Abstract
COVID-19 has infected 77.4 million people worldwide and has caused 1.7 million fatalities as of December 21, 2020. The primary cause of death due to COVID-19 is Acute Respiratory Distress Syndrome (ARDS). According to the World Health Organization (WHO), people who are at least 60 years old or have comorbidities that have primarily been targeted are at the highest risk from SARS-CoV-2.
Medical imaging provides a non-invasive, touch-free, and relatively safer alternative tool for diagnosis during the current ongoing pandemic. Artificial intelligence (AI) scientists are developing several intelligent computer-aided diagnosis (CAD) tools in multiple imaging modalities, i.e., lung computed tomography (CT), chest X-rays, and lung ultrasounds. These AI tools assist the pulmonary and critical care clinicians through (a) faster detection of the presence of a virus, (b) classifying pneumonia types, and (c) measuring the severity of viral damage in COVID-19-infected patients. Thus, it is of the utmost importance to fully understand the requirements of for a fast and successful, and timely lung scans analysis.
This narrative review first presents the pathological layout of the lungs in the COVID-19 scenario, followed by understanding and then explains the comorbid statistical distributions in the ARDS framework. The novelty of this review is the approach to classifying the AI models as per the by school of thought (SoTs), exhibiting based on segregation of techniques and their characteristics. The study also discusses the identification of AI models and its extension from non-ARDS lungs (pre-COVID-19) to ARDS lungs (post-COVID-19). Furthermore, it also presents AI workflow considerations of for medical imaging modalities in the COVID-19 framework. Finally, clinical AI design considerations will be discussed.
We conclude that the design of the current existing AI models can be improved by considering comorbidity as an independent factor. Furthermore, ARDS post-processing clinical systems must involve include (i) the clinical validation and verification of AI-models, (ii) reliability and stability criteria, and (iii) easily adaptable, and (iv) generalization assessments of AI systems for their use in pulmonary, critical care, and radiological settings.
Keywords: COVID-19, ARDS, Comorbidity, Medical imaging, CT, X-ray, US, Artificial intelligence, Deep learning, Machine learning, Transfer learning, Ultrasound
1. Introduction
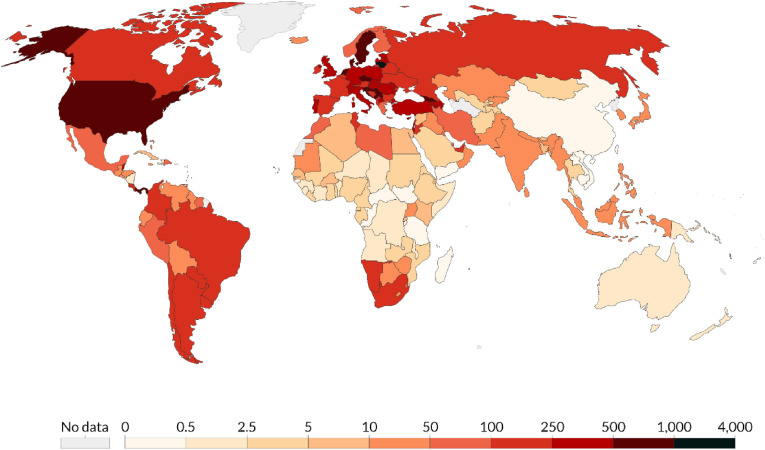
In December 2019, the novel coronavirus, severe acute respiratory distress syndrome coronavirus 2 (SARS-CoV-2) [1], appeared in Wuhan, in the Hubei province of the People's Republic of China. The disease caused by the virus was initially called novel coronavirus pneumonia (NCP) by the Chinese government [2]. Even though it has been argued that this disease is syndemic1 (has both biological and social factors) [3], it was subsequently renamed the coronavirus disease 2019 (COVID-19) by the World Health Organization (WHO). It is currently a global pandemic [4]. It is a respiratory disease that can lead to acute respiratory distress syndrome (ARDS) and eventually to death. As of December 21, 2020, more than 77.4 million COVID-19 cases have been reported, causing 1.7 million casualties [5]. COVID-19 has a Ro value between 2.43 and 3.10, meaning that it is highly contagious [6]. The colors in Fig. 1 (from white to dark red) show the number of cases reported per million of population worldwide. The eight countries with the highest mortality are the USA, Brazil, India, Mexico, the UK, Italy, France, and Spain [7].
Fig. 1.
Total confirmed cases per million as of December 21, 2020 [15]. (Source: Center for Systems Science and Engineering (CSSE) at Johns Hopkins University, Maryland, USA.).
Genetically, COVID-19 is closer to SARS-CoV-1 than to Middle East respiratory syndrome coronavirus (MERS-CoV). However, it differs from SARS-CoV-1 in the length of the incubation period, clinical severity, and transmissibility [8]. Despite government initiatives to implement social habits such as social distancing and wearing masks, and despite quarantining and non-pharmacological and preventive treatments for psychophysical wellbeing, the global spread of COVID-19 has increased [9,10].
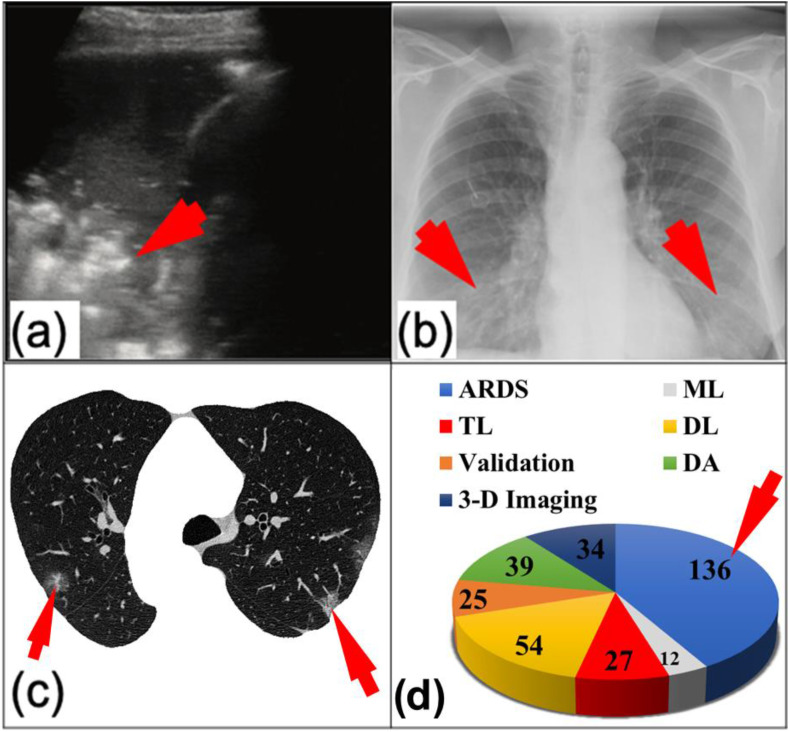
COVID-19 has unique imaging characteristics that constitute a visual identity when viewed through a radiologist's lens. Furthermore, COVID-19 adversely affects organs other than the lungs [11,12]. Due to these circumstances, research on this topic has exploded, with nearly 72,000 COVID-19-related articles published since December 2019 (see PubMed website [13]). This translates to an average of 2000 articles per week. Interestingly, more than 900 articles are on COVID-19 and artificial intelligence (AI); these articles cover the models of machine learning (ML), transfer learning (TL), and deep learning (DL) (see Fig. 2 (d)). It appears that AI can play a role in the characterization of ARDS in the lungs and the characterization and diagnosis of other parts of the body [14]. The topic of AI for ARDS characterization must be investigated carefully to help practitioners better manage viral COVID-19 pneumonia leading to ARDS.
Fig. 2.
Images of COVID-19 infection: (a) lung ultrasound (hyper-echoic region of the COVID-19 lung), (b) chest X-rays (the infected region in the lung), and (c) lung CT (segmented lung region; courtesy of Luca Saba, University of Cagliari, Italy). (d) The number of COVID-19 studies involving ARDS, ML, TL, DL, validation, data acquisition (DA), and 3-D imaging.
AI is involved in many facets of COVID-19 management, such as medical imaging, risk assessment, telemedicine, and patient follow-up [16]. Even though AI has penetrated into several fields of radiological imaging [17], such as classification of images into (a) controls, (b) community viral pneumonia, and (c) COVID-19 pneumonia [[18], [19], [20]], the question that remains unanswered is whether these models actually function the way the pulmonologists and critical care physicians want. For example, if the patient has a specific pre-existing disease, can an AI include this information into its knowledgebase and use it to more effectively correlate this comorbid condition, the age factor, and the grayscale patterns in lung scans with COVID-19 severity? Furthermore, there are other questions worth addressing, such as the following: (i) What kind of imaging modalities are useful for ARDS? (ii) What kind of image-based classifiers are most beneficial for detecting and classifying ARDS severity while also considering comorbidity, age group, and imaging characteristics? (iii) How can COVID-19 severity due to ARDS be measured? (iv) How should we estimate the survival of patients affected by ARDS? (v) How does a pre-existing disease or comorbidity affect the mortality from ARDS? (vi) What methods (new/hybrid) can be used to detect and classify the early stages of ARDS? All these issues must be addressed if valid diagnoses are to be given and if the therapeutic applications of the AI framework are to be evaluated accurately.
The most popular medical imaging tools used for lung imaging are ultrasound, X-rays, CT (see Fig. 2: (a) Hyper-echoic region of the COVID-19 lung, 2 (b) The infected region in the lung, and 2 (c) Segmented lung region), and a recently developed technique that combines PET and CT to visualize the lungs and their metabolic functions [[21], [22], [23]]. This study considers various schools of thought for AI-based solutions for COVID-19 lung severity classification. Specifically, this study considers machine learning (ML), transfer learning (TL), and deep learning (DL) techniques, and combination of these. It also discusses the crucial issues and concerns that ongoing AI-based COVID-19 research must tackle to help the medical community.
This review makes the following contributions. (i) It is the first review of its kind that explores the pathophysiological aspect of acute respiratory distress syndrome (ARDS) due to COVID-19. It explicitly discusses the deteriorating alveolar stages leading to the gas-exchange disorder. (ii) This study is the only one that links the derived comorbid conditions and ARDS from nearly fifty studies. These conditions include hypertension, diabetes, obesity, chronic kidney disease, cardiovascular diseases, liver disease, hyperlipidemia, renal dysfunction, cancer, human immunodeficiency virus (HIV), lung disease, and cerebrovascular disease. Furthermore, it computes the link between comorbidity, ARDS, and mortality. (iii) The review compares the AI techniques used to characterize lung diseases during non-ARDS times (pre-COVID-19) with those used in ARDS times (post-COVID-19). (iv) This is the first review of its kind that develops the seven different school of thought (SoT) types utilizing AI techniques to evaluate the COVID-19 severity in the ARDS framework. (v) This is the only review that exclusively compares different imaging modalities for evaluating the lung conditions in ARDS. (vi) This is the first review of its kind that shows the evolution of AI design for personalized medicine by linking AI with ARDS having comorbidity. This is very useful for evaluating ARDS conditions in the COVID-19 framework.
The layout of the study is as follows. The research strategy is presented in section 2. The pathophysiology of ARDS is described in section 3. The effect of comorbidity with COVID-19 is analyzed in section 4. In section 5, AI architectures are divided into schools of thought (SoT). The practical aspect of AI for COVID-19 is explained in section 6. The critical discussion and conclusion are given in sections 7, 8, respectively.
2. Research strategy
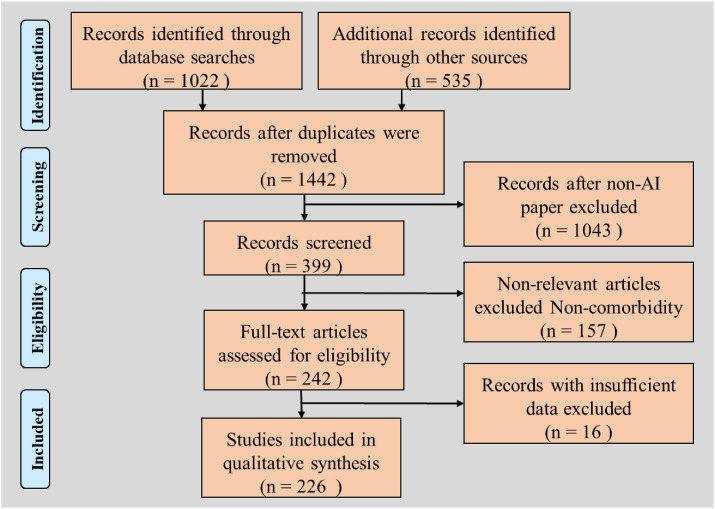
Fig. 3 shows the flowchart of our research strategy. We used four online research databases: PubMed, IEEE Xplore, ArXiv, and Web of Science. Initial screening used the keywords “COVID-19,” “coronavirus,” or “ARDS,” with the modality terms “lung CT,” “X-ray,” or “ultrasound.” The search was augmented with terms “artificial intelligence,” “machine learning,” or “transfer learning,” or “deep learning,” resulting in 1557 articles. We excluded 115 duplicate articles and articles that did not deal with COVID-19 severity (including classification), resulting in 1442 articles. Finally, we selected the articles based on our subjective assessment of their relevance and novelty, resulting in 399 articles. Excluding irrelevant articles, such as those that did not discuss comorbidity, resulted in 242 articles. Records with insufficient data were removed, leading to 230 resources. This was then used in our narrative study, which used AI-based, comorbid-based, and pathophysiology-based articles.
Fig. 3.
The flowchart showing the research strategy.
3. The pathophysiology of acute respiratory distress syndrome
The prime route of SARS-CoV-2 virus communicability is through the nasal droplets and saliva of an infected person [19]. The SARS-CoV-2 virus enters the alveolar type 2 cells (AT2 cells) by anchoring its viral spike proteins (S1 and S2) to the angiotensin-converting enzyme 2 (ACE2) receptor [24] (Fig. 4 ). Mossel et al. have reported that the previous coronavirus (CoV or SARS-CoV-1) replicates more aggressively in AT2 cells than in alveolar type 1 cells (AT1 cells) in the lung [25]. There is an 80% genetic resemblance between SARS-CoV-1 and SARS-CoV-2. It has been shown via molecular pathways [26] that SARS-CoV-2 has a high potential for binding with AT2 cells in the lungs [27].
Fig. 4.
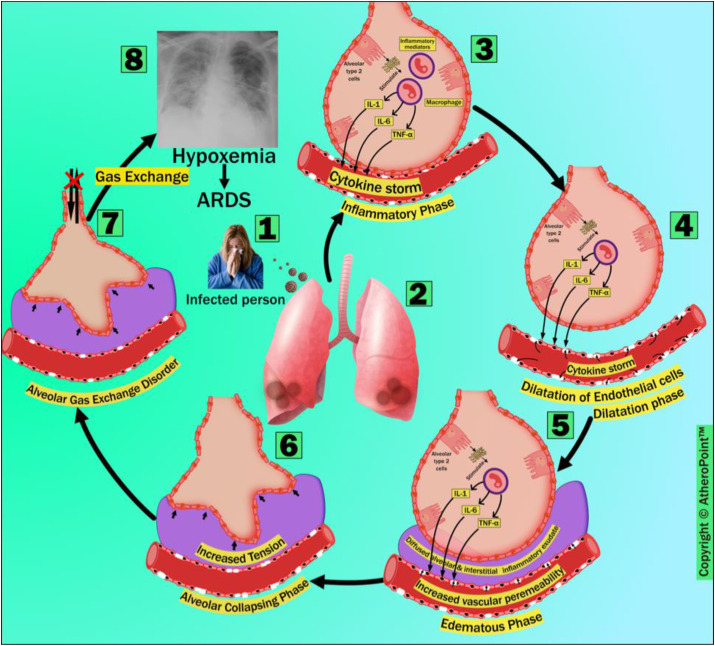
The pathophysiology of ARDS after COVID-19 infection, which consists of six phases: (i) inflammatory phase, (ii) dilatation phase, (iii) edematous phase, (iv) alveolar collapsing phase, (v) gas-exchange disorder, and (vi) hypoxemia. (Courtesy of AtheroPoint™, Roseville, CA, USA; reproduced with permission.)
In the inflammatory phase (marked as 3), inflammatory mediators are secreted as a systemic inflammatory response to the infection of AT2 cells by SARS-CoV-2 on the alveolar epithelium surface [28]. The secreted inflammatory mediators stimulate alveolar macrophage, producing cytokines like IL-1, IL-6, and TNF-α. In turn, the elevated production of cytokines and chemokines results in a condition called a cytokine storm. The sequence of the systemic inflammatory response, the cytokine storm, and the failure of multiple organs significantly influences ARDS's pathophysiology. These sequences can also be seen in all other viruses belonging to all CoV families (e.g., SARS-CoV-1 and MERS-CoV) [29,30]. This leads to the upregulation of trypsin.
Furthermore, it damages the endothelial tight junction protein and the zonula occludens, disorganizing and weakening endothelial cells and increasing these cells’ vascular permeability to intravascular fluids [31]. In the dilatation phase (marked as 4), the cytokine storm causes endothelial barrier dysfunction. This is caused by the release of the cytokines following a viral infection. In the edematous phase (marked as 5), intravascular permeability leads to the diffusion of intravascular fluid (proteins, neutrophils, erythrocytes, and platelets) to the sub-alveolar and interstitial space, thereby causing diffused alveolar and interstitial exudates, as well as alveolar edema. At this point, one can observe radiological consolidations using lung CT as a diagnostic approach, and monitor the treatment prognosis [32]. Diffused alveolar and interstitial exudates and increased sub-alveolar edema are the hallmark features of ARDS [33].
