Abstract
Escherichia coli Nissle 1917 (EcN) is a probiotic bacterium, commonly employed to treat certain gastrointestinal disorders. It is fast emerging as an important target for the development of therapeutic engineered bacteria, benefiting from the wealth of knowledge of E. coli biology and ease of manipulation. Bacterial synthetic biology projects commonly utilize engineered plasmid vectors, which are simple to engineer and can reliably achieve high levels of protein expression. However, plasmids typically require antibiotics for maintenance, and the administration of an antibiotic is often incompatible with in vivo experimentation or treatment. EcN natively contains plasmids pMUT1 and pMUT2, which have no known function but are stable within the bacteria. Here, we describe the development of the pMUT plasmids into a robust platform for engineering EcN for in vivo experimentation, alongside a CRISPR-Cas9 system to remove the native plasmids. We systematically engineered both pMUT plasmids to contain selection markers, fluorescent markers, temperature sensitive expression, and curli secretion systems to export a customizable functional material into the extracellular space. We then demonstrate that the engineered plasmids were maintained in bacteria as the engineered bacteria pass through the mouse GI tract without selection, and that the secretion system remains functional, exporting functionalized curli proteins into the gut. Our plasmid system presents a platform for the rapid development of therapeutic EcN bacteria.
Keywords: synthetic biology, microbiome, probiotic engineering
Escherichia coli Nissle 1917 (EcN) is a probiotic bacterium originally isolated from a particularly healthy soldier from World War I by the physician Alfred Nissle.1 Since then, this bacterium has found significant use as a probiotic therapy, outcompeting pathogens in the gut2 and thus protecting the host from infection. EcN has been at the forefront of probiotic genetic engineering,3 benefiting from the well-understood nature of E. coli biology, and from the many tools available to manipulate this organism. There are many projects working with engineered EcN,3 developing engineered therapeutic bacteria to tackle diseases in the gut like hyperammonemia,4 as well as outside the gut, such as for cancer detection and treatment.5,6
In recent years, the gut microbiome has emerged as a critical factor for human health;7 however, the gut ecosystem remains a poorly understood system. One important approach to probe the gut microbiome is the development of engineered microbes that can sense and report on the conditions in the gut,8 as well as deliver therapeutic molecules into the gut environment.9 Additionally, synthetic systems can provide insight into the behavior of engineered bacteria in the gut environment,10 aiding further engineering efforts. As such, genetic tools that simplify bacterial engineering both facilitate the study of gut health, and accelerate the development of sophisticated probiotic bacteria capable of sensing and treating gut disorders.
Synthetic biology projects typically utilize plasmid vectors, circular extrachromosomal DNA elements that can replicate within cells independently of the genome. Plasmids have many benefits: they are simple to manipulate, can be reliably transformed into E. coli cells, and can achieve high levels of gene expression due, in part, to a higher copy number than genomic DNA. Furthermore, several plasmids can be used in concert, allowing for modular assembly of complex synthetic genetic systems, as well as the simple independent testing of each plasmid in the system. An integral part of developing synthetic genetic systems is the iteration of prototypes in a design-build-test cycle,11 where during each cycle variants are tested to inform successive design iterations. Rapid and reliable genetic circuit construction and implementation is key for developing synthetic genetic systems, and plasmids offer an essential tool for this process. However, plasmid vectors also present a serious experimental limitation by requiring an antibiotic for selection and plasmid maintenance. In the context of in vivo therapeutic use in the gut, administration of an antibiotic is often incompatible with treatment and severely limiting to experiments as it induces drastic changes in the host microbiome.12
Synthetic plasmids have been employed to engineer bacteria for in vivo use; however, without antibiotic selection, significant plasmid loss has been observed.13 Several plasmid maintenance strategies have been developed,14 including toxin-antitoxin systems,15 microcin mediated postsegregational killing,16 and auxotrophy.17 However, such methods would require significant effort to optimize, and may themselves be burdensome to any engineered genetic system. Given the limitations of plasmids, EcN engineering projects that require stable transformants often insert DNA directly into the chromosome. However, genomic manipulations are typically limited by poor transformation efficiencies in EcN, and involve time-consuming and cumbersome protocols, impeding the iteration of genetic circuit designs. Furthermore, common genomic incorporation protocols such as Lambda Red based methods can be inefficient and have limitations on insert length,18,19 further slowing or outright preventing the development of large multicomponent synthetic genetic systems. Additionally, genomic incorporation limits recombinant DNA copy number to genomic copy number, making the achievement of high gene expression rates more difficult. Given the importance of rapid prototyping for the development of synthetic genetic systems, new paradigms are required to host synthetic DNA to facilitate the engineering of probiotic organisms.
Bacteria isolated from clinical samples often contain plasmids, including small cryptic plasmids that are maintained at high copy number despite containing little genetic information and conferring no apparent phenotype.20 Many of these plasmids have no known function, although one study linked the presence of such small cryptic plasmids to phage resistance.21 EcN contains two such cryptic plasmids, pMUT1 and pMUT2, which are stable within the bacteria, survive passage through the gut, and are used as targets to detect EcN in clinical PCR assays.22 The pMUT plasmids do not confer any detectable phenotype, are not essential to EcN and do little to affect growth.23 Furthermore, the pMUT plasmids do not present a metabolic burden to EcN, at least under laboratory conditions.24 While several projects have used pMUT plasmids to carry synthetic circuits,3 no systematic engineering attempt has been made to domesticate and characterize the efficacy of engineered pMUT plasmids in vivo.
In this work, we describe the systematic engineering of the E. coli Nissle 1917 cryptic plasmids pMUT1 and pMUT2 to create a series of plasmid vectors for use in the gut. We tested several sites on each plasmid to insert recombinant DNA cassettes containing selection and fluorescent markers, and characterized the gene expression in each case. We found that the native plasmids were not lost through transformation of an engineered variant, so we also developed a technique to remove the native plasmids through a CRISPR-Cas9 mechanism. We then added further functionality to these plasmid vectors: adapting and expanding a temperature sensitive expression system, as well as curli-based protein secretion to export proteins into the extracellular space. We then tested their use in vivo, finding that EcN retained the engineered pMUT plasmids during passage through the mouse GI tract, and that the plasmids were capable of secreting recombinant protein into the extracellular space of the gut.
Results
Revision of the pMUT Cryptic Plasmid Sequences
EcN’s cryptic plasmids were first documented by Hacker et al. in 2002,25 who published the sequences for pMUT1 and pMUT2 on the National Center for Biotechnology Information (NCBI) database with accession numbers A84793 and A95448, respectively. Since then, 3 whole genome-sequencing projects for EcN have been uploaded to NCBI, with 2 fully assembled genomes. The first assembly, ASM71459v1 (Reister et al.(26)), resulted in a single sequence containing the chromosome and both plasmids, with the plasmids erroneously inserted multiple times within the chromosomal sequence. A later assembly, ASM354697v1, has a genomic sequence separate from the 2 cryptic plasmid sequences (labeled pNissle1 and pMUT2). Here, the pNissle1 sequence contains both the sequence for pMUT1 and pMUT2 and is likely also an erroneous assembly. Also, the pMUT sequences from the whole genome sequencing projects differed from those originally uploaded, A84793 and A95448, which were sequenced using Sanger sequencing. As such, we could not find a correct pMUT1 sequence on NCBI and thus uploaded one for reference, NCBI accession number MW240712, and we refer to NCBI accession CP023342 for the correct pMUT2 sequence. We confirmed the pMUT sequences by Sanger sequencing the backbones of the pMUT-derived engineered vectors, finding the sequence traces aligned exactly with those derived from the whole genome sequencing efforts.
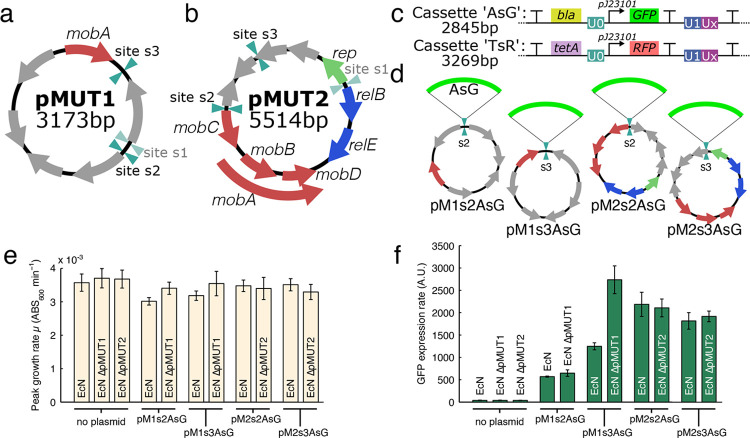
Figure 1a shows the plasmid maps and lengths of pMUT1 and pMUT2, and Figure S1 shows the annotations as determined by the NCBI automated prokaryotic annotation pipeline27 in greater detail. pMUT1 has a typical ColE1 origin of replication.22 pMUT2 is 96.9% homologous to the pUB6060 plasmid from Plesiomonas shigelloides, which has been described as having a ColE2-like replication and ColE1-like mobilization loci.28 Both plasmids contain mob genes involved in plasmid transfer; however, both plasmids lack the full gene complement necessary for conjugation, and have previously been described as nontransferable.29 Most of the putative proteins found on the plasmids have no known functions (Figure S1), except for the relB-relE toxin-antitoxin system on pMUT2.30 Toxin-antitoxin systems, often found on plasmids, are known to promote plasmid maintenance,15 so it is likely that these genes contribute to pMUT2 stability in EcN.
