Abstract
SARS-CoV-2, the causal agent of COVID 19, is a new human pathogen that appeared in Wuhan, late December 2019. SARS-CoV-2 is a positive sense RNA virus, having four structural and six accessory proteins including that encoded by ORF8 gene known to be one of the most hypervariable and rapidly evolving genes. Thus, global characterization of mutations in this gene is important for pathogenicity and diagnostics. 240 different nonsynonymous mutations and 2 deletions were identified in 45,400 ORF8 nucleotide sequences during six months pandemic with about half of these variants were deleterious for ORF8, and the quarter of them were located in conserved amino acids. Genetic diversity analysis showed two main regions that harbor L84S and S24L. L84S is by far the most predominant mutation, followed by S24L that appeared first in USA. Phylogenetic analysis of ORF8 variants revealed the appearance of small clades with that of L84S being closer to bats. This is the first study that revealed the global nonsynonymous mutations in ORF8 from January to June 2020.
Keywords: COVID-19, SARS-CoV-2, ORF8, Nonsynonymous mutations, Single nucleotide variations, Deletions, Phylogenetic analysis
1. Introduction
COVID-19 caused by pathogenic coronavirus SARS-Cov-2 appeared in Wuhan, China, late December 2019 (Wu et al., 2020; Zhou et al., 2020; Huang et al., 2020). More than 70 million persons have been affected leading to more than one million death by December 14th. SARS-Cov-2, like SARS-CoV (Drosten et al., 2003) and MERS (Middle East respiratory syndrome-related Coronavirus) (Zaki et al., 2012) belongs to the Betacoronavirus genus, of the Sarbecovirus subgenus, of the Nidovirales order, of the Coronaviridae family (Zhou et al., 2020). Viruses of this latter are positive sense, single stranded RNA and are characterized by their large genomes, up to 33,000 nucleotides (nts). The genome of SARS-CoV-2 is of 30,000 nts (Wu et al., 2020; Zhou et al., 2020) encoding, among others, four structural proteins: envelope (E), membrane (M), nucleocapsid (N) and spike (S) which is used by the virus to enter the human cells by engaging human angiotensin-converting enzyme 2 (ACE2) receptor (Li et al., 2003). Several nonstructural proteins are encoded by ORF1a and ORF1b, that constitute two thirds of the viral genome and are essential for replication since they are translated into polyproteins (Wu et al., 2020), generating 16 nonstructural proteins (nsp 1 to nsp16) (Cagliani et al., 2020). As for the remaining part of the genome, it concerns mainly accessory proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF8 and ORF10). ORF8, not shared by all members of the Sarbecovirus subgenus, is of 366 nts in length, encoding a protein of 121 amino acids (Wu et al., 2020), and constitute with orf3b, orf10 and S, the major difference between SARSr-CoV and SARS-CoV (Chan et al., 2020; Yang et al., 2020).
ORF8 and the RBD encoding domain of the S genes constitute the most hypervariable regions in the genome of SARS-CoV (Chan et al., 2020; Wu et al., 2016) which together with three Macro domains in ORF1a constitute the most evolving regions in SARS-CoV-2 genome (Tan et al., 2020). Single Nucleotide Variations (SNVs) in ORF8 at 28144T>C (251T>C, L84S) characterize the SARS-CoV-2 S clade (Tang et al., 2020), leading to divergence of phylogenetic group (Tang et al., 2020; Velazquez-Salinas et al., 2020). SARS-CoV-2 ORF8 protein shares 94,3% and 26% homology with that of Bat-SL-CoVZC45 and early phase containing a full-length ORF8ab SARS-CoV (SARS-CoV_GZ02) respectively (Wu et al., 2020; Chan et al., 2020; Zhang et al., 2020). However, mid and late phase SARS-CoV strains (SARS-CoV_BJ01) that have 29-nucleotide deletion responsible for the splitting of ORF8ab into ORF8a and ORF8b (Oostra et al., 2007), exhibit respectively only 10% and 16% homology with that of SARS-CoV-2 ORF8 protein (Zhang et al., 2020). The exact function of ORF8ab, as well as ORF8a and ORF8b is still elusive (Mohammad et al., 2020). Recently, it has been shown that SARS-CoV-2 ORF8 belongs to novel families of Immunoglobulin proteins (Tan et al., 2020; Flower et al., 2020), suggesting a role in modulating host immunity, and thus, providing support to the work of Zhang and colleagues that suggested its implication in downregulating MHC-1 (Zhang et al., 2020), or in inhibiting type I interferon (IFN-β) (Li et al., 2020).
The number of SARS-CoV-2 sequences is increasing (159 thousands sequences by October 23rd) from different countries, and are available on GISAID EpiCoV™ (Global Initiative on Sharing All Influenza Data; https://www.gisaid.org/) (Shu and M.J., 2017). Despite that, the mutation rate of SARS-CoV-2 is still low in comparison to other RNA viruses. However, mutations in the genome still occur as the virus propagates in humans. This will certainly lead to viral genomic diversity that is important for viral adaptation to different hosts and environments (Hufsky et al., 2020). Characterization of mutations, especially in fast evolving genes like ORF8 (Tan et al., 2020) is important not only for pathogenesis and immune modulation but also for drugs and diagnostic tests, especially ORF8 which is one of the viral proteins that has been shown to elicit strong and specific antibody response (Hachim et al., 2020; Wang et al., 2020a). In this context, ORF8 has been studied not only at the evolutionary level (Pereira, 2020; Chen et al., 2020) but also at the mutational one (Hassan et al., 2020), in which only 78 mutations in ORF8 were identified.
