereafter referred to as DhpsΔmye mice) was significantly reduced compared with cells from Dhpsfl/fl or Dhpsfl/fl;Lyz2cre/+ mice (Figure 1B). Similarly, the H. pylori-induced DHPS protein expression in BMmacs from Dhpsfl/fl and Dhpsfl/fl;Lyz2cre/+ mice was markedly attenuated in DhpsΔmye cells (Figure 1C). By densitometry, we found that the level of DHPS was reduced in H. pylori-stimulated macrophages from DhpsΔmye mice to the same level as unstimulated controls (Figure 1D). In addition, the level of Dhps mRNA, but not Dohh mRNA (Figure 1E), as well as DHPS protein (Figures 1F and 1G), was increased in the murine macrophage cell line RAW 264.7 infected with H. pylori. This induction was reduced by blocking phosphatidylinositol 3-kinase (PI3K) with LY294002 (Figures S1A and S1B); however, inhibitors of nuclear factor κB (NF-κB), mitogen-activated protein kinase 14 (MAPK14), MAPK1/3, or MAP2K1 had no effect on H. pylori-induced DHPS protein expression (Figures S1A and S1B), indicating that signaling through PI3K is involved in stimulation of DHPS in infected macrophages. In addition, expression of Dhps mRNA (Figure S1C) and DHPS protein (Figure S1D) was also induced by the colonic pathogen C. rodentium in BMmacs from Dhpsfl/fl and Dhpsfl/fl;Lyz2cre/+ mice and markedly reduced in DhpsΔmye BMmacs. Lastly, we observed that DHPS was also induced by H. pylori and C. rodentium in bone-marrow-derived dendritic cells (BMDCs; Figure S1E).
Using a specific antibody (Nishiki et al., 2013), we found that the level of EIF5AHyp was enhanced in BMmacs from Dhpsfl/fl mice infected with H. pylori compared with uninfected cells and was absent in uninfected and infected DhpsΔmye macrophages (Figures 1H and 1I). Similarly, H. pylori-induced hypusination in RAW 264.7 cells was significantly reduced by the DHPS inhibitor GC7 (Figures 1J and 1K). Moreover, we found hypusination of EIF5A was significantly induced by H. pylori in BMmacs from Odcfl/fl mice and from animals with specific Odc deletion in myeloid cells (OdcΔmye; Hardbower et al., 2017; Figures S1F and S1G). While EIF5AHyp was reduced in BMmacs from OdcΔmye mice compared with Odcfl/fl mice, more than 50% of the EIF5AHyp remained in infected cells (Figures S1F and S1G), explaining why increased immune effectors could still be identified in BMmacs from OdcΔmye mice lacking the transcriptional inhibitor putrescine (Hardbower et al., 2017).
These data demonstrate that bacterial infection of macrophages leads to enhanced hypusination, notably through the inducible expression of DHPS.
Increased Hypusination in H. pylori-Infected Patients
To determine whether DHPS is expressed in gastric macrophages in humans, we assessed gastric biopsies from uninfected individuals and H. pylori-infected patients with gastritis. The number of cells with double staining for EIF5AHyp and CD68 was significantly increased in tissues from infected patients compared with individuals without infection (Figures 2A and 2B).
Figure 2. Levels of Hypusinated EIF5A in Human Gastric Tissues during Hp Infection.

(A) The macrophage marker CD68 (red), EIF5AHyp (green), and nuclei (blue) were detected by immunofluorescence in patients with normal histology and with non-atrophic gastritis (NAG) caused by infection. Merged images are shown, with cells double positive for CD68 and EIF5AHyp depicted in yellow. Scale bars, 50 μm. These photomicrographs are representative images for five normal and nine infected patients.
(B) Quantification of EIF5AHyp staining in CD68+ cells. Bars represent mean ± SEM; ****p < 0.0001.
Hypusination Regulates Macrophage Function
To determine the role of hypusination in the innate response of macrophages to bacterial infection, we analyzed the proteome of BMmacs from three different Dhpsfl/fl and DhpsΔmye mice. These cells were stimulated or not with H. pylori, and an isobaric tag for relative and absolute quantification (iTRAQ)-based proteomics approach was utilized. We identified 2,994 proteins, comprising 2,895 murine proteins and 99 H. pylori proteins. There were 130 and 191 proteins induced by H. pylori infection in BMmacs from Dhpsfl/fl and DhpsΔmye mice, respectively (Table S1). The most induced proteins in Dhpsfl/fl BMmacs included (1) classical antibacterial effectors IRG1, NOS2, and ASSY; (2) the proteins IFIH1, ISG20, GBP2/4, and IFIT2/3 that are involved in the antiviral response; (3) the proinflammatory cytokines IL1A, IL1B, and CCL5; (4) the inflammasome constituent NLRP3; and (5) the mediators of autophagy IIGP1, CLEC4E, and SQSTM (Figure 3A; Table S1). Remarkably, there were also proteins less expressed in infected BMmacs from DhpsΔmye mice compared with Dhpsfl/fl animals; these included IRG1, NOS2, ASSY, IL1A, CCL5, IIGP1, CLEC4E, IRGM1, and SQSTM (Figure 3A; Table S1). We then used targeted approaches to verify our findings. We found by western blot that the level of the proteins NOS2, IRG1, and SQSTM were reduced in BMmacs from DhpsΔmye mice infected with H. pylori compared with infected Dhpsfl/fl-derived BMmacs (Figure 3B), thus validating the results obtained by proteomic analysis. We also confirmed that de novo translation of NOS2 was reduced in BMmacs from DhpsΔmye mice compared with Dhpsfl/fl macrophages (Figure S2). The production of NO2−, the stable metabolite of NO derived from NOS2 activity, was also reduced in infected DhpsΔmye BMmacs compared with cells from Dhpsfl/fl mice (Figure S3A). Importantly, the mRNA expression of the genes encoding these proteins was not affected by DHPS deletion (Figure 3C), further demonstrating that DHPS regulates only protein translation in infected macrophages. We also found that the expression of genes encoding for the chemokine CXCL1, the classical M1 marker IL-6, and the M2 marker arginase-1 was increased in response to H. pylori in both genotypes, but was not differentially expressed between them (Figure S3D), indicating that Dhps deletion in macrophages does not affect the inducible transcriptome in response to bacteria.
Figure 3. Regulation of the Inducible Proteome of Macrophages by Dhps Deletion.
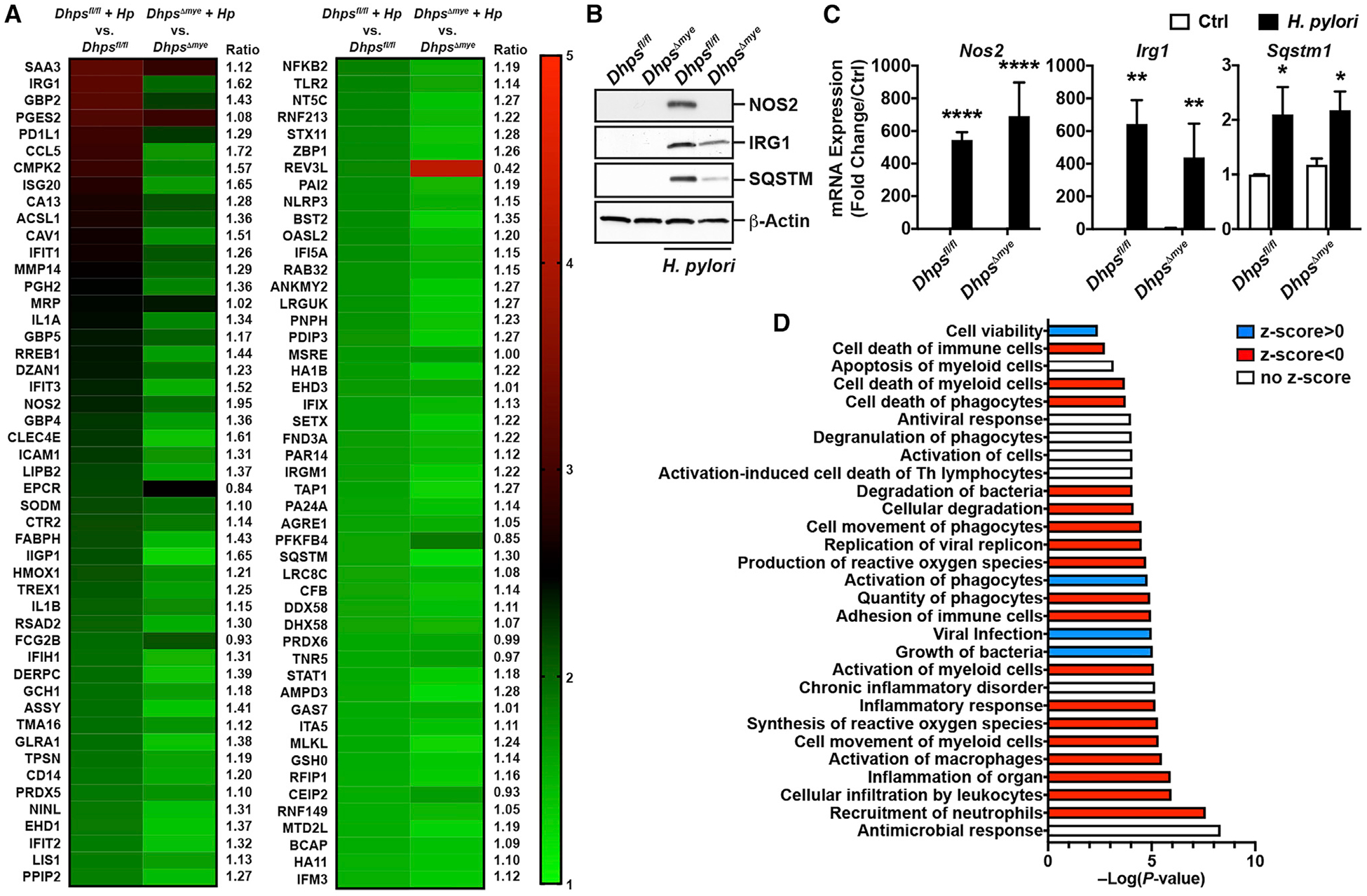
(A) BMmacs from Dhpsfl/fl (n = 3) and DhpsΔmye mice (n = 3) were infected with Hp PMSS1 for 24 h. Cells were lysed, and proteins were digested, labeled with iTRAQ reagents, and analyzed by liquid chromatography (LC)-coupled tandem mass spectrometry. The heatmap depicts the normalized fold change in the protein levels compared with uninfected cells; this panel represents the top 98 proteins induced by at least 1.3-fold in BMacs from Dhpsfl/fl and/or DhpsΔmye mice. For each protein, the ratio between the fold increase in infected Dhpsfl/fl and DhpsΔmye mice is indicated.