Figure 1.
Plasmid maps of the native cryptic pMUT1 (a) and pMUT2 (b) plasmids in EcN with known genes labeled; also shown are the sites where we inserted recombinant cassettes, with unsuccessful sites greyed out. (c) Insulated characterization cassettes inserted onto the pMUT plasmids to produce engineered pMUT vectors, with cassette “AsG” containing an ampicillin resistance gene and constitutively expressed GFP, and “TsR” containing tetracycline resistance and constitutive RFP (characterized in Figure S2). (d) For site characterization, we inserted the “AsG” cassette onto the 2 sites on each pMUT plasmid, and characterized both the (e) bacterial growth rate and (f) GFP expression rates. In each case, we characterized the performance of the engineered plasmids in both an unmodified EcN strain, and an EcN strain where the relevant native pMUT plasmid had been removed.
Engineering the pMUT Plasmids
We began pMUT engineering by selecting 3 sites, s1–s3, (Figure 1a,b) on each plasmid to insert an insulated cassette encoding antibiotic resistance and a fluorescent protein. The selected sites did not contain any known proteins and were away from the origin of replication in order to avoid disrupting any native functions. We kept the entire cryptic plasmid sequences as vector backbones, in order to maintain any features that may contribute to the stability of the plasmids.
We amplified the pMUT plasmids by PCR with primers (Table S1) to act as the vector backbone onto which the cassette was inserted. For both pMUT1 and pMUT2, we attempted cassette insertion on 3 sites on the plasmid, but in both cases we could not assemble a plasmid for site 1 (s1) as we could not amplify the backbone by PCR despite trying two primer pair variants.
We tested the 2 successful insertion cassettes for gene expression (Figure 2c): “AsG” which contained an ampicillin resistance gene (bla) and constitutively expressed superfolder GFP; and “TsR”, which contained a tetracycline resistance gene (tetA) and a constitutively expressed mCherry RFP. Both of these transcriptional elements were flanked by terminators to insulate the insertion cassette from transcription on the plasmid backbone and vice versa. Furthermore, we added Universal Nucleotide Sequences (UNS) from Torella et al.(31) to the cassettes in order to allow for the rapid assembly of modular genetic elements. The UNS are 40 bp genetic segments that act as spacer elements without significant DNA structure or function. UNS flanked each functional module we created, and are labeled as “U#” in genetic circuit diagrams. The UNS information can be found in Table S2.
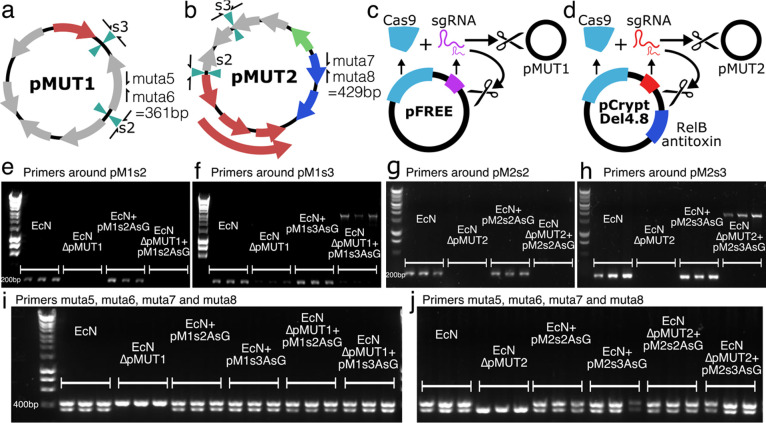
Figure 2.
Curing native pMUT plasmids. EcN pMUT plasmids were assessed with primers around the insertion sites and primers muta5 and muta6 on pMUT1 (a), and muta7 and muta8 on pMUT2 (b). (c) Plasmid pFREE cleaves pMUT1 through expression of Cas9 and gRNA targeting the colE1 origin of replication on pMUT1 and on pFREE itself. (d) Similarly, pCryptDel4.8 targets the origin of pMUT2 and itself, and also contains a RelB antitoxin to disrupt the RelE-RelB toxin-antitoxin system on pMUT2. (e−h) Agarose gels showing the results of colony PCRs around the insertion sites of the “AsG” cassette, revealing that transformation with an engineered plasmid does not displace the native plasmid. (i,j) pFREE and pCryptDel4.8 can cure EcN of native plasmids, and these can be replaced with engineered versions. In all agarose gel images, each condition is shown in triplicate, each lane representing a PCR result using a distinct bacterial colony.
We cloned the inserts “AsG” and “TsR” into the sites on either pMUT1 or pMUT2 to obtain plasmids pMXsYAsG and pMXsYTsR, where X is either 1 or 2, referring to pMUT1 or pMUT2, and where Y is the insertion site number (Figure 1d). We tested the gene expression from each insertion site by measuring the cell density and GFP fluorescence in a kinetic run for each insertion site with plasmids pMXsYAsG. We performed these assays with the engineered pMUT plasmid transformed into either an unmodified EcN, or EcN where the corresponding native pMUT had been knocked out.
As reported before,23 EcN pMUT plasmid knockouts did not grow differently under lab conditions (Figure 1e). Growth rates were broadly similar in all cases with the engineered plasmids (Figure 1e), although in the pMUT1 derived vectors, the presence of the native pMUT1 reduced growth slightly (p < 0.001 in both cases). We found that the insertion sites influenced GFP expression levels, with site 3 on pMUT1 and site 2 on pMUT2 giving the highest GFP expression levels (Figure 1f). The expression rates from pM1s3AsG further showed that the presence of the native plasmid altered gene expression from the engineered plasmid significantly. Since we would like our engineered vectors to be capable of high levels of gene expression, the high performing sites pM1s3 and pM2s2 were selected for further use. We also characterized RFP expression from pM1s3TsR and pM2s2TsR, finding a similar ratio of gene expression strengths to the GFP data (Figure S2), suggesting that their relative rate of gene expression is independent of the protein expressed.
We found that transforming with the engineered plasmids did not displace the native plasmids. We tested for pMUT plasmids with DNA primers muta5, muta6, muta7, and muta8 (Table S1), developed by Blum-Oehler et al.(22) to detect pMUT1 and pMUT2 in clinical samples. In a multiplexed PCR with these 4 muta primers, a 361 bp product is formed when pMUT1 is present, and a 429 bp product when pMUT2 is present (Figure 2a,b). Furthermore, we designed primers around the insertion sites on pMUT1 and pMUT2 to distinguish the native and engineered pMUT plasmids. We expected to find colonies in which the native pMUTs were knocked out through plasmid incompatibility—a process whereby two plasmids cannot stably coexist in the same bacterial cell line over multiple generations, typically occurring in plasmids containing similar or identical replication mechanisms. However, when unmodified EcN was transformed with an ampicillin resistant engineered pMUT plasmid (pM1s3AsG or pM2s2AsG), and grown on selective media, colony PCR with primers around the insertion sites (primers pMXsY_chk_F and R) produced a short 200 bp product, indicating the presence of native plasmid (Figure 2e–h). This was despite the fact that although the native and engineered plasmids had identical origins, only the engineered plasmids conferred antibiotic resistance. Since our data indicated that unmodified pMUT plasmid can impact the performance of the engineered plasmids, we developed a technique to rapidly remove the native plasmids prior to transformation with engineered pMUTs.
Curing the Native pMUT Plasmids
Since transforming EcN with engineered pMUTs did not displace the native plasmids, we required a strategy to remove them. While pMUT curing strategies exist, they rely on plasmid incompatibility to knock out native plasmids,23 which our data indicates is not an immediate process and requires multiple weeks of streaking onto selective media, based on our experience. We therefore used a CRISPR-Cas9 strategy to cleave both native pMUT plasmids, based on the pFREE system of Lauritsen et al.(32) The pFREE plasmid (Figure S3a) contains a Cas9 protein and 4 guide RNAs (gRNAs) that target two sites on two common origins of replication, ColE1 and pSC101, in order to rapidly cleave and cure plasmids with those origins. The pFREE plasmid also harbors a variant of the ColE1 origin, and therefore also cures itself during this process.