During the alveolar collapsing phase (marked as 6), increased tension (due to the drowning of alveoli in the fluid accumulated in the sub-alveolar and interstitial space) causes alveolar collapse, which results in the disruption of gas exchange from alveoli [34]. The alveolar gas-exchange disorder (marked as 7) occurs because of the ventilation-to-perfusion mismatch between carbon dioxide and oxygen. This condition leads to hypoxemia and ARDS (marked as 8). This leads to increased mortality if left untreated [35].
4. Comorbidity and ARDS
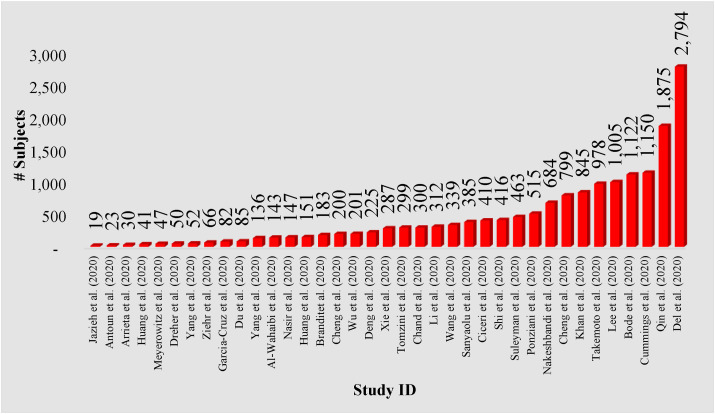
Comorbidities of COVID-19 in ARDS include (i) old age, (ii) ethnicity, (iii) hypertension, (iv) diabetes mellitus (DM), (v) elevated BMI, (vi) cardiac diseases, and (vii) respiratory disorders. Studies have shown that ARDS tends to be worse among older patients [[36], [37], [38]], African Americans (Blacks in general) [39], and those suffering from hypertension [36,40], diabetes [36,38,41], BMI [[42], [43], [44]], respiratory diseases [42], and myocarditis [[45], [46], [47]]. These comorbidities play a vital role in segregating AI models that can be independently trained for effective diagnosis and COVID-19 severity prediction. The selected comorbidity studies [[48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77], [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100], [101]] were taken from https://pubmed.ncbi.nlm.nih.gov/. Fig. 5 shows the number of subjects with comorbidities enrolled in the ARDS-based studies.
Fig. 5.
The number of subjects enrolled in the ARDS-based studies that consider comorbidities.
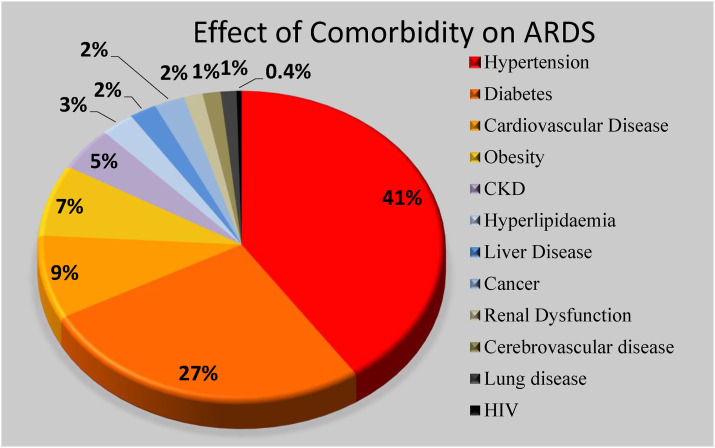
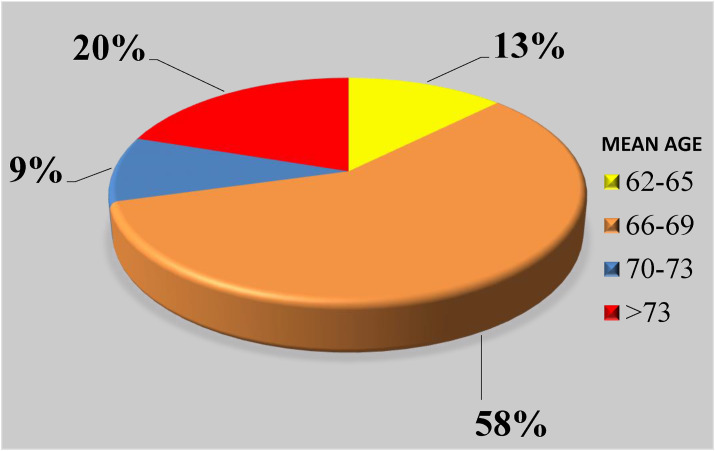
We used the following criteria during the selection process. The chosen keywords were hypertension, diabetes, obesity, chronic kidney disease, cardiovascular diseases, liver disease, hyperlipidemia, renal dysfunction, cancer, human immunodeficiency virus (HIV), lung disease, and cerebrovascular disease. The percentage of comorbidity subjects is shown in the pie chart as depicted in Fig. 6 . This was designed using the selected 48 studies. Note that these 48 studies were pure medical journals discussing the role of comorbidity (“pre-existing diseases”) and “age groups”. Therefore, we decided to only consider these studies as a rationale and motivation for statistical data collection, which was likely to be helpful in the design of the innovative AI solutions for COVID-19 severity diagnosis and monitoring. The two most important contributing factors to COVID-19 were diabetes and hypertension, which together account for about 68%. The rest of the comorbid predictors were nearly the same, in the range of 4%–13%. The selected studies stated that a total of 14% deaths were due to comorbidities in the ARDS framework. This pie chart demonstrates that comorbidity plays a vital role in the mortality of each cohort. Fig. 7 shows the distribution of the death due to the age factor with at least one comorbidity in the cohort. The most affected age range is 66–69, with a total coverage of 58% of the total cohort, followed by age >70 with total of 20% from the selected studies.
Fig. 6.
Depiction of comorbidities collected from 48 studies.
Fig. 7.
Mortality due to the age factor (in years) with comorbidities in the cohort from the selected studies.
All these subjects had COVID-19 with one or more of these comorbidities. These comorbidities all contributed to the worsening of COVID-19. Thus, each comorbidity represents a category of the cohort whose imaging characteristics (or grayscale features) are likely to be similar. Therefore, multiple knowledge-based AI systems can be developed for each comorbidity using independent training cohorts. Based on these independently trained systems corresponding to each comorbidity, lung scans be characterized and COVID-19 severity can be assessed.
5. Artificial Intelligence architectures for ARDS characterization by school of thought
Characterization Definition. The concept of the characterization of the diseases using AI has been adopted in nearly all medical imaging areas. This includes the AI's role in pinpointing the disease, extracting the ROI of the disease, and automatically classifying the disease against binary or multiclass events. We have chosen machine learning and deep learning characterization systems that overlap and synchronize with ARDS frameworks. The idea of using this characterization system is to share the origination and innovation spirits derived for different modalities and for different organs or applications. Note that some of these are taken from our own group intentionally to show the role of tissue characterization using AI or disease characterization using AI models. These systems are the crux solutions for diversification, which are unique and not available elsewhere. We specifically listed these as a “disease-specific portfolio” as a “one-stop-shop” to focus the search for the young scientists who are particularly interested in knowing how ARDS paradigms are related to other organ paradigms. Lastly, the third advantage of this cluster is to directly approach our group should more intricate details are necessary for such parallel characterization systems. Examples of AI-based characterization can be seen in several other body and disease applications, such as brain [[102], [103], [104]], stroke [[105], [106], [107]], plaque vascular wall [[108], [109], [110]], arrhythmia [111], liver [[112], [113], [114]], coronary artery [115,116], prostate [117], ovarian [118,119], diabetes [120], thyroid cancer [121], skin cancer [122,123], heart [[124], [125], [126]], gene expression [127], and rheumatoid arthritis [128]. This characterization framework can be extended to the ARDS framework.
Non-ARDS vs. ARDS. ARDS studies have started after December 2019. On the other hand, lung segmentation and classification techniques under the non-ARDS framework have existed for several years before the start of SARS-CoV-2. Some of the AI models used for non-ARDS lung data have been partially tried on ARDS lung data. For this reason, we have listed non-ARDS AI models against the ARDS AI models under three categories: segmentation, classification, and sub-regional applications (nodule, cancer, and tumor segmentation). This paper does not cover non-ARDS AI models or techniques. Table 1 shows the studies done in both non-ARDS and ARDS frameworks. The non-ARDS AI techniques were also applied to nodule, cancer, and tumor segmentation [[129], [130], [131], [132], [133], [134], [135], [136]]. Note that we excluded (a) lung cellular images and (b) animal studies by considering only human lungs. Further state-of-the-art 2017–2020 studies were only considered for non-ARDS models, and validation and inter- and intra-observer variability studies were also not considered in Table 1.
Table 1.
AI-based studies involved during Non-ARDS and ARDS periods. AI-based Non-ARDS: AI on ARDS lung data during pre-COVID-19. AI-based ARDS: AI on ARDS lung data postmarked December 2019 COVID-19 (post-COVID-19).
| Subsystems | AI-based Non-ARDS | AI-based ARDS |
|---|---|---|
| Segmentation | Characteristics | Characteristics |
| Watershed, region-based, contour-based, fusion-based, and model-based. | FC-Densenet103, Unet, DenseNet, and DenseNet121-FPN. | |
| References | References | |
| [129,[137], [138], [139], [140], [141], [142], [143], [144], [145], [146]] | [[147], [148], [149], [150], [151], [152], [153], [154], [155], [156], [157], [158], [159], [160], [161], [162], [163], [164], [165]] | |
| Classification | Characteristics | Characteristics |
| Gray scale feature extraction and ML classifier, and model-based techniques. | Resnet-50, CNN, SVM, ResNet101, VGG16, and VGG19. | |
| References | References | |
| [[166], [167], [168], [169], [170]] | [148,[150], [151], [152], [153],[157], [158], [159],[171], [172], [173], [174], [175]] | |
| Joint Segmentation and Classification | Characteristics | Characteristics |
| They use the same characteristics as adapted by segmentation and classification domain for AI-based Non-ARDS. | They use the same characteristics as adapted by segmentation and classification domain for AI-based ARDS. | |
| References | References | |
| [167,169,170] | [148,150,151,154,[156], [157], [158], [159],162] |
It is essential to note that there are three typical components of an AI-based ARDS system: (a) segmentation of the lung region, (b) classification component, and (c) COVID-19 severity measurement (see Table 2 ). For the ARDS framework in this narrative review, we have proposed and classified the literature based on architecture (or so-called school of thought (SoT)) (see Table 3 ).
Table 2.
Types of artificial intelligence architectures and severity index.
| SN | AI Components and Attributes |
||
|---|---|---|---|
| Lung Segmentation |
AI Component |
Severity Index |
|
| Auto/Semi-Auto | ML/DL/TL/DLaML/DLaTL | Categorical/Continuous/Categoricala Continuous | |
| 1 | Auto | DL | Categoricala Continuous |
| 2 | Auto | DLaML | Categoricala Continuous |
| 3 | Auto | DL | Categorical |
| 4 | Semi-Auto | DLaML | Categoricala Continuous |
| 5 | Auto | ML | Categoricala Continuous |
| 6 | Auto | DLaTL | Categorical |
| 7 | Auto | TL | Categorical |
Both technologies are present.
Table 3.
Clustering of multimodality artificial intelligence architectures and their salient features.
| SoT | Reference | Modality | Imaging | Highlight/Objective | Architecture Description | Performance Metrics |
|---|---|---|---|---|---|---|
| SoT-1 | [[147], [148], [149], [150], [151], [152], [153],229] | CT [229]: [147,148] X-ray [149]: [151] LUS [150]: | 3-D [229]: [147,148] 2-D [149]: [150,151] | Multiview fusion [229], Multi-view pyramid network with attention [147], training using human in loop [148], video-based real time prediction [150], end- to-end DL architecture for semi quantitative prediction COVID- 19 severity [151] | Resnet50 [229], Custom CNN with attention [147], VB-Net [148], commercial deep learning system by Lunit Inc [149], Spatial Transformer Net- work [150], ensemble of multiple networks (Backbone – ResNet, VGG, DenseNet, Inception; Segmentation- UNet, UNet++; Alignment- Spatial Transformer Network; Scoring Head-Feature Pyramid Network; Custom Network) [151] | ACC [229]: [147,148,150,151] |
| AUC [229]: [147] | ||||||
| Sensitivity [229]: [149,150] | ||||||
| Specificity [229]: [149] | ||||||
| Others [148]: [[149], [150], [151]] | ||||||
| SoT-2 | [152,153] | CT [152]: [153] | 3-D [153]: | Biomarker based model [152], model for severity in 3-D lung abnormalities [153] | Resnet34 with logistic regression [152], Dense UNet [153] | AUC [152]: |
| X-ray: LUS: | 2-D [152]: | Others [152]: [153] | ||||
| SoT-3 | [[154], [155], [156],171,173,174] |
3-D [154]: [173] 2-D [171]: [[155], [156], [157],174] |
ACC [171]: [155,156,173,174] | |||
| 3-D Convolution Network [154], | Resnet50 [154], Custom CNN | [157] | ||||
| CT [154] | multi-objective differential | [157,171,173], DenseNet [155] | AUC [154]: [155,156,173,174] | |||
| [155,171] | evolution based CNN [171], comparison of ten CNNs [156], | (AlexNet, VGG-16 | Sensitivity: [154,155,171] | |||
| [156,173] | weakly supervised DL model [173], truncated InceptionNet [174], modified DarkNet CNN [157] | VGG-19, SqueezeNet, | [156,157,173,174] | |||
| X-ray: | GoogleNet, MobileNet-V2, ResNet-18, ResNet-50, ResNet-101, and Xception) [156], InceptionNetV3 [174] | Specificity [154]: [155,171] | ||||
| [157,174] | [156,157,173,174] | |||||
| LUS: NA | Others [171]: [[155], [156], [157],173,174] | |||||
| SoT-4 | [157] | CT [157]: | 3-D [157]: | ML and DL hybrid network for classification and prognosis [157] | Resnet18 with Gradient Boosting [157] | ACC [157]: |
| X-ray: NA LUS: NA | 2-D: NA | AUC [157]: | ||||
| SoT-5 | [158,159] | CT [158]: | Pleural line identification using ML [158], automatic severity assessment and exploration of severity related features using ML [159] | Hidden Markov Model and Viterbi Algorithm combined with SVM [158], Random forest [159] | ACC [158]: [159] | |
| X-ray: NA LUS [158]: | 3-D:NA | AUC: [ [159] | ||||
| 2-D [158]: [159] | Sensitivity [158]: | |||||
| Specificity [158]: | ||||||
| Others [158]: [159] | ||||||
| SoT-6 | [160,172] | CT | Ensemble of DL and TL [160], multi-dilation CNN for extraction of COVID-19 features [172] | Custom CNN [160,172], (VGG- | ACC [160]: [172] | |
| X-ray [160]: [172] | 3-D:NA | 16, VGG-19, Inception-V3, Xception, InceptionResNet- V2, MobileNet- V2, DenseNet-201, NasNet- mobile) [160] | AUC [160]: [172] | |||
| LUS: | 2-D [160]: [172] | Sensitivity [160]: [172] | ||||
| Specificity [160]: [172] | ||||||
| Others [160]: [172] | ||||||
| SoT-7 | [[161], [162], [163], [164], [165],175] | CT [165]: | Explainable DL to provide explainability about the prediction | ACC [175]: [[161], [162], [163], [164], [165]] | ||
| X-ray: | 3-D:NA | [175], real time internet based COVID-19 detection service [161], | VGG-16 [161,175], Alexnet [165] | AUC [161]: [162,164] | ||
| [[163], [164], [165],175] | 2-D [175]: [[161], [162], [163], [164], [165]] | TL model trained on ensemble of two publicly available datasets [163], interpretable AI framework for COVID-19 classification [164], applying TL on comprehensive custom COVID- 19 CT and X-ray datasets [165] | DenseNet201 [162], Xception | Sensitivity [175]: [[161], [162], [163], [164], [165]] | ||
| LUS [161]: | [163], InceptionNetV3 [164] | Specificity [175]: [[161], [162], [163], [164], [165]] | ||||
| Others [175]: [[161], [162], [163], [164]] |
Definition of Schools of Thought and Their Relationship to AI Architectures. The classification of the AI architecture has been central to different engineering applications. Imaging has been the most prominent AI application. Recently, Biswas et al. [17] presented a class of AI architectures for medical imaging. In our current study we present several applications using different classes of architectures (SoT). Biswas also discussed the evolution of architectures in terms of the present and future of deep learning [16].
These architectures can use the manual or automated feature selection method. Typically, manual feature selection methods were used in the past, where the features were hand-picked and fed to the classification model during its learning and training process. These were older generation methods. In other words, this is an old school of thought. Since the evolution of the concept of mimicking the brain using a greater number of layers for filtering the features, deep learning emerged. The main characteristic of deep learning (DL) paradigms is the ability to compute millions of training parameters automatically. This is a more recently evolved subset of ML. These methods are another, more recent school of thought. Thus, the schools of thought are primarily defined by their foundational architecture.