(B) Levels of NOS2, IRG1, SQSTM, and β-actin proteins in BMmacs from Dhpsfl/fl and DhpsΔmye mice infected or not with Hp for 24 h.
(C) The expression of the genes encoding NOS2, IRG1, and SQSTM was analyzed in BMmacs infected or not for 6 h. In (B) and (C), n = 3–4 different mice per genotype. In all the panels, bars depict mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
(D) IPA was performed on the proteome dataset obtained from BMmacs from Dhpsfl/fl and DhpsΔmye mice, and the pathways related to diseases and functions are presented. The full lists of pathways are shown in Table S3.
See also Figures S3–S5 and Tables S1, S2, and S3.
To confirm this first proteomic analysis, we performed a second iTRAQ experiment using a new set of BMmacs from one Dhpsfl/fl mouse and one DhpsΔmye mouse. In this experiment, there were 248 proteins significantly upregulated by H. pylori in Dhpsfl/fl BMmacs (Table S2), and 78.5% of the induced proteins identified in the first experiment were also found in this independent study. Strikingly, the main H. pylori-stimulated immune mediators affected by Dhps deletion in the first experiment were also downregulated in BMmacs from DhpsΔmye mice in this second experiment (Figure S4A). Additionally, we confirmed by immunoblotting that the immune effectors NOS2, IRG1, and SQSTM were induced in Dhpsfl/fl BMmacs infected with C. rodentium compared with control cells and that their expression was reduced in infected DhpsΔmye BMmacs (Figure S4B), evidencing that the regulation of specific proteins by hypusination occurs in response to different pathogens. In this analysis, there were 27 and 13 proteins up- and downregulated, respectively, in uninfected DhpsΔmye BMmacs compared with Dhpsfl/fl cells (Table S2), but there was no major regulator of the antimicrobial activity of macrophages in the uninfected cells.
The differential proteome dataset from H. pylori-infected BMmacs from Dhpsfl/fl and DhpsΔmye mice was subjected to ingenuity pathway analysis (IPA) to determine biological functions and the important pathways regulated by hypusination in macrophages. The complete analysis is presented in Table S3. Strikingly, when we focused on the functions related to immune pathways, we found that the categories linked to macrophage activation and antimicrobial responses were significantly affected by Dhps deletion (Figure 3D); this was associated with an increase in “viral infection” and “growth of bacteria” pathways (Figure 3D). Overall, there was also a dampening of the pathways related to inflammation, adhesion, and death of immune cells (Figure 3D). In addition, we used IPA to identify the biochemical networks that were modulated in infected macrophages from DhpsΔmye versus Dhpsfl/fl mice. The highest scored networks were “hematological system development and function, inflammatory response, tissue morphology” (score 29; Figure S5A) and “antimicrobial response, connective tissue disorders, inflammatory response” (score 26; Figure S5B), further indicating that the specific alterations in protein expression regulated by DHPS lead to enhanced host defenses.
Next, we assessed the effect of hypusination on the macrophage proteome using a pharmacological approach. RAW 264.7 cells were treated or not with the DHPS inhibitor GC7 for 2 h prior to infection with H. pylori PMSS1. After 24 h, the proteome was similarly analyzed using the iTRAQ-based experimental approach. We quantified 3,556 proteins, comprising 3,507 murine proteins and 49 H. pylori proteins. There were 57 and 58 proteins significantly upregulated in response to H. pylori in cells without and with GC7 (Table S4). We found a series of proteins more highly expressed in H. pylori-infected macrophages compared with infected cells treated with GC7, including many of the same proteins affected by Dhps deletion in BMmacs, e.g., IRG1, NOS2, EHD1, and IRGM1 (Figure 4A; Table S4). Of note, 58 proteins were induced by the GC7 in uninfected cells, whereas 158 were downregulated by DHPS inhibition; the proteins that were the most inhibited were ribosomal proteins (Table S4). We confirmed by immunoblotting that the proteins NOS2, IRG1, and IRGM1 were less induced in response to H. pylori in RAW 264.7 cells treated with GC7 compared with infected macrophages without GC7 (Figure 4B), while their mRNA levels were not affected (Figure 4C). In addition, the NO metabolite NO2− was also produced in lower amounts by infected macrophages treated with GC7 (Figure S3B); GC7 also reduced H. pylori-induced autophagy in RAW 264.7 cells (Figure S3C). Lastly, the immune pathways involved in the activation and antimicrobial effects of macrophages were downregulated by GC7 in H. pylori-infected RAW 264.7 macrophages (Figure 4D; Table S5). Interestingly, the pathways predicted to impact cell death, including by autophagy, were also negatively affected by the inhibition of hypusination (Figure 4D; Table S5).
Figure 4. Effect of the Inhibition of DHPS on the Proteome of Activated Macrophages.

(A) Differential expression of proteins induced by Hp PMSS1 in RAW 264.7 cells ± GC7. The heatmap depicts the normalized fold change compared with uninfected cells of the 92 proteins induced significantly by at least 1.3-fold in infected RAW 264.7 macrophages, treated or not with GC7. For each protein, we calculated the ratio between the fold increase in infected macrophages and in infected macrophages + GC7.
(B) Levels of NOS2, IRG1, IRGM1, and β-actin proteins in RAW 264.7 cells ± GC7 infected or not with Hp for 24 h; these immunoblots are representative data of three to six independent experiments.
(C) Expression of the genes encoding NOS2, IRG1, and IRGM1 was determined in RAW 264.7 macrophages ± GC7 after a 6 h infection. Bars represent mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; n = 3–6 independent experiments.
(D) Diseases and functions pathways were determined by IPA using the differential proteome dataset obtained from RAW 264.7 cells. The full lists of pathways are shown in Table S5.
These data suggest that hypusination supports the antimicrobial protein expression profile of macrophages.
mRNA and Protein Sequence Analysis of the Effectors Regulated by Hypusination in Macrophages
Hypusinated EIF5A binds preferentially to mRNAs that contains an 5′-AAAUGU-3′ consensus sequence (Maier et al., 2010; Xu and Chen, 2001). For this in silico analysis, we focused on the 48 proteins downregulated by at least 1.3-fold in infected DhpsΔmye BMmacs and RAW 264.7 cells infected with H. pylori and treated with GC7. Among them, we found this binding site in the coding region of 23 of the proteins regulated by hypusination, and 20 more displayed the similar sequence with only one substitution (Table S6).
In addition, EIF5AHyp improves ribosome pausing during the translation of peptides enriched in diprolyl, diglycyl motifs and in basic and acidic amino acids (Gutierrez et al., 2013; Pelechano and Alepuz, 2017; Schuller et al., 2017). We identified 31 proteins with at least one diprolyl motif and 36 with one diglycyl motif (Table S6); 22 proteins exhibited at least one of each motif. Furthermore, we also identified 34 proteins containing at least one tripeptide whose translation is recognized to be facilitated by hypusinated EIF5A (Pelechano and Alepuz, 2017; Schuller et al., 2017). Only the proteins IFIT3, CCL5, and CLEC4E did not exhibit diprolyl, diglycyl, or EIF5A-regulated tripeptide motifs, but each of these did show the 5′-AAAUGU-3′ sequence (± 1 substitution).
Thus, all the innate effectors identified in this study to be affected by hypusination in macrophages exhibit at least one feature of EIF5AHyp-regulated proteins, validating our proteomic findings.
Regulation of the Mucosal Antimicrobial Response by DHPS
Our data indicate that the induction of DHPS in macrophages supports the translation of inducible proteins involved in the defense against pathogens. To verify this postulate, we tested the involvement of hypusination in the antimicrobial effect of macrophages. We observed increased H. pylori survival in BMmacs from DhpsΔmye mice compared with Dhpsfl/fl macrophages (Figure 5A) and in RAW 264.7 cells pre-treated with GC7 compared with untreated macrophages (Figure 5B).
Figure 5. Effect of Dhps Deletion on Hp Survival in Macrophage and in the Stomach.