In our present study, we were interested in a more global characterization of ORF8 mutations that could have occurred in the ORF8 gene during the pandemic period from January to June 2020. We had identified 240 mutations and 2 deletions in 45,400 sequences in ORF8 and analyzed the genetic diversity of mutations and their geographic distribution, in addition to making a phylogenetic tree of the ORF8 variants' sequences, together with that of bat and pangolin.
2. Materials and methods
2.1. Genome collection and variant analysis
The sequence of Wuhan-Hu-1 (NCBI, NC_045512.2), was used as a reference sequence for numbering, nucleotide location, and amino acid variations. ORF8 nonsynonymous variants and deletions were retrieved from CoV-GLUE (Singer et al., 2020). The total number of sequences that were analyzed by CoV-GLUE was 49,780. Only 45,400 sequences were taken into consideration, according to the exclusion criteria defined by CoV-GLUE (length between 29,000 and 35,000 nts, covers 95% of the coding sequence, and does not contain more than 10 unique variants, or frameshifting deletion or insertion in comparison to Wuhan-Hu-1).
Full genome viral DNA sequences were collected from GISAID EpiCoV™, from January to June 2020. ORF8 DNA and protein sequences were extracted manually and verified for the presence of variant, as well as for the presence of more than one variant in each sequence.
Sequences that contain N stretches were excluded from phylogenetic trees and nucleotide diversity analysis, in addition to the deletions of the latter. Those that contain symbols like K, R, M, Y, S, W at the DNA level were verified after Blast with the Wuhan-Hu-1 (NCBI, NC_045512.2) ORF8 DNA sequence and corrected manually for phylogenetic analysis and mutation counting.
The effect of each variant on ORF8 protein was analyzed by the Protein Variation Effect Analyzer (PROVEAN v1.1.3.) (Choi and Chan, 2015). The score threshold for prediction was −2.5.
2.2. DNA and protein sequence alignments
MAFFT (Katoh and Standley, 2013) was used for pairwise alignment. Alignment of DNA and protein sequences was accomplished on all sequence variants as well as on the followings: SARS-COV-2 Wuhan-Hu-1 (NC_045512.2, YP_009724396.1), Bat coronavirus RaTG13 (MN996532.1, QHR63307.1), Bat SARS-like coronavirus isolate Bat-SL-CoVZC45 (MG772933.1, AVP78036.1), bat SARS-like coronavirus WIV1 (KF367457.1, AGZ48840.1), Bat SARS-related coronavirus isolate F46 (KU973692.1, ARO76390.1), Pangolin coronavirus isolate MP789 (MT121216.1, QIG55952.1), SARS coronavirus GZ02 (AY390556.1, AAS00010.1) and Paguma Civet coronavirus (AY515512.1). ORF8 DNA sequences of different species were extracted manually after alignment.
Aligned sequences were seen by Seaview 5.0.4 (Gouy et al., 2010) and Biological Sequence Alignment Editor (BioEdit 7.2.5) (Hall, 1999).
ORF8 DNA and protein sequence identity were determined using Needleman-Wunsch Global Align Nucleotide/Protein Sequences.
2.3. Nucleotide diversity and population genetic analysis
The sequences downloaded from GISAID EpiCoV™ were subjected to cleaning, and only 1500 sequences were taken from North America, Oceania, Asia and Europe. As for Africa and South America, the available sequences that were taken were 344 from the former and 395 from the latter.
Pi which is the average number of nucleotide differences per site between two sequences was performed using DnaSP (DNA Sequence Polymorphism) 6.12.03 (Rozas et al., 2017). Files were converted by DnaSP in order to accomplish Fst analysis and Tajima's D test by Arlequin 3.5.2.2 (Excoffier and Lischer, 2010).
Phylogenetic tree construction.
Regarding phylogenetic tree construction, only one representative sequence of each variant was chosen.
DNA and protein phylogenetic trees were constructed with and without Bat coronavirus (RaTG13 MN996532.1, QHR63307.1), Bat SARS-like coronavirus isolate Bat-SL-CoVZC45 (MG772933.1, AVP78036.1), and Pangolin coronavirus isolate MP789 (MT121216.1, QIG55952.1). In addition, SARS-CoV-2 ORF8 DNA and protein sequences (gene ID: 43740577, YP_009724396.1,) and the 2 deletions' sequences, 269 and 259 of ORF8 DNA and protein sequences respectively, were included. After sequence alignment by MAFFT, RAxML-HPC-PTHREADS-SSE3 version 8.2.10 (Stamatakis, 2006), on raxmlGUI 2.0.0 for windows, was used for the inference of the phylogenetic trees using maximum likelihood approach, under GTRGAMMA and PROTGAMMA options for DNA and protein sequences respectively, with 1000 bootstrap replicates. Phylogenetic trees were visualized on Interactive Tree Of Life (iTol) web site (https://itol.embl.de/) (Letunic and Bork, 2007).