Although the AI industry is dominated by these two paradigms, it continues to look for alternatives. DL begins to look outdated, and more advanced architecture has started to emerge. For example, the challenges of DL during training have led to generating pre-trained weights to speed up and simplify the DL systems. This type of architecture is known as transfer learning. It is considered a more recent SoT and a classic in its category. In summary, the classification of the architectural has been synchronized more as a SoT based on the ingredients and components of the architecture. This is similar to the way segmentation of images many years ago was categorized into several architectures or SoT. For example, segmentation can be classified as region-based, contour-based, or knowledge-based. The categories were then fused together to generate intermediate architectures. These were known as fused architectures, and they combined region and contour or region with knowledge, generating another architectural paradigm or SoT. This can be seen in the classic papers by Suri [[176], [177], [178]].
Therefore, a SoT is a synonym for architectural design, where the individual and fused architectures are each considered a SoT. Fused architectures have been developed by different groups around the globe and are presented in the literature. Thus, the SoT is a more refined version of an architectural design. In summary, the idea of the School-of-Thought it to make it more granular rather than being too binary. Some of our previous studies have used the school of thought concept. Please note the SoT are technically the same as the generations of architectures; however, they give more subtle differences.
The AI models used in different SoT along with their salient features are listed below.
SoT-1 is useful for obtaining a quantitative measurement of the extent of COVID-19 lung damage and classifying the lung within the binary and multiclass frameworks. Lung segmentation is automated without human intervention, using either commercial software, such as XMedCon [179], or AI techniques, such as threshold segmentation [180] or UNet-based segmentation [146]. The AI component is state-of-the-art or custom deep learning (DL) architecture.
The purpose of SoT-2 is similar to that of SoT-1 (to quantitatively and categorically estimate COVID-19 severity). However, researchers have employed a hybrid architecture that is more complex than the architecture of SoT-1.
Nevertheless, by blending multiple AI architectures (DL and ML), SoT-2 has generally achieved comparable accuracy and accomplished additional sub-goals (e.g., prognosis analysis). The disadvantage of SoT-2 is the time and effort required for the amalgamation and fine-tuning of discrete AI components.
SoT-3 is used only to divide patients into two or more classes (e.g., COVID-19, non-COVID-19, and other kinds of pneumonia). The majority of the AI-based COVID-19 research follows SoT-3. Lung segmentation before classification is automated, just like in SoT-1. SoT-3 involves an end-to-end automated pipeline with the primary goal being classification in one of several categories of pneumonia, including COVID-19. Because of this, it is faster (and often easier) to carry out research using SoT-3. The primary disadvantage of SoT-3 is that it offers few insights that healthcare professionals can apply to treat COVID-19-infected patients. For example, SoT-3 does not help quantify biomarkers that represent the severity of the disease.
SoT-4 involves the semi-automatic segmentation of lungs from radiography images by professionals before use for AI-related processing. Multiple metrics are computed using an AI component that is a hybrid of various AI architectures. SoT-4 gives researchers the freedom to create a pipeline with the appropriate balance of accuracy, speed, and complexity, depending on the desired metrics. The disadvantage of SoT-4 is that designing the semi-automatic segmentation of lungs and the appropriate hybrid models takes a fairly long time.
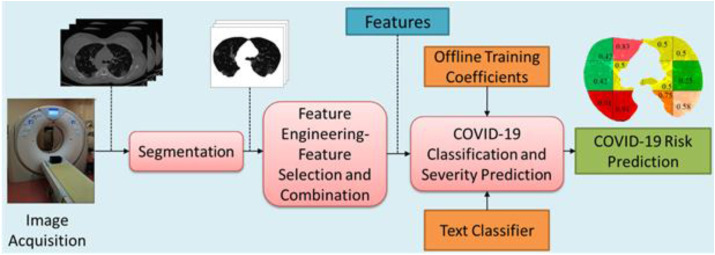
In SoT-5, researchers use automated lung segmentation, but they also use traditional machine learning (ML) models, hand-crafted feature engineering, and feature selection methods. The computed features are then used to compute biomarkers, which, in turn, are used to predict the severity of COVID-19 and to classify patients. Research belonging to SoT-6 uses automated segmentation with a hybrid model of DL and TL to produce a categorical metric for classifying patients. Finally, SoT-7 research is similar to SoT-6, with the sole difference being that the transfer learning (TL) model is the primary component [181]. It contains two components: an offline system and an online system. Relevant features are extracted manually by the researchers using radiography images. These features are then passed on to a classifier in an offline system for training. Once the offline system is trained, the training coefficients are transferred to the online classifier, which is then used for real-time classification and severity prediction. ML can also be effectively used to segment the lungs in medical images, as shown in Refs. [146,167]. An online ML-based COVID-19 risk prediction system is depicted in Fig. 8 . This is very much along the lines of previous ML systems published by our group [165,182,183].
Fig. 8.
An online ML-based COVID-19 risk prediction system. (Courtesy of AtheroPoint™, Roseville, CA, USA; reproduced with permission.)
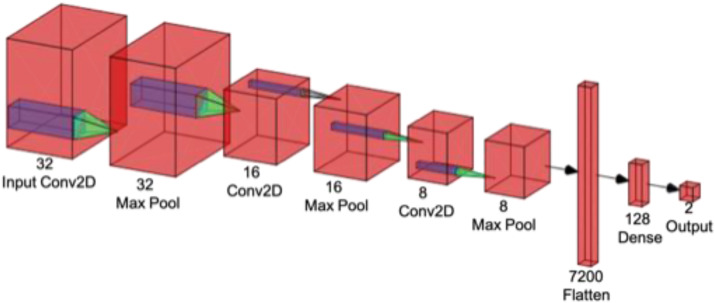
A reference DL architecture created using [184] is shown in Fig. 9 . The most commonly used type of DL architecture is the convolution neural network (CNN). This network consists of multiple convolutional filters that extract simple visual features from the image data. Multiple CNN filters are stacked together in layers so that complex visual features can be extracted from the image data. The CNN layers are often bundled together with pooling layers to reduce the spatial information contained in the intermediate representation and padding layers, thus maintaining the appropriate dimensions of the data.
Fig. 9.
A custom CNN-based DL architecture comprising different layers.
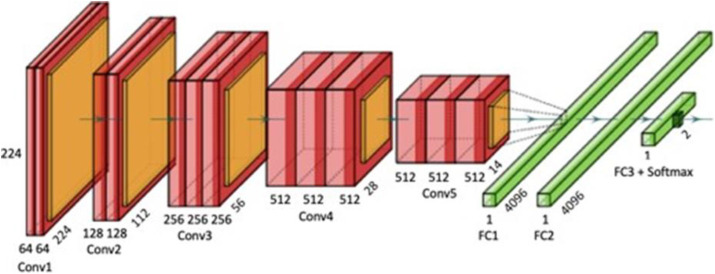
TL is an extension of state-of-the-art DL architectures that are pre-trained using massive datasets. TL techniques produce more accurate results when there is less training data, when the available hardware has fewer capabilities, or when little training time is available. The reference architecture of TL created using [185] (called VGG16) is shown in Fig. 10 .
Fig. 10.
An example of transfer learning (TL) architecture using VGG16.
Table 3 presents a comparison of multi-modality AI architectures for COVID-19 diagnostics, along with their salient features. The “Arch Type” refers to the seven SoT architectures that we have devised after clustering various AI research works based on their workflow and intent. The “Reference” and “Modality” attributes are self-explanatory. The “3-D/2-D Imaging” attribute shows whether the research has used two-dimensional or three-dimensional radiological imaging [186,187].
The “Highlight/Objective” attribute represents the distinguishing feature of the research. The “Architecture Description” attribute describes the underlying base AI architecture adapted in the study. The “Performance Metrics” attribute shows which metrics the researchers had used to validate their work. Readers are encouraged to look into dedicated AI-based reviews for a deeper understanding of AI and its applications.
6. Workflow considerations for COVID-19 lung characterization: CT vs. X-ray
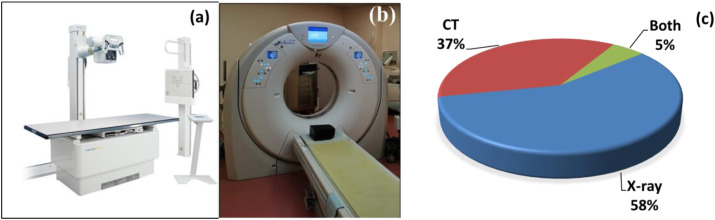
The three chest imaging modalities X-ray, CT, and ultrasound have been strongly recommended by WHO for the diagnosis of COVID-19 [90]. In their guidelines, the following observations were given for COVID-19 diagnosis. (i)) X-rays was found to be lower in sensitivity and higher specificity than chest CT imaging [188,189]. (ii) Chest X-rays are less expensive, have lower radiation, takes less acquisition time, and are less expensive to use for monitoring than CT. (iii) Chest CT was found to have higher sensitivity but lower specificity and emits more radiation than X-ray. (iv) Lung ultrasound was found to have very low diagnostic accuracy but offers an alternative for several other applications, such as the abdomen, carotid, urology, obstetrics, and gynecology imaging. On the other hand, ultrasound has a high risk of COVID-19 infection transmission due to close contact with the patient compared to other imaging modalities. Meanwhile, MRI is a popular imaging modality for diagnosing many severe diseases. However, according to the guidelines of the American College of Radiology (released on April 8, 2020) [190], MRI is not recommended for COVID-19 patients due to the high risk of infection [91]. The literature includes studies covering MRI and COVID-19 [[92], [93], [94]]; however, no relevant literature was found where MRI, COVID-19, and AI were all mentioned, mainly due to the high cost, long scanning time, and high risk of infection. FDG-PET/CT [191] imaging is another high-level imaging technique that was used in many studies to diagnose COVID-19 [95,96]. PET/CT is a more advanced imaging technique than CT alone; however, it is more expensive [23,192]. We did not find any relevant studies for the diagnosis of COVID-19 with FDG-PET/CT imaging that used AI. The suitability of X-ray and CT imaging for COVID-19 (see Fig. 11 ), based on recommendations by WHO, is shown in Table 4 , where we discuss the workflow and the practical implementation. The major studies for automated COVID-19 diagnosis using the AI paradigm with X-rays and CT are [[21], [22], [23],[192], [193], [194], [195], [196], [197], [198], [199], [200], [201], [202]]. Due to their popularity in the scientific community, there are several open-source COVID-19 datasets available for X-ray and CT imaging modalities, such as the one from RSNA (https://www.rsna.org). The percentages of studies using X-ray, CT, or both, shown in Fig. 11(c), are 58%, 37%, and 5%, respectively, with CT considered the gold standard. These studies were chosen because of the completeness of their AI models and the attributes governing them. The following attributes were considered for comparing X-rays and CT: (i) number of subjects, (ii) risk classes, (iii) 2-D vs. 3-D imaging, (iv) automated vs. non-automated ROI segmentation, (v) AI models, (vi) augmented vs. non-augmented, (vii) K-fold cross-validation, (vii) hardware and software, (ix) optimal models, and (x) performances. Based on these attributes, the studies on X-ray and CT imaging modality for automated COVID-19 diagnosis are compared, respectively, in Table 5, Table 6 . For the 1st attribute (number of subjects), some remarkable studies were found ([[203], [204], [205], [206], [207], [208]]) that used a cohort of over 1000 subjects. The 2nd attribute (number of classes) was broadly divided into binary and multiclass. More than half of the selected studies had binary classification [203,204,[209], [210], [211], [212], [213], [214], [215], [216]], while the rest had multiclass. For the 3rd attribute (2-D vs. 3-D), two studies ([210,212]) used a 3-D CT volume as input, while the rest used 2-D X-ray images. For the 4th attribute, ROI segmentation in chest images was an essential aspect prior to classification. Three types of ROI were observed; five studies ([203,211,212,217,218]) used automated ROI segmentation criteria, and only one study ([209]) adopted manual segmentation. Most studies used full-sized images for the classification.
Fig. 11.
(a) An X-ray scanner. (b) A CT-scanner (Courtesy of Luca Saba, University of Cagliari, Italy). (c) Studies using CT vs. X-ray.
Table 4.
Compatibility of imaging modality for COVID-19 and adaptability for AI [230].
| Imaging Modality | Suitable for COVID-19 as per WHO guidelines | Cost | Risk of radiation | Risk of infection due to close contact | Compatible with AI for COVID-19 diagnosis |
|---|---|---|---|---|---|
| PET/CT | High | Very High | Very High | Low | Low |
| CT | High | High | Very High | Low | Very High |
| X-ray | Medium | Low | High | Low | Medium |
| Ultrasound | Low | Low | No | High | Low |
| MRI | Low | High | No | Low | Low |
Table 5.
Artificial Intelligence-based studies for automatic COVID-19 detection using lung CT.
| SN | Reference | aSubj. | Risk Class | 2-D vs. 3-D | *ROI | AI Model | Augm. | CV | H/W-S/W | Optimal Model | Performances |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ardakani et al. (2020) [209] | 194 | 2 | 2-D | ✗ | CNN + TL | ✗ | NA | SPSS software (version 24, IBM) | ResNet101 | ACC:99.51% |
| SE:100% | |||||||||||
| SP:99.02% | |||||||||||
| AUC:0.994 | |||||||||||
| 2 | Wu et al. (2020) [217] | 495 | 3 | 2-D | ✓ | CNN + TL | ✗ | NA | GPU Python | ResNet50 | ACC:76.0% |
| SE:81.1% | |||||||||||
| SP:61.5% | |||||||||||
| AUC:0.819 | |||||||||||
| 3 | Zheng et al. (2020) [210] | 499 | 2 | 3-D | ✗ | 3-D CNN + TL | ✓ | NA | GPU PyTorch | UNet | ACC: 90.1% |
| SE: 90.7% | |||||||||||
| SP: 911% | |||||||||||
| AUC: 0.976 | |||||||||||
| 4 | Yang et al. (2020) [211] | 679 | 2 | 2-D | ✓ | CNN | ✓ | NA | GPU PyTorch | DenseNet169 | ACC:89% |
| FS:90% | |||||||||||
| AUC:0.98 | |||||||||||
| 5 | Gozes et al. (2020) [212] | 270 | 2 | 2-D & 3-D | ✓ | 3-D + 2-D CNN | ✓ | NA | NA | Resnet50 | SE: 98.2% |
| SP: 92.2% | |||||||||||
| AUC: 0.996 | |||||||||||
| 6 | Shi et al. (2020) [203] | 2685 | 2 | 2-D | ✓ | ML (RF) | ✗ | K5 | NA | RF | ACC:87.9% |
| SE: 90.7% | |||||||||||
| SP: 83.3% | |||||||||||
| AUC: 0.942 | |||||||||||
| 7 | Liu et al. (2020) [213] | 746 | 2 | 2-D | ✗ | LA-DNN + TL | ✗ | NA | NA | LA-DNN | ACC:88.8% |
| F1S: 94.7% | |||||||||||
| AUC: 0.88 | |||||||||||
| 8 | Panwar et al. (2020) [204] | 2482 | 2 | 2-D | ✗ | CNN + TL | ✗ | NA | NA | VGG19 | ACC:87.9% |
| SE: 90.7% | |||||||||||
| SP: 83.3% | |||||||||||
| AUC: 0.942 |
Subj.: Number of subjects in the study; Augm.: Augmentation; NA: Not available; ACC: Accuracy, SE: Sensitivity; SP: Specificity; AUC: Area under the curve; F-M: F-measure; KP: Kappa statistics; TL: Transfer Learning; FS: F1-Score LA-DNN: lesion-attention deep neural network; RF: Random forest; ML: machine learning; H/W: Hardware; S/W: Software; CV: Cross-Validation; K5: five-fold; ROI: Automated Region of Interest.
Table 6.
Artificial Intelligence-based studies for automatic COVID-19 detection using lung X-ray.
| SN | Reference | aSubj. | Risk Class | 2-D vs. 3-D | Auto ROI | AI Model | Augm. | CV | H/W-S/W | Optimal Model | Performances |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Narayan Das et al. (2020) [216] | NA | 3 | 2-D | ✗ | CNN + TL | ✗ | NA | NA | Proposed CNN | ACC: 97% |
| FM: 96% | |||||||||||
| SE: 97% SP:97% | |||||||||||
| KP:0.97 | |||||||||||
| 2 | Ouchicha et al. (2020) [205] | 2905 | 3 | 2-D | ✗ | CNN | ✗ | K5 | NA | CVDNet (proposed CNN) | ACC: 90% |
| PR: 96.72% | |||||||||||
| RC: 96.84% | |||||||||||
| FS: 96.68% | |||||||||||
| 3 | Hemdan et al. (2020) [214] | 50 | 2 | 2-D | ✗ | CNN | ✗ | NA | GPU Python | VGG19 DenseNet201 | ACC: 96.69% |
| PR: 83% | |||||||||||
| RC: 100% | |||||||||||
| FS: 91% | |||||||||||
| 4 | Zhang et al. (2020) [215] | 43,370 | 1, 2 | 2-D | ✗ | CNN | ✓ | K5 | NA | CAAD (proposed CNN) | ACC: 78.57% |
| SE: 71.70% | |||||||||||
| SP:79.40% | |||||||||||
| AUC: 0.83 | |||||||||||
| 5 | Togacar et al. (2020) [218] | 458 | 3 | 2-D | ✓ | CNN + TL | ✓ | K5 | CPU MATLAB | MobileNetV2 | ACC: 99.24% |
| SE: 100% | |||||||||||
| SP: 97.72% | |||||||||||
| FS: 99.43% | |||||||||||
| 6 | Farooq et al. (2020) [206] | 2839 | 4 | 2-D | ✗ | CNN + TL | ✓ | NA | NA | ResNet50 | ACC: 96.23% |
| RC: 100% | |||||||||||
| PR: 100% | |||||||||||
| FS: 100% | |||||||||||
| 7 | Cozzi et al. (2020) [207] | 1427 | 3 | 2-D | ✗ | CNN + TL | ✗ | K10 | NA | VGG19 | ACC: 96.78% |
| SE: 98.66% | |||||||||||
| SP: 96.46% | |||||||||||
| 8 | Pereira et al. (2020) [208] | 1144 | 7 | 2-D | ✗ | ML + DL + TL | ✗ | NA | NA | Multilayer Perceptron | FS: 89% |
Subj.: Number of subjects in the study; Augm: Augmentation; NA: Not available; ACC: Accuracy; SE: Sensitivity; SP: Specificity; AUC: Area under the curve; F-M: F-measure; KP: Kappa statistics; PR: precision; RC: recall; FS:F-Score; TL: Transfer Learning; LA-DNN: lesion-attention deep neural network; ML: machine learning; H/W: Hardware; S/W: Software; CAAD: confidence-aware anomaly detection model; ROI: Region of Interest.