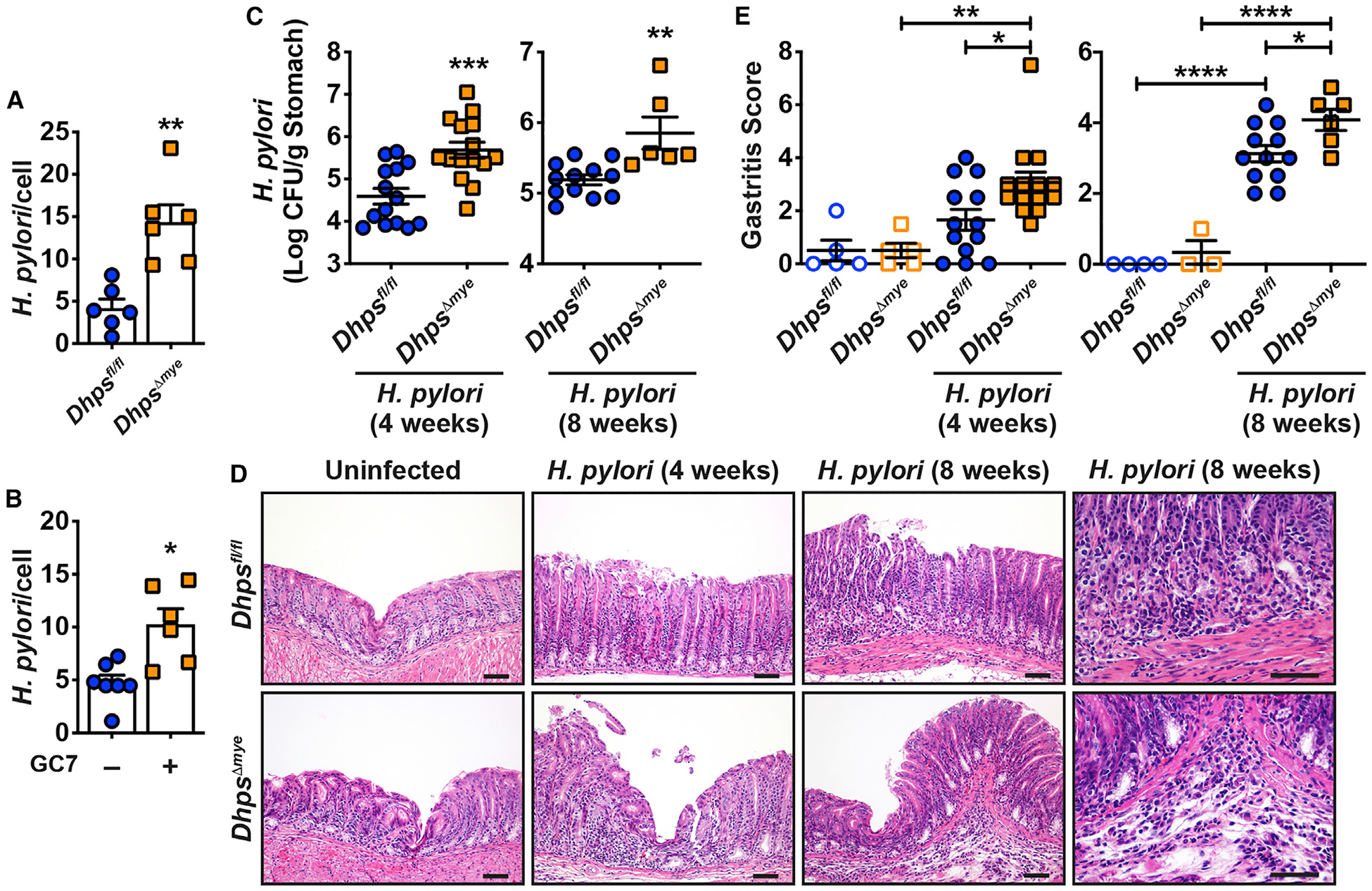
(A) BMmacs from Dhpsfl/fl and DhpsΔmye mice were infected with Hp at a multiplicity of infection of 100 for 24 h. Cocultures were washed and treated with gentamicin for 1 h. Cells were then harvested, counted, and lysed. The number of live bacteria was determined by counting colony-forming units (CFUs) after plating serial dilutions of cell lysates. Each dot represents BMmacs from one mouse.
(B) The same experiments were performed using RAW 264.7 macrophages ± GC7; each dot represents an individual well.
(C–E) Dhpsfl/fl and DhpsΔmye mice were infected or not with Hp PMSS1 for 4 and 8 weeks. Gastric colonization was determined by culture (C). H&E staining of the stomach (D) was used to generate the inflammation score of the gastric tissues (E). In all the panels, the mean ± SEM is shown; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Scale bars in (D), 50 μm.
See also Figures S9 and S10 and Table S7.
Next, to assess the effect of hypusination in infection in vivo, we used a well-established model of H. pylori infection in mice (Chaturvedi et al., 2010; Hardbower et al., 2016, 2017). The level of gastric colonization by H. pylori at 4 weeks and 8 weeks post-infection was significantly enhanced by 1.1 and 0.8 log, respectively, in animals lacking Dhps in myeloid cells compared with Dhpsfl/fl mice (Figure 5C). Concomitantly, hematoxylin and eosin (H&E) images showed more infiltration of immune cells in the gastric mucosa and more gastric hyperplasia in H. pylori-infected DhpsΔmye mice (Figure 5D). Scoring for acute and chronic inflammation in the gastric antrum and corpus (Chaturvedi et al., 2010; Hardbower et al., 2016, 2017) confirmed that gastritis was significantly higher in infected DhpsΔmye mice than Dhpsfl/fl mice at both time points (Figure 5E). This was confirmed by increased expression of the genes encoding for the chemokines CXCL1 and CXCL2 and for the T cell effectors IL-17 and IL-10 in the gastric mucosa of infected DhpsΔmye mice (Figure S6A). However, the inducible expression of the genes encoding for NOS2, IL-1β, and interferon (IFN)-γ was not affected by Dhps deletion (Figure S6A).
We then confirmed the role of DHPS in the antibacterial immunity of the gastrointestinal tract using the colonic pathogen C. rodentium. Similar to the result with H. pylori in the stomach, mice with specific deletion of Dhps in myeloid cells exhibited increased colonization by C. rodentium (Figure 6A). There was also a significant increase in colon weight (Figure 6B) and histologic colitis in DhpsΔmye mice (Figures 6C and 6D). Cxcl1, Il1b, Ifng, and Il17a mRNA levels were more abundant in the colon of C. rodentium-infected DhpsΔmye mice compared with Dhpsfl/fl mice (Figure S6B).
Figure 6. Outcome of C. rodentium Infection in Dhpsfl/fl and DhpsΔmye Mice.

(A–D) Animals were infected or not with C. rodentium for 14 days. Colonization of the colon was assessed by culture (A). Colon was weighed (B). H&E staining of the colon (C) was used to generate the score for colitis (D); the inflammation scores in uninfected Dhpsfl/fl and DhpsΔmye mice were 0 in stomach and colon. In all the panels, the mean ± SEM is depicted; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Scale bars in (C), 50 μm.
See also Figure S9.
Moreover, both H. pylori and C. rodentium infection of Dhpsfl/fl and DhpsΔmye mice led to the recruitment of macrophages in the gastric and colonic mucosa, respectively, compared with uninfected animals, as shown by immunofluorescence (IF) for the macrophage marker CD68 (Figures 7A and 7C). We did not observe noteworthy differences in the number of CD68+ cells between genotypes (Figures 7A and 7C). Remarkably, macrophages in H. pylori- and C. rodentium-infected Dhpsfl/fl mice exhibited high levels of EIF5AHyp (Figures 7A and 7C). By contrast, the level of EIF5AHyp was strikingly reduced in gastric and colonic CD68+ cells from DhpsΔmye mice infected with H. pylori (Figures 7A and 7B) or with C. rodentium (Figures 7C and 7D). Note that EIF5AHyp was also present in other cells from the gastric mucosa of H. pylori-infected mice and from the colon of animals infected with C. rodentium, but these levels were not altered by myeloid cell Dhps deletion (Figure 7C). Of interest, the levels of NOS2, which have been found to be regulated by hypusination, were also increased in CD68+ macrophages in H. pylori- and C. rodentium-infected Dhpsfl/fl mice and were markedly reduced in DhpsΔmye mice (Figures 7A–7D).
Figure 7. Expression of Immune Effectors Regulated by Hypusination in Infected Tissues.

(A–D) The gastric tissues of Dhpsfl/fl and DhpsΔmye mice infected for 8 weeks with Hp PMSS1 (A) or the colon of these animals infected with C. rodentium for 14 days (C) were immunostained for CD68 and EIF5AHyp, as well as CD68 and NOS2. Scale bars, 50 μm. Quantification of CD68+/EIF5AHyp + cells and CD68+/NOS2+ cells among the total number of CD68+ cells is shown in (B) and (D). The immunofluorescence images are representative of staining performed on two uninfected animals and five infected mice in each group, and the same number of animals was used for quantification. Bars represent mean ± SEM in all graphs; **p < 0.01 and ****p < 0.0001.
Lastly, we assessed the effect of Dhps deletion in macrophages on the gastric metabolomic signatures. There were 267 positively charged and 341 negatively charged metabolites significantly affected in infected DhpsΔmye mice compared with Dhpsfl/fl mice (Figure S7). As an example, the level of spermidine, the substrate for DHPS, was increased in the stomach of DhpsΔmye mice. Conversely, the NOS2 product citrulline, and the oxidized from of glutathione, which is a reliable indicator of oxidative stress, were less abundant in the DhpsΔmye animals (Figure S7). The analysis of the metabolomic pathways (Table S7) evidenced that the citrulline-NO cycle was significantly affected by infection in Dhpsfl/fl mice, but not in DhpsΔmye mice. We also found that the TCA cycle was affected by Dhps deletion in myeloid cells (Table S7).