We found that the pFREE plasmid cured pMUT1 (Figure 2c) with a success rate of around 60% (43 out of 73 colonies tested), but unsurprisingly did not cure pMUT2 as it does not have a ColE1 origin. To cure pMUT2, we redesigned guide RNAs to target locations on pMUT2 (Table S3) to make the pCryptDel range of plasmids. We tested 3 gRNA pairs designed to cure pMUT2, however, all designs failed until we included the antitoxin gene relE found on pMUT2 onto the plasmids (Figure 2d). At each iteration of the design, we tested many pCryptDel variants for their ability to remove pMUT2, and identified one, labeled pCryptDel4.8, that cured EcN of pMUT2. Upon sequencing, we found that this plasmid targeted two sites on pMUT2 for Cas9-mediated cleavage; however, it also contained an insertion mutation that altered one of the gRNAs targeting the ColE1 origin (Figure S3). Variants without this mutation could cure pMUT1, but did not cure pMUT2 in any of the colonies tested. pCryptDel4.8 self-cured in all colonies tested, and cured pMUT2 with an efficiency of around 21% (24 out of 117 colonies tested); however, it was very poor at curing pMUT1 (3 out of 97 colonies tested). In all cases of pMUT plasmid curing we tested, the pFREE and pCryptDel4.8 plasmids self-cured during the process. pFREE and pCryptDel4.8 can therefore be used to rapidly produce pMUT plasmid knockout strains through a single overnight growth step. Furthermore, the pFREE and pCryptDel plasmids can be used consecutively to create fully pMUT plasmid free EcN (Figure S4).
Incorporating Genetic Modules for in Vivo Use: Temperature Sensing
The goal of our work was to make plasmid vectors for bacterial protein secretion in the gut. As such, there are several experimental challenges to both controlling and assessing synthetic genetic systems within the bacteria. Since the bacteria are in the gut of the host organism, they cannot be readily interrogated, and due to the complex environment of the gut, it is unlikely that bacteria behave as they do under laboratory conditions. This sets a severe limitation on genetic induction systems that require exogenous chemical inducers in the gut due to the difficulty of supplying a steady inducer concentration. Inducers are normally provided in a concentrated form in the water for the animal, so the effective concentration in the gut is not clear.
However, inducible systems are desirable to simplify cloning and in vitro propagation of DNA and bacterial strains, especially for genes that encode products that are toxic or stress-inducing to the bacteria. Synthetic genetic circuits typically require the bacteria to express heterologous proteins, and these can impose significant metabolic burdens on their host.33 For constitutive high-expressing constructs, given a nonzero mutation rate, any defective mutants that relieve the metabolic burden will quickly come to dominate cultures due to faster growth. Therefore, for in vitro cloning and propagation it is desirable to use inducible systems to create an “off” state where the synthetic system does not significantly reduce fitness during culture propagation. Additionally, the uninduced state provides a further internal control in experiments that can provide valuable insight into the performance of the genetic system.
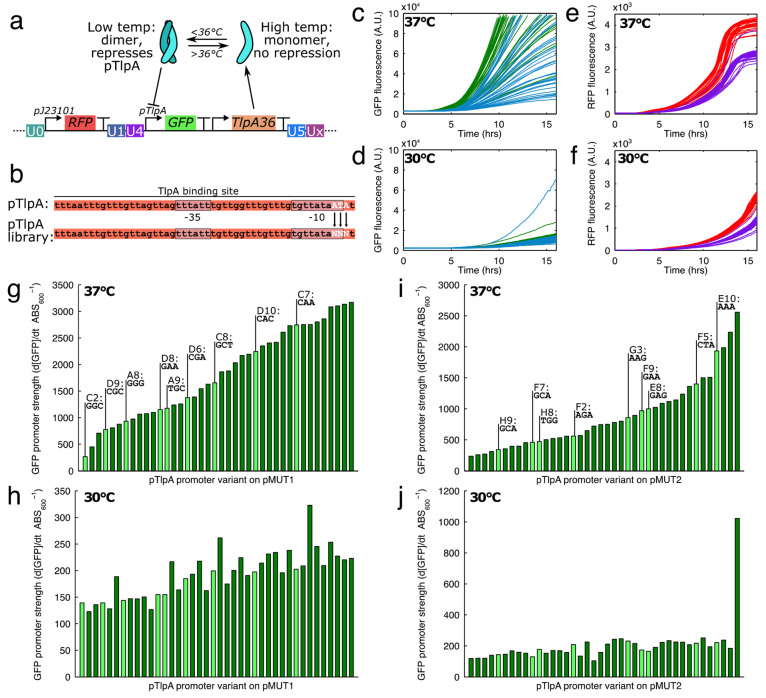
We implemented a temperature-sensitive gene expression system from Piraner et al.,34 based on the promoter pTlpA and repressor protein TlpA36. TlpA36 forms a dimer at temperatures below 36 °C, and this dimer binds pTlpA and prevents gene expression (Figure 3a). At temperatures above 36 °C, the repressor dimer is unstable and does not prevent gene expression. As such, this is an ideal system to provide constitutive high gene expression in vivo, as both human and mouse body temperatures are around 37 °C, while in vitro the bacteria can be grown at 30 °C. To characterize the temperature dependent gene expression, we designed a synthetic ratiometric construct containing pTlpA driving sfGFP, and the constitutive pJ23101 (BioBricks registry) driving mCherry, an RFP (Figure 3a).
Figure 3.
(a) Temperature sensitive expression was achieved with the TlpA36 protein, which dimerizes, binds, and represses the pTlpA promoter at temperatures below 36 °C. (b) To make a library with various promoter strengths, the pTlpA promoter was modified to contain 3 variable nucleotides near the −10 region of the promoter. Variable GFP expression strengths at 37 °C (c) and 30 °C (d) from the pTlpA library, with green curves showing 40 pM1s3AsR_TS* variants, and blue showing 40 pM2s2AsR_TS* variants. (e,f) In contrast, RFP was expressed by a constitutive J23101 promoter and RFP expression rates were not as variable as for GFP. When promoter strengths were quantified at 37 °C for the pMUT1 (g) and pMUT2 (i) engineered vectors, we found a range of strengths, and 9 promoters throughout the range were chosen for sequencing and further development (highlighted in lighter green). (h–j) In general, gene expression from the pTlpA* promoters was reduced at 30 °C by at least 10-fold.
Above a certain critical temperature, the pTlpA promoter is active and acts constitutively. We mutated the promoter to generate a library with varied expression strengths. The promoter variant could then be selected for a transcriptional unit of interest in order to optimize gene expression. TlpA, from which TlpA36 was derived, binds to the entire pTlpA promoter35 (Figure 3b), so to minimally disrupt the repressor-promoter interaction we limited our mutations to the edge of the promoter region. We found the −35 and −10 regions of the promoter with the BPROM software,36 and designed the 3 mutations in the −10 region of the pTlpA promoter, a region important to RNA polymerase binding and transcription strength. We therefore designed DNA oligos containing 3 freely varying nucleotides, and used them to assemble the “sR_TS” (i.e., “RFP, temperature sensitive”) circuit shown in Figure 3a on both the pM1s3A and pM2s2A backbones. The assembled plasmids were then transformed into an E. coli cloning strain, Mach1, for an initial screening and characterization. Transformants were initially screened for GFP on LB agar plates grown at 37 °C to select variants with a range of expression strengths, and 40 constructs were chosen each for pM1s3A and pM2s2A. Constructs were labeled by the microwell within the plate, with pM1s3AsR_TS-A1 to D10 for the engineered pMUT1 constructs and pM2s2AsR_TS-E1 to H10 for the pMUT2 constructs. The cells bearing the selected constructs were then monitored during growth in the plate reader, where we quantified the GFP, RFP fluorescence and absorbance at 600 nm at both 37 and 30 °C to identify their strength and inducibility (Figure 3c–f). We found that the GFP expression strengths varied significantly between constructs at 37 °C, whereas RFP expression remained similar in each case and did not vary as much with temperature.
We found the promoter strengths of the pTlpA variants (labeled pTlpA-A1 to pTlpA-H10) by calculating the amount of GFP produced per unit time and per cell (Figure 3g–j). We found that the promoters were weaker by around an order of magnitude at 30 °C compared to 37 °C, and when induced, covered a wide range of expression strengths on both pMUT vectors. We selected 9 of these constructs to cover the range of expression strengths each on the pM1s3A and pM2s2A backbones for sequencing and further use. The selected constructs are highlighted in light green on the Figure 3g–j, and the mutant nucleotides are shown as labels above. The promoter sequences and their associated expression strengths can be found in Table S4.
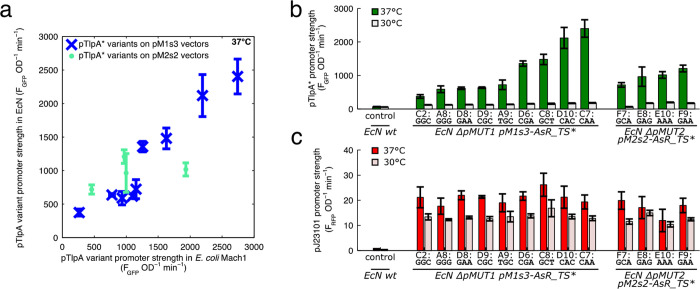
We then characterized the performance of the pM1s3AsR_TS* and pM2s2AsR_TS* constructs in EcN, in each case measuring the engineered pMUT construct performance in the absence of the native cryptic plasmid. We found that we could not transform some of the pM2s2AsR_TS* constructs into EcN ΔpMUT2 cells, and thus we could only characterize 4 of the pMUT2 derived constructs. The characterization data from the E. coli Mach1 cloning strain was broadly indicative of performance in EcN (Figure 4a), although the pM2s2A constructs in particular did not fully match their behavior in Mach1. The pTlpA* promoters covered a range of expression strengths when induced, and had significantly less expression at 30 °C (Figure 4b). By contrast, constitutive RFP expression from each construct was similar, and did not vary as much with temperature (Figure 4c).
