Abstract
Coronavirus disease (COVID-19) caused by SARS-CoV-2 has spread since the end of 2019 and has resulted in a pandemic with unprecedented socioeconomic consequences. This situation has created enormous demand for the improvement of current diagnostic methods and the development of new diagnostic methods for fast, low-cost and user-friendly confirmation of SARS-CoV-2 infection. This critical review focuses on viral electrochemical biosensors that are promising for the development of rapid medical COVID-19 diagnostic tools. The molecular biological properties of SARS-CoV-2 as well as currently known biochemical attributes of infection necessary for biosensor development are outlined. The advantages and drawbacks of conventional diagnostic methods, such as quantitative reverse-transcription polymerase chain reaction (qRT-PCR), are critically discussed. Electrochemical biosensors focusing on viral nucleic acid and whole viral particle detection are highlighted and discussed in detail. Finally, future perspectives on viral electrochemical biosensor development are briefly mentioned.
Keywords: Antibody, CRISPR/Cas9, Diagnostics, Electrochemistry, Electrode
1. Introduction
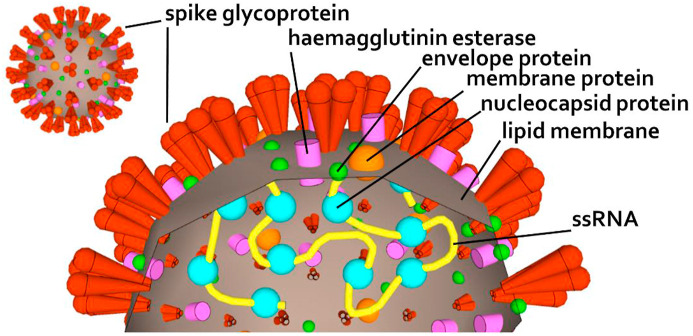
The new coronavirus SARS-CoV-2 first emerged in December 2019 in Wuhan, Hubei Province, China [1]. Due to its highly contagious nature, it has been rapidly spreading worldwide [2]. SARS-CoV-2 belongs to the order Nidovirales, family Coronaviridae, subfamily Orthocoronavirinae genus Betacoronavirus and subgenus Sarbecovirus, which also include severe acute respiratory syndrome-related coronavirus (SARS-CoV) and Middle East respiratory syndrome-related coronavirus (MERS-CoV) [[3], [4], [5]]. Virions of the Coronaviridae family are roughly spherical, 118–140 nm in diameter and notable for the large spike glycoprotein that extends from the virus envelope, which gave the name to the whole family (Fig. 1 ).
Fig. 1.
Schematic of the SARS-CoV-2 structure showing the surface structure (left) and cross-section (right).
In 2002, SARS-CoV emerged in Guangdong Province (China) and spread to five continents through air travel routes, infecting 8098 people and causing 774 deaths. No cases of SARS-CoV have been reported worldwide since 2004. MERS-CoV causing severe lower respiratory tract infection in people emerged in the Arabian Peninsula in 2012, where it remains a major public health concern, and was exported to 27 countries, infecting almost 2500 individuals and claiming 858 lives. By the end of August 2020, more than 21.2 million confirmed COVID-19 cases had been reported by the WHO, including 761,000 deaths.
SARS-CoV-2 mortality can significantly differ depending on the geographic area. Reports show that SARS-CoV-2 is rapidly moving across countries, and genomes with new mutation hotspots are emerging [6]. Thirteen variation sites in SARS-CoV-2 open reading frame 1 (ORF1) were recently characterized, with some positions showing mutation rates up to 30% [7]. The mutagenic process of the viral genome depends on viral enzymes that replicate nucleic acids, which is affected by little or no proofreading capacity and/or post-replicative nucleic acid repair. In most viruses, RNA polymerase lacks proofreading capability, with some exceptions, such as the Nidovirales order (to which the Coronavirus genus belongs). Eight new recurrent RNA-dependent RNA polymerase (RdRp) mutations in SARS-CoV-2 were characterized by Pachetti et al., with differences in occurrence in Europe, Asia and North America. Each of them possesses a different mutation pattern, showing the contribution of RdRp to maintaining its proofreading capability [6]. Due to the aforementioned mutations, early detection and monitoring of spread and disease development by standard RT-PCR might be challenging due to possible primer mismatches with targets [8]. Hence, focusing on conserved regions of the viral genome and multiple virus gene targets were recommended to minimize the probability of false-negative results [9].
Among quarantine, staying home and social distancing measures, rapid testing represents one of the cornerstones in slowing virus transmission and helps to understand SARS-CoV-2 epidemiology. Currently, qRT-PCR, antigen and serological (IgM and IgG antibodies) tests are used for COVID-19 diagnosis. PCR tests are considered the gold standard of COVID-19 testing, and together with antigen tests, PCR tests are able to determine active infection in patients who shed the virus. Although qRT-PCR possesses adequate sensitivity to determine early infection, false-negative or false-positive results in the range of tens of percent have been reported by several authors [10,11]. Although false-positive results have economic consequences of quarantine and contact tracing, in the case of false-negative results, infected people can spread the virus even without symptoms. Among the virus mutations mentioned above, Tahamran and Ardebili suggested that sample type, sampling and viral load kinetics can influence the results of PCR tests [12]. Serological tests determine blood anti-SARS-COV-2 IgM/IgG status and provide information about recent SARS-CoV-2 exposure (IgM and IgG positive) and past infection (IgM negative and IgG positive); however, IgM-positive and IgG-negative status can denote an active infectious state. Notably, no test can provide 100% sensitivity (the likelihood that the test will be positive for infected persons) or specificity (the likelihood that the test will be negative for non-infected persons), and all test results must be carefully interpreted with other clinical features, such as computed tomography images [13]. Woloshin et al. highlighted the urgent need for the determination of clinical and analytical test performance and for reference standards for measuring the sensitivity of SARS-CoV-2 tests in asymptomatic persons [14].
Electrochemical biosensors represent alternative approaches to detect viral nucleic acids or viral antigens and can contribute to the development of point-of care (POC) COVID-19 testing. Employment of “point-of-care-testing” (POCT) methods is worthwhile due to the prospect of immediate determination of the patient's negative/positive status at the site of testing within minutes or 1 h. Many POCT methods could be potentially useful for such purposes, but if the qRT-PCR cost, specificity and sensitivity requirements need to be fulfilled, the choice begins to be markedly limited.
The aim of this conceptual review is to discuss and conceptualize the use of electrochemical (bio)sensors for designing coronavirus POC analytical devices, which might be beneficial to maintain sensitivity, selectivity and costs at acceptable levels.
2. Analytical hallmarks of the viral target
2.1. Detection window
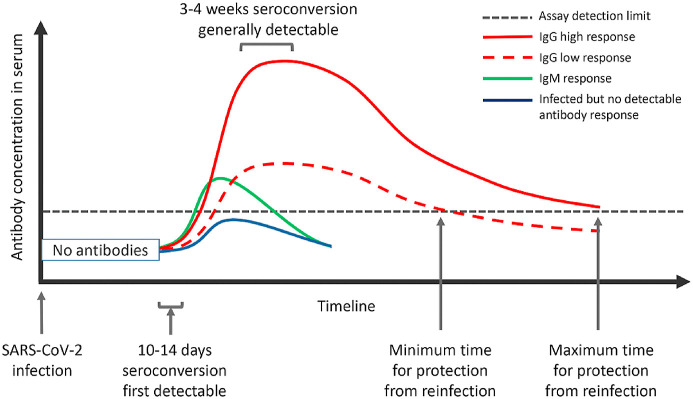
Many patients experiencing SARS-CoV-2 infection are not even hospitalized and have asymptomatic development. This fact causes many of these patients to not undergo qRT-PCR testing after interacting with their general practitioner. The critical point is that infectious persons are not determined and may still contribute to the worsening of the epidemiological situation. Fig. 2 describes the detection window when anti-SARS-CoV-2 antibodies cannot be detected and the different stages of immune response to infection (Fig. 2). In the first to second week after infection, nucleic acid-based POCT can represent an alternative to conventional qRT-PCR. However, after 10–14 days, when seroconversion occurs, antibody-based POCT can play an important role.