DISCUSSION
Hypusination of EIF5A is a unique mechanism that controls the translation of specific proteins. In this study, we have utilized an unbiased approach using specific genetic deletion of Dhps and a DHPS inhibitor to probe the role of the spermidine-EIF5AHyp axis in macrophage responses to pathogenic bacteria. Increased DHPS expression and levels of EIF5AHyp were observed in macrophages infected ex vivo with H. pylori and in gastric CD68+ cells from H. pylori- and C. rodentium-infected mice. This results in the translation of immune effectors principally involved in microbial clearance and cell death, as underlined by the IPA of the proteomic data. Importantly, these results were obtained with the human gastric pathogen H. pylori and the murine colon bacterial pathogen C. rodentium, providing strong evidence that our finding is not limited to one pathogen but concerns the global innate response of macrophages. Lastly, our data showing increased colonization of DhpsΔmye mice by both pathogens endorse the in vivo relevance of the paramount role of hypusination in the first line of defense of macrophages against bacterial infection of the gastrointestinal mucosa. Our present findings emphasize one more step in the complex regulation of the immune response by polyamines: first, putrescine regulates the transcription of innate effectors (Hardbower et al., 2017); and second, spermidine controls translation through hypusination, as shown herein. Of importance, we found that hypusination is reduced in infected BMmacs from OdcΔmye mice, but not completely suppressed, consistent with the partial reduction of spermidine concentration by Odc deletion in macrophages (Hardbower et al., 2017). This indicates that ODC-derived putrescine affects transcription, but does not sufficiently alter translation by modulation of hypusination. Taken together, our data herein and our prior study (Hardbower et al., 2017) illustrate a complex immunologic rheostat for macrophage innate immune effectors, where putrescine from ODC regulates transcription, and spermidine, acting through DHPS/hypusination, regulates translation, providing robust biological control.
Previous reports have evidenced that EIF5A knockdown using small hairpin RNA in cervical cancer cells (Mémin et al., 2014), pancreatic ductal adenocarcinoma cells (Fujimura et al., 2015), and HeLa cells (Mandal et al., 2016) regulates proteins involved in migration, invasion, proliferation, endoplasmic reticulum (ER) stress, and/or unfolded protein response. Moreover, mitochondrial enzymes and proteins involved in the TCA cycle are affected by GC7 in M0 and M2 macrophages and by Eif5a and Dhps silencing in murine embryonic fibroblasts (Puleston et al., 2019). We have identified a series of bacteria-induced proteins that are regulated by DHPS function in macrophages.
Interestingly, it has also been shown that hypusination of EIF5A is enhanced in alternatively activated macrophages, in which the arginase-ODC metabolic pathway is strongly activated, but not during the classical macrophage activation with lipopolysaccharide (LPS)/IFN-γ (Puleston et al., 2019). However, in the context of infected macrophages, we propose now that DHPS induction supports increased hypusination of EIF5A and translation of antimicrobial factors. Note that in this prior study (Puleston et al., 2019), the authors did not identify the proteins involved in the antimicrobial response since they were not induced by M2 stimulation. In this previous work, the authors demonstrated that the TCA cycle was dampened in M0, M1, and M2 macrophages treated with GC7 (Puleston et al., 2019). Accordingly, our proteomic analysis of RAW 264.7 cells ± GC7 evidenced that the enzymes SDHB, SUCB1, SUCB2, ODPA, and ODPB, which all belong to the TCA cycle, are downregulated by GC7 in unstimulated cells (Table S4). However, since these enzymes were also reduced by H. pylori infection, we suggest that the control of infection by hypusination is not dependent on the TCA cycle.
In our study, numerous immune effectors regulated by hypusination are involved in the host defense against pathogens. Notably, the protein levels of NOS2, ASSY, and IRG1 were markedly reduced during H. pylori infection in DhpsΔmye macrophages compared with Dhpsfl/fl and in GC7-treated macrophages. NOS2 expression is a hallmark of macrophage activation by pathogen-associated molecular patterns and proinflammatory cytokines (MacMicking et al., 1997) and has been already reported to be regulated by hypusination in pancreatic β cells (Maier et al., 2010) and in murine macrophages stimulated with the mycobacterial cell wall component trehalose-6,6′-dimycolate (Lee et al., 2016). NOS2-derived high-output NO production has been implicated in the killing of various parasites and bacteria (Duleu et al., 2004; El Kasmi et al., 2008; Qualls et al., 2012), including H. pylori (Gobert et al., 2001). Similarly, ASSY converts citrulline into arginine, the NOS2 substrate, to increase the NO-dependent killing of bacteria (Qualls et al., 2012). In addition, the innate effector IRG1 has been shown to be induced in RAW 264.7 cells and primary macrophages stimulated by LPS and IFN-γ (Strelko et al., 2011), tumor necrosis factor alpha, Toll-like receptor (TLR) 2, or TLR4 signaling (Degrandi et al., 2009) and leads to the synthesis of itaconic acid (Basler et al., 2006; Degrandi et al., 2009; Strelko et al., 2011). This metabolite inhibits isocitrate lyase, a key enzyme that is necessary for bacterial growth (Michelucci et al., 2013). Interestingly, it has been recently shown that the CRISPR-Cas9 deletion of both NOS2 and IRG1 reduced the killing of Legionella pneumophila by murine BMmacs stimulated by IFN-γ (Price et al., 2019). In addition, the maximal killing of L. pneumophila by macrophages required the expression of four other immune effectors: CASP11, NOX2, IRGM3, and IRGM1 (Price et al., 2019); this last protein was also found to be regulated by hypusination in our experiments. Thus, our data highlight that hypusination is a major regulator of host defense by affecting the translation of multiple antimicrobial effectors in macrophages; this postulate was supported by our finding that bacterial survival is increased in macrophages with DHPS knockout or inhibition. Of note, we also observed that hypusination regulates mediators involved in antiviral response, including the receptor of viral nucleic acids IFIH1, the antiviral exoribonuclease ISG20, the GTPases GBP2 and GBP4, and the antiviral proteins RRSAD2, IFIT2, and IFIT3, further indicating that hypusination is a general control mechanism of the overall innate response of macrophages.
The abundance of the proteins IIGP1, IRGM1, CLEC4E, and SQSTM, which are all implicated in autophagy, was affected by DHPS deletion or inhibition in infected macrophages, and we also found that GC7 reduced H. pylori-mediated autophagy. Remarkably, these effectors also play a fundamental role in the destruction of microorganisms (Al-Zeer et al., 2009; MacMicking et al., 2003; Pahari et al., 2019; Watson et al., 2012). Intriguingly, spermidine, the DHPS substrate, supports autophagy in various cells including human peripheral blood mononuclear cells (Eisenberg et al., 2009) or in a human colon cancer cell line (Morselli et al., 2011). In the same way, mice treated with spermidine exhibit prolonged lifespan and improved cardiomyocyte function due to increased cardiac autophagic flux (Eisenberg et al., 2016). Furthermore, it has been recently observed that hypusinated EIF5A controls the synthesis of the transcription factor EB, a master regulator of genes encoding mediators of autophagy, in naive B cells (Zhang and Simon, 2020). This transcription factor was not evidenced to be regulated by infection and/or inhibition of DHPS in our study, suggesting that EIF5AHyp may regulate autophagy by various pathways. Nonetheless, we contend that hypusination is also involved in the autophagy-dependent clearance of pathogenic bacteria by macrophages.
Polyamine metabolism plays a fundamental role in the outcome of bacterial infections, including those of the gastrointestinal tract (Chaturvedi et al., 2010; Gobert et al., 2018; Hardbower et al., 2017). This report highlights that the spermidine-hypusine metabolic pathway represents an unrecognized foundation of the antimicrobial armature. It is important to underline that reduced levels of EIF5AHyp in tissue macrophages led not only to increased colonization by H. pylori and C. rodentium but also to enhanced inflammation. We therefore propose that the loss of innate host defense in DhpsΔmye macrophages led to the uncontrolled development of infection, which results in increased inflammation. Epithelial responses and chronic inflammation are also critical for the outcome of gastrointestinal infections. Hypusination has been shown to support the development of cancerous cells (Nakanishi and Cleveland, 2016); in contrast, it has also been recently evidenced that the polyamine-EIF5AHyp metabolic pathway exhibits tumor suppressor functions in a mouse lymphoma model (Scuoppo et al., 2012). Given that H. pylori is the main causative agent for the development of gastric cancer (Parsonnet et al., 1991), the relative functional contribution of hypusination in epithelial, myeloid, and T cells in host defense against carcinogenetic events deserves further investigation and is underway in our laboratory.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Keith T. Wilson (keith.wilson@vumc.org).
Materials availability
Dhpsfl/fl;Lyz2cre/+ and Dhpsfl/fl;Lyz2cre/cre mice were generated for this study and are available upon request.
Data and code availability
The mass spectrometry proteomics data regarding BMmacs (Tables S1 and S2) and RAW 264.7 cells (Table S4) have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Perez-Riverol et al., 2019), and the accession numbers are ProteomeXchange Consortium: PXD018187 and ProteomeXchange Consortium: PXD010082, respectively. The accession number for the metabolomics data reported in this paper is EMBL-EBI MetaboLights: MTBLS607.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human tissues
Biopsies from gastric tissues were obtained from human subjects in Colombia as described (Mera et al., 2018), under protocols approved by the ethics committees of the local hospitals and of the Universidad del Valle in Cali, Colombia, as well as the Institutional Review Board at Vanderbilt University. We used biopsies from 5 uninfected subjects (4 females and 1 male) aged 26–64 years (mean ± standard deviation: 47.2 ± 13.6 years) and 9 H. pylori-infected patients (5 females and 4 males) aged 26–45 years (mean ± standard deviation: 34.6 ± 6.1 years).