3. Results
3.1. Sequence alignment between SARS-CoV-2 ORF8 with other animal viral species
Sequence alignment between SARS-CoV-2 ORF8 DNA and protein sequences with other animal viral species are shown in Table 1 in order to determine the degree of conservation at the DNA and protein levels.
Table 1.
ORF8 DNA and protein sequence homology between SARS-CoV-2 and other animal viral species.
| SARS-CoV2 ORF8 | Bat CoV RaTG13 | Bat-SL-CoV WIV1 | Bat-SL-CoVZC45 | Bat SARSr CoV KU | Pangolin CoV | Paguma civet | SARS-CoV |
|---|---|---|---|---|---|---|---|
| DNA homology | 91% | 68% | 89% | 54% | 80% | 56% | 56% |
| Protein homology | 95% | 58% | 94% | 27% | 82% | 30% | 30% |
The Conserved amino acids among different animal viral species are shown in Fig. 1 .
Fig. 1.
ORF8 Protein sequence alignments of SARS-CoV-2 ORF8 with other animal viral species.
ORF8 Protein sequence alignments of Human SARS-CoV-2 (YP_009724396.1), Bat coronavirus RaTG13 (QHR63307.1), Bat-SL-CoVZC45 (AVP78036.1), Bat SARS-like coronavirus WIV1 (AGZ48840.1), Bat SARS-related coronavirus isolate F46 (ARO76390.1), Pangolin coronavirus (QIG55952.1), SARS CoV GZ02 (AAS00010.1), and Paguma Civet coronavirus using MAFFT.
3.2. Nonsynonymous variants
240 ORF8 different nonsynonymous mutations were identified in 6337 sequences retrieved from COV GLUE (Singer et al., 2020) and downloaded from GISAID EpiCoV™. 573 sequences contained more than one variant, and 5165 sequences had only one variant, thus reducing the total number of sequences with variants in ORF8 to 5738 sequences (≈13% of 45,400 sequences have mutation in ORF8). It was found that 17 out of 240 mutations were nonsense, thus representing 7%, whilst missense mutations represented around 93% of the nonsynonymous mutations. Fig. S1 shows the aligned variants' sequences with SARS-CoV-2 ORF8 (YP_009724396.1).
The effect of each mutation on ORF8 protein was predicted by PROVEAN. Table S1 shows the list of nonsynonymous mutations including their effects on ORF8 protein and their locations and conservations, as well as the number of sequences of each one.
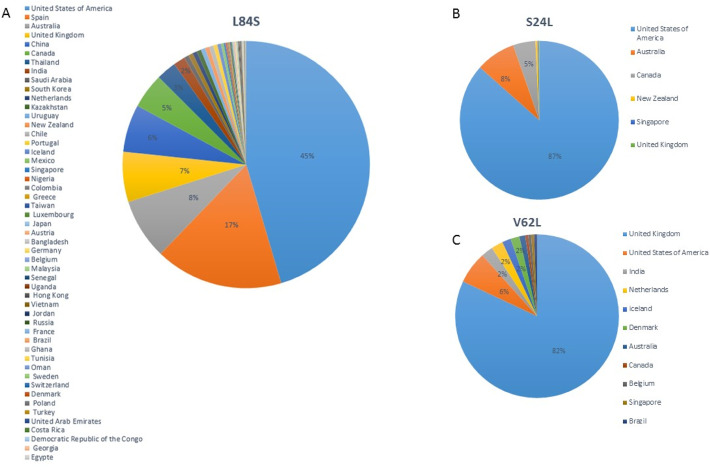
Concerning nonsynonymous mutations, the number of sequences with L84S worldwide is 3233, followed by S24L in 879 sequences mostly from United States of America (USA), V62L in 172 (Fig. 2 ), A65S in 83 sequences, A51V in 44 sequences, S67F in 38 sequences, Q18* in 29 sequences, and E19D and T26I were each found in 27 different sequences (Table S2).
Fig. 2.
Geographic distribution of most frequent SARS-CoV-2 ORF8 variants.
Representation and distribution of the three most frequent SARS-CoV-2 ORF8 variants. A: L84S; B: S24L and C: V62L. L84S is highly present in USA, Spain, Australia and United Kingdom. S24L is highly present in USA, followed by Australia and Canada. United Kingdom has the highest V62L representation worldwide. This representation is relative, because it depends on the number of SARS-CoV-2 genome sequencing that is variable between countries, and does not necessarily reflects the reality. Less than 2% are not indicated.
74 different double co-mutations were also observed, with L84S, V62L being the most frequent, and were found in 330 sequences, followed by L84S,A65S and L84S,E19D that were found in 28 and 27 different sequences respectively, and finally S24L, S69L that were found in 17 different sequences.
Twelve triple co-mutations were also observed, where for example, L84S,V62L,P36S was found in 4 sequences, whilst each of L84S,V62L,I88V and V62A,R48G,V49L co-mutation was observed in 2 different sequences.