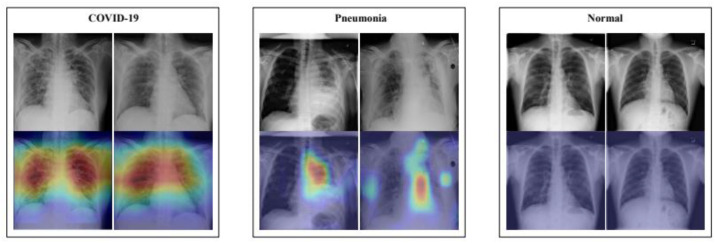
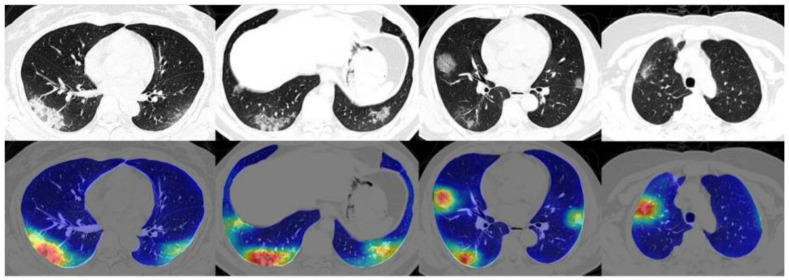
The 5th attribute (type of AI models) includes two main categories, ML- and DL-based studies. In one study on chest X-rays, images were classified as normal, pneumonia, other diseases, or COVID-19 using a DL-based network, with an accuracy of 90.13% [219]. Some of the images and their corresponding color maps are shown in Fig. 12 . In another study, COVID-19 abnormalities were detected in lung CT scans using a DL-based network, and a heatmap of corresponding severity levels was generated [220]. The resultant images are shown in Fig. 13 .
Fig. 12.
X-ray scans of COVID-19, pneumonia, and normal lungs (Reproduced with permission [219]).
Fig. 13.
CT scans classified as positive for coronavirus abnormalities and their corresponding color heatmaps (Reproduced with permission [220]).
For the 6th attribute (image augmentation), data limitation is always a big challenge for medical studies. Therefore, most of the studies used TL with DL models in such scenarios, especially when data is limited (i.e., in the thousands). Due to limited data, augmentation was applied to increase the data synthetically [[210], [211], [212],215,218]. For this method, the images are synthetically increased using operations like image transformation (rotation, translation, and scaling), blurring, color jiggling, etc. There have been studies that use a patch-based framework with a relatively small number of trainable parameters for COVID-19 diagnosis, and use multi-scaling for spatial features [221,222]. For the cross-validation (the 7th attribute) of the data, the K-fold strategy was used to evaluate the performance of the AI technique. In this technique, K is the number of folds (or combinations) of training and testing samples.
In each round, the training is performed, and the model predicts the outcome based on the test sample. These predictions are compared against the gold standard to compute the ROI and performance of the AI system. Some authors ([203,205,215], and [218]) used five-fold CV, and one author [207] used ten-fold CV. The hardware constraints (the 8th attribute) is discussed further in the critical discussion section. During optimization (the 9th attribute) of the AI model, it is important to avoid overfitting. CV and TL were used to suppress over-fitting in supervised learning. Overfitting is a situation where the model can predict known data accurately, but is unable to predict unknown data. There are many ways to avoid overfitting; one of them is to use more data in the training model. The performance (the 10th attribute) of the classifier depends on various factors, such as the number of subjects, the sample size, the training iterations, the training time, etc. Therefore, we cannot identify a single best-performing model or method at this stage. However, over time, as more studies and trials are conducted, the superior AI models will emerge.
7. Critical discussion
Clinical Requirements for COVID-19 AI Systems. An ideal AI system for COVID-19 must be robust and stable, and its output must vary within the acceptable limits with changes in the demographics or other patient-related characteristics. It must be reliable and reproducible; i.e., it must yield similar results across multiple trials. Furthermore, when used in an operating room, it must be reasonably fast and cost-effective compared to traditional COVID-19 diagnostic techniques (e.g., real-time polymerase chain reaction (RT-PCR) tests). The AI system must be generalized by being trained and tested on an equal percentage of data. COVID-19 severity is a critical metric for doctors treating COVID-19 patients, and it is a desirable requirement of any AI diagnostic system.
Radiologists, pulmonologists, and doctors must validate the results of the AI system before its effectiveness can be determined. Finally, the AI system should segment the impact of COVID-19 on different organs using 3-D imaging to further assist clinicians. There is a vast potential for improvement for existing SoT using the AI-assisted diagnostics of COVID-19 patients by incorporating the above clinical requirements.
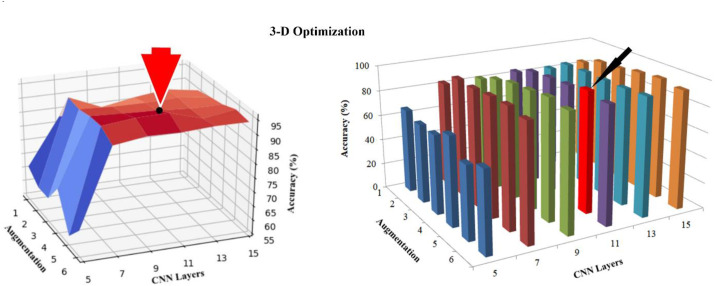
System Optimization. The majority of AI-based systems for COVID-19 detection and classification are based on DL. Several researchers have used data augmentation to increase the volume of COVID-19 data for training. The number of convolution layers is a hyperparameter that researchers need to adjust based on their intuition. However, DL-focused AI systems for COVID-19 need to be optimized based on the relationship between augmentation, the number of convolution layers, and classification accuracy (Fig. 14).
Fig. 14.
A 3-D graph representing the relationship between CNN layers, data augmentation, and accuracy. (Courtesy of AtheroPoint™, Roseville, CA, USA; reproduced with permission [14].)
Scientific Validation. The behavior of an AI-based COVID-19 diagnostics system should be observed and validated under various comorbidity conditions. There are numerous scenarios in which the underlying assumptions may change. For example, the CT scan's thickness can be altered, or a different view of a CT scan (coronal, sagittal, and axial) can be given to the system for diagnostic purposes. Furthermore, the stability of a system must be validated by changing the combination of data that is used (e.g., K10, K5, K2, the so-called partition protocol). A stable AI system will yield a minimum standard deviation value across different data combinations [165]. The system should also be validated for patients from different age groups and with different comorbidities.
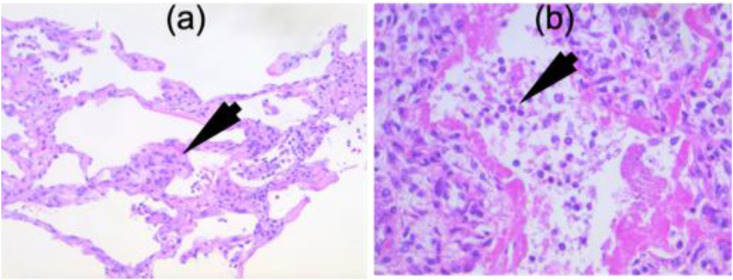
Clinical Validation. In order for the research to be accepted by the regulatory authorities, a gold standard validation should be performed through a physical examination of the body organs against the results predicted by the AI system. One way of doing this is through the microscopic validation of the body tissues for the severity of ARDS from COVID-19, as shown in Fig. 15 . Inter-observer and intra-observer variability analysis [223] should also be performed to minimize any human bias in the system.
Fig. 15.
Microscopic views of (a) interstitial pneumonia and (b) COVID-19 pneumonia. (Courtesy of Luca Saba, A.O.U., Cagliari, Italy.)
AI for COVID-19 Comorbidity and Age-Group Frameworks. The AI system must be designed to accommodate the patients’ comorbidities (e.g., diabetes, cardiovascular diseases, obesity, retinopathy, pancreatic diseases, blood vessel diseases, and angiography [224]), along with their age groups. In Ref. [225], an AI system was developed to predict cardiovascular diseases in a multi-ethnic patient scenario. The AI systems designed in Refs. [226,227] can be extended to COVID-19 diagnosis by including comorbidities and age groups.
The primary AI system can be broken down into multiple subsystems for various comorbidity classes, with each comorbidity class having further subclasses for different age groups. For instance, if there are five comorbidities and three age groups, there will be 15 subsystems. Each subsystem should be trained separately using a different gold standard database [228]. The appropriate AI subsystem for a new patient can be determined by feeding additional input information about the patient's comorbidities and age group into the system.
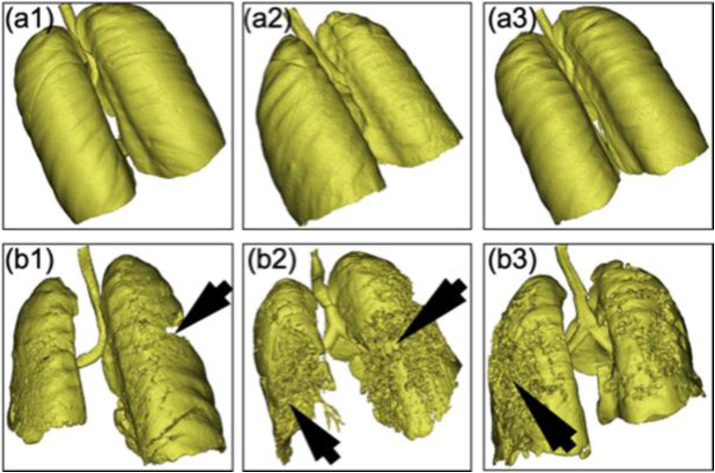
3-D Image Acquisition and Processing. Most AI research for COVID-19 shows the severity of the lung infection in 2-D, which is undesirable because the disease progression cannot be depicted accurately in 2-D. It is therefore helpful to evaluate the COVID-19 data in 3-D (see Fig. 16). Scans of the Patients can be acquired during the contrast enhancement of a lung ultrasound [231]. A contrast agent, if included in the bloodstream, aids the early management and prognosis of COVID-19. In Ref. [232], the authors used 3-D CT for diagnosis and classification of lung lesions affected by COVID-19. Recently, the authors in Ref. [233] used DensNet121-FPN for segmentation of the lung and classification using deep learning strategy.
Fig. 16.
Three lungs with non-COVID-19 pneumonia (a1, a2, and a3). Three lungs with COVID-19 pneumonia with different COVID-19 severities (b1, b2, and b3). (Courtesy of AtheroPoint™, Roseville, CA, USA; reproduced with permission.)
Multi-Modality Data. Radiography provides many solutions for diagnosing COVID-19, like lung CT, chest X-rays, lung ultrasound, and PET/CT scans. The choice of modality depends on the patient's comorbidities, age, and pre-test probabilities (PTPs) [18]. A PTP test on troponin levels (which is an indirect indicator of hypoxia) can be conducted to identify the severity of a COVID-19 infection. During the initial phase of the COVID-19 infection, X-rays and lung ultrasounds can be useful owing to their low cost and easy obtainability, and to the small footprint of the medical device. However, if a patient has comorbidities, is elderly, or has COVID-19-induced ARDS (which leads to hypoxia), lung CT is likely the best modality due to its higher resolution and robust diagnostic capabilities. Any AI system for COVID-19 must be adaptable and generalizable to multiple modalities according to each patient's requirements.
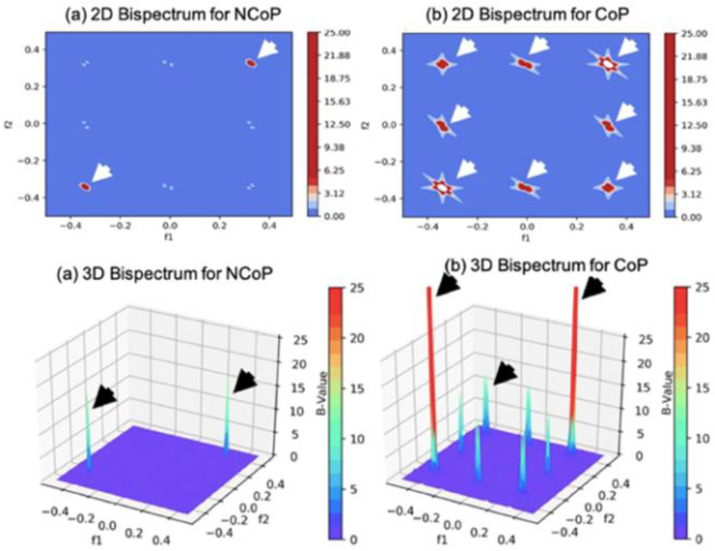
Tissue Characterization of Lung Scans. In our previous work [3], we performed a bispectrum (B) analysis on COVID-19 lung tissues based on a higher order spectrum (HoS) to validate the survey results without using AI. The results (Fig. 17 ) confirm that COVID-19-infected samples possess much higher B-values than non-infected samples. Thus, AI can effectively exploit the underlying patterns to distinguish COVID-19 samples from non-COVID-19 samples.
Fig. 17.
Bispectrum analysis of non-COVID-19 pneumonia (NCoP) and COVID-19 pneumonia (CoP). (Courtesy of AtheroPoint™, Roseville, CA, USA; reproduced with permission.)
Data Collection. It is desirable to collect data during a sizeable temporal window and from geographically distant locations, because this reduces biases related to age, comorbidity, ethnicity, and other factors among COVID-19 patients. Patient data must also be multiclass, consisting of a control class and various types of pneumonia classes (e.g., COVID-19, viral, bacterial, HIV, and MERS). Meeting these conditions helps to create a more generalized and robust AI system that can be used for COVID-19 diagnostics in any medical setting without the need to perform localized validations or retraining.
AI and Hardware Constraints. GPUs are essential in DL studies [234,235]. Most of the studies (training and testing) are performed on GPUs using open-source platforms like Google-Colab, with Python as the language and TensorFlow or PyTorch as the framework [14,102].
Strengths, Limitations, and Extensions. This study provides an insight into acute respiratory distress syndrome; into its relationship to comorbidity; into medical imaging modalities for imaging COVID-19 subjects in an ARDS framework; into the workflow and practical aspects of premier imaging tools; into the design of AI architectures and their adaptation to handle ARDS-based lung severity diagnoses; and finally, into the role of AI-based solutions for comorbid conditions. The study also highlights the critical components needed for a safe and effective AI-based paradigm for risk assessment of COVID-19 severity.
Even though the study offers several new directions through the amalgamation of comorbidity-based AI designs, it is missing several other facets that could be included in a more compressive review. These include the biological processes that inherit comorbidities. This omission is mainly due to limitations of manuscript length. More stringent comparisons of deep learning paradigms can be adapted; we have provided leads to previously published dedicated reviews on AI.
Future research should look for a way to quickly determine the COVID-19 severity using a global positioning system in target lung regions in 2-D and 3-D images, while considering comorbidity and time constraints [[236], [237], [238]]. This fundamental study can also be further developed in several other directions, covering the fields of neurology [239], cardiology, diabetology [240], and ophthalmology [241]. This includes the applications of statistical tools for more systematic review and meta-analysis (SRMA). Since this is a narrative review, the SRMA is beyond its scope.
8. Conclusion
This study is one of the latest contributions that investigated different kinds of comorbidities, its contributions to the ARDS-based framework, its effect on mortality, and finally, the study proposed the AI-based solutions using comorbidity as an independent factor in their design. The salient features of the seven types of school-of-thought were presented, and their architectural characteristics were highlighted. We conclude that there are critical components of the AI system that must be thoroughly investigated before it can be applied clinically for diagnosis of COVID-19 severity. These include (a) scientific and clinical validation, (b) optimization of layers vs. augmentation for best AI optimized architecture design, (c) application of imaging modality based on COVID-19 symptoms severity and troponin release. Lastly, we conclude that the most powerful paradigm where the future holds for ARDS characterization is by incorporating the comorbidity and age-group as one of the distinguishing and novel features in the AI design [[242], [243], [244]].
Declaration of competing interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome. We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.
We confirm that we have given due consideration to the protection of intellectual propertyassociated with this work and that there are no impediments to publication, including thetiming of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
We understand that the Corresponding Author is the sole contact for the Editorial process. He is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs. We confirm that we have provided a current, correct email address which is accessible by the Corresponding Author and which has been configured to accept email from.”