Animals
Mice were used under protocols M/05/176 and M/08/124 approved by the Institutional Animal Care and Use Committee at Vanderbilt University and Institutional Biosafety Committee, and the Research and Development Committee of the Veterans Affairs Tennessee Valley Healthcare System. Procedures were performed in accordance with institutional policies, AAALAC guidelines, the AVMA Guidelines on Euthanasia, NIH regulations (Guide for the Care and Use of Laboratory Animals), and the United States Animal Welfare Act (1966). To achieve specific deletion of Dhps in myeloid cells, we crossed C57BL/6J Dhpsfl/fl mice, in which exons 2 to 7 of the Dhps gene were flanked by Cre recombinase recognition sequences (Levasseur et al., 2019), to mice with the myeloid driver Lyz2cre. Dhpsfl/fl;Lyz2cre/+ mice were then crossed to each other to generate littermate Dhpsfl/fl;Lyz2cre/cre, Dhpsfl/fl;Lyz2cre/+ and Dhpsfl/fl mice in expected proportions (25%, 52% and 23%, respectively). Thereafter, we used littermates, namely Dhpsfl/fl mice as controls, Dhpsfl/fl;Lyz2cre/+ mice, and Dhpsfl/fl;Lyz2cre/cre mice, which we termed DhpsΔmye mice. We also used Odcfl/fl and OdcΔmye mice that are bred in our animal facility (Hardbower et al., 2017). Adult mice (8–12 weeks) of both sexes were used for isolation of BMmacs. Only male mice (8–12 weeks) were used for infection with H. pylori and C. rodentium.
Isolation of myeloid cells
BMmacs and BMDCs from Dhpsfl/fl, Dhpsfl/fl;Lyz2cre/+, and Dhpsfl/fl;Lyz2cre/cre mice were isolated and differentiated as described (Hardbower et al., 2016).
Bacteria
The cagA+ H. pylori strains PMSS1 (Arnold et al., 2011) and C. rodentium DBS100 (Barthold et al., 1976) were grown on Tryptic Soy agar plates containing 10% sheep blood and MacConkey agar plates, respectively. Bacteria were harvested directly from plates to infect the cells. From the plates, H. pylori and C. rodentium were also grown in Brucella broth containing 10% FBS and in Luria-Bertani broth liquid medium overnight, respectively; they were then diluted to an A600 nm of 0.1 in these media, grown, and harvested at the exponential phase to infect mice.
METHOD DETAILS
Infections
C57BL/6J, Dhpsfl/fl, Dhpsfl/fl;Lyz2cre/+, Dhpsfl/fl;Lyz2cre/cre, Odcfl/fl, and OdcΔmye mice were bred in our animal facility. Mice were fed ad libitum with regular 5L0D chow (LabDiet). For infections, male mice at 8–12 weeks of age were randomly distributed in the different groups and infected by oral gavage with i) 109 H. pylori PMSS1 in 0.2 mL Brucella broth, two times every 2 days (Sierra et al., 2018), or ii) 5×108 C. rodentium DBS100 in in 0.2 mL Luria-Bertani broth (Gobert et al., 2018; Hardbower et al., 2016). Control mice were gavaged with the corresponding broth alone. In both models, mice were monitored daily and those that showed extreme distress, became moribund, or lost more than 20% of initial body weight were euthanized. Animals were sacrificed after 4 or 8 weeks (H. pylori) and 14 days (C. rodentium). Stomachs and colons were harvested and analyzed as described (Chaturvedi et al., 2010; Gobert et al., 2018; Hardbower et al., 2016, 2017). The number of bacteria in gastric and colon tissues was determined by counting the CFUs after plating serial dilutions of homogenized tissues.
Histopathology
Histologic assessments were performed by a gastrointestinal pathologist (M.B.P.) in a blinded manner. At necropsy, stomach and colon segments were fixed in 10% neutral buffered formalin and stained with H&E. For H. pylori-infected mice, inflammation was assessed on a longitudinal strip using the following histopathological features: acute and chronic inflammation of the antrum and the corpus regions of the stomach (0–3 for each), leading to a final 0–12 score (Chaturvedi et al., 2010; Hardbower et al., 2016, 2017). For C. rodentium infection, the histologic colitis score (0–18) was the sum of acute and chronic inflammation scores (0–3 for each) multiplied by extent of inflammation (0–3), as reported (Chaturvedi et al., 2010; Gobert et al., 2018; Hardbower et al., 2016, 2017).
Immunostaining
Immunofluorescent staining for EIF5AHyp, NOS2, and the macrophage marker CD68 was performed on murine and human gastric tissues as described (Hardbower et al., 2016, 2017) using the following antibodies (see the Key Resources Table): Rabbit polyclonal anti-EIF5AHyp, 1/2000; rabbit polyclonal anti-NOS2, 1/1000; rabbit polyclonal anti-mouse CD68, 1/100; mouse monoclonal anti-Human CD68, ready to use; donkey anti-mouse IgG, Alexa fluor 555-labeled, 1/500; goat anti-rabbit IgG, Alexa fluor 488-labeled, 1/400; goat anti-rabbit IgG, Alexa fluor 555-labeled, 1/500. Quantification was performed by two blinded observers and the scoring was averaged.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-EIF5AHyp antibody | Nishiki et al., 2013 | N/A |
| Rabbit polyclonal anti-EIF5A antibody | Abcam | Cat#ab227537 |
| Rabbit polyclonal anti-DHPS | Abcam | Cat#ab202133 |
| Rabbit polyclonal anti-NOS2 | Millipore | Cat#ABN26; RRID: AB_10805939 |
| Rabbit monoclonal anti-IRG1 | Abcam | Cat#ab222411; RRID: AB_2868510 |
| Rabbit polyclonal anti-IRGM1 | Abcam | Cat# ab118569; RRID: AB_10901765 |
| Rabbit polyclonal anti-SQSTM | Proteintech | Cat#55274–1-AP; RRID: AB_11182278 |
| Rabbit polyclonal anti-mouse CD68 | Boster Biological | Cat#A1518 |
| Mouse monoclonal anti-human CD68 | Biocare Medical | Cat#PM033AA |
| Mouse monoclonal anti-β-actin | Sigma | Cat#A5316 |
| Goat anti-rabbit IgG, HRP-labeled | Jackson ImmunoResearch | Cat#111-035-003; RRID: AB_2313567 |
| Goat anti-mouse IgG, HRP-labeled | Jackson ImmunoResearch | Cat#115-035-003; RRID: AB_10015289 |
| Donkey anti-mouse IgG, Alexa fluor 555-labeled | Life Technologies | Cat#A-31570; RRID: AB_2536180 |
| Goat anti-rabbit IgG, Alexa fluor 488-labeled | Life Technologies | Cat#A-11008; RRID: AB_143165 |
| Goat anti-rabbit IgG, Alexa fluor 555-labeled | Life Technologies | Cat#A-21429; RRID: AB_2535850 |
| Bacterial and Virus Strains | ||
| Helicobacter pylori strain PMSS1 | Arnold et al., 2011 | N/A |
| Citrobacter rodentium strain DBS100 | Barthold et al., 1976 | N/A |
| Biological Samples | ||
| Human gastric biopsies | Universidad del Valle, Cali, Colombia | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Bay 11–7082 | Calbiochem | Cat#196870 |
| ERKi | Calbiochem | Cat#3328006 |
| LY294002 | Calbiochem | Cat#440206 |
| PD98059 | Calbiochem | Cat#513000 |
| SB203580 | Calbiochem | Cat#559395 |
| GC7 | Biosearch Technologies | Cat#G-1000 |
| Oligo dT | Invitrogen | Cat#18418020 |
| 4plus Streptavidin HRP Label | Biocare Medical | Cat#HP604H |
| Critical Commercial Assays | ||
| RNeasy Mini kit | QIAGEN | Cat#74106 |
| Superscript II Reverse Transcriptase | Invitrogen | Cat#18064022 |
| iQ™ SYBR Green kit | Bio-Rad | Cat#1708880 |
| BCA Protein Assay Kit | Pierce | Cat#23225 |
| Griess Reagent System | Promega | Cat#G2930 |
| Protease Inhibitor Cocktail Set III | Calbiochem | Cat#539134 |
| Phosphatase Inhibitor Cocktail Set I | Calbiochem | Cat#539131 |
| CYTO-ID® Autophagy Detection Kit | Enzo | Cat#ENZ-51031–0050 |
| Click-IT L-Homopropargylglycine | Invitrogen | Cat#C10186 |
| Biotin azide | Invitrogen | Cat#B10184 |
| Click-iT Protein Reaction Buffer Kit | Invitrogen | Cat#C10276 |
| PureProteome Protein A Magnetic Bead System | Millipore | Cat#LSKMAGA02 |
| Lookout Mycoplasma detection kit | Sigma-Aldrich | Cat#D9307 |
| Deposited Data | ||
| Proteomics dataset from BMmacs from Dhpsfl/fl and DhpsΔmye ± H. pylori or C. rodentium | This paper | ProteomeXchange Consortium (http://www.proteomexchange.org): PXD018187 |
| Proteomics dataset from RAW 264.7 cells ± H. pylori ± GC7 | This paper | ProteomeXchange Consortium (http://www.proteomexchange.org): PXD010082 |
| Metabolomics dataset | This paper | EMBL-EBI MetaboLights (https://www.ebi.ac.uk/metabolights/): MTBLS607 |
| Experimental Models: Cell Lines | ||
| Mus musculus RAW 264.7 cells | ATCC | Cat#TIB-71 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J Dhpsfl/fl | Levasseur et al., 2019 | N/A |
| C57BL/6J Lyz2cre/cre | The Jackson Laboratory | Cat#004781 |
| C57BL/6J Dhpsfl/fl;Lyz2cre/+ | This paper | N/A |
| C57BL/6J Dhpsfl/fl;Lyz2cre/cre | This paper | N/A |
| C57BL/6J Odcfl/fl | Hardbower et al., 2017 | N/A |
| C57BL/6J OdcΔmye | Hardbower et al., 2017 | N/A |
| Oligonucleotides | ||
| Murine Dhps (F) | This paper | CTTCCAGGCTACCAACTTCG |
| Murine Dhps (R) | This paper | GAGTCAGGTCTGCGTGATGA |
| Murine Dohh (F) | This paper | ACATGTGCAGGACCCTACCT |
| Murine Dohh (R) | This paper | GCACGGTATCGCTCAAAGA |
| Murine Nos2 (F) | This paper | CACCTTGGAGTTCACCCAGT |
| Murine Nos2 (R) | This paper | ACCACTCGTACTTGGGATGC |
| Murine Irg1 (F) | This paper | ATCTTGGACCTGGGGTCAG |