Figure 4.
(a) Comparison of the temperature sensitive gene expression circuit in EcN and in E. coli Mach1 strains. (b) GFP expression from the pTlpA library in EcN, showing a range of expression strengths. (c) Constitutive RFP from the “AsR_TS*” cassettes from the engineered pMUT plasmids in EcN.
Curli Secretion from Engineered pMUT Vectors
Many proteins and peptides have therapeutic potential in the gut,37 and as such the secretion of such peptides into the extracellular space from EcN inhabiting the gut is an attractive approach to therapy. Curli are well-characterized bacterial extracellular matrix proteins, secreted natively by E. coli using dedicated machinery38 to form robust fibers. Engineered curli systems are emerging as a versatile platform for custom protein materials, as they are capable of tolerating mutations and fusions to functional protein domains, and consequently they are being developed as gut therapeutics.39
To express curli, we used a synthetic csgBACEFG operon,40 which encodes the major and minor curli fiber subunits, csgA and csgB, and the secretion machinery necessary for transport from the periplasm to the extracellular space in csgEFG. CsgC prevents intracellular CsgA polymerization, which would be toxic to the bacterium.41 We fused the CsgA monomer to an E-tag epitope tag in a 37 aa flexible linker (Figure 5a) to enable detection with anti-E-tag antibodies, and this cassette was “csg-Etag”. In order to further demonstrate a functional curli variant, we also produced constructs where CsgA was fused to a GFP nanobody (NbGFP).42 Nanobodies, also known as VHH domains, are single chain antibody fragments,43 capable of binding tightly to a specific antigen. In this case, the csgA:NbGFP fusion should bind GFP, and due to the insolubility of the curli material, should remove purified GFP from solution. The csgA:NbGFP fusion in cassette “csg-Etag-NbGFP” also encoded an E-tag in a 37 aa flexible linker between the csgA and NbGFP sequences, and the NbGFP was followed by a 6xHis tag (Figure 5a).
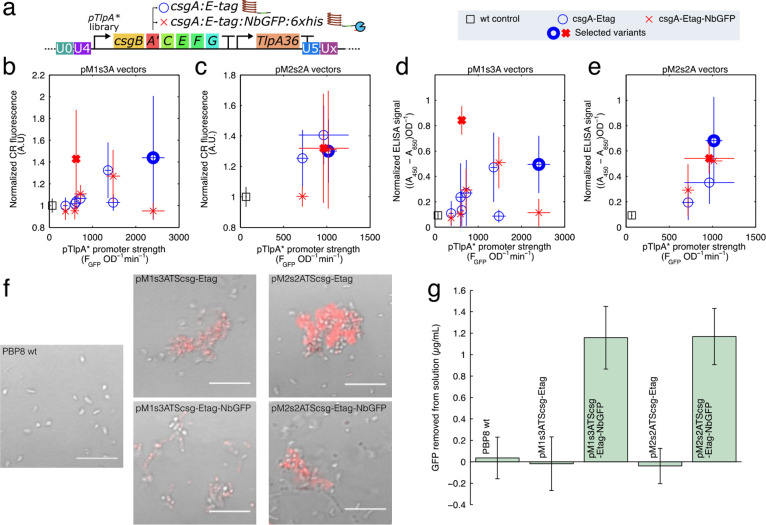
Figure 5.
(a) Diagram of the temperature sensitive curli production construct, containing a pTlpA promoter variant driving the expression of a synthetic curli operon csgBACEFG, where the csgA sequence is appended with an E-tag epitope tag (labeled cassette “csg-Etag”), or an E-tag and a GFP nanobody sequence with 6xHis (cassette “csg-Etag-NbGFP”). The temperature sensitive pTlpA* promoter variants were all used to generate pM1s3ATScsg-# variants (b,d) and pM2s2ATScsg-# (c,e) variants. These variants were assayed with a Congo Red (CR) curli assay (b,c), where CR dye stains the curli proteins and becomes fluorescent, and an anti-E-tag filtration ELISA (d,e). In panels b–e, thick markers represent the variants selected from the promoter library for subsequent use. (f) Representative confocal micrographs of bacterial cultures harboring the selected plasmids with temperature inducible curli grown at 37 °C in the presence of CR, with the red CR fluorescence overlaying a brightfield image. Scale is 10 μm. (g) Curli fused to GFP nanobodies (NbGFP) was able to remove a significant amount of sfGFP from a 4 μg/mL solution of purified sfGFP in PBS.
Overexpression of the csgBACEFG operon can be toxic to cells, and as such the expression strength requires significant tuning to obtain a high yield of curli fibers. We therefore used the pTlpA promoter library to express a synthetic curli operon to identify an optimal promoter strength. In total, we were able to generate 8 of each “csg-Etag” and “csg-Etag-NbGFP” on pM1s3A vectors, and 3 each on pM2s2A vectors. For each pTlpA*-curli construct, we characterized the curli production using the Congo Red (CR) fluorescence method44 (Figure 5b,c), as well as by a filtration ELISA method with anti-E-tag antibodies (Figure 5d,e). Both methods showed similar results, with certain combinations of promoter and csgA variant producing significant yields of curli material. For the csgA:Etag constructs, higher promoter strength generated the higher curli yields, and we chose the highest expressing constructs to make plasmids pM1s3ATScsg-Etag and pM2s2ATScsg-Etag, which used the pTlpA-C7 and pTlpA-E8 promoters, respectively. By contrast, the “csg-Etag-NbGFP” constructs had peak expression at intermediate promoter strengths, with the chosen plasmids pM1s3ATScsg-NbGFP and pM2s2ATScsg-NbGFP containing the pTlpA-D8 and pTlpA-E10 promoters. In all cases, the temperature sensitive curli constructs expressed poorly at 30 °C (Table S4).
At 37 °C, the selected temperature sensitive curli expression constructs produced curli, which caused the bacterial cultures to form aggregates that were fluorescent upon the addition of CR (Figure 5f). Additionally, at 37 °C, bacteria with the csgA:NbGFP fusions successfully bound and removed purified GFP from a solution of purified GFP, demonstrating the desired function of the nanobody (Figure 5g).
Engineered pMUTs Performance in the Mouse Gut
One of the original motivations for this work was to address and improve retention rates of synthetic plasmids in bacteria within the mouse gut. In a preliminary experiment testing engineered EcN in the mouse gut, we found that EcN harboring engineered synthetic plasmids experienced plasmid loss during passage through the gut without selection. In this experiment, we fed mice PBP8 cells (EcN ΔcsgBACEFG::cat(CamR)) transformed with either plasmid pKAG,45 a pSB4K5 based plasmid containing constitutively expressed sfGFP, or pL6FO,44 a similar synthetic plasmid with an IPTG inducible csgBACEFG operon (Figure S5a). The engineered bacteria were administered to the mice on day 0 of the experiment (Figure S5b), and we tracked both the overall PBP8 population and the plasmid bearing population in fecal samples over the subsequent days. As we were selecting for the PBP8 bacteria by treating with chloramphenicol, we found that the PBP8 persisted in the gut in all cases after administration (Figure S5c). However, we found significant plasmid loss for all synthetic plasmids (Figure S5d), which we tested by challenging with kanamycin. On day 1 after administration, pL6FO was only present in only around 15% of the population when the curli operon was in the IPTG induced state. Furthermore, for plasmid pKAG, and pL6FO with IPTG induced curli operon, the plasmid-bearing bacteria were not found in the gut after 5 days.
We sought to test the plasmid retention of our engineered pMUTs after passing through the mouse gut. Additionally, we were interested in determining the ability of our plasmid system to produce and secrete proteins in an in vivo context, as this feature is key to therapeutic peptide delivery in the gut. Bacterial gene expression in a mammalian gut significantly differs from expression under laboratory conditions,46 and as such in vitro characterization is unlikely to be representative of in vivo functionality.
Typically, it is difficult to assess the gene expression of engineered bacteria in the gut, because the bacteria must usually be grown in vitro after isolation from fecal samples, which disrupts any measurement of in vivo gene expression. Direct detection of heterologously produced proteins in fecal samples is similarly challenging. For most proteins and affinity tags, proteolytic degradation by intestinal proteases is likely to significantly reduce any measurable signal. This is particularly problematic considering the high background signal one can expect from a complex biological medium such as feces. These experimental limitations were, in large part, what motivated us to test the pMUT system using curli fibers and VHH domains. In addition to the potential utility of these proteins, both curli and nanobodies are known for their resistance to the harsh conditions,47,48 thereby increasing the likelihood of their detection in fecal pellets.