Variants, such as V62L,R115S (EPI_ISL_469002), V62L,K53N (EPI_ISL_459067), L84S,V62L,C102Y (EPI_ISL_436434), I121L,F120V,L118* (EPI_ISL_454578), C102F,L60F (EPI_ISL_451832), A65S,V62F,Q72H (EPI_ISL_456596), and A65T,C83Y (EPI_ISL_430806) were excluded from protein and DNA phylogenetic trees construction because of the uncertainty of their associations (presence of letters, like K, R, Y, M, S, W reflecting the presence of virus quasi-species). Others such as P70S (EPI_ISL_425132), I39V,P38V (EPI_ISL_434511), E64V (EPI_ISL_455774), H112R (EPI_ISL_465538), I74L (EPI_ISL_455577), Q29R (EPI_ISL_440153), Q91H (EPI_ISL_428962), A14S (EPI_ISL_462306) and S24L,D34E,D35E, N89S (EPI_ISL_427171), were not included in the protein and in the DNA phylogenetic trees construction due to the presence of N stretches in ORF8 sequence. L7-I121del was excluded from the protein tree because it was of the same sequence as L7*.
P85T and F86S were not found after sequence verification. However, V62F was found in our analysis (EPI_ISL_456596) but not documented by COV-GLUE.
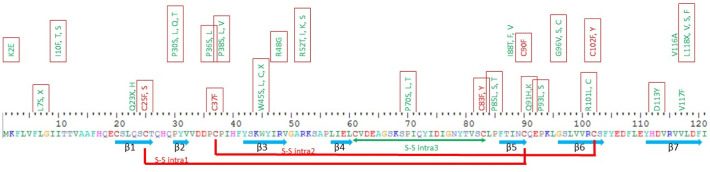
Thus, 125 mutations, that are deleterious to ORF8 protein, were found in SARS –CoV-2 ORF8, with 59 of them were found to be at conserved amino acids among 8 viral species (Fig. 3 ).
Fig. 3.
ORF8 Mutations at conserved amino acids between 8 viral species.
Protein sequence of SARS-CoV-2 ORF8 (YP_009724396.1), β-strands and disulfide bridges are indicated below the protein sequence (Flower et al., 2020). 59 mutations that were found at conserved amino acids between 8 viral species are indicated above. Mutations at Cysteines 25, 37, 83, 90 and 102 that form disulfide bridges are indicated in red. No mutation was found at C61. Some variants were found at conserved amino acids and were located in β-strands like Q23X(H), C25F(S), P30S (L,Q,T), W45S(L,C,X), R48G, I88T (F,V), C90F, G96V(S,C), R101(L,C) and C102F(Y), D113Y, V116A, V117F and L118X(V,S,F). Red squares indicate deleterious mutations. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. ORF8 diversity evaluation and evolution at population level
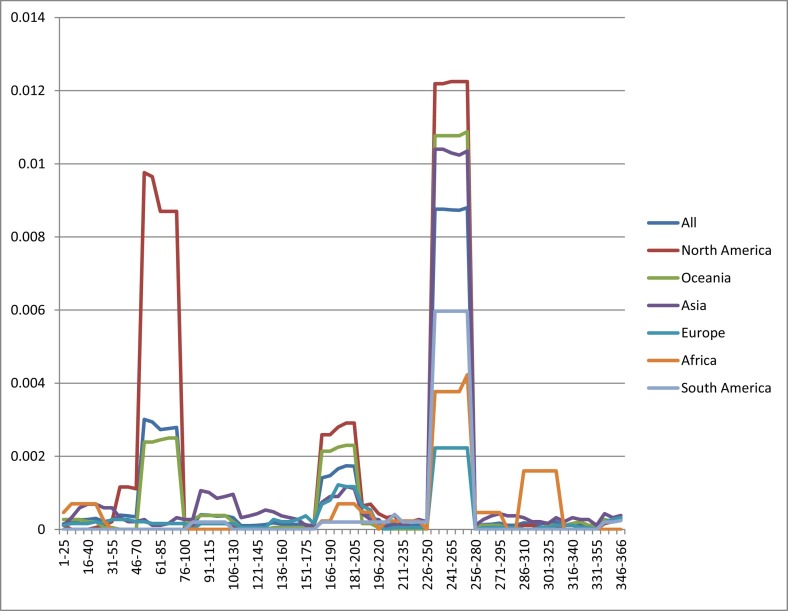
The number of sequences obtained post-cleaning included 19,000 from Europe, 6000 from Asia, 9000 from North America, 6000 from Oceania, 344 from Africa, and 395 from South America. The analysis of Pi distribution over the length of ORF8 in six continental sequences revealed four regions of diversity including 51–95, 161–205, 245–260 and 286–316 (Fig. 4 ). Actually, the region from 51 to 95 was found to be predominant in North America and Oceania but not in the other continents whilst that from 161 to 205 was found mainly in North America and Oceania, but in low number. Moreover, the region from 245 to 260 was found to be abundant in all continents, whilst that from 286 to 316 appeared mainly in Africa.