Fig. 2.
A schematic representation of the immune response following SARS-CoV-2 infection (adapted from Ref. [15]).
The main factor influencing the practitioner's choice of analytical method is the cost of the qRT-PCR procedure, which consists of 3 general parts: i) acquiring the sample (personal time and collection material), ii) transport to a certified laboratory and iii) testing in the certified laboratory (pre-analytical phase costs such as biosafety level certification, qRT-PCR kit costs, and administrative costs). Next, the POCT can benefit from decreased expenses to only i) acquire the sample, ii) kit costs, and iii) send the data via a network (e.g., smart city platforms) to the nearest epidemiologic authority.
If captured within the correct timeframe after disease onset, SARS-CoV-2-specific antibody responses can be helpful for the rapid determination of active infections and past infections. Hence, the added value of serological testing is the determination of past infection, where qRT-PCR fails. Seroconversion occurs from approximately 10 days after symptom onset, but the exact timing of IgM (green line) and IgG (red line) appearance is currently unclear. The IgG antibody titres rise from day 10 onwards to reach a peak whose height is likely to be influenced, on a case by case basis, by disease severity and virus load (difference between the solid red, and dashed red and blue lines). Currently, the exact level of antibody protection against reinfection (black dotted line), the duration of the total humoral immune response above this level and the rate of decline of mild or severe infection-induced antibody levels are not known. Similarly, the proportion of infected individuals who do not mount a protective immune response (green line) has not been determined [15].
2.2. SARS-CoV-2 PCR detection
The order Nidovirales contains positive-sense, single-stranded RNA (ssRNA) genomes that are capped and polyadenylated. The Nidovirales order contains enveloped viruses with the largest RNA genomes among currently known RNA viruses in the range of 12.7–33.5 kb, which can infect a variety of vertebrate and invertebrate hosts [16,17]. Unique among positive-sense ssRNA viruses, large nidoviruses encode a 3′-to-5′ exoribonuclease (ExoN) that is implicated in controlling RNA replication fidelity. Nidovirus genomes are typically organized into many ORFs occupying over 90% of the genome. The products of these regions predominantly control genome expression, replication, virus assembly and dissemination [16,18].
Most of the molecular diagnostics being developed for the diagnosis of COVID-19 infections involve qRT-PCR test methods similar to those developed for the diagnosis of SARS-CoV. Coronaviruses contain molecular targets within their genome that can be used for PCR assays, including genes of structural proteins, the envelope spike glycoprotein (S protein), envelope protein (E protein), membrane protein (M protein), helicase (Hel), and nucleocapsid protein (N protein), as well as genes required for viral replication, such as RdRp, haemagglutinin esterase (HE), and open reading frames ORF1a and ORF1b [[19], [20], [21]].
Centers for Disease Control and Prevention (CDC) recommends two N protein targets (N1 and N2) while the WHO recommends first line screening with the E gene assay followed by a confirmatory assay using the RdRp gene [22,23]. Many laboratories have developed assays independently of the CDC or WHO. Chan et al. developed three novel qRT-PCR assays targeting the RdRp/Hel, S, and N genes of SARS-CoV-2 [24]. Chu et al. described two 1-step qRT-PCR assays to detect two different regions (ORF1b and N) of the viral genome separately [23]. Two molecular targets (E and RdRp) were selected for testing in the study by Corman et al. [20]. The nucleocapsid and ORF1ab genes were chosen in other studies [[24], [25], [26]]. Despite a considerable number of recent studies there has been no indication that any one of these sequence regions used offers a significant advantage for clinical diagnostic testing. Ideal design would include at least one conserved region as well as a specific region to mitigate the effects of genetic drift, especially as the virus evolves within new populations [22].
3. Design of coronavirus biosensors
Undoubtedly, electrochemical biosensing of viruses is challenging and ambitious. However, electrochemistry provides several well-known advances for biosensor design, such as inherent high sensitivity, relatively low-cost sensors and equipment, user-friendly manipulation, fast analysis, and suitability for miniaturization, hence creating POC devices. Current state-of-the-art electrochemical biosensors and related technologies are critically discussed in subsequent paragraphs.
3.1. Recognition element
SARS-CoV-2 possesses several suitable targets for biological recognition elements. First, viral RNA is one. Viral nucleic acids are definitely the ideal target since they are suitable for early stage-detection when the IgM and IgG immune responses are still too low. Myriad studies have reported hybridization electrochemical biosensors aimed at viral nucleic acids and have claimed their reliable detection [[27], [28], [29], [30], [31], [32]]. However, most of these studies have used synthetic oligonucleotides as analytes, which are often just slightly longer than the corresponding probes. Such biosensors are suitable for the detection of PCR products where the quantity of amplicons is relatively high and the only interferents are polymerases, primers and nucleoside triphosphates, but they can hardly be used for the detection of viral genomic nucleic acids in real biological samples. Notably, regarding genetic complexity and genome size, the Coronaviridae family is the largest RNA virus identified to date (29.7 kb) [33].
Antibodies are accepted as the gold standard for biorecognition, and they are broadly used in medical diagnostic and therapeutic practice. Antibody affinity towards antigens (reaching the nanomolar range in the case of good antibodies) has resulted in various electrochemical biosensors ranging from enzyme-amplified ELISA-like to label-free formats. However, there are numerous challenges that antibody-based research faces. Many researchers have highlighted the insufficient characterization and low quality of antibodies used in biomedical research, causing their poor reliability by binding to a plethora of irrelevant antigens [34,35]. Engineering recombinant antibodies provided extensive possibilities for modifications with tags, labels or reporters and the production or creation of antibody fragments. In contrast to heterogeneous polyclonal antibodies, monoclonal and recombinant antibodies are homogeneous molecules that bind to single antigen epitopes. This attribute makes them more sensitive to pH, salt concentration or temperature change. Especially in the field of biosensors, where multiple washing steps are inherent parts of working procedures, such conditions can induce uncertainties in the results, which further highlights the need for antibody validation [36]. Recent advances in this field were reviewed by Park [37].
Regarding antibodies focusing on whole viral particles, the most problematic issue is their long-term reliability; the viral genome changes under immune system pressure and can result in epitope changes. Approximately 15 amino acids (discontinuous in the protein primary sequence) of the epitope come into spatial contact with the antibody paratope [38]. Of these 15 amino acids, 5 strongly influence antibody-antigen interactions, and their point mutation can decrease the relative binding constant by two or three orders of magnitude [39]. Currently, researchers focus on antibodies against the SARS-CoV-2 S protein of the viral envelope. By binding to the S protein, these antibodies prevent viral particles from interacting with cellular angiotensin-converting enzyme 2 (ACE2) receptors; hence, they are able to neutralize (neutralizing antibodies, nAbs) infection and should help with vaccine design [40]. Such antibodies are suitable for the development of whole viral particle biosensors since they focus on highly exposed parts of viral antigens [41]. Although there is overall structural homology between SARS-CoV and SARS-CoV-2 S proteins, Wrapp et al. reported that none of three SARS-CoV S protein receptor-binding domain (RBD) binding monoclonal antibodies bind to the SARS-CoV-2 S protein [42]. Although several commercially available antibodies against the S protein and other SARS-CoV-2 structural proteins (E, M and N protein) are available, their specificity against individual serotypes of the virus has not been determined. Notably, slow and long epidemics allow selection pressure to drift further and possibly create novel mutated antigens [43]. A well-reported case of an S protein point mutation (D614G), which appeared in early April 2020 and as of the end of the year has become the dominant variant in the pandemic, has been reported [44,45]. Hence the performance of biorecognition elements of viral biosensors and POC devices needs to be frequently re-evaluated.