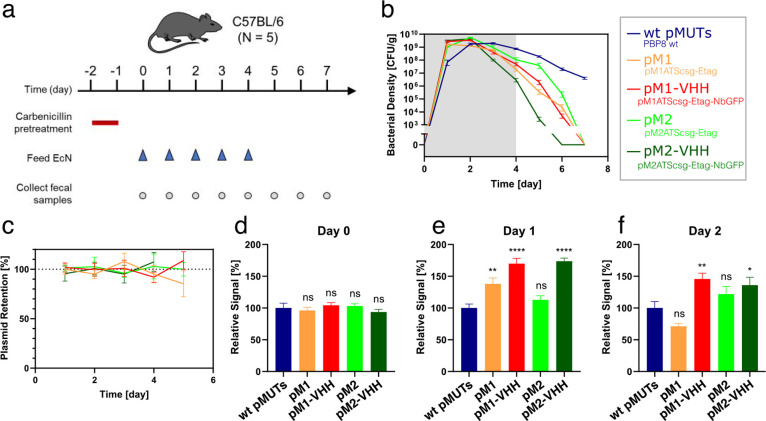
We designed an experiment to test the retention of the engineered pMUTs in vivo, as well as the expression of protein through the plasmid system within the mouse gut. Four plasmids were tested, expressing either cassette “csg-Etag” or “csg-Etag-NbGFP” on pM1s3ATS* or pM2s2TAS* vectors. In each case, PBP8 cells (EcN ΔcsgBACEFG::cat(CamR)) were used, with the native pMUT knocked out whenever the corresponding engineered version was present. As a negative control, we used PBP8 harboring both wild-type pMUTs with no engineered plasmids, making for a total of 5 experimental cohorts. We labeled the conditions: “wt pMUTs” for the control; “pM1” for PBP8 ΔpMUT1 pM1s3ATScsg-Etag; “pM1-VHH” for PBP8 ΔpMUT1 pM1s3ATScsg-Etag-NbGFP; “pM2” for PBP8 ΔpMUT2 pM2s2ATScsg-Etag; “pM2-VHH” for PBP8 ΔpMUT2 pM2s2ATScsg-Etag-NbGFP. Each of the five bacterial strains were administered to C57BL/6 mice (n = 5).
The mice were fed bacterial suspension daily for 5 days and monitored for 3 additional days after cessation of bacterial administration (Figure 6a). Each day, fecal pellets were collected for colony counting and protein detection. Like most human E. coli isolates, EcN is a poor colonizer of the mouse gut,49 though it can transiently persist in mice pretreated with antibiotics. Therefore, carbenicillin was given 2 days prior to bacterial feeding, in order to allow the engineered EcN to reach high density, with the antibiotic administration lasting 24 h to avoid artificially selecting for the engineered plasmids. As a result, EcN density gradually dropped over the course of the experiment, likely due to the recovery of native mouse microbiome (Figure 6b). All the engineered EcN were cleared from the mice by day 7, unlike the wt control, and we hypothesized that this was due to the increased fitness of the unmodified bacteria. When characterized in vitro (Figure S6), the unmodified EcN grew significantly faster than any engineered pMUT variant, suggesting that the burden of the recombinant gene expression reduced fitness.
Figure 6.
Engineered pMUTs in the mouse gut. (a) Timeline of in vivo study. (b) Bacterial density of PBP8 over time, as measured by CFU counts from fecal samples plated on LB agar with Cm. (c) Plasmid retention over time. (d–f) Relative in vivo protein expression levels from fecal filtration ELISA on days 0, 1, and 2 (d, e, and f, respectively). At each day, engineered pMUT conditions were tested against a WT pMUT control by one-way ANOVA, followed by pairwise Welch’s t test. ns, not significant; *p < 0.05, **p < 0.01, ****p < 0.0001. All data are represented as mean ± SEM.
Each fecal pellet was plated on two types of selective plates: chloramphenicol (Cm), selecting for PBP8 irrespective of plasmid presence or identity; and chloramphenicol with carbenicillin (Cm+Carb), which selected specifically for PBP8 with an engineered plasmid. Plasmid retention rates were calculated as the ratio of Cm+Carb to Cm colony counts. All four engineered pMUT cohorts showed no plasmid loss during GI transit, with none of the retention rates differing significantly from 100% (Figure 6c).
Protein expression was tested via fecal filtration ELISA, modified from a previous protocol.39 In both engineered pMUT1 and pMUT2, significant levels of E-tagged curli fibers were detected (Figure 6d). Since the mice were fed EcN grown at 30 °C, there was no curli expression prior to feeding, so this result demonstrated the ability of the engineered pMUT system to express and secrete proteins in vivo. Interestingly, in both plasmids, the nanobody-containing constructs produced a higher signal than their nanobody-free counterparts. We suspect this may be due to the robust CsgA and NbGFP protein domains flanking and protecting the E-tag from proteolysis when in the gut lumen.
Discussion
In this work we developed plasmid vectors based on the E. coli Nissle 1917 pMUT cryptic plasmids, and characterized their performance in the mouse gut. Our work developed a simple method to remove the native pMUT plasmids, and generated reliable pMUT plasmid vectors capable of secreting a functional curli material within the mouse gut without plasmid loss. Furthermore, our pMUT-based plasmid vectors simplified in vivo experiments by forgoing the need for antibiotics for plasmid maintenance or inducers for gene expression through temperature sensitive circuits.
The pMUT plasmids have no known function, but are stable within EcN during passage through the gut, and can thus act as vectors for recombinant DNA. While previous studies have used the pMUT plasmids,3 and shown their high plasmid retention in vitro,24 their in vivo efficacy had never been systematically characterized. Through our attempts to cure the native pMUT plasmids, our results suggest that pMUT2 stability in EcN is improved by the RelB/RelE toxin-antitoxin system, as we could not cure pMUT2 without expressing the antitoxin gene from our pCryptDel4.8 plasmid. This demonstrates how the approach of building and testing genetic tools not only creates useful systems, but also provides insight into the underlying biology.
Despite the common use of plasmids in the development of engineered microbes, they are not typically utilized in clinical applications, where exogenous genetic sequences are instead incorporated into the chromosome of the chassis organism. A major reason for this is plasmid loss, and this phenomenon severely limits the efficacy of plasmid based genetic systems in vivo. Antibiotics, commonly used for plasmid maintenance in vitro, are incompatible with many in vivo assays, as they disrupt the native microbiota as well as any pathogens, and additionally raise significant questions regarding the effective antibiotic dose. Engineered plasmid maintenance strategies could address this issue, but such methods have so far not fully overcome plasmid loss,16 and would currently require significant development and optimization. The addition of engineered plasmid retention would also create a further metabolic burden from the plasmid, potentially reducing the efficacy of other synthetic genetic circuits. The pMUT plasmids, by contrast, are in some sense already optimized for EcN, as they are stable and do not impose any noticeable burden. While it is possible that some recombinant inserts may hinder their stability, we did not observe any loss of the engineered pMUT plasmids, despite adding synthetic circuits that imposed a significant reduction in growth when induced.
A further concern for plasmid based engineering is horizontal gene transfer (HGT), whereby a plasmid with an antibiotic resistance gene or virulence factor would run the risk of being introduced into the host microbiome.50 While such concerns are valid for most synthetic plasmids, the unique features of the engineered pMUT system could address these limitations. Most prominently, the absence of antibiotic selection could eliminate the possibility of spreading resistance genes, as the resistance gene can be excised from any engineered bacterium through a recombinase. Furthermore, the presence of these plasmids in wild-type EcN suggests that the risk posed by any sequence found natively on the plasmid is negligible. Indeed, the safety profile of EcN over decades of probiotic use implies that HGT of pMUT-encoded genes is either exceedingly rare, relatively harmless or both. Lastly, some have proposed utilizing HGT as a tool for in situ microbiome engineering.51 As a selection-free, probiotic-derived plasmid system, the pMUT platform could prove a valuable addition to the toolbox of this emerging microbial intervention strategy. Thus, while the pMUT plasmids could indeed be utilized for the research and development of engineered strains, they could also open the door to plasmid-based production of therapeutics in vivo, in both clinical and preclinical settings.
There are several benefits to using engineered pMUT plasmid vectors compared to genomic incorporation. The first is speed and reliability, since plasmid assembly and transformation are the only steps required for the production of an engineered EcN strain, and this can be done in several days. This can facilitate the rapid construction and development of probiotic bacteria, speeding the development and optimization of prototypes. A further benefit is the ability to incorporate relatively large recombinant genetic constructs with ease. Indeed, one of the largest constructs we made was around 13 Kbp (pM2s2ATScsg-NbGFP), incorporating over 7 Kbp of recombinant DNA. Furthermore, both engineered pMUT1 and pMUT2 plasmids could be used simultaneously to house synthetic DNA, allowing for the incorporation of even larger and more complex synthetic DNA systems.
A major benefit to simplifying the process of bacterial engineering is the ability to rapidly and reliably generate multiple variant strains, and thus screen and optimize genetic circuits of interest. The pTlpA promoter library in this case demonstrated how even a relatively small functional change, such as the addition of a fusion protein, can require the redesign of regulatory elements within genetic circuits for optimal function. Here, the addition of an anti-GFP nanobody required a weaker promoter for curli expression compared to unmodified CsgA-Etag, suggesting that the nanobody reduced secretion efficacy, likely through the toxicity of expression and secretion. However, the weaker expression did not reduce overall curli production in the nanobody constructs, suggesting that curli production was not limited by the expression of the other genes in the csgBACEFG operon.