I, Mainak Biswas on behalf of all authors of the manuscript “A Narrative Review on Characterization of Acute Respiratory Distress Syndrome in COVID-19 Lungs using Artificial Intelligence” hereby declare that the details furnished above are true and correct to the best of my knowledge and belief. In case any of the above information is found to be false or untrue or misleading or misrepresenting, I am aware that I may be held liable for it.
Symbol Table
- 2-D
Two dimensions
- 3-D
Three dimensions
- ACC
Accuracy
- AI
Artificial intelligence
- ARDS
Acute respiratory distress syndrome
- AUC
Area-under-the-curve
- CNN
Convolution neural network
- CT
Computed tomography
- DL
Deep learning
- FS
F1-score
- GPU
Graphics Processing Unit
- H/W and S/W
Hardware and software
- LUS
Lung ultrasound
- ML
Machine learning
- MRI
Magnetic resonance imaging
- RF
Random forest
- RSNA
Radiological Society of North America
- SE
Sensitivity
- SP
Specificity
- TL
Transfer learning
- US
Ultrasound
- X-ray
Röntgen radiation
Footnotes
Quotation from Wikipedia [https://en.wikipedia.org/wiki/Syndemic]: “A syndemic or synergistic epidemic is the aggregation of two or more concurrent or sequential epidemics or disease clusters in a population with biological interactions, which exacerbate the prognosis and burden of disease.”
References
- 1.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7 doi: 10.1186/s40779-020-00240-0. 11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horton R.J.L. Offline: COVID-19 is not a pandemic. 2020;396:874. doi: 10.1016/S0140-6736(20)32000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D. Cucinotta, M. Vanelli, WHO Declares COVID-19 a Pandemic. [DOI] [PMC free article] [PubMed]
- 5.https://www.worldometers.info/coronavirus/
- 6.D'Arienzo M., Coniglio A. Assessment of the SARS-CoV-2 basic reproduction number, R (0), based on the early phase of COVID-19 outbreak in Italy. Biosaf Health. 2020;2:57–59. doi: 10.1016/j.bsheal.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravalli S., Musumeci G. Multidisciplinary Digital Publishing Institute; 2020. Coronavirus Outbreak in Italy: Physiological Benefits of Home-Based Exercise during Pandemic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilder-Smith A., Chiew C.J., Lee V.J. Can we contain the COVID-19 outbreak with the same measures as for SARS? Lancet Infect. Dis. 2020;20:e102–e107. doi: 10.1016/S1473-3099(20)30129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maugeri G., Castrogiovanni P., Battaglia G., Pippi R., D'Agata V., Palma A., Di Rosa M., Musumeci G.J.H. vol. 6. 2020. (The Impact of Physical Activity on Psychological Health during Covid-19 Pandemic in Italy). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesser I.A., Nienhuis C.P., Health P. vol. 17. 2020. p. 3899. (The Impact of COVID-19 on Physical Activity Behavior and Well-Being of Canadians). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.V. Viswanathan, A. Puvvula, A.D. Jamthikar, A Pathophysiological Bidirectional Association between Diabetes Mellitus and COVID-19 Leading to Heart and Brain Injury: A Mini-Review.
- 12.Saba L., Gerosa C., Wintermark M., Hedin U., Fanni D., Suri J.S., Balestrieri A., Faa G. Can COVID19 trigger the plaque vulnerability—a Kounis syndrome warning for “asymptomatic subjects”. Cardiovasc. Diagn. Ther. 2020;10:1352–1355. doi: 10.21037/cdt-20-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pubmed COVID-19 Publicatons.
- 14.Skandha S.S., Gupta S.K., Saba L., Koppula V.K., Johri A.M., Khanna N.N., Mavrogeni S., Laird J.R., Pareek G., Miner M., Sfikakis P.P., Protogerou A., Misra D.P., Agarwal V., Sharma A.M., Viswanathan V., Rathore V.S., Turk M., Kolluri R., Viskovic K., Cuadrado-Godia E., Kitas G.D., Nicolaides A., Suri J.S. 3-D optimized classification and characterization artificial intelligence paradigm for cardiovascular/stroke risk stratification using carotid ultrasound-based delineated plaque: atheromatic™ 2.0. Comput. Biol. Med. 2020;125:103958. doi: 10.1016/j.compbiomed.2020.103958. [DOI] [PubMed] [Google Scholar]
- 15.https://ourworldindata.org/grapher/total-confirmed-cases-of-covid-19-per-million-people
- 16.Saba L., Biswas M., Kuppili V., Cuadrado Godia E., Suri H.S., Edla D.R., Omerzu T., Laird J.R., Khanna N.N., Mavrogeni S., Protogerou A., Sfikakis P.P., Viswanathan V., Kitas G.D., Nicolaides A., Gupta A., Suri J.S. The present and future of deep learning in radiology. Eur. J. Radiol. 2019;114:14–24. doi: 10.1016/j.ejrad.2019.02.038. [DOI] [PubMed] [Google Scholar]
- 17.Biswas M., Kuppili V., Saba L., Edla D.R., Suri H.S., Cuadrado-Godia E., Laird J.R., Marinhoe R.T., Sanches J.M., Nicolaides A., Suri J.S. State-of-the-art review on deep learning in medical imaging. Front. Biosci. 2019;24:392–426. doi: 10.2741/4725. [DOI] [PubMed] [Google Scholar]
- 18.Suri J.S., Puvvula A., Biswas M., Majhail M., Saba L., Faa G., Singh I.M., Oberleitner R., Turk M., Chadha P.S., Johri A.M., Sanches J.M., Khanna N.N., Viskovic K., Mavrogeni S., Laird J.R., Pareek G., Miner M., Sobel D.W., Balestrieri A., Sfikakis P.P., Tsoulfas G., Protogerou A., Misra D.P., Agarwal V., Kitas G.D., Ahluwalia P., Kolluri R., Teji J., Maini M.A., Agbakoba A., Dhanjil S.K., Sockalingam M., Saxena A., Nicolaides A., Sharma A., Rathore V., Ajuluchukwu J.N.A., Fatemi M., Alizad A., Viswanathan V., Krishnan P.R., Naidu S. COVID-19 pathways for brain and heart injury in comorbidity patients: a role of medical imaging and artificial intelligence-based COVID severity classification: a review. Comput. Biol. Med. 2020;124 doi: 10.1016/j.compbiomed.2020.103960. 103960-103960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatto M., Bertuzzo E., Mari L., Miccoli S., Carraro L., Casagrandi R., Rinaldo A. Spread and dynamics of the COVID-19 epidemic in Italy: effects of emergency containment measures. Proc. Natl. Acad. Sci. U. S. A. 2020;117:10484–10491. doi: 10.1073/pnas.2004978117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.rekha Hanumanthu S.J.C. 2020. Solitons, Fractals, Role of Intelligent Computing in COVID-19 Prognosis: A State-Of-The-Art Review; p. 109947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng Y., Lei L., Chen Y., Zhang W. The potential added value of FDG PET/CT for COVID-19 pneumonia. Eur. J. Nucl. Med. Mol. Imag. 2020;47:1634–1635. doi: 10.1007/s00259-020-04767-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C., Zhou J., Xia L., Cheng X., Lu D. 18F-FDG PET/CT and serial chest CT findings in a COVID-19 patient with dynamic clinical characteristics in different period. Clin. Nucl. Med. 2020;45:495–496. doi: 10.1097/RLU.0000000000003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurea S., Mainolfi C.G., Bombace C., Annunziata A., Attanasio L., Petretta M., Del Vecchio S., Cuocolo A. FDG-PET/CT imaging during the Covid-19 emergency: a southern Italian perspective. Eur. J. Nucl. Med. Mol. Imag. 2020;47:2691–2697. doi: 10.1007/s00259-020-04931-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mossel E.C., Wang J., Jeffers S., Edeen K.E., Wang S., Cosgrove G.P., Funk C.J., Manzer R., Miura T.A., Pearson L.D., Holmes K.V., Mason R.J. SARS-CoV replicates in primary human alveolar type II cell cultures but not in type I-like cells. Virology. 2008;372:127–135. doi: 10.1016/j.virol.2007.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saba Luca, Clara Gerosa D.F., Marongiu Francesco, La Nasa Giorgio, Caocci Giovanni, Barcellona Doris, Balestrieri Antonella, Coghe Ferdinando, Orru Germano, Jasjit S Suri. 2020. Riccardo Cau, Massimo Castagnola, Gavino Faa, Molecular Pathways Triggered by COVID19 in Different Organs: ACE2 Receptor-Expressing Cells under Attack? A Review. [DOI] [PubMed] [Google Scholar]
- 27.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian Z., Travanty E.A., Oko L., Edeen K., Berglund A., Wang J., Ito Y., Holmes K.V., Mason R.J. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. Am. J. Respir. Cell Mol. Biol. 2013;48:742–748. doi: 10.1165/rcmb.2012-0339OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding Y., Wang H., Shen H., Li Z., Geng J., Han H., Cai J., Li X., Kang W., Weng D., Lu Y., Wu D., He L., Yao K. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 2003;200:282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Zheng X., Tong Q., Li W., Wang B., Sutter K., Trilling M., Lu M., Dittmer U., Yang D. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J. Med. Virol. 2020;92:491–494. doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S., Le T.Q., Kurihara N., Chida J., Cisse Y., Yano M., Kido H. Influenza virus-cytokine-protease cycle in the pathogenesis of vascular hyperpermeability in severe influenza. J. Infect. Dis. 2010;202:991–1001. doi: 10.1086/656044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matthay M.A., Ware L.B., Zimmerman G.A. The acute respiratory distress syndrome. J. Clin. Invest. 2012;122:2731–2740. doi: 10.1172/JCI60331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katzenstein A.L., Bloor C.M., Leibow A.A. Diffuse alveolar damage--the role of oxygen, shock, and related factors. A review. Am. J. Pathol. 1976;85:209–228. [PMC free article] [PubMed] [Google Scholar]
- 35.Nuckton T.J., Alonso J.A., Kallet R.H., Daniel B.M., Pittet J.-F., Eisner M.D., Matthay M.A. Pulmonary dead-space fraction as a risk factor for death in the acute respiratory distress syndrome. N. Engl. J. Med. 2002;346:1281–1286. doi: 10.1056/NEJMoa012835. [DOI] [PubMed] [Google Scholar]
- 36.Wu C., Chen X., Cai Y., Xia J.a., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y., Song J., Wang S., Chao Y., Yang Z., Xu J., Zhou X., Chen D., Xiong W., Xu L., Zhou F., Jiang J., Bai C., Zheng J., Song Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lian J., Jin X., Hao S., Cai H., Zhang S., Zheng L., Jia H., Hu J., Gao J., Zhang Y., Zhang X., Yu G., Wang X., Gu J., Ye C., Jin C., Lu Y., Yu X., Yu X., Ren Y., Qiu Y., Li L., Sheng J., Yang Y. Analysis of epidemiological and clinical features in older patients with coronavirus disease 2019 (COVID-19) outside wuhan. Clin. Infect. Dis. 2020;71:740–747. doi: 10.1093/cid/ciaa242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Sun W., Li J., Chen L., Wang Y., Zhang L., Yu L. Cold Spring Harbor Laboratory; 2020. Clinical Features and Progression of Acute Respiratory Distress Syndrome in Coronavirus Disease 2019. [Google Scholar]
- 39.Khan A., Chatterjee A., Singh S. Cold Spring Harbor Laboratory; 2020. Comorbidities and Disparities in Outcomes of COVID-19 Among African American and White Patients. [Google Scholar]
- 40.Zhang P., Zhu L., Cai J., Lei F., Qin J.-J., Xie J., Liu Y.-M., Zhao Y.-C., Huang X., Lin L., Xia M., Chen M.-M., Cheng X., Zhang X., Guo D., Peng Y., Ji Y.-X., Chen J., She Z.-G., Wang Y., Xu Q., Tan R., Wang H., Lin J., Luo P., Fu S., Cai H., Ye P., Xiao B., Mao W., Liu L., Yan Y., Liu M., Chen M., Zhang X.-J., Wang X., Touyz Rhian M., Xia J., Zhang B.-H., Huang X., Yuan Y., Loomba R., Liu Peter P., Li H. Association of inpatient use of angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers with mortality among patients with hypertension hospitalized with COVID-19. Circ. Res. 2020;126:1671–1681. doi: 10.1161/CIRCRESAHA.120.317134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maniruzzaman M., Kumar N., Menhazul Abedin M., Shaykhul Islam M., Suri H.S., El-Baz A.S., Suri J.S. Comparative approaches for classification of diabetes mellitus data: machine learning paradigm. Comput. Methods Progr. Biomed. 2017;152:23–34. doi: 10.1016/j.cmpb.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 42.Dreher M., Kersten A., Bickenbach J., Balfanz P., Hartmann B., Cornelissen C., Daher A., Stöhr R., Kleines M., Lemmen S.W., Brokmann J.C., Müller T., Müller-Wieland D., Marx G., Marx N. The characteristics of 50 hospitalized COVID-19 patients with and without ARDS. Dtsch Arztebl Int. 2020;117:271–278. doi: 10.3238/arztebl.2020.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palaiodimos L., Kokkinidis D.G., Li W., Karamanis D., Ognibene J., Arora S., Southern W.N., Mantzoros C.S. Severe obesity, increasing age and male sex are independently associated with worse in-hospital outcomes, and higher in-hospital mortality, in a cohort of patients with COVID-19 in the Bronx, New York. Metabolism. 2020;108 doi: 10.1016/j.metabol.2020.154262. 154262-154262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu T., Cai S., Zheng Z., Cai X., Liu Y., Yin S., Peng J., Xu X. Association between clinical manifestations and prognosis in patients with COVID-19. Clin. Therapeut. 2020;42:964–972. doi: 10.1016/j.clinthera.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bandyopadhyay D., Akhtar T., Hajra A., Gupta M., Das A., Chakraborty S., Pal I., Patel N., Amgai B., Ghosh R.K., Fonarow G.C., Lavie C.J., Naidu S.S. COVID-19 pandemic: cardiovascular complications and future implications. Am. J. Cardiovasc. Drugs: Drugs, Devices, and other Interventions. 2020;20:311–324. doi: 10.1007/s40256-020-00420-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doyen D., Moceri P., Ducreux D., Dellamonica J. Myocarditis in a patient with COVID-19: a cause of raised troponin and ECG changes. Lancet. 2020;395:1516. doi: 10.1016/S0140-6736(20)30912-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madjid M., Safavi-Naeini P., Solomon S.D., Vardeny O. Potential effects of coronaviruses on the cardiovascular system: a review. JAMA Cardiol. 2020;5:831–840. doi: 10.1001/jamacardio.2020.1286. [DOI] [PubMed] [Google Scholar]
- 48.Suleyman G., Fadel R.A., Malette K.M., Hammond C., Abdulla H., Entz A., Demertzis Z., Hanna Z., Failla A., Dagher C., Chaudhry Z., Vahia A., Abreu Lanfranco O., Ramesh M., Zervos M.J., Alangaden G., Miller J., Brar I. Clinical characteristics and morbidity associated with coronavirus disease 2019 in a series of patients in metropolitan detroit. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyerowitz E.A., Kim A.Y., Ard K.L., Basgoz N., Chu J.T., Hurtado R.M., Lee C.K., He W., Minukas T., Nelson S., Ojikutu B.O., Robbins G., Sanchez S., Triant V.A., Zachary K., Gandhi R.T. Disproportionate burden of coronavirus disease 2019 among racial minorities and those in congregate settings among a large cohort of people with HIV. AIDS. 2020;34:1781–1787. doi: 10.1097/QAD.0000000000002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandran M., Chan Maung A., Mithal A., Parameswaran R. Vitamin D in COVID - 19: dousing the fire or averting the storm? – a perspective from the Asia-Pacific. Osteoporosis and Sarcopenia. 2020;6:97–105. doi: 10.1016/j.afos.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang T.S., Ding Y., Freund M.K., Johnson R., Schwarz T., Yabu J.M., Hazlett C., Chiang J.N., Wulf A., Geschwind D.H., Butte M.J., Pasaniuc B. medRxiv; 2020. Prior Diagnoses and Medications as Risk Factors for COVID-19 in a Los Angeles Health System. [Google Scholar]
- 52.Sanyaolu A., Okorie C., Marinkovic A., Patidar R., Younis K., Desai P., Hosein Z., Padda I., Mangat J., Altaf M. Comorbidity and its impact on patients with COVID-19. SN Compr Clin Med. 2020:1–8. doi: 10.1007/s42399-020-00363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takemoto M., Menezes M.O., Andreucci C.B., Knobel R., Sousa L., Katz L., Fonseca E.B., Nakamura-Pereira M., Magalhaes C.G., Diniz C., Melo A., Amorim M., C. Brazilian Group for Studies of, Pregnancy . BJOG; 2020. Clinical Characteristics and Risk Factors for Mortality in Obstetric Patients with Severe COVID-19 in Brazil: a Surveillance Database Analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gudipati S., Brar I., Murray S., McKinnon J.E., Yared N., Markowitz N.J. 2020. Descriptive Analysis of Patients Living with HIV Affected by COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bornstein S.R., Rubino F., Khunti K., Mingrone G., Hopkins D., Birkenfeld A.L., Boehm B., Amiel S., Holt R.I.G., Skyler J.S., DeVries J.H., Renard E., Eckel R.H., Zimmet P., Alberti K.G., Vidal J., Geloneze B., Chan J.C., Ji L., Ludwig B. Practical recommendations for the management of diabetes in patients with COVID-19. Lancet Diabet. Endocrinol. 2020;8:546–550. doi: 10.1016/S2213-8587(20)30152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guzik T.J., Mohiddin S.A., Dimarco A., Patel V., Savvatis K., Marelli-Berg F.M., Madhur M.S., Tomaszewski M., Maffia P., D'Acquisto F., Nicklin S.A., Marian A.J., Nosalski R., Murray E.C., Guzik B., Berry C., Touyz R.M., Kreutz R., Wang D.W., Bhella D., Sagliocco O., Crea F., Thomson E.C., McInnes I.B. COVID-19 and the cardiovascular system: implications for risk assessment, diagnosis, and treatment options. Cardiovasc. Res. 2020;116:1666–1687. doi: 10.1093/cvr/cvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bassendine M.F., Bridge S.H., McCaughan G.W., Gorrell M.D. COVID-19 and comorbidities: a role for dipeptidyl peptidase 4 (DPP4) in disease severity? J. Diabetes. 2020;12:649–658. doi: 10.1111/1753-0407.13052. [DOI] [PubMed] [Google Scholar]