Aptamers, short ssRNA or DNA molecules, are able to bind target molecules with high affinity and selectivity. Many aptamers focusing on inorganic molecules, proteins, cells and whole viral particles have been reported. Aptamer degradation can play an important role in the field of biosensors [40]. In particular, RNA aptamers are prone to degradation by RNases. The ubiquitous occurrence of RNases due to their secretion by human skin and air-dispersed microorganisms can lead to surface contamination. In addition, the presence of RNases in real samples, such as body fluids such as tears, saliva or sweat, can hamper the performance of aptasensors [46]. Hence, RNA aptamers need to be modified to protect against nuclease activity [47].
The development of antiviral aptamers is a difficult task. If whole viral particles are considered, membrane proteins are targeted first. However, full-length membrane protein purification is a rather complex issue. Natural protein structures cannot be mimicked by recombinant extracellular parts of proteins that are subject to vector-dependent post-translational modification and do not oligomerize. Cell-SELEX (systematic evolution of ligands by exponential enrichment), which uses whole cells instead of target proteins, was developed to overcome these drawbacks. Recently, an advanced SELEX method (Viro-SELEX) suitable for the development of membrane protein aptamers was developed [48]. It uses enveloped viruses, such as the baculovirus expression system, to incorporate membrane proteins, such as influenza haemagglutinin (HA), into the surrogate viral envelope and for whole viral particle aptamer enrichment through SELEX [49]. Recently, Song et al. reported two SARS-CoV-2 DNA aptamers targeting the RBD of the S protein [50]. The dissociation constants (Kd) of CoV2-RBD-1 (51-base) and CoV2-RBD-4 (67-base) were 5.8 and 19.9 nM, respectively. Based on previously selected SARS-CoV aptamers, Chen et al. designed an aptamer for the SARS-CoV-2 N protein, which was shown to be a promising early-stage diagnostic marker in serum [51,52].
There are other biorecognition elements that have been proven to be beneficial for electrochemical viral biosensor development. This advance can be demonstrated using the typical case of influenza. Sialic acid (SA), covering a broad range of neuraminic acid derivatives, is a part of host cell surface glycoproteins and glycolipids that are recognized by the viral glycoprotein HA and mediate endocytosis and infection. Notably, SA serves as a receptor for many other viruses [53]. Recognition of various SA derivatives and conformations by HA genotypes determines influenza virus transmission. Immobilized SA can serve as a recognition element of viral biosensors. For example, Hiroguchi et al. used various SA molecules immobilized on electrodes to distinguish between human and avian influenza A [54]. Analogous to the case for influenza, ACE2 which mediates SARS-CoV-2 entry into cells, can serve for biosensor biorecognition purposes [55].
In recent years, mimicking the immunological interaction between antibodies and antigens with molecular imprinting strategies has been trending [56]. Although the technology was originally designed for small molecules, molecular imprinting has extended towards supramolecular complexes including viruses. Still facing many challenges, such as virus size, fragility or instability in organic solvents, the most recent publications in the field address these challenges. For example, Gast et al. imprinted hexon protein (the most abundant and accessible protein of human adenovirus type 5 capsid) and reported high selectivity of this solution towards entire viral particles [57]. Successful imprinting of the SARS-CoV-2 structure on a polymer was demonstrated in a reprint by Parisi et al. [58]. They fabricated “monoclonal-type” plastic antibodies (nanoparticles using MIP technology) specific for the RBD of the SARS-CoV-2 S protein with no cross reactivity towards the RBD of the SARS-CoV S protein. Possession of the mentioned antibodies was also declared by the commercial company MIP Diagnostics Ltd. (Bedford, UK).
3.2. Sensitivity challenges of viral biosensors
The main challenge of viral electrochemical biosensors is their sensitivity. SARS-CoV-2 shares epidemiological attributes with the previous epidemic agents SARS-CoV and MERS-CoV. In the case of many infections, specific IgM antibodies can be detected after 3 days of non-immunodeficient patient infection. IgM represents the first line of organism defence against viral infections and hence can indicate recent exposure to viruses. In contrast, IgG can be detected with a delay of several days. Although the detailed immune system response to SARS-CoV-2 virus has still not been described, we can expect similar serological responses, such as in the case of SARS-CoV. Anti-SARS-CoV IgM and IgG antibodies were reported to be detectable after 3–6 and 8 days, respectively [59]. Zhou et al. showed that anti-SARS-CoV-2 IgM and IgG peak at 9 days and within a second week after disease onset, respectively [2]. It is important to note that the immune response of individual patients is variable, making serological testing prone to errors. For example, Long et al. showed that plateaued IgG and IgM levels can vary widely (by more than 20-fold), IgG and IgM titres differ between severe and non-severe patients, seroconversion shows distinct patterns, and even hospitalization with IgG- and IgM-negative status is rare [51]. Taking into account the fact that symptoms can occur even after 24 days (3 days median) and occurrence of patients without any symptoms, tracking virus spread is a difficult task, and combined IgG/IgM-based-rapid tests are beneficial but do not ensure reliable SARS-CoV-2 confirmation in the early stage after onset [52]. The field of serological rapid diagnostics of SARS-CoV-2 infection is dominated by lateral flow test strips, which were reviewed in detail by Quesada et al. [60]. Currently, commercial companies are selling corresponding rapid diagnostic kits or serological ELISA kits enabling high throughput and multiple formats of assays (electrochemiluminescence assay, enzyme immunoassay or fluorescent immunoassay). Sinawang et al. demonstrated that integration of lateral flow technology and electrochemical detection is beneficial and can substantially decrease demands for operators’ manual operations [61]. However, such an approach can hardly compete with user-friendly optical evaluation of common lateral-flow strips.
In our opinion, early detection of viral infection is the field where electrochemistry can prove its potential. However, we will discuss the obstacles of such an approach first. The important topic is definitely analytical parameters, which biosensors need to fulfil. The viral RNA loads are as similarly variable as antibodies. They are not constant during the infection and change by several orders of magnitude. Pan et al. reported that the viral load of throat swabs and sputum peaked approximately 5–6 days after symptom onset and reached 104 - 107 copies per ml during this time [62]. However, He et al. observed the highest viral loads in throat swabs at the time of symptom onset and concluded that infectiousness peaked on or before onset [63]. Recent findings declare that viral loads of asymptomatic and symptomatic patients are similar, suggesting that identification of patients with no or mild symptoms is possible [64]. By simple calculation, the target concentrations of SARS-CoV-2 biosensors should be in the range from tens of attomolar to tens of femtomolar. Although the viral load of COVID-19 patients will be more specified in the future, it is evident that most of the currently published viral hybridization biosensors do not meet these analytical parameter demands. Hence we can conclude that various advanced signal amplification strategies need to be adopted.