In our in vivo experiments, we observed slower clearance of the WT pMUT control strain compared to those expressing proteins through engineered pMUTs (Figure 6b). We believe this reflects the added metabolic burden imposed on the cells by overexpression of heterologous protein, rather than any feature of the engineered plasmids, as our previous work yielded similar trends with different plasmid systems.39 In addition, as was the case in the aforementioned study, bacterial density and protein expression varied between the different conditions, indicating these factors depend on the specific proteins being expressed. Praveschotinunt et al.(39) also demonstrated that PBP8 and WT EcN do not differ substantially in their in vivo behavior, and that strains expressing wild-type curli fibers can exhibit similar bacterial densities to those producing GFP. These findings suggest that the performance observed by the strains in this work is unlikely to be specific to curli. Taken together, such observations support the compatibility of engineered pMUTs with in vivo expression of a variety of proteins, though the expression strength would have to be adjusted to achieve optimal results for each desired application—as would be the case for any other expression platform, be it genomic integration or plasmid-based.
Conclusion
In this work we have harnessed native cryptic plasmids to create a robust genetic platform for engineering probiotic E. coli Nissle 1917 bacteria. While E. coli is not a major component of the human or mouse microbiome, it is often present at sites of inflammation.52 As such, E. coli is capable of playing an important therapeutic role, both by competing with pathogenic bacteria as well as being able to deliver therapeutic compounds to where they are needed.
It is becoming increasingly clear that the state of the gut microbiome has important ramifications for human health, and there are many unanswered questions about the role of the microbes. In this work we have developed a reliable genetic platform to host synthetic DNA for E. coli Nissle. Our platform simplifies research, facilitating new experiments to investigate the gut microbiome, and speeds the development of therapeutic engineered bacteria that can be deployed clinically.
Materials and Methods
DNA Cloning
All plasmid assembly was performed using Gibson Assembly, with the exception of the pCryptDel# plasmids, where the gRNA array was assembled using Golden Gate assembly due to many repeats in the DNA sequence. Custom DNA oligos were ordered from Integrated DNA Technologies (IDT) and used in PCR with Q5 polymerase (New England Biolabs) to create amplicons for subsequent Gibson assembly. DNA purification from PCR was done with ZymoClean Gel DNA Recovery Kit (ZymoGen). DNA assembly products were transformed into chemically competent E. coli Mach1 cells (Thermo Fisher Scientific) and plated onto LB Agar plates with appropriate antibiotics.
DNA libraries were generated by designing degenerate bases at selected locations on DNA oligos, flanked by 25 bp of the unmodified sequence. The resulting DNA was synthesized (IDT) used as primers to make amplicons for plasmid assembly. The resulting pool of assembled plasmid variants was transformed into Mach1 cells and plated onto 10 plates. After overnight incubation at 37 °C, the plates were imaged for GFP and RFP fluorescence in a FluorChem M Imager (Protein Simple), and colonies were selected.
Colony PCR
Assessment of cryptic plasmids was done by colony PCR using 25 μL reactions with Quick-Load Taq PCR mix (New England Biolabs) following the manufacturer’s instructions. After the PCR, the reactions were added to a 1% agarose TAE gel with SybrSafe DNA stain and ran in a gel electrophoresis setup (constant 120 V, 35 min). Gels were then imaged in FluorChem M Imager (Protein Simple).
Bacterial Culture
E. coli bacteria were grown in LB Miller media during plasmid preparation and genetic circuit characterization. For characterization assays, starter cultures of the appropriate bacterial cultures were grown overnight in LB media in a shaking incubator. For all temperature sensitive constructs, started cultures were grown at 30 °C, whereas we used 37 °C for all other constructs. Unless explicitly indicated otherwise, all characterization was done at 37 °C.
Kinetic plate reader assays were performed by diluting starter culture 1:1000 into the appropriate selective media. We then added 200 μL of the inoculated culture into the wells of black, clear-bottom, 96-well plates (655090, Greiner Bio-One). The plates were then grown in a Synergy HT plate reader (BioTek), reading absorbance (600 nm), GFP (excitation: 485/20 nm, emission: 528/20 nm), RFP (ex: 590/20 nm, em: 645/40 nm). Reads were taken every 10 min for 16 h, and plates were shaken continuously outside of reading (Double Orbital, 548 cpm (2 mm)).
Plasmid Curing
In order to cure Nissle and any derived strains of cryptic plasmids, they were transformed with plasmids pFREE or pCryptDel4.8, in order to cure pMUT1 or pMUT2 respectively. After transformation, cells were grown overnight in liquid LB media with 50 μg/mL kanamycin. Then, the overnight culture was diluted 1:1000 into fresh LB media supplemented with 50 μg/mL kanamycin, 0.2% rhamnose and 0.43 μM anhydrotetracycline (ATC), and grown overnight at 37 °C. After 24 h, the culture was streaked out onto several LB agar plates without antibiotics and these were left to grow overnight. Then, the colonies were assessed by colony PCR with primers muta5, muta6, muta7, and muta8 to find colonies that had been cured of cryptic plasmids.
Growth and Gene Expression Characterization
Data from kinetic plate reader runs was initially cleaned by subtracting the background signal and smoothing the time courses for all fluorescence and absorbance data. Growth rates were found by fitting the absorbance curves to a Gompertz model, and subsequently extracting the peak growth rate. Promoter strength was quantified from kinetic fluorescence data by first finding the gradient of the fluorescence signal, normalizing this to the absorbance signal, resulting in a per cell measure of fluorescent protein production per unit time. Promoter strength was then quoted to be the average of this term for an hour around peak exponential phase.
Curli Measurement
Curli was measured by the CR method outlined in Kan et al.(44) Bacterial starter cultures were grown overnight in LB and the relevant antibiotics at 30 °C. Then, we inoculated selective LB media 1:1000 with starter culture, and placed 300 μL into 1 mL deep well plates (780210, Greiner Bio-One) in a shaking incubator (900 rpm (1 mm)) at either 30 or 37 °C. 0.025% CR was added to the media upon inoculation. After 24 h of growth, 200 μL of each well was transferred into black, clear bottom plates and the absorbance (600 nm) and CR fluorescence (ex: 525 nm, em: 625 nm) was read in a Synergy HT plate reader. The results were then normalized to the host strain without engineered plasmids.
Curli production was also measured by whole cell filtration ELISA to measure the E-tagged CsgA proteins. 80 μL bacterial overnight cultures were added into each well of a 96-well filter plate in triplicate (0.22 μm pore size, Multiscreen-GV, Merck/Millipore Sigma). Samples were vacuum-filtered, and washed in TBST (TBS, 0.1% Tween-20), and blocked for 1.5 h at 37 °C with 1% bovine serum albumin (BSA) and 0.01% H2O2 in TBST. After additional washing, samples were incubated with HRP-conjugated goat polyclonal E-tag epitope antibody (Novus Biologicals) for 1.5 h at room temperature (1:5000 in TBST). Following additional washes in TBST, 100 μL Ultra-TMB ELISA reagent (Thermo Scientific) was added to each well and covered with aluminum foil to protect from light. After approximately 15–25 min of incubation at room temperature, the reaction was stopped using 50 μL per well of 2 M sulfuric acid. 100 μL were transferred from each well into a 96-well plate and absorbance was measured at 450 and 650 nm. The signal was calculated by subtracting the 650 nm absorbance value from the 450 nm absorbance value.
GFP Sequestration Assay
To test the function of the GFP nanobody, bacterial cultures were first grown overnight at stated temperatures in LB media with appropriate antibiotics. They were then pelleted at 3000g for 10 min, washed once in PBS, and resuspended in a solution of PBS containing 4 μg/mL purified sfGFP. The solutions were then left in a rotating mixer for an hour at room temperature, then centrifuged again at 3000g for 10 min. The supernatant GFP signal was then measured in the plate reader, and compared to the fluorescence of the initial sfGFP solution. In order to prevent nonspecific GFP protein adsorption to the plasticware used in the experiment, a sterile solution of 1% BSA (bovine serum albumin) in PBS was used to block the plastic tubes prior to the experiment. To do this, we filled the 1.5 mL tubes with 1 mL of the BSA solution and left in a rotating mixer for an hour.
In Vivo Study of Engineered pMUT Plasmid Retention and Protein Expression
The protocol described below was reviewed and approved by the Harvard Medical Area Standing Committee on Animals (HMA IACUC, ref. No. IS00000516–3). Twenty-five female 8- to 9-week-old C57BL/6NCrl mice were randomly split into five experimental cohorts: WT pMUTs, pM1, pM1-VHH, pM2, and pM2-VHH (N = 5). Bacterial suspensions were prepared in advance by growing to mid-exponential phase (OD600 of 0.5) at 30 °C (shaking at 225 rpm), pelleting the cells, resuspending to OD600 of 10 in PBS supplemented with 20% sucrose and 10% glycerol, and flash-freezing in liquid nitrogen. Aliquots of these bacterial suspensions were stored at −80 °C and allowed to thaw immediately preceding daily feeding, in order to maintain consistent bacterial density of the inoculum.