- 58.Bode B., Garrett V., Messler J., McFarland R., Crowe J., Booth R., Klonoff D.C. Glycemic characteristics and clinical outcomes of COVID-19 patients hospitalized in the United States. J Diabet. Sci Technol. 2020;14:813–821. doi: 10.1177/1932296820924469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akram J., Azhar S., Shahzad M., Latif W., Khan K.S. Pakistan Randomized and Observational Trial to Evaluate Coronavirus Treatment (PROTECT) of Hydroxychloroquine, Oseltamivir and Azithromycin to treat newly diagnosed patients with COVID-19 infection who have no comorbidities like diabetes mellitus: a structured summary of a study protocol for a randomized controlled trial. Trials. 2020;21:702. doi: 10.1186/s13063-020-04616-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Q., Xie L., Zhang W., Zhao L., Wu H., Jiang J., Zou J., Liu J., Wu J., Chen Y., Wu J. Analysis of the clinical characteristics, drug treatments and prognoses of 136 patients with coronavirus disease 2019. J. Clin. Pharm. Therapeut. 2020;45:609–616. doi: 10.1111/jcpt.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Halaji M., Farahani A., Ranjbar R., Heiat M., Dehkordi F.J. vol. 28. 2020. pp. 6–17. (Emerging Coronaviruses: First SARS, Second MERS and Third SARS-CoV-2: Epidemiological Updates of COVID-19). [PubMed] [Google Scholar]
- 62.Grimaldi D., Aissaoui N., Blonz G., Carbutti G., Courcelle R., Gaudry S., Gaultier A., D'Hondt A., Higny J., Horlait G., Hraiech S., Lefebvre L., Lejeune F., Ly A., Piagnerelli M., Sauneuf B., Serck N., Soumagne T., Szychowiak P., Textoris J., Vandenbunder B., Vinsonneau C., Lascarrou J.B., group C.s. Characteristics and outcomes of acute respiratory distress syndrome related to COVID-19 in Belgian and French intensive care units according to antiviral strategies: the COVADIS multicentre observational study. Ann. Intensive Care. 2020;10:131. doi: 10.1186/s13613-020-00751-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zaim S., Chong J.H., Sankaranarayanan V., Harky A. COVID-19 and multiorgan response. Curr. Probl. Cardiol. 2020;45:100618. doi: 10.1016/j.cpcardiol.2020.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ponziani F.R., Del Zompo F., Nesci A., Santopaolo F., Ianiro G., Pompili M., Gasbarrini A., C.-g. Gemelli against Liver involvement is not associated with mortality: results from a large cohort of SARS-CoV-2 positive patients. Aliment. Pharmacol. Ther. 2020;52(6):1060–1068. doi: 10.1111/apt.15996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brandt J.S., Hill J., Reddy A., Schuster M., Patrick H.S., Rosen T., Sauer M.V., Boyle C., Ananth C.V. Epidemiology of coronavirus disease 2019 in pregnancy: risk factors and associations with adverse maternal and neonatal outcomes. Am. J. Obstet. Gynecol. 2020 doi: 10.1016/j.ajog.2020.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng L.L., Guan W.J., Duan C.Y., Zhang N.F., Lei C.L., Hu Y., Chen A.L., Li S.Y., Zhuo C., Deng X.L., Cheng F.J., Gao Y., Zhang J.H., Xie J.X., Peng H., Li Y.X., Wu X.X., Liu W., Peng H., Wang J., Xiao G.M., Chen P.Y., Wang C.Y., Yang Z.F., Zhao J.C., Zhong N.S. Effect of recombinant human granulocyte colony-stimulating factor for patients with coronavirus disease 2019 (COVID-19) and lymphopenia: a randomized clinical trial. JAMA Intern Med. 2021;181(1):71–78. doi: 10.1001/jamainternmed.2020.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cummings M.J., Baldwin M.R., Abrams D., Jacobson S.D., Meyer B.J., Balough E.M., Aaron J.G., Claassen J., Rabbani L.E., Hastie J., Hochman B.R., Salazar-Schicchi J., Yip N.H., Brodie D., O'Donnell M.R. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet. 2020;395:1763–1770. doi: 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakeshbandi M., Maini R., Daniel P., Rosengarten S., Parmar P., Wilson C., Kim J.M., Oommen A., Mecklenburg M., Salvani J., Joseph M.A., Breitman I. The impact of obesity on COVID-19 complications: a retrospective cohort study. Int. J. Obes. 2020;44:1832–1837. doi: 10.1038/s41366-020-0648-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao J., Li X., Gao Y., Huang W.J. Risk factors for the exacerbation of patients with 2019 Novel Coronavirus. A Meta Analysis. 2020;17:1744. doi: 10.7150/ijms.47052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H., Wang T., Guo W., Chen J., Ding C., Zhang X., Huang J., Han M., Li S., Luo X., Zhao J., Ning Q. 2020. Clinical Characteristics of 113 Deceased Patients with Coronavirus Disease 2019: Retrospective Study, Bmj. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qin C., Zhou L., Hu Z., Yang S., Zhang S., Chen M., Yu H., Tian D.S., Wang W. Clinical characteristics and outcomes of COVID-19 patients with a history of stroke in wuhan, China. Stroke. 2020;51:2219–2223. doi: 10.1161/STROKEAHA.120.030365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi S., Qin M., Shen B., Cai Y., Liu T., Yang F., Gong W., Liu X., Liang J., Zhao Q., Huang H., Yang B., Huang C. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in wuhan, China. JAMA Cardiol. 2020;5:802–810. doi: 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deng Y., Liu W., Liu K., Fang Y.Y., Shang J., Zhou L., Wang K., Leng F., Wei S., Chen L., Liu H.G. Clinical characteristics of fatal and recovered cases of coronavirus disease 2019 in Wuhan, China: a retrospective study. Chin Med J (Engl) 2020;133:1261–1267. doi: 10.1097/CM9.0000000000000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang F., Shi S., Zhu J., Shi J., Dai K., Chen X. Clinical characteristics and outcomes of cancer patients with COVID-19. J. Med. Virol. 2020;92(10):2067–2073. doi: 10.1002/jmv.25972. [DOI] [PubMed] [Google Scholar]
- 75.Del Sole F., Farcomeni A., Loffredo L., Carnevale R., Menichelli D., Vicario T., Pignatelli P., Pastori D. Features of severe COVID-19: a systematic review and meta-analysis. Eur. J. Clin. Invest. 2020;50 doi: 10.1111/eci.13378. e13378-e13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ciceri F., Castagna A., Rovere-Querini P., De Cobelli F., Ruggeri A., Galli L., Conte C., De Lorenzo R., Poli A., Ambrosio A., Signorelli C., Bossi E., Fazio M., Tresoldi C., Colombo S., Monti G., Fominskiy E., Franchini S., Spessot M., Martinenghi C., Carlucci M., Beretta L., Scandroglio A.M., Clementi M., Locatelli M., Tresoldi M., Scarpellini P., Martino G., Bosi E., Dagna L., Lazzarin A., Landoni G., Zangrillo A. Early predictors of clinical outcomes of COVID-19 outbreak in Milan, Italy. Clin. Immunol. 2020;217:108509. doi: 10.1016/j.clim.2020.108509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kutluhan M.A., Taş A., Şahin A., Ürkmez A., Topaktas R., Ataç Ö., Verit A.J. 2020. Assessment of Clinical Features and Renal Functions in Coronavirus Disease‐19: A Retrospective Analysis of 96 Patients. [DOI] [PubMed] [Google Scholar]
- 78.Derespina K.R., Kaushik S., Plichta A., Conway E.E., Jr., Bercow A., Choi J., Eisenberg R., Gillen J., Sen A.I., Hennigan C.M.J. 2020. Clinical Manifestations and Outcomes of Critically Ill Children and Adolescents with Coronavirus Disease 2019 in New York City. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blumfield E., Levin T.L.J.P.r. vol. 50. 2020. pp. 1369–1374. (COVID-19 in Pediatric Patients: a Case Series from the Bronx, NY). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jazieh A.-R., Alenazi T.H., Alhejazi A., Al Safi F., Al Olayan A.J.J.G.O. vol. 6. 2020. pp. 471–475. (Outcome of Oncology Patients Infected with Coronavirus). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Al-Wahaibi K., Al-Wahshi Y., Mohamed Elfadil O. Myocardial injury is associated with higher morbidity and mortality in patients with 2019 novel coronavirus disease (COVID-19) SN Compr Clin Med. 2020:1–7. doi: 10.1007/s42399-020-00569-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chand S., Kapoor S., Orsi D., Fazzari M.J., Tanner T.G., Umeh G.C., Islam M., Dicpinigaitis P.V. COVID-19-Associated critical illness-report of the first 300 patients admitted to intensive care units at a New York city medical center. J. Intensive Care Med. 2020;35:963–970. doi: 10.1177/0885066620946692. [DOI] [PubMed] [Google Scholar]
- 83.Lee Y.R., Kang M.K., Song J.E., Kim H.J., Kweon Y.O., Tak W.Y., Jang S.Y., Park J.G., Lee C., Hwang J.S., Jang B.K., Suh J.I., Chung W.J., Kim B.S., Park S.Y. Clinical outcomes of coronavirus disease 2019 in patients with pre-existing liver diseases: a multicenter study in South Korea. Clin. Mol. Hepatol. 2020;26:562–576. doi: 10.3350/cmh.2020.0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang L., He W., Yu X., Hu D., Bao M., Liu H., Zhou J., Jiang H. Coronavirus disease 2019 in elderly patients: characteristics and prognostic factors based on 4-week follow-up. J. Infect. 2020;80:639–645. doi: 10.1016/j.jinf.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du Y., Tu L., Zhu P., Mu M., Wang R., Yang P., Wang X., Hu C., Ping R., Hu P., Li T., Cao F., Chang C., Hu Q., Jin Y., Xu G. Clinical features of 85 fatal cases of COVID-19 from wuhan. A retrospective observational study. Am. J. Respir. Crit. Care Med. 2020;201:1372–1379. doi: 10.1164/rccm.202003-0543OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ziehr D.R., Alladina J., Petri C.R., Maley J.H., Moskowitz A., Medoff B.D., Hibbert K.A., Thompson B.T., Hardin C.C. Respiratory pathophysiology of mechanically ventilated patients with COVID-19: a cohort study. Am. J. Respir. Crit. Care Med. 2020;201:1560–1564. doi: 10.1164/rccm.202004-1163LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Garcia-Cruz E., Manzur-Sandoval D., Rascon-Sabido R., Gopar-Nieto R., Barajas-Campos R.L., Jordan-Rios A., Sierra-Lara Martinez D., Jimenez-Rodriguez G.M., Murillo-Ochoa A.L., Diaz-Mendez A., Lazcano-Diaz E., Araiza-Garaygordobil D., Cabello-Lopez A., Melano-Carranza E., Bucio-Reta E., Gonzalez-Ruiz F.J., Cota-Apodaca L.A., Santos-Martinez L.E., Fernandez-de la Reguera G., Ramos-Enriquez A., Rojas-Velasco G., Alvarez-Alvarez R.J., Baranda-Tovar F. Critical care ultrasonography during COVID-19 pandemic: the ORACLE protocol. Echocardiography. 2020;37(9):1353–1361. doi: 10.1111/echo.14837. [DOI] [PubMed] [Google Scholar]
- 88.Huang Y., Guo H., Zhou Y., Guo J., Wang T., Zhao X., Li H., Sun Y., Bian X., Fang C. The associations between fasting plasma glucose levels and mortality of COVID-19 in patients without diabetes. Diabetes Res. Clin. Pract. 2020;169:108448. doi: 10.1016/j.diabres.2020.108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arrieta J., Galwankar S., Lattanzio N., Ray D., Agrawal A. Studying the clinical data of COVID positive patients admitted to a tertiary care academic hospital. J. Emergencies, Trauma, Shock. 2020;13:131–134. doi: 10.4103/JETS.JETS_67_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oltean M., Søfteland J.M., Bagge J., Ekelund J., Felldin M., Schult A., Magnusson J., Friman V., Karason K.J.I.D. vol. 52. 2020. pp. 830–837. (Covid-19 in Kidney Transplant Recipients: a Systematic Review of the Case Series Available Three Months into the Pandemic). [DOI] [PubMed] [Google Scholar]
- 91.Marinaki S., Tsiakas S., Korogiannou M., Grigorakos K., Papalois V., Boletis I.J. A systematic review of COVID-19 infection in kidney transplant recipients: a universal effort to preserve patients'. Lives and Allografts. 2020;9:2986. doi: 10.3390/jcm9092986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rajpal A., Rahimi L., Ismail‐Beigi F.J.J.o.d. 2020. Factors Leading to High Morbidity and Mortality of COVID‐19 in Patients with Type 2 Diabetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang I., Lim M.A., Pranata R. Diabetes mellitus is associated with increased mortality and severity of disease in COVID-19 pneumonia - a systematic review, meta-analysis, and meta-regression. Diabet Metab Syndr. 2020;14:395–403. doi: 10.1016/j.dsx.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Salerno M., Sessa F., Piscopo A., Montana A., Torrisi M., Patanè F., Murabito P., Volti G.L., Pomara C. No autopsies on COVID-19 deaths: a missed opportunity and the lockdown of science. J. Clin. Med. 2020;9:1472. doi: 10.3390/jcm9051472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arrieta J., Galwankar S., Lattanzio N., Ray D., Agrawal A. Common clinical characteristics and complications determining the outcome in a COVID-Positive predominantly geriatric population. J. Emergencies, Trauma, Shock. 2020;13 doi: 10.4103/JETS.JETS_67_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tomazini B.M., Maia I.S., Cavalcanti A.B., Berwanger O., Rosa R.G., Veiga V.C., Avezum A., Lopes R.D., Bueno F.R., Silva M.V.A.O., Baldassare F.P., Costa E.L.V., Moura R.A.B., Honorato M.O., Costa A.N., Damiani L.P., Lisboa T., Kawano-Dourado L., Zampieri F.G., Olivato G.B., Righy C., Amendola C.P., Roepke R.M.L., Freitas D.H.M., Forte D.N., Freitas F.G.R., Fernandes C.C.F., Melro L.M.G., Junior G.F.S., Morais D.C., Zung S., Machado F.R., Azevedo L.C.P., Investigators C.C.-B.I. Effect of dexamethasone on days alive and ventilator-free in patients with moderate or severe acute respiratory distress syndrome and COVID-19: the CoDEX randomized clinical trial. J. Am. Med. Assoc. 2020;324:1307–1316. doi: 10.1001/jama.2020.17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nasir N., Farooqi J., Mahmood S.F., Jabeen K. COVID-19-associated pulmonary aspergillosis (CAPA) in patients admitted with severe COVID-19 pneumonia: an observational study from Pakistan. Mycoses. 2020;63:766–770. doi: 10.1111/myc.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Antoun L., Taweel N.E., Ahmed I., Patni S., Honest H. Maternal COVID-19 infection, clinical characteristics, pregnancy, and neonatal outcome: a prospective cohort study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020;252:559–562. doi: 10.1016/j.ejogrb.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Khan M., Khan H., Khan S., Nawaz M. Epidemiological and clinical characteristics of coronavirus disease (COVID-19) cases at a screening clinic during the early outbreak period: a single-centre study. J. Med. Microbiol. 2020;69:1114–1123. doi: 10.1099/jmm.0.001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xie J., Zu Y., Alkhatib A., Pham T.T., Gill F., Jang A., Radosta S., Chaaya G., Myers L., Zifodya J.S., Bojanowski C.M., Marrouche N.F., Mauvais-Jarvis F., Denson J.L. Metabolic syndrome and COVID-19 mortality among adult black patients in new orleans. Diabetes Care. 2020;44(1):188–193. doi: 10.2337/dc20-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li T., Lu L., Zhang W., Tao Y., Wang L., Bao J., Liu B., Duan J. Clinical characteristics of 312 hospitalized older patients with COVID-19 in Wuhan, China. Arch. Gerontol. Geriatr. 2020;91:104185. doi: 10.1016/j.archger.2020.104185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saba L., Agarwal M., Sanagala S., Gupta S., Sinha G., Johri A., Khanna N., Mavrogeni S., Laird J., Pareek G.J.E.L. 2020. Brain MRI-Based Wilson Disease Tissue Classification: an Optimised Deep Transfer Learning Approach. [Google Scholar]