Fig. 4.
ORF8 nucleotide diversity analysis in six continents.
Pi analysis of ORF8 sequence at level 25, step 5, in six continents, reveals the presence of 3 pics: from 51 to 95, 161 to 205, 245 to 260 and 286 to 316. X axis represents the level of DNA sequence whilst Y axis represents the Pi value.
Pi on whole sequence showed that the highest diversity is in North America, whilst the lowest one is in Europe. The low diversity in Africa and South America could be attributed to the low number of available sequences.
Tajima's D test demonstrated that the population expansion in Africa and South America is weak, whereas, in North America it is slightly weak and hardly significant. The highest value was observed in Asia and Europe.
Fst analysis revealed that most of the value was significant. The highest value was between North America and Europe, with 9% of differentiation; however, low value was obtained between North America and Oceania and between South America and the other continents.
3.4. Deletions
A deletion of 6 bases (Refseq: 28090-28095del (GTTCTA)) was reported in 16 different sequences, mostly from England, from the GR, GH, G and O clades leading to G66-S67del and K68E substitution in ORF8 protein. The other deletion is of 344 nts from 27,910 to 28,254 causing a deletion of amino acids from Leucine at position 7 to Isoleucine at position 121 (L7-I121del), was found in two sequences (Table 2). This deletion was observed in two sequences from Bangladesh. Other deletions were not included in our study, like 62 nt deletion (EPI_ISL_452497) and 138-nt deletion (EPI_ISL_426967) due to the presence of N stretches at the beginning of ORF8 gene in the former, and the presence of N in the codon of the fourth amino acid in the latter. However, the 382 deletion (EPI_ISL_414378) (Su et al., 2020) was not included because it causes the complete loss of ORF8 protein.
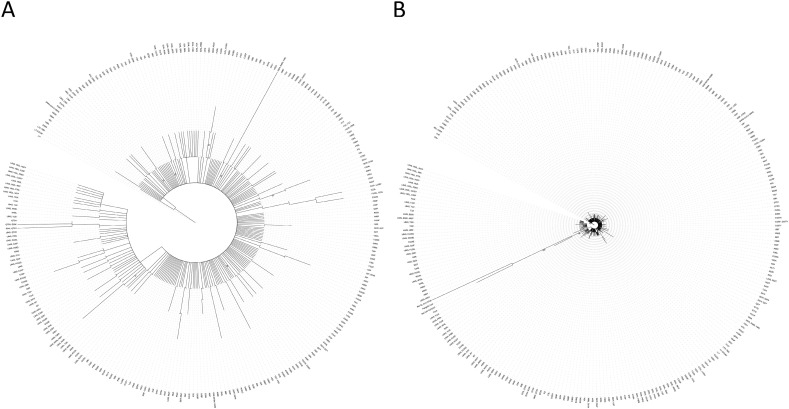
3.5. Phylogenetic trees
Phylogenetic trees' construction using maximum likelihood approach with 1000 bootstrap replicates of ORF8 DNA sequences without bat and pangolin ORF8 revealed the presence of clades. Besides the L84S which is the largest clade, others, such as S24L, V62L, Q72H, R52, and I121L were also observed. L84S and V62L associated with other nonsynonymous mutations constituted a subclade inside the L84S clade in the DNA tree (Fig. 5A) showing a distinct clade in the protein tree (Fig. S2). The maximum bootstrap value was 52 and 57 for Protein and DNA phylogenetic trees, respectively.
Fig. 5.
Phylogenetic tree of SARS-CoV-2 ORF8 DNA variants.
A: ORF8 DNA of Human SARS-CoV-2 (SARS CoV2 ORF8) (gene ID: 43740577), and the 269 variants sequences and 2 deletions' sequences (G66-S67del, K68E, L7-I121del) were analyzed. B: Bat coronavirus RaTG13 (MN996532.1), Bat SARS-like coronavirus isolate bat-SL-CoVZC45 (MG772933.1), and Pangolin coronavirus isolate MP789 (MT121216.1), were added to the phylogenetic analysis. Phylogenetic construction was performed using maximum likelihood approach, under GTRGAMMA option, with 1000 bootstrap replicates. Bootstrap value from 50 and above were indicated. Maximum bootstrap value in A is 57, and 100 in B.
A phylogenetic tree with Bat CoV RaTG13, Bat-SL-CoVZC45, and Pangolin Coronavirus reveals that L84S is closer to them than to the SARS-COV-2 Wuhan-Hu-1 reference sequence (indicated as SARS COV2 ORF8) (Fig. 5B). The maximum bootstrap value was 78 and 100, between Bat CoV RaTG3 and the Bat-SL-CoVZC45 and pangolin CoV group at the protein and DNA trees, respectively.
4. Discussion
As the epidemic is growing, and the number of genome sequencing is increasing, new viral mutations will appear creating a genomic diversity of SARS-CoV-2. Despite the fact that the mutation's rate in SARS-CoV-2 is less than that of other RNA viruses, like influenza (Nextstrain.org), 240 different nonsynonymous mutations, from January to June 2020, were detected in the ORF8 gene leading to nonsense and missense mutations of 7 and 93%, respectively. These results go in the same direction as for the missense and nonsense mutations in the ORF8 gene (Koyama et al., 2020) and SARS-CoV-2 genome (Pathan et al., 2020), in which they show that the missense mutations were higher than the nonsense ones.