| Murine Irg1 (R) | This paper | TAAAGGCCACATCCTGCTG |
| Murine Irgm1 (F) | This paper | GACTCTGGCAATGGCATGT |
| Murine Irgm1 (R) | This paper | ACAGCACCACATTGGGAAA |
| Murine Sqstm1 (F) | This paper | aagaacgcgtgctgatacct |
| Murine Sqstm1 (R) | This paper | ttcctccttggctttgtctc |
| Murine Cxcl1 (F) | This paper | GCTGGGATTCACCTCAAGAA |
| Murine Cxcl1 (R) | This paper | CTTGGGGACACCTTTTAGCA |
| Murine Cxcl2 (F) | This paper | GCCAAGGGTTGACTTCA |
| Murine Cxcl2 (R) | This paper | TGTCTGGGCGCAGTG |
| Murine Il6 (F) | This paper | AGTTGCCTTCTTGGGACTGA |
| Murine Il6 (R) | This paper | TCCACGATTTCCCAGAGAAC |
| Murine Arg1 (F) | This paper | AAGAAAAGGCCGATTCACCT |
| Murine Arg1 (R) | This paper | CACCTCCTCTGCTGTCTTCC |
| Murine Il1b (F) | This paper | ACCTGCTGGTGTGTGACGTTCC |
| Murine Il1b (R) | This paper | GGGTCCGACAGCACGAGGCT |
| Murine Ifng (F) | This paper | GGCCATCAGCAACAACATAAGCgT |
| Murine Ifng (R) | This paper | TGGGTTGTTGACCTCAAACTTGGC |
| Murine Il17 (F) | This paper | ATCCCTCAAAGCTCAGCGTGTC |
| Murine Il17 (R) | This paper | GGGTCTTCATTGCGGTGGAGAG |
| Murine Il10 (F) | This paper | CCAAGCCTTATCGGAAATGA |
| Murine Il10 (R) | This paper | TCACTCTTCACCTGCTCCAC |
| Murine Actb (F) | This paper | CCAGAGCAAGAGAGGTATCC |
| Murine Actb (R) | This paper | CTGTGGTGGTGAAGCTGTAG |
| Recombinant DNA | ||
| N/A | N/A | N/A |
| Software and Algorithms | ||
| GraphPad Prism 8, version 8.4.3 | Scientific Software | https://www.graphpad.com/scientific-software/prism/ |
| Ingenuity Pathway Analysis | QIAGEN | https://digitalinsights.qiagen.com/?promo=qiagen-ipa |
| ImageJ 1.53a | National Institutes of Health | https://imagej.nih.gov |
Co-culture of myeloid cells and bacteria
We used BMmacs, BMDCs, and the murine macrophage cell line RAW 264.7 that was maintained in DMEM containing 10% FBS, 10 mM HEPES and 1 mM sodium pyruvate. Cells were stimulated with H. pylori at a multiplicity of infection of 10–100 for 6 or 24 h. For C. rodentium, macrophages were infected for 3 h with a multiplicity of infection of 1–10; then, cocultures were washed and a fresh media containing gentamycin, penicillin, and streptomycin was added to each well for 3 or 21 more h. In some experiments, cells were pre-treated with GC7 (Biosearch Technologies; 10 μM), Bay 11–7082 (Calbiochem; 5 μM), ERK inhibitor (Calbiochem; 20 μM), LY294002 (Calbiochem; 10 μM), PD98059 (Calbiochem; 10 μM), or SB203580 (Calbiochem; 2 μM) 1 h before infection. RAW264.7 cells tested negative for mycoplasma contamination using the Lookout Mycoplasma detection kit (Sigma-Aldrich).
To determine the survival of bacteria in macrophages, cocultures were washed thoroughly with PBS after 24 h, incubated 1 h with 200 μg/ml gentamicin, and lysed in 0.1% saponin for 30 min at 37°C (Gobert et al., 2014). The number of bacteria in each lysate was determined by counting the CFUs after plating serial dilutions on blood agar plates.
Analysis of mRNA levels
Total RNA was isolated using the RNeasy Mini kit (QIAGEN). Reverse transcription was performed using Superscript II Reverse Transcriptase and Oligo dT (Invitrogen). mRNAs were amplified by real-time PCR using the iQ™ SYBR Green kit (Bio-Rad) (Gobert et al., 2014) and the primers listed in Key Resources Table.
Proteomics and IPA
Proteomic analysis was performed as described (Gobert et al., 2019; Noto et al., 2019). After infections, macrophages were lysed in 50 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, and 2 mM EDTA and protein concentrations were determined by BCA. Protein samples were precipitated with ice-cold acetone overnight at −20°C. Precipitates were washed with cold acetone, dried, and reconstituted in 8 M urea in 250 mM TEAB (pH 8.0). Samples were reduced with 50 mM TCEP, alkylated with 200 mM MMTS, and diluted with TEAB to obtain a final solution containing 2 M urea, and digested with sequencing-grade trypsin overnight. Quantitative proteomics analysis was performed using iTRAQ. For 25 μg of protein, 1 unit of 4-plex iTRAQ labeling reagent (SCIEX) was used. Labeling reagent was reconstituted in ethanol such that each protein sample was labeled at a final concentration of 90% ethanol, and labeling was performed for 2 h. Four-plex iTRAQ experiments were conducted. The resulting labeled peptides were then desalted by a modified Stage-tip method. Following desalting, peptides were reconstituted in 0.1% formic acid, and peptides were loaded onto a self-packed biphasic C18/SCX MudPIT column using a helium-pressurized cell. Using a Dionex Ultimate 3000 nanoLC and autosampler, MudPIT analysis was performed with a 15-step salt pulse gradient. Following each salt pulse delivered by the autosampler, peptides were gradient-eluted from the reverse analytical column at a flow rate of 350 nl/min. Mobile phase solvents consisted of 0.1% formic acid, 99.9% water (solvent A) and 0.1% formic acid, 99.9% acetonitrile (solvent B). For the peptides from the first 13 SCX fractions, the reverse phase gradient consisted of 2%–50% B in 83 min, followed by a 10 min equilibration at 2% B. For the last 2 SCX-eluted peptide fractions, the peptides were eluted from the reverse phase analytical column using a gradient of 2%–98%B in 83 min, followed by a 10 min equilibration at 2% B. A Q Exactive Plus or HF mass spectrometer (Thermo Scientific), equipped with a nanoelectrospray ionization source, was used to mass analyze the eluting peptides. The Q Exactive instrument was operated in data-dependent mode acquiring HCD MS/MS after each MS1 scan on the 15 most abundant ions using an MS2 target of 105 ions. The HCD-normalized collision energy was set to 30, dynamic exclusion was set to 30 s, and peptide match and isotope exclusion were enabled.
Peptide/protein identifications and quantitative analysis were performed using Spectrum Mill (Agilent). MS/MS spectra were searched against a subset of the UniProt KB protein database containing Mus musculus and Helicobacter pylori 26695 proteins. Autovalidation procedures in Spectrum Mill were used to filter the data to < 1% false discovery rates at the protein and peptide level. Log2 protein ratios were fit to a normal distribution using non-linear (least-squares) regression. The calculated mean derived from the Gaussian fit was used to normalize individual log2 ratios for each quantified protein. The normalized log2 ratios were then fit to a normal distribution, and the mean and standard deviation values derived from the Gaussian fit of the normalized ratios were used to calculate p values. Subsequently, p values were corrected for multiple comparisons by the Benjamini-Hochberg method.
IPA software (QIAGEN) was used for the functional interpretation of differential expression results obtained from the aforementioned proteomic analyses. The pathways related to Diseases and Functions and the protein interaction networks were generated. The statistical significance for each assignment was expressed by a corresponding p values calculated using Fisher’s exact test.
Untargeted Metabolomics
Gastric tissues were homogenized by sonication in water:methanol (9:1) containing 50 mM ammonium acetate (pH~6) to yield a tissue density of 50 mg/ml. A portion of the homogenate (100 μL) was combined with HPLC-grade methanol (300 μL), vortexed vigorously, and centrifuged at 10,000 × g. An aliquot of the supernatant (100 μL) was diluted with an equal volume of HPLC-grade acetonitrile. Discovery metabolomics data were acquired using a Vanquish ultrahigh performance liquid chromatography (UHPLC) system interfaced to a Q Exactive HF quadrupole/orbitrap mass spectrometer (Thermo Fisher Scientific). Each sample was injected twice. The first injection was made in positive ESI mode; the second injection was made in negative mode. A Zic-cHILIC analytical column (3 μm, 2.1 × 150 mm; Merck SeQuant) was used for all chromatographic separations. Mobile phases were made up of 0.2% acetic acid and 15 mM ammonium acetate in (A) H2O:CH3CN (9:1) and in (B) CH3CN:CH3OH:H2O (90:5:5). The flow rate was maintained at 300 μl/min, the total chromatographic run time was 20 min, and the sample injection volume was 10 μl. Mass spectra were acquired over a precursor ion scan range of m/z 100 to 1,200 at a resolving power of 30,000 using the following ESI source parameters: spray voltage 5 kV (3 kV in negative mode); capillary temperature 300°C; S-lens RF level 60 V; N2 sheath gas 40; N2 auxiliary gas 10; auxiliary gas temperature 100°C. MS/MS spectra were acquired for the top-five most abundant precursor ions with an MS/MS AGC target of 105, a maximum MS/MS injection time of 100 ms, and a normalized collision energy of 30. Chromatographic alignment, peak picking, and statistical comparisons were performed using XCMS (https://xcmsonline.scripps.edu).