- 103.Tandel G.S., Biswas M., Kakde O.G., Tiwari A., Suri H.S., Turk M., Laird J.R., Asare C.K., Ankrah A.A., Khanna N.J.C. A review on a deep learning perspective in brain cancer classification. 2019;11:111. doi: 10.3390/cancers11010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saba L., Tiwari A., Biswas M., Gupta S.K., Godia-Cuadrado E., Chaturvedi A., Turk M., Suri H.S., Orru S., Sanches J.M. Wilson's disease: a new perspective review on its genetics. Diagnosis Treatment. 2019;11:166–185. doi: 10.2741/E854. [DOI] [PubMed] [Google Scholar]
- 105.Acharya U.R., Mookiah M.R.K., Sree S.V., Afonso D., Sanches J., Shafique S., Nicolaides A., Pedro L.M., e Fernandes J.F., Suri J.S.J.M. b. engineering, computing, Atherosclerotic plaque tissue characterization in 2D ultrasound longitudinal carotid scans for automated classification. Paradigm Stroke Risk Assess. 2013;51:513–523. doi: 10.1007/s11517-012-1019-0. [DOI] [PubMed] [Google Scholar]
- 106.Sharma A.M., Gupta A., Kumar P.K., Rajan J., Saba L., Nobutaka I., Laird J.R., Nicolades A., Suri J.S. vol. 17. 2015. p. 55. (A Review on Carotid Ultrasound Atherosclerotic Tissue Characterization and Stroke Risk Stratification in Machine Learning Framework). [DOI] [PubMed] [Google Scholar]
- 107.Biswas M., Kuppili V., Saba L., Edla D.R., Suri H.S., Sharma A., Cuadrado-Godia E., Laird J.R., Nicolaides A., Suri J.S.J.M. b. engineering, computing, Deep learning fully convolution network for lumen characterization in diabetic patients using carotid ultrasound: a tool for stroke risk. 2019;57:543–564. doi: 10.1007/s11517-018-1897-x. [DOI] [PubMed] [Google Scholar]
- 108.Saba L., Jain P.K., Suri H.S., Ikeda N., Araki T., Singh B.K., Nicolaides A., Shafique S., Gupta A., Laird J.R. vol. 41. 2017. p. 98. (Plaque Tissue Morphology-Based Stroke Risk Stratification Using Carotid Ultrasound: a Polling-Based PCA Learning Paradigm). [DOI] [PubMed] [Google Scholar]
- 109.Acharya U., Sree S.V., Mookiah M., Saba L., Gao H., Mallarini G., Suri J.J. Part H: journal of Engineering in Medicine, Computed tomography carotid wall plaque characterization using a combination of discrete wavelet transform and texture features. A pilot Study. 2013;227:643–654. doi: 10.1177/0954411913480622. [DOI] [PubMed] [Google Scholar]
- 110.Acharya U.R., Molinari F., Saba L., Nicolaides A., Shafique S., Suri J.S. 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, IEEE. 2012. Carotid ultrasound symptomatology using atherosclerotic plaque characterization: a class of Atheromatic systems; pp. 3199–3202. [DOI] [PubMed] [Google Scholar]
- 111.Prasad H., Martis R.J., Acharya U.R., Min L.C., Suri J.S. 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), IEEE. 2013. Application of higher order spectra for accurate delineation of atrial arrhythmia; pp. 57–60. [DOI] [PubMed] [Google Scholar]
- 112.Saba L., Dey N., Ashour A.S., Samanta S., Nath S.S., Chakraborty S., Sanches J., Kumar D., Marinho R., Suri J.S.J.C.m., biomedicine p.i. vol. 130. 2016. pp. 118–134. (Automated Stratification of Liver Disease in Ultrasound: an Online Accurate Feature Classification Paradigm). [DOI] [PubMed] [Google Scholar]
- 113.Acharya U.R., Sree S.V., Ribeiro R., Krishnamurthi G., Marinho R.T., Sanches J., Suri J.S.J.M.p. vol. 39. 2012. pp. 4255–4264. (Data Mining Framework for Fatty Liver Disease Classification in Ultrasound: a Hybrid Feature Extraction Paradigm). [DOI] [PubMed] [Google Scholar]
- 114.Biswas M., Kuppili V., Edla D.R., Suri H.S., Saba L., Marinhoe R.T., Sanches J.M., Suri J.S.J.C.m. vol. 155. 2018. pp. 165–177. (p.i. Biomedicine, Symtosis: A Liver Ultrasound Tissue Characterization and Risk Stratification in Optimized Deep Learning Paradigm). [DOI] [PubMed] [Google Scholar]
- 115.Boi A., Jamthikar A.D., Saba L., Gupta D., Sharma A., Loi B., Laird J.R., Khanna N.N., Suri J.S. vol. 20. 2018. p. 33. (A Survey on Coronary Atherosclerotic Plaque Tissue Characterization in Intravascular Optical Coherence Tomography). [DOI] [PubMed] [Google Scholar]
- 116.Acharya U.R., Faust O., Kadri N.A., Suri J.S., Yu W.J.C.i.b. vol. 43. 2013. pp. 1523–1529. (Medicine, Automated Identification of Normal and Diabetes Heart Rate Signals Using Nonlinear Measures). [DOI] [PubMed] [Google Scholar]
- 117.Pareek G., Acharya U.R., Sree S.V., Swapna G., Yantri R., Martis R.J., Saba L., Krishnamurthi G., Mallarini G., El-Baz A.J. vol. 12. 2013. t; pp. 545–557. (Reatment, Prostate Tissue Characterization/classification in 144 Patient Population Using Wavelet and Higher Order Spectra Features from Transrectal Ultrasound Images). [DOI] [PubMed] [Google Scholar]
- 118.Acharya U.R., Molinari F., Sree S.V., Swapna G., Saba L., Guerriero S., Suri J.S. vol. 14. 2015. pp. 251–261. (Treatment, Ovarian Tissue Characterization in Ultrasound: a Review). [DOI] [PubMed] [Google Scholar]
- 119.Acharya U.R., Sree S.V., Kulshreshtha S., Molinari F., Koh J.E.W., Saba L., Suri J.S. vol. 13. 2014. pp. 529–539. (Treatment, GyneScan: an Improved Online Paradigm for Screening of Ovarian Cancer via Tissue Characterization). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Viswanathan V., Jamthikar A.D., Gupta D., Shanu N., Puvvula A., Khanna N.N., Saba L., Omerzum T., Viskovic K., Mavrogeni S.J.F.i.B. vol. 25. 2020. pp. 1132–1171. (Low-cost Preventive Screening Using Carotid Ultrasound in Patients with Diabetes). [DOI] [PubMed] [Google Scholar]
- 121.Acharya U.R., Swapna G., Sree S.V., Molinari F., Gupta S., Bardales R.H., Witkowska A., Suri J.S. vol. 13. 2014. pp. 289–301. (Treatment, A Review on Ultrasound-Based Thyroid Cancer Tissue Characterization and Automated Classification). [DOI] [PubMed] [Google Scholar]
- 122.Shrivastava V.K., Londhe N.D., Sonawane R.S., Suri J.S.J.C.m. vol. 126. 2016. pp. 98–109. (p.i. Biomedicine, Computer-Aided Diagnosis of Psoriasis Skin Images with HOS, Texture and Color Features: a First Comparative Study of its Kind). [DOI] [PubMed] [Google Scholar]
- 123.Shrivastava V.K., Londhe N.D., Sonawane R.S., Suri J.S.J.C.m. vol. 150. 2017. pp. 9–22. (p.i. Biomedicine, A Novel and Robust Bayesian Approach for Segmentation of Psoriasis Lesions and its Risk Stratification). [DOI] [PubMed] [Google Scholar]
- 124.Acharya U.R., Joseph K.P., Kannathal N., Lim C.M., Suri J.S.J.M. vol. 44. 2006. pp. 1031–1051. (B. Engineering, Computing, Heart Rate Variability: a Review). [DOI] [PubMed] [Google Scholar]
- 125.Corrias G., Cocco D., Suri J.S., Meloni L., Cademartiri F., Saba L.J.C.D. Therapy, Heart applications of 4D flow. 2020;10:1140. doi: 10.21037/cdt.2020.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Acharya U.R., Sree S.V., Krishnan M.M.R., Krishnananda N., Ranjan S., Umesh P., Suri J.S.J.C.m. vol. 112. 2013. pp. 624–632. (p.i. Biomedicine, Automated Classification of Patients with Coronary Artery Disease Using Grayscale Features from Left Ventricle Echocardiographic Images). [DOI] [PubMed] [Google Scholar]
- 127.Maniruzzaman M., Rahman M.J., Ahammed B., Abedin M.M., Suri H.S., Biswas M., El-Baz A., Bangeas P., Tsoulfas G., Suri J.S.J.C.m., biomedicine p.i. vol. 176. 2019. pp. 173–193. (Statistical Characterization and Classification of Colon Microarray Gene Expression Data Using Multiple Machine Learning Paradigms). [DOI] [PubMed] [Google Scholar]
- 128.Khanna N.N., Jamthikar A.D., Gupta D., Piga M., Saba L., Carcassi C., Giannopoulos A.A., Nicolaides A., Laird J.R., Suri H.S.J. vol. 21. 2019. p. 7. (Rheumatoid Arthritis: Atherosclerosis Imaging and Cardiovascular Risk Assessment Using Machine and Deep Learning–Based Tissue Characterization). [DOI] [PubMed] [Google Scholar]
- 129.Jiang J., Hu Y.C., Tyagi N., Zhang P., Rimner A., Deasy J.O., Veeraraghavan H. Cross-modality (CT-MRI) prior augmented deep learning for robust lung tumor segmentation from small MR datasets. Med. Phys. 2019;46:4392–4404. doi: 10.1002/mp.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Aresta G., Jacobs C., Araujo T., Cunha A., Ramos I., van Ginneken B., Campilho A. iW-Net: an automatic and minimalistic interactive lung nodule segmentation deep network. Sci. Rep. 2019;9:11591. doi: 10.1038/s41598-019-48004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Anthimopoulos M., Christodoulidis S., Ebner L., Geiser T., Christe A., Mougiakakou S. Semantic segmentation of pathological lung tissue with dilated fully convolutional networks. IEEE J Biomed Health Inform. 2019;23:714–722. doi: 10.1109/JBHI.2018.2818620. [DOI] [PubMed] [Google Scholar]
- 132.Weikert T., Akinci D'Antonoli T., Bremerich J., Stieltjes B., Sommer G., Sauter A.W. Evaluation of an AI-powered lung nodule algorithm for detection and 3D segmentation of primary lung tumors. Contrast Media Mol. Imaging. 2019:1545747. doi: 10.1155/2019/1545747. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu H., Cao H., Song E., Ma G., Xu X., Jin R., Jin Y., Hung C.C. A cascaded dual-pathway residual network for lung nodule segmentation in CT images. Phys. Med. 2019;63:112–121. doi: 10.1016/j.ejmp.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 134.Wong Yuzhen N., Barrett S. A review of automatic lung tumour segmentation in the era of 4DCT. Rep. Practical Oncol. Radiother. 2019;24:208–220. doi: 10.1016/j.rpor.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Park B., Park H., Lee S.M., Seo J.B., Kim N. Lung segmentation on HRCT and volumetric CT for diffuse interstitial lung disease using deep convolutional neural networks. J. Digit. Imag. 2019;32:1019–1026. doi: 10.1007/s10278-019-00254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nasrullah N., Sang J., Alam M.S., Mateen M., Cai B., Hu H. Automated lung nodule detection and classification using deep learning combined with multiple strategies. Sensors. 2019;19 doi: 10.3390/s19173722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu M., Qi S., Yue Y., Teng Y., Xu L., Yao Y., Qian W. Segmentation of lung parenchyma in CT images using CNN trained with the clustering algorithm generated dataset. Biomed. Eng. Online. 2019;18:2. doi: 10.1186/s12938-018-0619-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Baek S., He Y., Allen B.G., Buatti J.M., Smith B.J., Tong L., Sun Z., Wu J., Diehn M., Loo B.W., Plichta K.A., Seyedin S.N., Gannon M., Cabel K.R., Kim Y., Wu X. Deep segmentation networks predict survival of non-small cell lung cancer. Sci. Rep. 2019;9:17286. doi: 10.1038/s41598-019-53461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pang T., Guo S., Zhang X., Zhao L. Automatic lung segmentation based on texture and deep features of HRCT images with interstitial lung disease. BioMed Res. Int. 2019:2045432. doi: 10.1155/2019/2045432. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chen G., Xiang D., Zhang B., Tian H., Yang X., Shi F., Zhu W., Tian B., Chen X. Automatic pathological lung segmentation in low-dose CT image using eigenspace sparse shape composition. IEEE Trans. Med. Imag. 2019;38:1736–1749. doi: 10.1109/TMI.2018.2890510. [DOI] [PubMed] [Google Scholar]
- 141.Senthil Kumar K., Venkatalakshmi K., Karthikeyan K. Lung cancer detection using image segmentation by means of various evolutionary algorithms. Comput Math Methods Med. 2019:4909846. doi: 10.1155/2019/4909846. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu C., Zhao R., Pang M. A fully automatic segmentation algorithm for CT lung images based on random forest. Med. Phys. 2020;47:518–529. doi: 10.1002/mp.13939. [DOI] [PubMed] [Google Scholar]
- 143.Geng L., Zhang S., Tong J., Xiao Z. Lung segmentation method with dilated convolution based on VGG-16 network. Comput Assist Surg (Abingdon) 2019;24:27–33. doi: 10.1080/24699322.2019.1649071. [DOI] [PubMed] [Google Scholar]
- 144.Sousa A.M., Martins S.B., Falcao A.X., Reis F., Bagatin E., Irion K. ALTIS: a fast and automatic lung and trachea CT-image segmentation method. Med. Phys. 2019;46:4970–4982. doi: 10.1002/mp.13773. [DOI] [PubMed] [Google Scholar]
- 145.Souza J.C., Bandeira Diniz J.O., Ferreira J.L., Franca da Silva G.L., Correa Silva A., de Paiva A.C. An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Comput. Methods Progr. Biomed. 2019;177:285–296. doi: 10.1016/j.cmpb.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 146.Noor N.M., Than J.C.M., Rijal O.M., Kassim R.M., Yunus A., Zeki A.A., Anzidei M., Saba L., Suri J.S. Automatic lung segmentation using control feedback system: morphology and texture paradigm. J. Med. Syst. 2015;39 doi: 10.1007/s10916-015-0214-6. [DOI] [PubMed] [Google Scholar]
- 147.Ni Q., Sun Z.Y., Qi L., Chen W., Yang Y., Wang L., Zhang X., Yang L., Fang Y., Xing Z., Zhou Z., Yu Y., Lu G.M., Zhang L.J. A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest CT images. Eur. Radiol. 2020;30:6517–6527. doi: 10.1007/s00330-020-07044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shi Y.J.a.p.a. 2020. Lung Infection Quantification of Covid-19 in Ct Images with Deep Learning. [Google Scholar]
- 149.Hwang E.J., Kim H., Yoon S.H., Goo J.M., Park C.M. Implementation of a deep learning-based computer-aided detection system for the interpretation of chest radiographs in patients suspected for COVID-19. Korean J. Radiol. 2020;21:1150–1160. doi: 10.3348/kjr.2020.0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Roy S., Menapace W., Oei S., Luijten B., Fini E., Saltori C., Huijben I., Chennakeshava N., Mento F., Sentelli A., Peschiera E., Trevisan R., Maschietto G., Torri E., Inchingolo R., Smargiassi A., Soldati G., Rota P., Passerini A., van Sloun R.J.G., Ricci E., Demi L. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Trans. Med. Imag. 2020;39:2676–2687. doi: 10.1109/TMI.2020.2994459. [DOI] [PubMed] [Google Scholar]
- 151.Signoroni A., Savardi M., Benini S., Adami N., Leonardi R., Gibellini P., Vaccher F., Ravanelli M., Borghesi A., Maroldi R.J.a.p.a. 2020. End-to-end Learning for Semiquantitative Rating of COVID-19 Severity on Chest X-Rays. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li Z., Zhong Z., Li Y., Zhang T., Gao L., Jin D., Sun Y., Ye X., Yu L., Hu Z., Xiao J., Huang L., Tang Y. From community-acquired pneumonia to COVID-19: a deep learning-based method for quantitative analysis of COVID-19 on thick-section CT scans. Eur. Radiol. 2020;30:6828–6837. doi: 10.1007/s00330-020-07042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chaganti S., Grenier P., Balachandran A., Chabin G., Cohen S., Flohr T., Georgescu B., Grbic S., Liu S., Mellot F., Murray N., Nicolaou S., Parker W., Re T., Sanelli P., Sauter A.W., Xu Z., Yoo Y., Ziebandt V., Comaniciu D. Automated quantification of CT patterns associated with COVID-19 from chest CT. Radiology: Artif. Intell. 2020;2 doi: 10.1148/ryai.2020200048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296:E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang S., Jiang L., Cao Z., Wang L., Cao J., Feng R., Zhang Z., Xue X., Shi Y., Shan F. Deep learning for detecting corona virus disease 2019 (COVID-19) on high-resolution computed tomography: a pilot study. Ann. Transl. Med. 2020;8 doi: 10.21037/atm.2020.03.132. 450-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hu S., Gao Y., Niu Z., Jiang Y., Li L., Xiao X., Wang M., Fang E.F., Menpes-Smith W., Xia J., Ye H., Yang G. Weakly supervised deep learning for COVID-19 infection detection and classification from CT images. IEEE Access. 2020;8:118869–118883. [Google Scholar]
- 157.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K., Ye L., Gao M., Zhou Z., Li L., Wang J., Yang Z., Cai H., Xu J., Yang L., Cai W., Xu W., Wu S., Zhang W., Jiang S., Zheng L., Zhang X., Wang L., Lu L., Li J., Yin H., Wang W., Li O., Zhang C., Liang L., Wu T., Deng R., Wei K., Zhou Y., Chen T., Lau J.Y.-N., Fok M., He J., Lin T., Li W., Wang G. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181:1423–1433. doi: 10.1016/j.cell.2020.04.045. e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Carrer L., Donini E., Marinelli D., Zanetti M., Mento F., Torri E., Smargiassi A., Inchingolo R., Soldati G., Demi L., Bovolo F., Bruzzone L. Automatic pleural line extraction and COVID-19 scoring from lung ultrasound data. IEEE Trans. Ultrason. Ferroelectrics Freq. Contr. 2020;67:2207–2217. doi: 10.1109/TUFFC.2020.3005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tang Z., Zhao W., Xie X., Zhong Z., Shi F., Liu J., Shen D.J.a.p.a. 2020. Severity Assessment of Coronavirus Disease 2019 (COVID-19) Using Quantitative Features from Chest CT Images. [Google Scholar]