Among the 240 nonsynonymous mutations, L84S, the most frequent is located between C83 and P85 (Flower et al., 2020). It seems that this variant has no effect on protein structure (Flower et al., 2020). Likewise, S24L which is part of the β1 strand (Tan et al., 2020; Flower et al., 2020) and V62L come next in the order. Those analyses are in accordance with the results that were obtained by Pi analysis since these variants are located in the three major regions. To note, the highest value of the second and third pics that refer to North America is predominant in the United States of America (Data not shown) because of the double co mutation L84S and V62L.
The lowest value of Tajima's D test was in Asia and Europe indicating a large expansion in the viral population. In Asia, Tajima's D test high value could reflect the large number of affected patients and the low number of sequenced viruses, whilst in Europe, it could be related to the high number of infected patients, as was the case in Italy and Spain, or to the so-called herd immunity that was the objective of United Kingdom. Indeed herd immunity strategy allows a rapid expansion of the virus population from different sources allowing the virus population to accumulate an excess of rare mutations. Kim and colleagues (Kim et al., 2017) showed that Tajima's D values are a good indication of the epidemiological dynamic. D values decrease rapidly with the increase of infected individuals and increase with the decrease in the number of infected patients. Our study is a good example of the possible usage of D values, to assess the virus dynamics in a population.
Mutations could be dynamic, as it has been proposed that some mutations like L84S are disappearing, and there is emergence of new ones that render the virus more infective, like S24L that was first sequenced in the USA (Wang et al., 2020b). This could explain the weak signal of population expansion whose distribution of S24L, on a large number of sequences, and, at the same time, of L84S increased the Pi value and added some equilibrium with the low frequency of new mutations.
The absence of S24L (except one case in the United Kingdom) is probably the reason behind high Fst value between North America and Europe. As for low Fst value between North America and Oceania, it could refer to high host travel between these continents, especially that the appearance of this variant appeared simultaneously, being earlier in USA (03-03-2020 first submitted).
Mutations in other SARS-CoV-2 genes like S gene (Korber et al., 2020), ORF3a (Issa et al., 2020), and others were reported (Koyama et al., 2020). However, ORF8 and S gene constitute the most hypervariable regions in the genome of SARS-CoV, and were shown to be recombination hot spots (Chan et al., 2020; Wu et al., 2016).
In addition to the deletions reported here, a 382 nt deletion (27,848:28229) of the ORF7b and ORF8 that includes its transcriptional regulator sequence (TRS) was documented from Singapore (Su et al., 2020) and Taïwan (Gong et al., 2020). It was found that this deletion was associated with higher replication fitness (Su et al., 2020). Other deletions in SARS-CoV-2 genome were observed in ORF7a (81 nts) from Arizona (Holland et al., 2020), apparently of the same origin (Su et al., 2020). This deletion could be a way of immune evasion since ORF8 elicits specific strong antibody response (Hachim et al., 2020; Su et al., 2020). This also could explain the presence of deletions and mutations like M1I (T,V) and nonsense ones.
Deletions were also observed in other Coronaviruses. SARS-CoV ORF8 deletions of 29, 82 and 415 nts were also observed (Consortium, 2004). The deletion of 29 nts in SARS CoV ORF8ab was found in strains of mid and late phase whose effect has been a matter of debate. It was suggested that it is involved in adaptation of SARS-CoV to inter human transmission (Liu et al., 2014) leading to a protein of new function adequate to later stages of epidemic (Consortium, 2004; Guan et al., 2003). Whilst Muth and colleagues (Muth et al., 2018) observed a reduction in viral replication, for them, this deletion provide viral survival despite reduced fitness due to a founder effect. According to Worobey and colleagues (Worobey and Holmes, 1999), deletions are a way to get rid of accumulated mutations in some DNA fragments.
SARS-CoV-2 ORF8 protein has been shown to be implicated in immune evasion by downregulating surface expression of MHC-1 (Zhang et al., 2020). Wang and colleagues (Wang et al., 2020b) showed that L84S decreases the function of ORF8, and thus is beneficial for human immune system, whilst S24L increases its function since it reinforces protein folding stability. As for deletions, 382 nts deletion was shown to cause milder infection (Young et al., 2020).
It would be of great importance to decipher the effects of variants and deletions experimentally on ORF8 protein structure and function, and to analyze the effects of these mutations and their relations to viral infectivity and their impacts on disease severity and clinical features since D614G in the S protein seems to enhance infectivity but not disease severity (Korber et al., 2020), and the deletions close to the spike S1/S2 cleavage were more related to mild symptoms (Andres et al., 2020). In addition, It would be interesting to track these mutations in order to see if the virus is fixing some variants that could help in tracking the spread of the disease (Grubaugh et al., 2020) and could be important for pathogenicity.