3.3. Sample treatment
Antibodies are produced by B cell lymphocytes and are secreted into the bloodstream hence free antibodies can be found within bodily fluids such as blood, serum, plasma, saliva and tears. However, the sensing of viral nucleic acids is completely different. As declared several times, real sample preparation represents a critical bottleneck of biosensor translation from research laboratories to clinical practice or even personal analytical devices [65]. The sample treatment procedure starts with obtaining the sample. Most POC tests use swabs for sampling. During the COVID-19 pandemic, nasopharyngeal swabs, oropharyngeal swabs and sputum are common clinical samples. No discernible differences were observed between naso- and oropharyngeal swabs showing viral load on the order of magnitude of 105 RNA copies per swab even in the early stages of disease [66]. As Zasada et al. reported, the role of swab properties should not be underestimated [67]. They showed that swab material was related to the adsorbed volumes of the specimen and elution buffers to the amount of recovered nucleic acid. To detect virus infection by the interaction of the electrochemical biosensor surface with viral RNA/DNA, nucleic acids need to be released and recovered from viral particles by disruption of the viral capsid or optionally the viral envelope. Here, several approaches can be adopted. Occasionally, simple heating (45–100 °C) of the sample can be sufficient for some applications. In the case of possible nucleases within the sample, the addition of ethylenediamine-tetraacetic acid (EDTA) or glycol-bis(2-aminoethylether)-N,N,N,N-tetraacetic acid (EGTA) is recommended to chelate divalent and trivalent cations required as cofactors for several nucleases. However, neither EDTA nor EGTA inactivates RNases hence alternative approaches such as the use of diethyl pyrocarbonate (DEPC)-treated or polyvinyl sulfonic acid-treated solutions, have been developed [68,69]. Although such chemical treatment of samples can interfere with subsequent PCR amplification, hybridization of nucleic acids and application of electrochemical biosensors should not be influenced. Chaotropic salts (sodium iodide or guanidinium thiocyanate) or detergents (sodium dodecyl sulfate, SDS) are used in combination with heating to disrupt capsids by denaturing and minimizing inter- and intraprotein hydrophobic interactions. In addition, guanidinium thiocyanate is beneficial and hence inhibits RNases.
The biology of SARS-CoV-2 simplifies further sample treatment. SARS-CoV-2 belongs to the group of positive-sense ssRNA viruses. First, nucleoproteins are not strongly bound to RNA and do not need to be removed before analysis. Second, most DNA viruses and some RNA viruses possess double-stranded nucleic acids. In these cases, biorecognition of nucleic acids by hybridization targets with probes is not possible because even after denaturation, long single strands possess higher affinity towards each other than short oligonucleotide probes. However, the possibility of sequence-specific targeting of dsDNA via triplex-creating or duplex/triplex invasion with protein nucleic acids, locked nucleic acids or bridged nucleic acids has been reported [63,70,71]. Notably, long sequences of viral ssRNA/ssDNA are prone to create thermodynamically stable but flexible secondary structures caused by intramolecular base pairing, in which groups of nucleotides are not accessible for complementary strands and can prevent target nucleic acids from hybridizing to probes. ssRNAs of exact secondary structures are functional products that play regulatory roles in essential processes such as replication, repair, transcription, translocation and recombination. Heat denaturation of ssDNA/ssRNA is necessary before target-to-probe hybridization and hybridization at increased temperatures and salt concentrations is desired.
3.4. Practical consideration
In this chapter, we will focus on practical examples of viral infection biosensors, which, in our opinion, have demonstrated innovative and unique approaches. In this field, we have focused on only biosensors of viral nucleic acids and whole viral particles, hence representing the only possibility for early stage diagnostics and being suitable for the design of point-of-need devices (see Fig. 3 ).
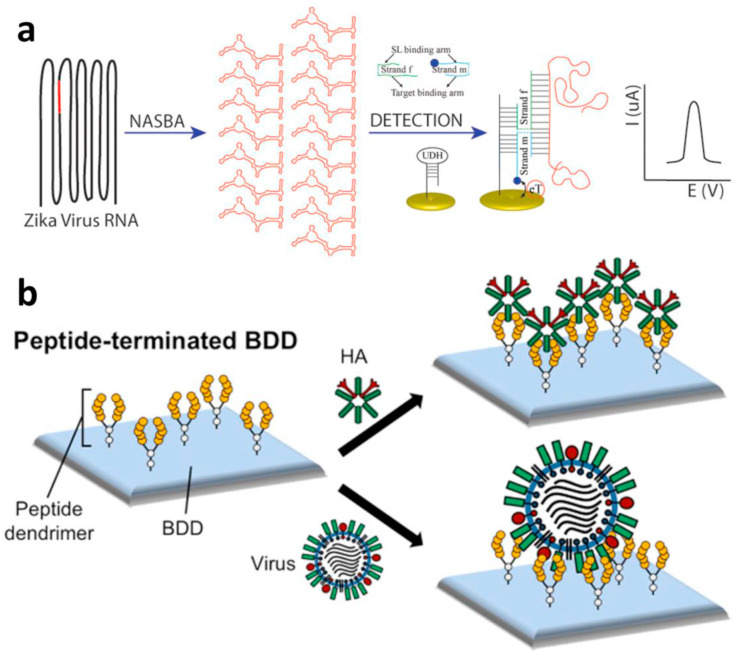
Fig. 3.
Schematic representation of (a) HA and influenza virus binding to peptide-terminated boron-doped diamond (BDD) electrodes (adapted from Ref. [72]), and (b) schematic representation of HA and virus binding to peptide-terminated BDD (adapted from Ref. [79]).
The first study we decided to mention is on Zika virus detection demonstrated by Lynch et al. [72]. Zika virus is, as along with SARS-CoV-2, a positive-sense ssRNA virus (Fig. 3a). They used nucleic acid sequence-based amplification (NASBA) for viral RNA fragment amplification. Further, they immobilized a DNA stem loop on the electrode and used two DNA adaptor strands partially complementary to both the target viral amplicon and stem loop to perform biorecognition. One DNA adaptor is labelled with electroactive methylene blue and responsible for the target recognition (blue probe), and the other is responsible for target RNA secondary structure unwinding (green probe). In the presence of the target, a 4-way junction structure comprising an unwinded stem loop, both DNA adaptors and target RNA is created and brings redox markers in close proximity to the electrode surface, hence providing a detectable signal. They reached biologically relevant concentrations of the target in unpurified amplicons in less than 1 h. Notably, NASBA is a highly suitable tool for ssRNA rapid diagnostic test development because no denaturation steps are required here for RNA fragment amplification, and the whole process is isothermal (approximately 40 °C). Hence, expensive thermocyclers are not necessary.
Another alternative method for isothermal nucleic acid amplification is loop-mediated isothermal amplification (LAMP). The combination of LAMP with magnetic separation and electrochemical detection of human papillomavirus DNA fragments (HPV16 and HPV18) was reported by Bartosik et al. [73]. Magnetic particles were shown to be useful in the analytical preconcentration of samples and hence are being used to enhance the sensitivity of electrochemical viral biosensors [74,75]. They mixed a LAMP-obtained solution with magnetic particles modified with a capture probe targeting the LAMP amplicon. Washed particles were subsequently incubated with an anti-digoxigenin antibody conjugated with horseradish peroxidase (HRP); hence, digoxigenin-labelled deoxyuridine triphosphate was a part of the LAMP mastermix. They utilized magnetic attraction of the construct on the surface of electrode and amperometric sensing of the HRP oxidation product (benzoquinone).
An interesting approach for various influenza A subtypes was reported by Matsubara et al. [76]. They demonstrated that biorecognition based on receptor-ligand binding can be beneficial. More precisely, they targeted influenza viral particles via membrane fusion glycoprotein HA affinity to a sialyloligosaccharide receptor-mimicking peptide. On a boron-doped diamond electrode, they showed a linear response between 3 and 400 pfu·ml−1, as reported by the charge transfer resistance induced by adhered viral particles. However, the biosensor was not tested using a real sample.