Western blot analysis
Macrophages were lysed using RIPA buffer containing the Protease Inhibitor Cocktail (Set III, Calbiochem) and the Phosphatase Inhibitor Cocktail (Set I, Calbiochem). Protein concentrations were determined using the BCA Protein Assay (Pierce). Western blotting was performed using 2 μg of protein per lane for all the blots, except for the EIF5A and EIF5AHyp blots for which 10 μg was used. The concentration of the primary and secondary Abs (see the Key Resources Table) were as follows: Rabbit polyclonal anti-DHPS, 1/2000; rabbit polyclonal anti-EIF5AHyp, 1/10000; rabbit monoclonal anti-EIF5A, 1/2000; rabbit polyclonal anti-NOS2, 1/2000; rabbit monoclonal anti-IRG1, 1/2000; rabbit polyclonal anti-IRGM1, 1/2000; rabbit polyclonal anti-SQSTM, 1/10000; mouse monoclonal anti-β-actin, 1/20000; goat anti-rabbit IgG, HRP-labeled, 1/5000; goat anti-mouse IgG, HRP-labeled, 1/5000. Densitometric analysis of western blots was performed with ImageJ 1.53a software.
NOS2 translation assay
BMmacs from Dhpsfl/fl and DhpΔmye mice were infected or not with H. pylori for 18 h. Cells were then washed with PBS and methionine-free, cysteine-free DMEM (GIBCO) was added to the cells for 2 h. This medium was then replaced by methionine-free, cysteine-free DMEM supplemented with 50 μM Click-IT homopropargylglycine (Invitrogen) for 4 h. Proteins were extracted using 1% SDS in 50 mM Tris-HCl containing the Protease Inhibitor Cocktail (Set III, Calbiochem) and the Phosphatase Inhibitor Cocktail (Set I, Calbiochem). Proteins (50 μg) were labeled with biotin azide (Invitrogen) using the Click Chemistry Reagents (Invitrogen), precipitated by methanol, resuspended in 50 mM Tris-HCl, and immunoprecipitated using a NOS2 Ab (Millipore) and the PureProteome Protein A Magnetic Bead System (Millipore). The resulting precipitates were used for western blotting using 4plus Streptavidin HRP Label (Biocare Medical).
Determination of autophagy
We determined autophagy in RAW 264.7 cells infected or not with H. pylori, in the presence or absence of GC7, by flow cytometry using the CYTO-ID® Autophagy Detection Kit (Enzo).
Measurement of NO2−
NO2− concentration was determined by the standard Griess assay (Promega)
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed with GraphPad Prism 8.4.3 and the significance level was set as p < 0.05. All the data shown represent the mean ± SEM. Data that were not normally distributed according to the D’Agostino & Pearson normality test were log or square root transformed. Student’s t test or ANOVA with the Tukey test were used to determine significant differences between two groups or to analyze significant differences among multiple test groups, respectively. All statistical tests were two-sided.
Supplementary Material
Highlights.
Pathogenic bacteria induce DHPS and hypusination of EIF5A in macrophages
Hypusinated EIF5A controls the translation of numerous antimicrobial effectors
Macrophage DHPS is required to clear gastrointestinal pathogens
ACKNOWLEDGMENTS
This work was supported by NIH grants R01CA190612, P01CA116087, P01CA028842, and R21AI142042 (K.T.W.), and R01 DK060581 (R.G.M.); Veterans Affairs Merit Review grants I01BX001453 and I01CX002171 (K.T.W.); Department of Defense grant W81XWH-18-1-0301 (K.T.W.); the Thomas F. Frist Sr. Endowment (K.T.W.); and the Vanderbilt Center for Mucosal Inflammation and Cancer (K.T.W.). Y.L.L. was supported by NIH grant T32AI138932. Proteomics analyses were supported in part by Core Scholarships from the Vanderbilt University Medical Center Digestive Disease Research Center funded by NIH grant P30DK058404 and the Vanderbilt Ingram Cancer Center support grant P30CA068485.
Footnotes
DECLARATION OF INTERESTS
K.T.W. serves as an associate editor for the journal Gastroenterology. He receives compensation for this work from the American Gastroenterological Association. This position is not related to this study.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108510.
REFERENCES
- Aksu M, Trakhanov S, and Görlich D (2016). Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A. Nat. Commun 7, 11952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zeer MA, Al-Younes HM, Braun PR, Zerrahn J, and Meyer TF (2009). IFN-gamma-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS ONE 4, e4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold IC, Lee JY, Amieva MR, Roers A, Flavell RA, Sparwasser T, and Müller A (2011). Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterology 140, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold SW, Coleman GL, Bhatt PN, Osbaldiston GW, and Jonas AM (1976). The etiology of transmissible murine colonic hyperplasia. Lab. Anim. Sci 26, 889–894. [PubMed] [Google Scholar]
- Basler T, Jeckstadt S, Valentin-Weigand P, and Goethe R (2006). Mycobacterium paratuberculosis, Mycobacterium smegmatis, and lipopolysaccharide induce different transcriptional and post-transcriptional regulation of the IRG1 gene in murine macrophages. J. Leukoc. Biol 79, 628–638. [DOI] [PubMed] [Google Scholar]
- Bussière FI, Chaturvedi R, Cheng Y, Gobert AP, Asim M, Blumberg DR, Xu H, Kim PY, Hacker A, Casero RA Jr., and Wilson KT (2005). Spermine causes loss of innate immune response to Helicobacter pylori by inhibition of inducible nitric-oxide synthase translation. J. Biol. Chem 280, 2409–2412. [DOI] [PubMed] [Google Scholar]
- Chaturvedi R, Asim M, Hoge S, Lewis ND, Singh K, Barry DP, de Sablet T, Piazuelo MB, Sarvaria AR, Cheng Y, et al. (2010). Polyamines impair immunity to Helicobacter pylori by inhibiting L-arginine uptake required for nitric oxide production. Gastroenterology 139, 1686–1698, 1698.e1–1698.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrandi D, Hoffmann R, Beuter-Gunia C, and Pfeffer K (2009). The proinflammatory cytokine-induced IRG1 protein associates with mitochondria. J. Interferon Cytokine Res 29, 55–67. [DOI] [PubMed] [Google Scholar]
- Duleu S, Vincendeau P, Courtois P, Semballa S, Lagroye I, Daulouède S, Boucher JL, Wilson KT, Veyret B, and Gobert AP (2004). Mouse strain susceptibility to trypanosome infection: an arginase-dependent effect. J. Immunol 172, 6298–6303. [DOI] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. (2009). Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol 11, 1305–1314. [DOI] [PubMed] [Google Scholar]
- Eisenberg T, Abdellatif M, Schroeder S, Primessnig U, Stekovic S, Pendl T, Harger A, Schipke J, Zimmermann A, Schmidt A, et al. (2016). Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med 22, 1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kasmi KC, Qualls JE, Pesce JT, Smith AM, Thompson RW, Henao-Tamayo M, Basaraba RJ, König T, Schleicher U, Koo MS, et al. (2008). Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat. Immunol 9, 1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Choi S, Wyse M, Strnadel J, Wright T, and Klemke R (2015). Eukaryotic translation initiation factor 5A (EIF5A) regulates pancreatic cancer metastasis by modulating RhoA and Rho-associated kinase (ROCK) protein expression levels. J. Biol. Chem 290, 29907–29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathi M, Padgett LR, Yamada K, Devinsky O, Willaert R, Person R, Au PB, Tagoe J, McDonald M, Karlowicz D, et al. (2019). Recessive rare variants in deoxyhypusine synthase, an enzyme involved in the synthesis of hypusine, are associated with a neurodevelopmental disorder. Am. J. Hum. Genet 104, 287–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, Cheng Y, Mobley HL, and Wilson KT (2001). Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. USA 98, 13844–13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert AP, Verriere T, Asim M, Barry DP, Piazuelo MB, de Sablet T, Delgado AG, Bravo LE, Correa P, Peek RM Jr., et al. (2014). Heme oxygenase-1 dysregulates macrophage polarization and the immune response to Helicobacter pylori. J. Immunol 193, 3013–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert AP, Al-Greene NT, Singh K, Coburn LA, Sierra JC, Verriere TG, Luis PB, Schneider C, Asim M, Allaman MM, et al. (2018). Distinct immunomodulatory effects of spermine oxidase in colitis induced by epithelial injury or infection. Front. Immunol 9, 1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert AP, Latour YL, Asim M, Finley JL, Verriere TG, Barry DP, Milne GL, Luis PB, Schneider C, Rivera ES, et al. (2019). Bacterial pathogens hijack the innate immune response by activation of the reverse transsulfuration pathway. MBio 10, e02174–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, and Dever TE (2013). eIF5A promotes translation of polyproline motifs. Mol. Cell 51, 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardbower DM, Singh K, Asim M, Verriere TG, Olivares-Villagómez D, Barry DP, Allaman MM, Washington MK, Peek RM Jr., Piazuelo MB, and Wilson KT (2016). EGFR regulates macrophage activation and function in bacterial infection. J. Clin. Invest 126, 3296–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardbower DM, Asim M, Luis PB, Singh K, Barry DP, Yang C, Steeves MA, Cleveland JL, Schneider C, Piazuelo MB, et al. (2017). Ornithine decarboxylase regulates M1 macrophage activation and mucosal inflammation via histone modifications. Proc. Natl. Acad. Sci. USA 114, E751–E760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins ZA, Hååg PG, and Johansson HE (2001). Human eIF5A2 on chromosome 3q25-q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics 71, 101–109. [DOI] [PubMed] [Google Scholar]
- Joe YA, Wolff EC, and Park MH (1995). Cloning and expression of human deoxyhypusine synthase cDNA. Structure-function studies with the recombinant enzyme and mutant proteins. J. Biol. Chem 270, 22386–22392. [DOI] [PubMed] [Google Scholar]