Phylogenetic analysis of protein and DNA sequences of SARS-CoV-2 ORF8 variants, together with that of Bat and Pangolin, revealed that L84S (L84S, A65V) is closer to both of them. The highest bootstrap value was obtained between Bat RaTG3 and Bat-SL-CoVZC45 and pangolin CoV group due to the high level of divergence compared to those among the sequences of ORF8 variants. In these variants, the low value of bootstrap could be due to the small size of protein sequence (121 aa) and to the low level of divergence between sequences which, in most cases, is only a single amino acid (i.e. one base at the level of DNA sequence). The exception to this, is the sequences that have double or triple co mutations in which we observed the presence of clades and subclades. To note that, L84S clade is also dependent on another variant located in the ORF1ab at 8782C>T, and this makes the possibility that L84S clade could be more ancestral than the L clade especially with the presence of COVID19 asymptomatic patients. This could also make the possibility that human SARS-CoV-2 was propagated before the sequenced virus of Wuhan.
Last but not least, this work revealed the effectiveness of COV-GLUE and its accuracy in detecting nonsynonymous mutations and deletions in ORF8 gene because only V62F was not documented, and P85T and F86S were not found among the 240 mutations.
5. Conclusion
ORF8 gene is one of the coronaviruses accessory proteins that offer flexibility and that are prone to mutations and one of the most hypervariable regions in their genomes. 240 different nonsynonymous mutations and 2 deletions were found in SARS-CoV-2 ORF8 gene in 45,400 sequences. About half of these mutations are deleterious to ORF8 protein, and the quarter of them are among animal viruses' conserved aminoacids. Those mutations, regardless of their effects on ORF8 itself, might have an effect on SARS-CoV-2 biology, and as a result, might curb the discovery of antiviral drugs and vaccine as well, especially that SARS-CoV-2 ORF8 is one of the viral antigens that elicit host immune response. Further analysis of these mutations at the genetic and functional levels together with the clinical symptoms of patients will help understand host-virus interaction and might speed the eradication of the virus.
The following are the supplementary data related to this article.
ORF8 variant's sequence alignments. ORF8 protein sequence alignments of Human SARS-CoV-2 (YP_009724396.1) and ORF8 variants are shown. The sequences are the same as those that are used for phylogenetic trees, in addition to L7-l121del. Identical sequences are indicated as dots, whilst deletions and truncated sequences are shown as dashes.
Phylogenetic tree of SARS-CoV-2 ORF8 protein variants. A: ORF8 proteins of Human SARS-CoV-2 (SARS CoV2 ORF8) (YP_009724396.1), and the 259 variants' sequences and 1 deletion sequence (G66-S67del, K68E) were analyzed (L7X was the same as L7-I121del at the protein level, one of them L7X was chosen for phylogenetic construction). B: Bat Coronavirus RaTG13 (QHR63307.1), Bat-SL-CoVZC45 (AVP78036.1) and Pangolin Coronavirus (QIG55952.1) were added to the analysis. Phylogenetic construction was performed using maximum likelihood approach, under PROTGAMMA option, with 1000 bootstrap replicates. Bootstrap value from 50 and above were indicated. Maximum bootstrap value in A is 52, and 78 for B.
Supplementary tables
CRediT authorship contribution statement
AAK performed sequences downloading and bioinformatics analysis, as well as revision of the manuscript, GL participated in bioinformatics analysis, discussion and writing of the manuscript, LEZ conceived the project, participated in bioinformatics analysis, Writing- Original draft preparation.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
Acknowledgments
The authors like to thank GISAID team, for its extraordinary work, without them, we are unable to do such work. CoV-GLUE web site team, are grateful for permitting us the use of its Data. Sincere thanks to Dr. Letunic, the responsible of iTol website, for offering a free subscription, due to the economic crisis in Lebanon. Many thanks to Dr. Abbas Hijazi. A sincere thanks to Dr Hussein Fayyad Kazan and Iman EL ZEIN for their English correction of this paper.
Funding
No funding.