A common approach for enhancing electrochemical biosensor sensitivity, which is highly desirable, is to increase the surface of working electrodes by various nanostructures or attach biorecognition elements to nanomaterials or engineered biomaterials. For example, Baek et al. attached norovirus-targeting peptides to WS2 nanoflower composites with gold nanoparticles [77]. This construct was used to bind norovirus particles from real sample extracts and was drop-cast on carbon screen-printed electrodes (SPE), where the norovirus signal was recognized as an increased charge transfer resistance of electrochemical impedance spectroscopy plots (LOD 2.37 copies ∙ ml−1). An alternative approach to improve biosensor performance was used by Chavan et al. [78]. They increased the amounts of antibodies bound on the electrode by bioengineered apoferritin, which resulted in 24 protein-G units and 6 histidine tags per apoferritin molecule. Histidine tags served for apoferritin immobilization on electrodes via nickel nitrilotriacetic acid (Ni-NTA) linkers and protein-G for anti-infectious pancreatic necrosis virus antibody attachment.
An electrochemical biosensor of influenza reaching an appealing LOD of 5–10 viral particles per sample in 5 min was reported by Nidzworski et al. [79]. They targeted the influenza M protein, which forms a coat within the viral capsid (Fig. 3b). The biosensing mechanism was based on immobilization of anti-M antibody on a boron-doped diamond working electrode via 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide/N-hydroxysuccinimide (EDC/NHS) chemistry on electrografted 4-aminobenzoic acid. The binding of the M protein on the electrode increased the charge transfer resistance against Fe(CN)6 3−/4−. To analyse real samples, induced nasal swabs were collected in buffer containing 0.5% Triton X-100, allowing M1 protein to be released. They obtained stable impedimetric signals of electrochemical impedance spectroscopy (EIS) even in the presence of bacteria and yeast within the sample. The analytical parameters of the mentioned biosensors are summarized within Table 1 .
Table 1.
Summary table of viral biosensors.
| analyte | target type | amplification | reporter | method | LOD | linear range | electrode | time (min) | reference |
|---|---|---|---|---|---|---|---|---|---|
| zika virus | NAa | NASBA | methylene blue | SWVb | 1.11 fg μl−1 1.8·105 copies·ml−1 |
1–75 nM | GDEc | 60 | [72] |
| HPVd | NA | LAMP | hydroquinone | AMPe | 0.1 ng | 0.1–50 ng | SPE chip | 150 | [73] |
| norovirus | NA | – | Fe [(CN)6]3–/4– | EISf | 2.37 copies·ml−1 in spiked sample; 6.21 copies·ml−1 in real sample | up to 104 copies·ml−1 | SPE | 60 | [77] |
| influenza | WVPg | – | Fe [(CN)6]3–/4– | EIS | 0.33–0.91 pfuh | 20–400 pfu | BDD electrode | – | [76] |
| pancreatic necrosis virus | WVP | – | Fe [(CN)6]3–/4– | EIS | 2.69 TCID50·ml−1i | 100–100000 TCID50·ml−1 | GDE | – | [78] |
| influenza | WVP | – | Fe [(CN)6]3–/4– | EIS | 1 fg ·ml−1 5–10 WVP per sample |
up to 100 fg | BDD electrode | 5 | [79] |
| SARS-CoV-2 | WVP | – | 1-naphthol | DPV | 6.5 pfu·ml−1 | – | SPE | 30 | [80] |
| SARS-CoV-2 | WVP | – | – | DPV | 1.68·10−22 μg ·ml−1 | – | SPE | 1 | [81] |
| SARS-CoV-2 | NA | – | toluidine blue | DPV | 200 copies·ml−1 | 1 pM - 10 aM | SPE | 181 | [82] |
Nucleic acid.
Square wave voltammetry.
Gold disc electrode.
Human papillomavirus.
Amperometry.
Electrochemical impedance spectroscopy.
Whole viral particles.
Plaque forming unit.
Fifty-percent tissue culture infective dose.
An electrochemical immunoassay for SARS-CoV-2 was reported by Fabiani et al. [80]. They focused on the S protein and N protein and reached LODs of 19 and 8 ng.ml−1, respectively. Their workflow comprised i) pre-coating of magnetic particles (not included in the total analysis time), ii) immunoassay on magnetic particles as a solid support with a reporting antibody labelled with alkaline phosphatase (ALP), and iii) electrochemical analysis. The product of enzymatic cleavage of 1-naphthyl phosphate by ALP, 1-naphthol, was analysed using carbon black-modified screen-printed electrodes and differential pulse voltammetry (DPV) [83]. The report addresses several current challenges, such as total analysis time (below 30 min here), easy sampling (untreated saliva) and portability (printed sensor and portable potentiostat). An interesting sensor (no biorecognition element) for various viruses based on the differentiable fingerprint of their glycoproteins in voltammograms was reported by Hashemi et al. [81]. They benefited from the properties of graphene oxide modified with 8-hydroxyquinoline, EDC and NHS composites with gold nanostars, which have high adsorptive capability for viral S proteins due to the presence of various functional groups. Label-free DPV performed on glassy carbon electrodes modified with the composite provided peaks at different potentials for infectious bronchitis virus, avian influenza, Newcastle disease virus and even SARS-CoV-2 in untreated blood and saliva; hence, the potentials of peaks served for virus identification and the peak intensities served for virus quantification. Compared to RT-PCR they declared only 5% miss rate in the case of SARS-CoV-2 in just 1 min of analysis. A complex nanotechnological approach to the SARS-CoV-2 genome using a supersandwich-type biosensor was developed by Zhao et al. [82]. First, they selectively isolated clinical RNA samples using magnetic particles modified with thiolated capture DNA probe (CP) via affinity of thiols to gold. Second, the fabricated p-sulfocalix [8] arene-functionalized graphene was loaded with gold nanoparticles and toluidine blue (electrochemical label). To gold nanoparticles in the composite, a partially hybridized labelling probe (LP) and auxiliary probe (AP) were attached using a labelling probe thiol group. Probes were designed such that the 5′- and 3′-termini of the target sequence were complementary to the CP and LP, respectively, and the 5′- and 3′-regions of AP had complementary sequences with two different LP areas. Therefore, sequence-specific detection can be achieved by using CPs and LPs, and AP hybridizes many times with the LP to produce long concatamers, resulting in high biosensor sensitivity. The reported biosensor provided a remarkable LOD of 200 RNA copies · ml−1 in clinical specimens, and the detectable positive rate was 85.5% in confirmed patients.