48 h prior to initial administration of bacteria (day −2), the drinking water was supplemented with 2 g/L carbenicillin (Teknova). Antibiotic-free drinking water was restored 24 h later (day −1). Starting on day 0, each cohort was fed 50 μL of its respective bacterial suspension by allowing the mice to lap the liquid from a pipet tip (as previously described by Mohawk et al.(53)). Bacterial administration was carried out daily from day 0 to day 4. Fecal pellets were collected and weighed daily from day 0 to day 7.
Mice
Female 8- to 9-week-old C57BL/6NCrl mice were obtained from Charles River Laboratories. Mice were housed in sterile vinyl isolators within the Harvard Medical School animal facility, and kept under specific-pathogen-free (SPF) conditions. Both sterile food (JL Rat and Mouse/Auto 6F 5K67, LabDiet) and water were provided ad libitum. All mice were allowed 1 week to acclimate prior to any experimental procedure. To further minimize impact of living environment on experimental outcomes, mice were randomized between housing isolators at the beginning of the experiment. All experiments were conducted in compliance with the US National Institutes of Health guidelines and approved by the Harvard Medical Area Standing Committee on Animals.
Plasmid Retention Analysis
Immediately following daily collection of fecal pellets, each sample was homogenized in 1 mL of PBS, serially diluted, and plated in quadruplicate to enumerate colony forming units (CFU). Samples were plated on two types of LB agar plates: 25 μg/mL chloramphenicol-only plates (Cm) and 100 μg/mL carbenicillin and 25 μg/mL chloramphenicol plates (Cm+Carb). While all PBP8-derived strains carried a chromosomal Cm resistance gene, only the engineered pMUT plasmids harbored a Carb resistance marker. Plasmid retention rate was therefore estimated by calculating the Cm+Carb to Cm ratio of sample weight-normalized CFU counts. Following the plating procedure, fecal homogenates were flash-frozen and stored at −80 °C for subsequent analysis.
Fecal Filtration ELISA
To detect E-tagged curli fibers in fecal samples, a filtration ELISA protocol was adapted from Praveschotinunt et al.(39) Fecal homogenate was centrifuged at 1000g for 1 min to separate large insoluble material, and the supernatant was transferred onto a 96-well filter plate in triplicate (0.22 μm pore size, Multiscreen-GV, Merck/Millipore Sigma). For each sample, the homogenate volume dispensed was normalized to 1.25 mg of fecal pellet per well. After samples were added to the filter plate, the procedure to detect E-tagged material proceeded as described above in the Curli Measurement section. In each assay, the signal was normalized by dividing by the WT pMUTs control, such that the WT pMUTs control corresponded to 100%.
Acknowledgments
This work is grateful for and made use of the facilities of the Harvard Center for Comparative Medicine. pFREE was a gift from Morten Norholm (Addgene plasmid # 92050). This work was supported by the National Institutes of Health (1R01DK110770), NSF Grant 1410751 (Division of Materials Research) and the Wyss Institute for Biologically Inspired Engineering.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.0c00466.
Table S1: DNA primers used in this work; Table S2: Sequence of UNS sequences; Table S3: gRNA sequences used in the pFREE and pCryptDel plasmids; Table S4: Variant TlpA* promoter sequences, and their properties; Figure S1: Detailed plasmid maps of pMUT1 and pMUT2; Figure S2: GFP and RFP characterization of engineered pMUT plasmids; Figure S3: Plasmid vectors to cure EcN cryptic plasmids; Figure S4: Curing native pMUT plasmids; Figure S5: In vivo synthetic plasmid loss; Supplementary methods for Figure S5; Figure S6: In vitro growth characterization of bacteria used in vivo (PDF)
Dataset S1: All data used in the figures (XLSX)
Dataset S2: In vivo data (XLSX)
Author Contributions
⊥ A.K. and I.G. gave equal contributions.
Author Contributions
A.K., I.G., N.S.J. conceived and designed experiments. A.K., I.G., S.E., P.P. performed research. The manuscript was written through contributions of all authors. The final version of the manuscript has been approved by all the authors.
The authors declare the following competing financial interest(s): Authors have filed a provisional patent application based on the work presented in this paper.
Notes
As described in the Results section, the pMUT1 sequence was deposited onto the NCBI database (accession MW240712). All data discussed in the paper are available in the Supporting Information. Useful plasmid DNA produced over the course of this work will be available on AddGene, alongside their sequence files.
Supplementary Material
References
- Sonnenborn U. (2016) Escherichia coli strain Nissle 1917—from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett. 363 (19), fnw212. 10.1093/femsle/fnw212. [DOI] [PubMed] [Google Scholar]
- Altenhoefer A.; Oswald S.; Sonnenborn U.; Enders C.; Schulze J.; Hacker J.; Oelschlaeger T. A (2004) The probiotic Escherichia coli strain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens. FEMS Immunol. Med. Microbiol. 40 (3), 223. 10.1016/S0928-8244(03)00368-7. [DOI] [PubMed] [Google Scholar]
- Ou B.; Yang Y.; Tham W. L.; Chen L.; Guo J.; Zhu G. (2016) Genetic engineering of probiotic Escherichia coli Nissle 1917 for clinical application. Appl. Microbiol. Biotechnol. 100 (20), 8693. 10.1007/s00253-016-7829-5. [DOI] [PubMed] [Google Scholar]
- Kurtz C. B.; Millet Y. A.; Puurunen M. K.; Perreault M.; Charbonneau M. R.; Isabella V. M.; Kotula J. W.; Antipov E.; Dagon Y.; Denney W. S.; Wagner D. A.; West K. A.; Degar A. J.; Brennan A. M.; Miller P. F. (2019) An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci. Transl. Med. 11 (475), eaau7975. 10.1126/scitranslmed.aau7975. [DOI] [PubMed] [Google Scholar]
- Danino T.; Prindle A.; Kwong G. A.; Skalak M.; Li H.; Allen K.; Hasty J.; Bhatia S. N. (2015) Programmable probiotics for detection of cancer in urine. Sci. Transl. Med. 7 (289), 289ra84. 10.1126/scitranslmed.aaa3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventhal D. S.; Sokolovska A.; Li N.; Plescia C.; Kolodziej S. A.; Gallant C. W.; Christmas R.; Gao J.-R.; James M. J.; Abin-Fuentes A.; Momin M.; Bergeron C.; Fisher A.; Miller P. F.; West K. A.; Lora J. M. (2020) Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat. Commun. 11 (1), 2739. 10.1038/s41467-020-16602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh K.; Meyer K. M.; Aagaard K. M.; Wilmes P. (2016) The human gut microbiome in health: establishment and resilience of microbiota over a lifetime. Environ. Microbiol. 18 (7), 2103. 10.1111/1462-2920.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riglar D. T.; Giessen T. W.; Baym M.; Kerns S. J.; Niederhuber M. J.; Bronson R. T.; Kotula J. W.; Gerber G. K.; Way J. C.; Silver P. A. (2017) Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol. 35 (7), 653. 10.1038/nbt.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker W. R.; Shepherd E. S.; Sonnenburg J. L. (2017) Tunable Expression Tools Enable Single-Cell Strain Distinction in the Gut Microbiome. Cell 169 (3), 538. 10.1016/j.cell.2017.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riglar D. T.; Richmond D. L.; Potvin-Trottier L.; Verdegaal A. A.; Naydich A. D.; Bakshi S.; Leoncini E.; Lyon L. G.; Paulsson J.; Silver P. A. (2019) Bacterial variability in the mammalian gut captured by a single-cell synthetic oscillator. Nat. Commun. 10 (1), 1. 10.1038/s41467-019-12638-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarczyk A. L.; Lin A.; Weiss R. (2012) Foundations for the design and implementation of synthetic genetic circuits. Nat. Rev. Genet. 13 (6), 406. 10.1038/nrg3227. [DOI] [PubMed] [Google Scholar]
- Theriot C. M.; Koenigsknecht M. J.; Carlson P. E.; Hatton G. E.; Nelson A. M.; Li B.; Huffnagle G. B.; Li J. Z.; Young V. B. (2014) Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5 (1), 3114. 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M.; Watzl S.; Oelschlaeger T. A.; Rath H. C.; Göttl C.; Lehn N.; Schölmerich J.; Linde H.-J. (2005) Green fluorescent protein for detection of the probiotic microorganism Escherichia coli strain Nissle 1917 (EcN) in vivo. J. Microbiol. Methods 61 (3), 389. 10.1016/j.mimet.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Yano H.; Shintani M.; Tomita M.; Suzuki H.; Oshima T. (2019) Reconsidering plasmid maintenance factors for computational plasmid design. Comput. Struct. Biotechnol. J. 17, 70. 10.1016/j.csbj.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F. (2003) Toxins-Antitoxins: Plasmid Maintenance, Programmed Cell Death, and Cell Cycle Arrest. Science 301 (5639), 1496. 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- Fedorec A. J. H.; Ozdemir T.; Doshi A.; Ho Y.-K.; Rosa L.; Rutter J.; Velazquez O.; Pinheiro V. B.; Danino T.; Barnes C. P. (2019) Two New Plasmid Post-segregational Killing Mechanisms for the Implementation of Synthetic Gene Networks in Escherichia coli. iScience 14, 323. 10.1016/j.isci.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. W.; Lim H. G.; Yang J.; Noh M. H.; Seo S. W.; Jung G. Y. (2018) Synthetic auxotrophs for stable and tunable maintenance of plasmid copy number. Metab. Eng. 48, 121. 10.1016/j.ymben.2018.05.020. [DOI] [PubMed] [Google Scholar]
- Kuhlman T. E.; Cox E. C. (2010) Site-specific chromosomal integration of large synthetic constructs. Nucleic Acids Res. 38 (6), e92. 10.1093/nar/gkp1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A.; Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97 (12), 6640. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burian J.; Guller L.; Mačor M.; Kay W. W. (1997) Small Cryptic Plasmids of Multiplasmid, ClinicalEscherichia coli. Plasmid 37 (1), 2. 10.1006/plas.1996.1273. [DOI] [PubMed] [Google Scholar]
- Feldgarden M.; Golden S.; Wilson H.; Riley M. A. (1995) Can phage defence maintain colicin plasmids in Escherichia coli. Microbiology 141 (11), 2977. 10.1099/13500872-141-11-2977. [DOI] [PubMed] [Google Scholar]
- Blum-Oehler G.; Oswald S.; Eiteljörge K.; Sonnenborn U.; Schulze J.; Kruis W.; Hacker J. (2003) Development of strain-specific PCR reactions for the detection of the probiotic Escherichia coli strain Nissle 1917 in fecal samples. Res. Microbiol. 154 (1), 59. 10.1016/S0923-2508(02)00007-4. [DOI] [PubMed] [Google Scholar]
- Hacker J., Oelschlaeger T., Oswald S., Sonnenborn U., and Proppert H. (2011) US7993902B2.