References
- Andres C., et al. Naturally occurring SARS-CoV-2 gene deletions close to the spike S1/S2 cleavage site in the viral quasispecies of COVID19 patients. Emerg. Microbes Infect. 2020;9(1):1900–1911. doi: 10.1080/22221751.2020.1806735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R., et al. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect. Genet. Evol. 2020;83:104353. doi: 10.1016/j.meegid.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., et al. Extended ORF8 gene region is valuable in the epidemiological investigation of severe acute respiratory syndrome-similar coronavirus. J. Infect. Dis. 2020;222(2):223–233. doi: 10.1093/infdis/jiaa278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Chan A.P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31(16):2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium C.S.M.E. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303(5664):1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Drosten C., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Excoffier L., Lischer H.E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Flower T.G., Buffalo C.Z., Hooy R.M., Allaire M., Ren X., Hurley JH. Structure of SARS-CoV-2 ORF8, a rapidly evolving coronavirus protein implicated in immune evasion. bioRxiv [Preprint] 2020 Aug 27 doi: 10.1101/2020.08.27.270637. Update in: Proc Natl Acad Sci U S A. 2021 Jan 12;118(2): PMID: 32869027; PMCID: PMC7457612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y.N., et al. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microbes Infect. 2020;9(1):1457–1466. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Grubaugh N.D., Petrone M.E., Holmes E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020;5(4):529–530. doi: 10.1038/s41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Hachim A., et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020 Oct;21(10):1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hassan S.S., et al. A unique view of SARS-CoV-2 through the lens of ORF8 protein. bioRxiv. 2020 doi: 10.1016/j.compbiomed.2021.104380. p. 2020.08.25.267328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L.A., et al. An 81-nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (January to March 2020) J. Virol. 2020;94(14) doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufsky F. Computational strategies to combat COVID-19: useful tools to accelerate SARS-CoV-2 and coronavirus research. Brief. Bioinform. 2020 Nov doi: 10.1093/bib/bbaa232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa E., et al. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. mSystems. 2020;5(3) doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Omori R., Ito K. Inferring epidemiological dynamics of infectious diseases using Tajima's D statistic on nucleotide sequences of pathogens. Epidemics. 2017;21:21–29. doi: 10.1016/j.epidem.2017.04.004. [DOI] [PubMed] [Google Scholar]
- Korber B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. (e19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23(1):127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Li W., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.X., et al. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad S., et al. SARS-CoV-2 ORF8 and SARS-CoV ORF8ab: genomic divergence and functional convergence. Pathogens. 2020:9(9). doi: 10.3390/pathogens9090677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth D., et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018;8(1):15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra M., de Haan C.A.M., Rottier P.J.M. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 2007;81(24):13876–13888. doi: 10.1128/JVI.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan R.K., Biswas M., Khandaker M.U. Time series prediction of COVID-19 by mutation rate analysis using recurrent neural network-based LSTM model. Chaos, Solitons Fractals. 2020;138:110018. doi: 10.1016/j.chaos.2020.110018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F. Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene. Infect. Genet. Evol. 2020;85:104525. doi: 10.1016/j.meegid.2020.104525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J., et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34(12):3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- Shu Y., M.J. GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22(13):30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J., et al. CoV-GLUE: a web application for tracking SARS-CoV-2 genomic variation. Preprints.org. 2020 doi: 10.20944/preprints202006.0225.v1. 2020060225. [DOI] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Su Y.C.F., et al. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio. 2020;11(4) doi: 10.1128/mBio.01610-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., et al. Novel immunoglobulin domain proteins provide insights into evolution and pathogenesis of SARS-CoV-2-related viruses. mBio. 2020;11(3) doi: 10.1128/mBio.00760-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Duan Y., Zhang H., Wang Y., Qian Z., Cui J., Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Salinas L., et al. Positive selection of ORF3a and ORF8 genes drives the evolution of SARS-CoV-2 during the 2020 COVID-19 pandemic. Front. Microbiol. 23 October 2020;11 doi: 10.3389/fmicb.2020.550674. 2020.04.10.035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. Accurate siagnosis of COVID-19 by a novel immunogenic secreted SARS-CoV-2 orf8 protein. mBio. 2020;11(5) doi: 10.1128/mBio.02431-20. p. e02431-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Chen J., Gao K., Hozumi Y., Yin C., Wei G. Characterizing SARS-CoV-2 mutations in the United States. Res. Sq. [Preprint] 2020 Aug 11 doi: 10.21203/rs.3.rs-49671/v1. PMID: 32818213; PMCID: PMC7430589. [DOI] [Google Scholar]
- Worobey M., Holmes E.C. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 1999;80(Pt 10):2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]
- Wu Z., et al. ORF8-related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2016;213(4):579–583. doi: 10.1093/infdis/jiv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., et al. SARS-CoV-2: characteristics and current advances in research. Virol. J. 2020;17(1):117. doi: 10.1186/s12985-020-01369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young B.E., et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396(10251):603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang Y., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. Preprint at bioRxiv. 24 May 2020 doi: 10.1101/2020.05.24.111823. PPR: PPR166823. [DOI] [Google Scholar]
- Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ORF8 variant's sequence alignments. ORF8 protein sequence alignments of Human SARS-CoV-2 (YP_009724396.1) and ORF8 variants are shown. The sequences are the same as those that are used for phylogenetic trees, in addition to L7-l121del. Identical sequences are indicated as dots, whilst deletions and truncated sequences are shown as dashes.
Phylogenetic tree of SARS-CoV-2 ORF8 protein variants. A: ORF8 proteins of Human SARS-CoV-2 (SARS CoV2 ORF8) (YP_009724396.1), and the 259 variants' sequences and 1 deletion sequence (G66-S67del, K68E) were analyzed (L7X was the same as L7-I121del at the protein level, one of them L7X was chosen for phylogenetic construction). B: Bat Coronavirus RaTG13 (QHR63307.1), Bat-SL-CoVZC45 (AVP78036.1) and Pangolin Coronavirus (QIG55952.1) were added to the analysis. Phylogenetic construction was performed using maximum likelihood approach, under PROTGAMMA option, with 1000 bootstrap replicates. Bootstrap value from 50 and above were indicated. Maximum bootstrap value in A is 52, and 78 for B.
Supplementary tables