3.5. Perspectives of viral electrochemical biosensors
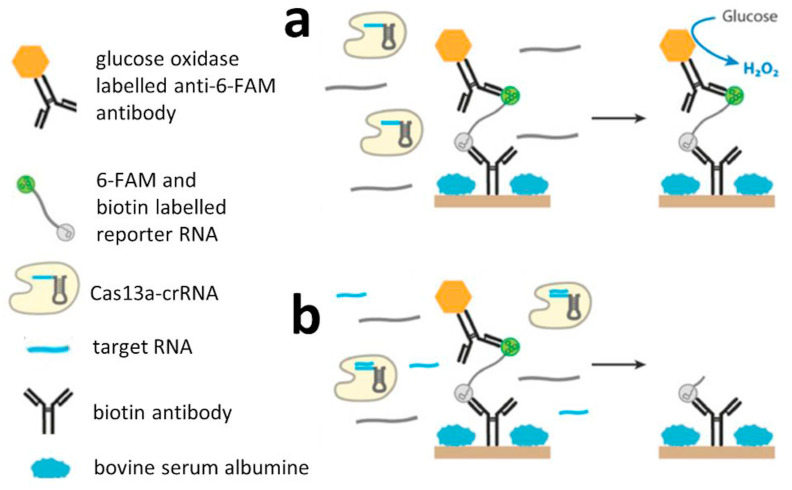
The versatility of the systems, which would enable us to easily detect new analytes, paves the way for new pathways in the strategy of designing such platforms. In this field, CRISPR systems in combination with bioengineered enzymes with cleavage activity and nucleic acid amplification are of great interest. Approaches such as CRISPR/Cas9 triggered isothermal exponential amplification reaction (CAS-EXARP) and NASBA-CRISPR cleavage (NASBACC) can be used to genotype pathogens and differentiate single nucleotide polymorphisms (SNPs) [84,85]. In 2016, a comprehensive study of the type VI CRISPR/Cas system demonstrated the collateral cleavage enzymatic activity of Cas13a after its target-specific binding [86]. This approach was used by Bruch et al. to develop an electrochemical nucleic acid (miRNA) biosensor (Fig. 4 ) [87]. Collateral cleavage-based CRISPR/Cas technology has the potential to significantly affect the field of biosensors by offering a much more rapid and precise method for ultrasensitive nucleic acid detection. Even CRISPR/Cas-based deployable POC paper devices may be possible [88,89]. Due to the great potential offered by CRISPR/Cas-based biosensing, several articles have been published as reviews, news, comments, or insights on CRISPR diagnostics since 2017 [90], which represent evidence for great potential in this field of biosensor development.
Fig. 4.
Collateral cleavage-based electrochemical RNA biosensor. (a) Without the presence of the target RNA, the Cas13-crRNA complex exhibits no cleavage activity towards the reporter RNA; hence, the glucose oxidase-labelled 6-FAM antibody remains immobilized on the electrode via affinity of the biotin antibody and reporter RNA labelled with 6-FAM and biotin. Hence, glucose oxidase can mediate amperometric detection of H2O2. (b) However, if the target RNA is present within the sample, Cas13a-crRNA cleaves the RNA reporter, and glucose oxidase is washed out and provides no (or lower) current response (modified from Ref. [87]).
4. Conclusion
In summary, the presented review summarizes current state-of-the-art approaches to viral electrochemical biosensors, with a focus on SARS-CoV-2. We provide a brief overview of qRT-PCR and serological tests, show their advantages and drawbacks and consider them regarding advances in electrochemical tests. Individual parameters of electrochemical biosensors are critically discussed, promising works are described and we provide future perspectives of the field. We expect that the SARS-CoV-2 pandemic will accelerate the transformation of many analytical approaches from proof-of-concept status to applied technologies.
We demonstrate that electrochemistry brings several advantages among conventional SARS-CoV-2 diagnostic methods. First, it can provide solutions for portable analytical devices composed of miniaturized sensors and minipotentiostats compatible with smartphones. Fast on-site and accurate analysis helps to reduce the economic damage of infection spread and possible quarantine. Furthermore, in comparison with qRT-PCR, electrochemical biosensors can offer attractive price per analysis. Sampling and sample treatment represent critical attributes of POC devices, and even minimal manual operations mean disqualifying drawbacks. It is evident that neither qRT-PCR and serological tests, nor electrochemical SARS-CoV-2 biosensors are flawless tools for COVID-19 diagnostics, but these techniques should complement each other. As we demonstrated, electrochemical biosensors can focus either on viral nucleic acids or whole viral particles. We believe that electrochemistry can provide solutions for rapid and inexpensive POC tests, which should be the first diagnostic method a potentially infected person uses prior to clinical diagnosis. The serious socioeconomic consequences of false-negative tests must be considered, and we suggest that attention should be paid to the sensitivity, selectivity and positive predictive value of published and especially commercial SARS-CoV-2 diagnostic tools.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was financially supported by IGA TP (AF-IGA-2018-tym005) and CEITEC 2020 (LQ1601).
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H.W., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 4.Fan Y., Zhao K., Shi Z.-L., Zhou P. Bat coronaviruses in China. Viruses. 2019;11:210. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu H., Chu D.K., Peiris M., Poon L.L. Multivariate analyses of codon usage of SARS-CoV-2 and other betacoronaviruses. Virus Evol. 2020;6 doi: 10.1093/ve/veaa032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:1–9. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C., Liu Z., Chen Z., Huang X., Xu M., He T., Zhang Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020;92:667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kupferschmidt K. INFECTIOUS DISEASES Genome analyses help track coronavirus' moves. Science. 2020;367:1176–1177. doi: 10.1126/science.367.6483.1176. [DOI] [PubMed] [Google Scholar]
- 9.Perchetti G.A., Nalla A.K., Huang M.L., Jerome K.R., Greninger A.L. Multiplexing primer/probe sets for detection of SARS-CoV-2 by qRT-PCR. J. Clin. Virol. 2020;129:3. doi: 10.1016/j.jcv.2020.104499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y., Yang M., Shen C., Wang F., Yuan J., Li J., Zhang M., Wang Z., Xing L., Wei J., Peng L., Wong G., Zheng H., Liao M., Feng K., Li J., Yang Q., Zhao J., Zhang Z., Liu L., Liu Y. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv. 2020;2020 2002.2011.20021493. [Google Scholar]
- 11.Arevalo-Rodriguez I., Buitrago-Garcia D., Simancas-Racines D., Zambrano-Achig P., del Campo R., Ciapponi A., Sued O., Martinez-Garcia L., Rutjes A., Low N., Bossuyt P.M., Perez-Molina J.A., Zamora J. FALSE-NEGATIVE results OF initial RT-PCR assays for COVID-19: a systematic review. medRxiv. 2020;2020 doi: 10.1371/journal.pone.0242958. 2004.2016.20066787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y.C., Zhang H.Q., Xie J.C., Lin M.J., Ying L.J., Pang P.P., Ji W.B. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection — challenges and implications. N. Engl. J. Med. 2020;383:e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 15.Kellam P., Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J. Gen. Virol. 2020 doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saberi A., Gulyaeva A.A., Brubacher J.L., Newmark P.A., Gorbalenya A.E. A planarian nidovirus expands the limits of RNA genome size. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nga P.T., del Carmen Parquet M., Lauber C., Parida M., Nabeshima T., Yu F., Thuy N.T., Inoue S., Ito T., Okamoto K. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeffelholz M.J., Tang Y.-W. Laboratory diagnosis of emerging human coronavirus infections–the state of the art. Emerg. Microb. Infect. 2020;9:747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu D.K., Pan Y., Cheng S.M., Hui K.P., Krishnan P., Liu Y., Ng D.Y., Wan C.K., Yang P., Wang Q. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan J.F.-W., Yip C.C.-Y., To K.K.-W., Tang T.H.-C., Wong S.C.-Y., Leung K.-H., Fung A.Y.-F., Ng A.C.-K., Zou Z., Tsoi H.-W. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv D.-f., Ying Q.-m., Weng Y.-s., Shen C.-b., Chu J.-g., Kong J.-p., Sun D.-h., Gao X., Weng X.-b., Chen X.-q. Clinica Chimica Acta; 2020. Dynamic Change Process of Target Genes by RT-PCR Testing of SARS-Cov-2 during the Course of a Coronavirus Disease 2019 Patient. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu H., Wu J., Hong L., Luo Y., Song Q., Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect. Dis. 2020;20:689. doi: 10.1016/S1473-3099(20)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cajigas S., Alzate D., Orozco J. Gold nanoparticle/DNA-based nanobioconjugate for electrochemical detection of Zika virus. Microchimica Acta. 2020;187:594. doi: 10.1007/s00604-020-04568-1. [DOI] [PubMed] [Google Scholar]