- Zainuddin H. S.; Bai Y.; Mansell T. J. (2019) CRISPR-based curing and analysis of metabolic burden of cryptic plasmids in Escherichia coli Nissle 1917. Eng. Life Sci. 19 (6), 478. 10.1002/elsc.201900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J., Sonnen-Born U., Schulze J., Blum-Oehler G., Malinka J., and Proppert H. (2002) US6391631B1.
- Reister M.; Hoffmeier K.; Krezdorn N.; Rotter B.; Liang C.; Rund S.; Dandekar T.; Sonnenborn U.; Oelschlaeger T. A. (2014) Complete genome sequence of the Gram-negative probiotic Escherichia coli strain Nissle 1917. J. Biotechnol. 187, 106. 10.1016/j.jbiotec.2014.07.442. [DOI] [PubMed] [Google Scholar]
- Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Ciufo S., and Li W. (2013) Prokaryotic Genome Annotation Pipeline, National Center for Biotechnology Information. [Google Scholar]
- Avison M. B.; Walsh T. R.; Bennett P. M. (2001) pUB6060: A Broad-Host-Range, DNA Polymerase-I-Independent ColE2-like Plasmid. Plasmid 45 (2), 88. 10.1006/plas.2000.1511. [DOI] [PubMed] [Google Scholar]
- Sonnenborn U.; Schulze J. (2009) The non-pathogenic Escherichia coli strain Nissle 1917 – features of a versatile probiotic. Microb. Ecol. Health Dis. 21 (3–4), 122. 10.3109/08910600903444267. [DOI] [Google Scholar]
- Gotfredsen M.; Gerdes K. (1998) The Escherichia coli relBE genes belong to a new toxin–antitoxin gene family. Mol. Microbiol. 29 (4), 1065. 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- Torella J. P.; Lienert F.; Boehm C. R.; Chen J.-H.; Way J. C.; Silver P. A. (2014) Unique nucleotide sequence–guided assembly of repetitive DNA parts for synthetic biology applications. Nat. Protoc. 9 (9), 2075. 10.1038/nprot.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritsen I.; Porse A.; Sommer M. O. A.; Nørholm M. H. H. (2017) A versatile one-step CRISPR-Cas9 based approach to plasmid-curing. Microb. Cell Fact. 10.1186/s12934-017-0748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorgy A.; Jimenez J. I.; Yazbek J.; Huang H.-H.; Chung H.; Weiss R.; Del Vecchio D. (2015) Isocost Lines Describe the Cellular Economy of Genetic Circuits. Biophys. J. 109 (3), 639. 10.1016/j.bpj.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piraner D. I.; Abedi M. H.; Moser B. A.; Lee-Gosselin A.; Shapiro M. G. (2017) Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nat. Chem. Biol. 13 (1), 75. 10.1038/nchembio.2233. [DOI] [PubMed] [Google Scholar]
- Hurme R.; Berndt K. D.; Namork E.; Rhen M. (1996) DNA Binding Exerted by a Bacterial Gene Regulator with an Extensive Coiled-coil Domain. J. Biol. Chem. 271 (21), 12626. 10.1074/jbc.271.21.12626. [DOI] [PubMed] [Google Scholar]
- Salamov V. S. A., and Solovyevand A. (2011) Automatic annotation of microbial genomes and metagenomic sequences. In Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, p 61, Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- Muheem A.; Shakeel F.; Jahangir M. A.; Anwar M.; Mallick N.; Jain G. K.; Warsi M. H.; Ahmad F. J. (2016) A review on the strategies for oral delivery of proteins and peptides and their clinical perspectives. Saudi Pharm. J. 24 (4), 413. 10.1016/j.jsps.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart M. M.; Chapman M. R. (2006) Curli Biogenesis and Function. Annu. Rev. Microbiol. 60 (1), 131. 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveschotinunt P.; Duraj-Thatte A. M.; Gelfat I.; Bahl F.; Chou D. B.; Joshi N. S. (2019) Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat. Commun. 10 (1), 1. 10.1038/s41467-019-13336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveschotinunt P.; Dorval Courchesne N.-M.; den Hartog I.; Lu C.; Kim J. J.; Nguyen P. Q.; Joshi N. S. (2018) Tracking of Engineered Bacteria In Vivo Using Nonstandard Amino Acid Incorporation. ACS Synth. Biol. 7 (6), 1640. 10.1021/acssynbio.8b00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M. L.; Chorell E.; Taylor J. D.; Åden J.; Götheson A.; Li F.; Koch M.; Sefer L.; Matthews S. J.; Wittung-Stafshede P.; Almqvist F.; Chapman M. R. (2015) The Bacterial Curli System Possesses a Potent and Selective Inhibitor of Amyloid Formation. Mol. Cell 57 (3), 445. 10.1016/j.molcel.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U.; Zolghadr K.; Tillib S.; Nowak D.; Schermelleh L.; Gahl A.; Backmann N.; Conrath K.; Muyldermans S.; Cardoso M. C.; Leonhardt H. (2006) Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3 (11), 887. 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- Ghahroudi M. A.; Desmyter A.; Wyns L.; Hamers R.; Muyldermans S. (1997) Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 414 (3), 521. 10.1016/S0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- Kan A.; Birnbaum D. P.; Praveschotinunt P.; Joshi N. S. (2019) Congo Red Fluorescence for Rapid In Situ Characterization of Synthetic Curli Systems. Appl. Environ. Microbiol. 85 (13), e00434. 10.1128/AEM.00434-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge T. J.; Federici F.; Steiner P. J.; Kan A.; Haseloff J. (2013) Cell shape-driven instability generates self-organised, fractal patterning of cell layers. ACS Synth. Biol. 2 (12), 705. 10.1021/sb400030p. [DOI] [PubMed] [Google Scholar]
- Heithoff D. M.; Conner C. P.; Hanna P. C.; Julio S. M.; Hentschel U.; Mahan M. J. (1997) Bacterial infection as assessed by in vivo gene expression. Proc. Natl. Acad. Sci. U. S. A. 94 (3), 934. 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson S. K.; Emödy L.; Müller K. H.; Trust T. J.; Kay W. W. (1991) Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J. Bacteriol. 173 (15), 4773. 10.1128/JB.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden R. H. J.; Frenken L. G. J.; de Geus B.; Harmsen M. M.; Ruuls R. C.; Stok W.; de Ron L.; Wilson S.; Davis P.; Verrips C. T. (1999) Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1431 (1), 37. 10.1016/S0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- Saito K. (1961) [Studies on the habitation of pathogenic Escherichia coli in the intestinal tract of mice. I. Comparative experiments on the habitation of each type of resistant pathogenic Escherichia coli under an administration of streptomycin]. Paediatr. Jpn. 65, 385. [PubMed] [Google Scholar]
- Lerner A.; Matthias T.; Aminov R. (2017) Potential Effects of Horizontal Gene Exchange in the Human Gut. Front. Immunol. 10.3389/fimmu.2017.01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth R. U.; Cabral V.; Chen S. P.; Wang H. H. (2016) Manipulating Bacterial Communities by in situ Microbiome Engineering. Trends Genet. 32 (4), 189. 10.1016/j.tig.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes J. M. (2007) The role of Escherichia coli in inflammatory bowel disease. Gut 56 (5), 610. 10.1136/gut.2006.111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk K. L.; Melton-Celsa A. R.; Zangari T.; Carroll E. E.; O’Brien A. D. (2010) Pathogenesis of Escherichia coli O157:H7 strain 86–24 following oral infection of BALB/c mice with an intact commensal flora. Microb. Pathog. 48 (3), 131. 10.1016/j.micpath.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.