- Latour YL, Gobert AP, and Wilson KT (2020). The role of polyamines in the regulation of macrophage polarization and function. Amino Acids 52, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Starkey PM, and Gordon S (1985). Quantitative analysis of total macrophage content in adult mouse tissues. Immunochemical studies with monoclonal antibody F4/80. J. Exp. Med 161, 475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WB, Kang JS, Choi WY, Zhang Q, Kim CH, Choi UY, Kim-Ha J, and Kim YJ (2016). Mincle-mediated translational regulation is required for strong nitric oxide production and inflammation resolution. Nat. Commun 7, 11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur EM, Yamada K, Piñeros AR, Wu W, Syed F, Orr KS, Anderson-Baucum E, Mastracci TL, Maier B, Mosley AL, et al. (2019). Hypusine biosynthesis in β cells links polyamine metabolism to facultative cellular proliferation to maintain glucose homeostasis. Sci. Signal 12, eaax0715.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J, Xie QW, and Nathan C (1997). Nitric oxide and macrophage function. Annu. Rev. Immunol 15, 323–350. [DOI] [PubMed] [Google Scholar]
- MacMicking JD, Taylor GA, and McKinney JD (2003). Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science 302, 654–659. [DOI] [PubMed] [Google Scholar]
- Madeo F, Eisenberg T, Pietrocola F, and Kroemer G (2018). Spermidine in health and disease. Science 359, eaan2788.. [DOI] [PubMed] [Google Scholar]
- Maier B, Ogihara T, Trace AP, Tersey SA, Robbins RD, Chakrabarti SK, Nunemaker CS, Stull ND, Taylor CA, Thompson JE, et al. (2010). The unique hypusine modification of eIF5A promotes islet beta cell inflammation and dysfunction in mice. J. Clin. Invest 120, 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal A, Mandal S, and Park MH (2016). Global quantitative proteomics reveal up-regulation of endoplasmic reticulum stress response proteins upon depletion of eIF5A in HeLa cells. Sci. Rep 6, 25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mémin E, Hoque M, Jain MR, Heller DS, Li H, Cracchiolo B, Hanauske-Abel HM, Pe’ery T, and Mathews MB (2014). Blocking eIF5A modification in cervical cancer cells alters the expression of cancer-related genes and suppresses cell proliferation. Cancer Res. 74, 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mera RM, Bravo LE, Camargo MC, Bravo JC, Delgado AG, Romero-Gallo J, Yepez MC, Realpe JL, Schneider BG, Morgan DR, et al. (2018). Dynamics of Helicobacter pylori infection as a determinant of progression of gastric precancerous lesions: 16-year follow-up of an eradication trial. Gut 67, 1239–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelucci A, Cordes T, Ghelfi J, Pailot A, Reiling N, Goldmann O, Binz T, Wegner A, Tallam A, Rausell A, et al. (2013). Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA 110, 7820–7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli E, Mariño G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Bénit P, Rustin P, Criollo A, et al. (2011). Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol 192, 615–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, and Cleveland JL (2016). Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids 48, 2353–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, et al. (2005). CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307, 254–258. [DOI] [PubMed] [Google Scholar]
- Nishiki Y, Farb TB, Friedrich J, Bokvist K, Mirmira RG, and Maier B (2013). Characterization of a novel polyclonal anti-hypusine antibody. Spring-erplus 2, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Murozumi K, Shirahata A, Park MH, Kashiwagi K, and Igarashi K (2005). Independent roles of eIF5A and polyamines in cell proliferation. Biochem. J 385, 779–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto JM, Rose KL, Hachey AJ, Delgado AG, Romero-Gallo J, Wroblewski LE, Schneider BG, Shah SC, Cover TL, Wilson KT, et al. (2019). Carcinogenic Helicobacter pylori strains selectively dysregulate the in vivo gastric proteome, which may be associated with stomach cancer progression. Mol. Cell. Proteomics 18, 352–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahari S, Negi S, Aqdas M, Arnett E, Schlesinger LS, and Agrewala JN (2019). Induction of autophagy through CLEC4E in combination with TLR4: an innovative strategy to restrict the survival of Mycobacterium tuberculosis. Autophagy 16, 1021–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, and Wolff EC (2018). Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J. Biol. Chem 293, 18710–18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Cooper HL, and Folk JE (1981). Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. USA 78, 2869–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Lee YB, and Joe YA (1997). Hypusine is essential for eukaryotic cell proliferation. Biol. Signals 6, 115–123. [DOI] [PubMed] [Google Scholar]
- Park JH, Aravind L, Wolff EC, Kaevel J, Kim YS, and Park MH (2006). Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc. Natl. Acad. Sci. USA 103, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, and Sibley RK (1991). Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med 325, 1127–1131. [DOI] [PubMed] [Google Scholar]
- Peek RM Jr., Fiske C, and Wilson KT (2010). Role of innate immunity in Helicobacter pylori-induced gastric malignancy. Physiol. Rev 90, 831–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE (2009). Mammalian polyamine metabolism and function. IUBMB Life 61, 880–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE (2016). Functions of polyamines in mammals. J. Biol. Chem 291, 14904–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, and Alepuz P (2017). eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res. 45, 7326–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JV, Russo D, Ji DX, Chavez RA, DiPeso L, Lee AY, Coers J, and Vance RE (2019). IRG1 and inducible nitric oxide synthase act redundantly with other interferon-gamma-induced factors to restrict intracellular replication of Legionella pneumophila. MBio 10, e02629–e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston DJ, Buck MD, Klein Geltink RI, Kyle RL, Caputa G, O’Sullivan D, Cameron AM, Castoldi A, Musa Y, Kabat AM, et al. (2019). Polyamines and eIF5A hypusination modulate mitochondrial respiration and macrophage activation. Cell Metab. 30, 352–363.e358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Qiu Q, Gu X, Tian Y, and Zhang Y (2016). ATM mediates spermi-dine-induced mitophagy via PINK1 and Parkin regulation in human fibroblasts. Sci. Rep 6, 24700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualls JE, Subramanian C, Rafi W, Smith AM, Balouzian L, DeFreitas AA, Shirey KA, Reutterer B, Kernbauer E, Stockinger S, et al. (2012). Sustained generation of nitric oxide and control of mycobacterial infection requires argininosuccinate synthase 1. Cell Host Microbe 12, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Wu CC, Dever TE, Buskirk AR, and Green R (2017). eIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 66, 194–205.e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, Yoon S, Krasnitz A, Teruya-Feldstein J, Pappin D, et al. (2012). A tumour suppressor network relying on the polyamine-hypusine axis. Nature 487, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra JC, Asim M, Verriere TG, Piazuelo MB, Suarez G, Romero-Gallo J, Delgado AG, Wroblewski LE, Barry DP, Peek RM Jr., et al. (2018). Epidermal growth factor receptor inhibition downregulates Helicobacter pylori-induced epithelial inflammatory responses, DNA damage and gastric carcinogenesis. Gut 67, 1247–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert H, Pällmann N, Miller KK, Hermans-Borgmeyer I, Venz S, Sendoel A, Preukschas M, Schweizer M, Boettcher S, Janiesch PC, et al. (2014). A novel mouse model for inhibition of DOHH-mediated hypusine modification reveals a crucial function in embryonic development, proliferation and oncogenic transformation. Dis. Model. Mech 7, 963–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelko CL, Lu W, Dufort FJ, Seyfried TN, Chiles TC, Rabinowitz JD, and Roberts MF (2011). Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc 133, 16386–16389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, and Cox JS (2012). Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150, 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerås C, Ave P, Huerre M, Arondel J, Ulevitch RJ, Mathison JC, and Sansonetti P (2000). Blockade of CD14 increases Shigella-mediated invasion and tissue destruction. J. Immunol 164, 3214–3221. [DOI] [PubMed] [Google Scholar]
- Xu A, and Chen KY (2001). Hypusine is required for a sequence-specific interaction of eukaryotic initiation factor 5A with postsystematic evolution of ligands by exponential enrichment RNA. J. Biol. Chem 276, 2555–2561. [DOI] [PubMed] [Google Scholar]
- Xu A, Jao DL, and Chen KY (2004). Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem. J 384, 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, and Simon AK (2020). Polyamines reverse immune senescence via the translational control of autophagy. Autophagy 16, 181–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data regarding BMmacs (Tables S1 and S2) and RAW 264.7 cells (Table S4) have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Perez-Riverol et al., 2019), and the accession numbers are ProteomeXchange Consortium: PXD018187 and ProteomeXchange Consortium: PXD010082, respectively. The accession number for the metabolomics data reported in this paper is EMBL-EBI MetaboLights: MTBLS607.