- 28.Fani M., Rezayi M., Meshkat Z., Rezaee S.A., Makvandi M., Angali K.A. A novel electrochemical DNA biosensor based on a gold nanoparticles-reduced graphene oxide-polypyrrole nanocomposite to detect human T-lymphotropic virus-1. IEEE Sensor. J. 2020;20:10625–10632. [Google Scholar]
- 29.Srisomwat C., Teengam P., Chuaypen N., Tangkijvanich P., Vilaivan T., Chailapakul O. Pop-up paper electrochemical device for label -free hepatitis B virus DNA detection. Sensor. Actuator. B Chem. 2020;316:8. [Google Scholar]
- 30.Shariati M., Sadeghi M. Ultrasensitive DNA biosensor for hepatitis B virus detection based on tin-doped WO3/In(2)O(3)heterojunction nanowire photoelectrode under laser amplification. Anal. Bioanal. Chem. 2020;412:5367–5377. doi: 10.1007/s00216-020-02752-z. [DOI] [PubMed] [Google Scholar]
- 31.Ilkhani H., Farhad S. A novel electrochemical DNA biosensor for Ebola virus detection. Anal. Biochem. 2018;557:151–155. doi: 10.1016/j.ab.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Manzano M., Viezzi S., Mazerat S., Marks R.S., Vidic J. Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron. 2018;100:89–95. doi: 10.1016/j.bios.2017.08.043. [DOI] [PubMed] [Google Scholar]
- 33.Family - Coronaviridae . In: Virus Taxonomy. King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Elsevier; San Diego: 2012. pp. 806–828. [Google Scholar]
- 34.Baker M. Antibody anarchy: a call to order. Nature. 2015;527:545–551. doi: 10.1038/527545a. [DOI] [PubMed] [Google Scholar]
- 35.Bradbury A., Pluckthun A. Standardize antibodies used in research. Nature. 2015;518:27–29. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- 36.O'Kennedy R., Fitzgerald S., Murphy C. Don't blame it all on antibodies - the need for exhaustive characterisation, appropriate handling, and addressing the issues that affect specificity. Trac. Trends Anal. Chem. 2017;89:53–59. [Google Scholar]
- 37.Park M. Orientation control of the molecular recognition layer for improved sensitivity: a review. BioChip J. 2019;13:82–94. [Google Scholar]
- 38.Benjamin D.C., Perdue S.S. Site-directed mutagenesis in epitope mapping. Methods. 1996;9:508–515. doi: 10.1006/meth.1996.0058. [DOI] [PubMed] [Google Scholar]
- 39.Dougan D.A., Malby R.L., Gruen L.C., Kortt A.A., Hudson P.J. Effects of substitutions in the binding surface of an antibody on antigen affinity. Protein Eng. 1998;11:65–74. doi: 10.1093/protein/11.1.65. [DOI] [PubMed] [Google Scholar]
- 40.Sago C.D., Kalathoor S., Fitzgerald J.P., Lando G.N., Djeddar N., Bryksin A.V., Dahlman J.E. Barcoding chemical modifications into nucleic acids improves drug stability in vivo. J. Mater. Chem. B. 2018;6:7197–7203. doi: 10.1039/c8tb01642a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 42.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boni M.F., Gog J.R., Andreasen V., Feldman M.W. Epidemic dynamics and antigenic evolution in a single season of influenza A. Proc. R. Soc. B-Biol. Sci. 2006;273:1307–1316. doi: 10.1098/rspb.2006.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta A.M., Chakrabarti J., Mandal S. Non-synonymous mutations of SARS-CoV-2 leads epitope loss and segregates its variants. Microb. Infect. 2020;22:598–607. doi: 10.1016/j.micinf.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., McDanal C., Perez L.G., Tang H.L., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C., Sheffield C.-G.G. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen C.-H. In: Diagnostic Molecular Biology. Shen C.-H., editor. Academic Press; 2019. Chapter 6 - extraction and purification of nucleic acids and proteins; pp. 143–166. [Google Scholar]
- 47.Wang R.E., Wu H., Niu Y., Cai J. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011;18:4126–4138. doi: 10.2174/092986711797189565. [DOI] [PubMed] [Google Scholar]
- 48.Kwon J., Narayan C., Kim C., Han M.J., Kim M., Jang S.K. Development of a subtype-specific diagnostic system for influenza virus H3N2 using a novel virus-based systematic evolution of ligands by exponential enrichment (Viro-SELEX) J. Biomed. Nanotechnol. 2019;15:1609–1621. doi: 10.1166/jbn.2019.2789. [DOI] [PubMed] [Google Scholar]
- 49.Narayan C., Kwon J., Kim C., Kim S.-J., Jang S.K. Virus-based SELEX (viro-SELEX) allows development of aptamers targeting knotty proteins. Analyst. 2020;145:1473–1482. doi: 10.1039/c9an01943j. [DOI] [PubMed] [Google Scholar]
- 50.Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020;92:9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 51.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 52.Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D.S.C., Du B., Li L.-j., Zeng G., Yuen K.-Y., Chen R.-c., Tang C.-l., Wang T., Chen P.-y., Xiang J., Li S.-y., Wang J.-l., Liang Z.-j., Peng Y.-x., Wei L., Liu Y., Hu Y.-h., Peng P., Wang J.-m., Liu J.-y., Chen Z., Li G., Zheng Z.-j., Qiu S.-q., Luo J., Ye C.-j., Zhu S.-y., Zhong N.-s. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv. 2020;2020 2002.2006.20020974. [Google Scholar]
- 53.Matrosovich M., Herrler G., Klenk H.D. In: SialoGlyco Chemistry and Biology II: Tools and Techniques to Identify and Capture Sialoglycans. Gerardy-Schahn R., Delannoy P., von Itzstein M., editors. Springer International Publishing; Cham: 2015. Sialic acid receptors of viruses; pp. 1–28. [Google Scholar]
- 54.Horiguchi Y., Goda T., Matsumoto A., Takeuchi H., Yamaoka S., Miyahara Y. Direct and label-free influenza virus detection based on multisite binding to sialic acid receptors. Biosens. Bioelectron. 2017;92:234–240. doi: 10.1016/j.bios.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang X., Qi Q., Jing Q., Ao S., Zhang Z., Ding M., Wu M., Liu K., Wang W., Ling Y., Zhang Z., Fu W. arXiv. 2020:1–20. [Google Scholar]
- 56.Gast M., Sobek H., Mizaikoff B. Advances in imprinting strategies for selective virus recognition a review. Trac. Trends Anal. Chem. 2019;114:218–232. [Google Scholar]
- 57.Gast M., Sobek H., Mizaikoff B. Selective virus capture via hexon imprinting. Mater. Sci. Eng. C. 2019;99:1099–1104. doi: 10.1016/j.msec.2019.02.037. [DOI] [PubMed] [Google Scholar]
- 58.Parisi O.I., Dattilo M., Patitucci F., Malivindi R., Pezzi V., Perrotta I., Ruffo M., Amone F., Puoci F. “Monoclonal-type” plastic antibodies for SARS-CoV-2 based on molecularly imprinted polymers. bioRxiv. 2020;2020 2005.2028.120709. [Google Scholar]
- 59.Lee H.K., Lee B.H., Seok S.H., Baek M.W., Lee H.Y., Kim D.J., Na Y.R., Noh K.J., Park S.H., Kumar D.N., Kariwa H., Nakauchi M., Heo S.J., Park J.H. Production of specific antibodies against SARS-coronavirus nucleocapsid protein without cross reactivity with human coronaviruses 229E and OC43. J. Vet. Sci. 2010;11:165–167. doi: 10.4142/jvs.2010.11.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quesada-González D., Merkoçi A. Nanoparticle-based lateral flow biosensors. Biosens. Bioelectron. 2015;73:47–63. doi: 10.1016/j.bios.2015.05.050. [DOI] [PubMed] [Google Scholar]
- 61.Prima D., Sinawang V., Rai R.E., Ionescu R.S. Marks, Electrochemical lateral flow immunosensor for detection and quantification of dengue NS1 protein. Biosens. Bioelectron. 2016;77:400–408. doi: 10.1016/j.bios.2015.09.048. [DOI] [PubMed] [Google Scholar]
- 62.Pan Y., Zhang D.T., Yang P., Poon L.L.M., Wang Q.Y. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emehiser R.G., Hall E., Guenther D.C., Karmakar S., Hrdlicka P.J. Head-to-head comparison of LNA, MPγPNA, INA and Invader probes targeting mixed-sequence double-stranded DNA. Org. Biomol. Chem. 2020;18:56–65. doi: 10.1039/c9ob02111f. [DOI] [PubMed] [Google Scholar]
- 64.Zou L.R., Ruan F., Huang M.X., Liang L.J., Huang H.T., Hong Z.S., Yu J.X., Kang M., Song Y.C., Xia J.Y., Guo Q.F., Song T., He J.F., Yen H.L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ritzi-Lehnert M. Development of chip-compatible sample preparation for diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2012;12:189–206. doi: 10.1586/erm.11.98. [DOI] [PubMed] [Google Scholar]
- 66.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 67.Zasada A.A., Zacharczuk K., Woźnica K., Główka M., Ziółkowski R., Malinowska E. The influence of a swab type on the results of point-of-care tests. AMB Express. 2020;10:46. doi: 10.1186/s13568-020-00978-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Earl C.C., Smith M.T., Lease R.A., Bundy B.C. Polyvinylsulfonic acid: a Low-cost RNase inhibitor for enhanced RNA preservation and cell-free protein translation. Bioengineered. 2018;9:90–97. doi: 10.1080/21655979.2017.1313648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yakovlev G.I., Mitkevich V.A., Makarov A.A. Ribonuclease inhibitors. Mol. Biol. 2006;40:867–874. [Google Scholar]
- 70.Chen Y., Murayama K., Kashida H., Kamiya Y., Asanuma H. A triplex-forming linear probe for sequence-specific detection of duplex DNA with high sensitivity and affinity. Chem. Commun. 2020;56:5358–5361. doi: 10.1039/d0cc01865a. [DOI] [PubMed] [Google Scholar]
- 71.Emehiser R.G., Hrdlicka P.J. Chimeric γPNA–Invader probes: using intercalator-functionalized oligonucleotides to enhance the DNA-targeting properties of γPNA. Org. Biomol. Chem. 2020;18:1359–1368. doi: 10.1039/c9ob02726b. [DOI] [PubMed] [Google Scholar]
- 72.Lynch C.A., Foguel M.V., Reed A.J., Balcarcel A.M., Calvo-Marzal P., Gerasimova Y.V., Chumbimuni-Torres K.Y. Selective determination of isothermally amplified Zika virus RNA using a universal DNA-hairpin probe in less than 1 hour. Anal. Chem. 2019;91:13458–13464. doi: 10.1021/acs.analchem.9b02455. [DOI] [PubMed] [Google Scholar]
- 73.Bartosik M., Jirakova L., Anton M., Vojtesek B., Hrstka R. Genomagnetic LAMP-based electrochemical test for determination of high-risk HPV16 and HPV18 in clinical samples. Anal. Chim. Acta. 2018;1042:37–43. doi: 10.1016/j.aca.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 74.Kudr J., Haddad Y., Richtera L., Heger Z., Cernak M., Adam V., Zitka O. Magnetic nanoparticles: from design and synthesis to real world applications. Nanomaterials. 2017;7 doi: 10.3390/nano7090243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kudr J., Klejdus B., Adam V., Zitka O. Magnetic solids in electrochemical analysis. Trac. Trends Anal. Chem. 2018;98:104–113. [Google Scholar]
- 76.Matsubara T., Ujie M., Yamamoto T., Einaga Y., Daidoji T., Nakaya T., Sato T. Avian influenza virus detection by optimized peptide termination on a boron-doped diamond electrode. ACS Sens. 2020;5:431–439. doi: 10.1021/acssensors.9b02126. [DOI] [PubMed] [Google Scholar]
- 77.Baek S.H., Park C.Y., Nguyen T.P., Kim M.W., Park J.P., Choi C., Kim S.Y., Kailasa S.K., Park T.J. Novel peptides functionalized gold nanoparticles decorated tungsten disulfide nanoflowers as the electrochemical sensing platforms for the norovirus in an oyster. Food Contr. 2020;114 [Google Scholar]
- 78.Chavan S.G., Yagati A.K., Mohammadniaei M., Min J., Lee M.H. Robust bioengineered apoferritin nanoprobes for ultrasensitive detection of infectious pancreatic necrosis virus. Anal. Chem. 2019;91:5841–5849. doi: 10.1021/acs.analchem.9b00187. [DOI] [PubMed] [Google Scholar]
- 79.Nidzworski D., Siuzdak K., Niedzialkowski P., Bogdanowicz R., Sobaszek M., Ryl J., Weiher P., Sawczak M., Wnuk E., Goddard W.A., Jaramillo-Botero A., Ossowski T. A rapid-response ultrasensitive biosensor for influenza virus detection using antibody modified boron-doped diamond. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-15806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fabiani L., Saroglia M., Galatà G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hashemi S.A., Golab Behbahan N.G., Bahrani S., Mousavi S.M., Gholami A., Ramakrishna S., Firoozsani M., Moghadami M., Lankarani K.B., Omidifar N. Ultra-sensitive viral glycoprotein detection NanoSystem toward accurate tracing SARS-CoV-2 in biological/non-biological media. Biosens. Bioelectron. 2021;171:112731. doi: 10.1016/j.bios.2020.112731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., Li H., Zhao C.-Y., Zhang L., Wei J., Zhang Y.-P., Li C.-P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kudr J., Zhao L., Nguyen E.P., Arola H., Nevanen T.K., Adam V., Zitka O., Merkoci A. Inkjet-printed electrochemically reduced graphene oxide microelectrode as a platform for HT-2 mycotoxin immunoenzymatic biosensing. Biosens. Bioelectron. 2020;156:8. doi: 10.1016/j.bios.2020.112109. [DOI] [PubMed] [Google Scholar]
- 84.Huang M.Q., Zhou X.M., Wang H.Y., Xing D. Clustered regularly interspaced short palindromic repeats/cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal. Chem. 2018;90:2193–2200. doi: 10.1021/acs.analchem.7b04542. [DOI] [PubMed] [Google Scholar]
- 85.Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W., Ferrante T., Ma D., Donghia N., Fan M., Daringer N.M., Bosch I., Dudley D.M., O'Connor D.H., Gehrke L., Collins J.J. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 86.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., Severinov K., Regev A., Lander E.S., Koonin E.V., Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:9. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bruch R., Baaske J., Chatelle C., Meirich M., Madlener S., Weber W., Dincer C., Urban G.A. CRISPR/Cas13a-Powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater. 2019;31:1905311. doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- 88.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A., Garcia K.F., Barnes K.G., Chak B., Mondini A., Nogueira M.L., Isern S., Michael S.F., Lorenzana I., Yozwiak N.L., MacInnis B.L., Bosch I., Gehrke L., Zhang F., Sabeti P.C. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Y., Li S.Y., Wang J., Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]