- 160.Rajaraman S., Siegelman J., Alderson P.O., Folio L.S., Folio L.R., Antani S.K. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access. 2020;8:115041–115050. doi: 10.1109/ACCESS.2020.3003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Born J., Brändle G., Cossio M., Disdier M., Goulet J., Roulin J., Wiedemann N.J.a.p.a. POCUS; 2020. POCOVID-net: Automatic Detection of COVID-19 from a New Lung Ultrasound Imaging Dataset. [Google Scholar]
- 162.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2020:1–8. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 163.Tsiknakis N., Trivizakis E., Vassalou E.E., Papadakis G.Z., Spandidos D.A., Tsatsakis A., Sánchez-García J., López-González R., Papanikolaou N., Karantanas A.H., Marias K. Interpretable artificial intelligence framework for COVID-19 screening on chest X-rays. Exp Ther Med. 2020;20:727–735. doi: 10.3892/etm.2020.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Maghdid H.S., Asaad A.T., Ghafoor K.Z., Sadiq A.S., Khan M.K. 2020. Diagnosing COVID-19 Pneumonia from X-Ray and CT Images Using Deep Learning and Transfer Learning Algorithms. [Google Scholar]
- 165.Shrivastava V.K., Londhe N.D., Sonawane R.S., Suri J.S. Computer-aided diagnosis of psoriasis skin images with HOS, texture and color features: a first comparative study of its kind. Comput. Methods Progr. Biomed. 2016;126:98–109. doi: 10.1016/j.cmpb.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 166.Chen A., Karwoski R.A., Gierada D.S., Bartholmai B.J., Koo C.W. Quantitative CT analysis of diffuse lung disease. Radiographics. 2020;40:28–43. doi: 10.1148/rg.2020190099. [DOI] [PubMed] [Google Scholar]
- 167.Than J.C.M., Saba L., Noor N.M., Rijal O.M., Kassim R.M., Yunus A., Suri H.S., Porcu M., Suri J.S. Lung disease stratification using amalgamation of Riesz and Gabor transforms in machine learning framework. Comput. Biol. Med. 2017;89:197–211. doi: 10.1016/j.compbiomed.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 168.Hattori A., Takamochi K., Oh S., Suzuki K. New revisions and current issues in the eighth edition of the TNM classification for non-small cell lung cancer. Jpn. J. Clin. Oncol. 2019;49:3–11. doi: 10.1093/jjco/hyy142. [DOI] [PubMed] [Google Scholar]
- 169.Saba T. Automated lung nodule detection and classification based on multiple classifiers voting. Microsc. Res. Tech. 2019;82:1601–1609. doi: 10.1002/jemt.23326. [DOI] [PubMed] [Google Scholar]
- 170.Zhang G., Yang Z., Gong L., Jiang S., Wang L. Classification of benign and malignant lung nodules from CT images based on hybrid features. Phys. Med. Biol. 2019;64:125011. doi: 10.1088/1361-6560/ab2544. [DOI] [PubMed] [Google Scholar]
- 171.Singh D., Kumar V., Vaishali, Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution-based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1379–1389. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mahmud T., Rahman M.A., Fattah S.A. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020;122 doi: 10.1016/j.compbiomed.2020.103869. 103869-103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Das D., Santosh K.C., Pal U. 2020. Truncated Inception Net: COVID-19 Outbreak Screening Using Chest X-Rays, Research Square. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. 103792-103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Progr. Biomed. 2020;196:105608. doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Suri J.S., Singh S., Reden L.J.P.A. vol. 5. 2002. pp. 77–98. (Applications, Fusion of Region and Boundary/surface-Based Computer Vision and Pattern Recognition Techniques for 2-D and 3-D MR Cerebral Cortical Segmentation (Part-II): A State-Of-The-Art Review). [Google Scholar]
- 177.Suri J.S., Singh S., Reden L.J.P.A. Applications, Computer vision and pattern recognition techniques for 2-D and 3-D MR cerebral cortical segmentation (Part I): a state-of-the-art review. 2002;5:46–76. [Google Scholar]
- 178.Suri J.S., Liu K., Reden L., Laxminarayan S.J., Society B. A review on MR vascular image processing: skeleton versus nonskeleton approaches: part II. 2002;6:338. doi: 10.1109/titb.2002.804136. [DOI] [PubMed] [Google Scholar]
- 179.https://xmedcon.sourceforge.io/
- 180.Sahu S.P., Kamble B., Doriya R. Springer Singapore; 2020. 3D Lung Segmentation Using Thresholding and Active Contour Method, Advances in Intelligent Systems and Computing; pp. 369–380. [Google Scholar]
- 181.Taylor M.E. Springer Berlin Heidelberg; 2009. Transfer between Different Reinforcement Learning Methods, Transfer in Reinforcement Learning Domains; pp. 139–179. [Google Scholar]
- 182.Acharya U.R., Swapna G., Sree S.V., Molinari F., Gupta S., Bardales R.H., Witkowska A., Suri J.S. A review on ultrasound-based thyroid cancer tissue characterization and automated classification. Technol. Canc. Res. Treat. 2014;13:289–301. doi: 10.7785/tcrt.2012.500381. [DOI] [PubMed] [Google Scholar]
- 183.Shih A.R., Uruga H., Bozkurtlar E., Chung J.H., Hariri L.P., Minami Y., Wang H., Yoshizawa A., Muzikansky A., Moreira A.L., Mino-Kenudson M. Problems in the reproducibility of classification of small lung adenocarcinoma: an international interobserver study. Histopathology. 2019;75:649–659. doi: 10.1111/his.13922. [DOI] [PubMed] [Google Scholar]
- 184.LeNail A. Nn-SVG: Publication-ready neural network architecture schematics. J Open Source Software. 2019;4:747. [Google Scholar]
- 185.Iqbal H. 2018. HarisIqbal88/PlotNeuralNet v1.0.0. [Google Scholar]
- 186.Ye F., Pu J., Wang J., Li Y., Zha H. 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE; 2017. Glioma grading based on 3D multimodal convolutional neural network and privileged learning. [Google Scholar]
- 187.Wang S., Zha Y., Li W., Wu Q., Li X., Niu M., Wang M., Qiu X., Li H., Yu H., Gong W., Bai Y., Li L., Zhu Y., Wang L., Tian J. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yoo S.H., Geng H., Chiu T.L., Yu S.K., Cho D.C., Heo J., Choi M.S., Choi I.H., Cung Van C., Nhung N.V., Min B.J., Lee H. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Front. Med. 2020;7:427. doi: 10.3389/fmed.2020.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhu J., Shen B., Abbasi A., Hoshmand-Kochi M., Li H., Duong T.Q. Deep transfer learning artificial intelligence accurately stages COVID-19 lung disease severity on portable chest radiographs. PloS One. 2020;15 doi: 10.1371/journal.pone.0236621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Guidance Statement by the American College of Radiology.
- 191.Yasar H., Ceylan M. A novel comparative study for detection of Covid-19 on CT lung images using texture analysis, machine learning, and deep learning methods. Multimed. Tool. Appl. 2020:1–25. doi: 10.1007/s11042-020-09894-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Setti L., Kirienko M., Dalto S.C., Bonacina M., Bombardieri E. FDG-PET/CT findings highly suspicious for COVID-19 in an Italian case series of asymptomatic patients. Eur. J. Nucl. Med. Mol. Imag. 2020;47:1649–1656. doi: 10.1007/s00259-020-04819-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Alonso Sanchez J., García Prieto J., Galiana Morón A., Pilkington-Woll J.P. PET/CT of COVID-19 as an organizing pneumonia. Clin. Nucl. Med. 2020;45:642–643. doi: 10.1097/RLU.0000000000003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Castanheira J., Mascarenhas Gaivão A., Mairos Teixeira S., Pereira P.J., Costa D.C. Asymptomatic COVID-19 positive patient suspected on FDG-PET/CT. Nucl. Med. Commun. 2020;41:598–599. doi: 10.1097/MNM.0000000000001221. [DOI] [PubMed] [Google Scholar]
- 195.Cohen J.P., Dao L., Roth K., Morrison P., Bengio Y., Abbasi A.F., Shen B., Mahsa H.K., Ghassemi M., Li H., Duong T. Predicting COVID-19 pneumonia severity on chest X-ray with deep learning. Cureus. 2020 doi: 10.7759/cureus.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Galougahi M.K., Ghorbani J., Bakhshayeshkaram M., Naeini A.S., Haseli S.J.A.R. 2020. Olfactory Bulb Magnetic Resonance Imaging in SARS-CoV-2-Induced Anosmia: the First Report. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gunraj H., Wang L., Wong A.J.a.p.a. 2020. Covidnet-ct: A Tailored Deep Convolutional Neural Network Design for Detection of Covid-19 Cases from Chest Ct Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ismael A.M., Sengur A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021;164:114054. doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kandemirli S.G., Dogan L., Sarikaya Z.T., Kara S., Akinci C., Kaya D., Kaya Y., Yildirim D., Tuzuner F., Yildirim M.S., Ozluk E., Gucyetmez B., Karaarslan E., Koyluoglu I., Demirel Kaya H.S., Mammadov O., Kisa Ozdemir I., Afsar N., Citci Yalcinkaya B., Rasimoglu S., Guduk D.E., Kedir Jima A., Ilksoz A., Ersoz V., Yonca Eren M., Celtik N., Arslan S., Korkmazer B., Dincer S.S., Gulek E., Dikmen I., Yazici M., Unsal S., Ljama T., Demirel I., Ayyildiz A., Kesimci I., Bolsoy Deveci S., Tutuncu M., Kizilkilic O., Telci L., Zengin R., Dincer A., Akinci I.O., Kocer N. Brain MRI findings in patients in the intensive care unit with COVID-19 infection. Radiology. 2020;297:E232–e235. doi: 10.1148/radiol.2020201697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kay F., Abbara S. Radiological Society of North America; 2020. The Many Faces of COVID-19: Spectrum of Imaging Manifestations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., van der Laak J.A.W.M., van Ginneken B., Sánchez C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 202.Minaee S., Kafieh R., Sonka M., Yazdani S., Jamalipour Soufi G. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Shi F., Xia L., Shan F., Wu D., Wei Y., Yuan H., Jiang H., Gao Y., Sui H., Shen D.J.a.p.a. 2020. Large-scale Screening of Covid-19 from Community Acquired Pneumonia Using Infection Size-Aware Classification. [DOI] [PubMed] [Google Scholar]
- 204.Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Singh V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos, Solit. Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110190. 110190-110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ouchicha C., Ammor O., Meknassi M. CVDNet: a novel deep learning architecture for detection of coronavirus (Covid-19) from chest x-ray images. Chaos, Solit. Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110245. 110245-110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Farooq M., Hafeez A.J.a.p.a. 2020. Covid-resnet: A Deep Learning Framework for Screening of Covid19 from Radiographs. [Google Scholar]
- 207.Cozzi D., Albanesi M., Cavigli E., Moroni C., Bindi A., Luvarà S., Lucarini S., Busoni S., Mazzoni L.N., Miele V. Chest X-ray in new Coronavirus Disease 2019 (COVID-19) infection: findings and correlation with clinical outcome. Radiol. Med. 2020;125:730–737. doi: 10.1007/s11547-020-01232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pereira R.M., Bertolini D., Teixeira L.O., Silla C.N., Jr., Costa Y.M.G. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput. Methods Progr. Biomed. 2020;194 doi: 10.1016/j.cmpb.2020.105532. 105532-105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103795. 103795-103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. Cold Spring Harbor Laboratory; 2020. Deep Learning-Based Detection for COVID-19 from Chest CT Using Weak Label. [Google Scholar]
- 211.Yang X., He X., Zhao J., Zhang Y., Zhang S., Xie P.J.A.e.-p. 2020. Covid-CT-dataset: A CT Scan Dataset about Covid-19. arXiv: 2003.13865. [Google Scholar]
- 212.Gozes O., Frid-Adar M., Sagie N., Kabakovitch A., Amran D., Amer R., Greenspan H. Springer International Publishing; 2020. A Weakly Supervised Deep Learning Framework for COVID-19 CT Detection and Analysis, Thoracic Image Analysis; pp. 84–93. [Google Scholar]
- 213.Liu B., Gao X., He M., Liu L., Yin G. A fast online COVID-19 diagnostic system with chest CT scans. Proc KDD. 2020;2020 [Google Scholar]
- 214.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-net: A Framework of Deep Learning Classifiers to Diagnose Covid-19 in X-Ray Images. [Google Scholar]
- 215.Zhang J., Xie Y., Liao Z., Pang G., Verjans J., Li W., Sun Z., He J., Yi Li C.S. 2020. Viral Pneumonia Screening on Chest X-Ray Images Using Confidence-Aware Anomaly Detection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Narayan Das N., Kumar N., Kaur M., Kumar V., Singh D. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. Ing Rech Biomed. 2020 doi: 10.1016/j.irbm.2020.1007.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wu Y.-H., Gao S.-H., Mei J., Xu J., Fan D.-P., Zhao C.-W., Cheng M. 2020. JCS: an Explainable COVID-19 Diagnosis System by Joint Classification and Segmentation. [DOI] [PubMed] [Google Scholar]
- 218.Toğaçar M., Ergen B., Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103805. 103805-103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Basu S., Mitra S.J.a.p.a. 2020. Deep Learning for Screening COVID-19 Using Chest X-Ray Images. [Google Scholar]
- 220.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E.J.a.p.a. 2020. Rapid Ai Development Cycle for the Coronavirus (Covid-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring Using Deep Learning Ct Image Analysis. [Google Scholar]
- 221.Yan T., Wong P.K., Ren H., Wang H., Wang J., Li Y.J.C. Solitons, Fractals, Automatic distinction between covid-19 and common pneumonia using multi-scale convolutional neural network on chest ct scans. 2020;140:110153. doi: 10.1016/j.chaos.2020.110153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Oh Y., Park S., Ye J.C. 2020. Deep Learning Covid-19 Features on Cxr Using Limited Training Data Sets. [DOI] [PubMed] [Google Scholar]
- 223.Saba L., Than J.C.M., Noor N.M., Rijal O.M., Kassim R.M., Yunus A., Ng C.R., Suri J.S. Inter-observer variability analysis of automatic lung delineation in normal and disease patients. J. Med. Syst. 2016;40 doi: 10.1007/s10916-016-0504-7. [DOI] [PubMed] [Google Scholar]
- 224.Liu K., Suri J.S. Google Patents; 2005. Automatic Vessel Indentification for Angiographic Screening. [Google Scholar]
- 225.Ambale-Venkatesh B., Yang X., Wu C.O., Liu K., Hundley W.G., McClelland R., Gomes A.S., Folsom A.R., Shea S., Guallar E., Bluemke D.A., Lima J.A.C. Cardiovascular event prediction by machine learning: the multi-ethnic study of atherosclerosis. Circ. Res. 2017;121:1092–1101. doi: 10.1161/CIRCRESAHA.117.311312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tandel G.S., Balestrieri A., Jujaray T., Khanna N.N., Saba L., Suri J.S. Multiclass magnetic resonance imaging brain tumor classification using artificial intelligence paradigm. Comput. Biol. Med. 2020;122:103804. doi: 10.1016/j.compbiomed.2020.103804. [DOI] [PubMed] [Google Scholar]
- 227.Shrivastava V.K., Londhe N.D., Sonawane R.S., Suri J.S. A novel and robust Bayesian approach for segmentation of psoriasis lesions and its risk stratification. Comput. Methods Progr. Biomed. 2017;150:9–22. doi: 10.1016/j.cmpb.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 228.Jamthikar A., Gupta D., Saba L., Khanna N.N., Viskovic K., Mavrogeni S., Laird J.R., Sattar N., Johri A.M., Pareek G.J.C.i.B. 2020. Medicine, Artificial Intelligence Framework for Predictive Cardiovascular and Stroke Risk Assessment Models: A Narrative Review of Integrated Approaches Using Carotid Ultrasound; p. 104043. [DOI] [PubMed] [Google Scholar]
- 229.Wu X., Hui H., Niu M., Li L., Wang L., He B., Yang X., Li L., Li H., Tian J., Zha Y. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Eur. J. Radiol. 2020;128 doi: 10.1016/j.ejrad.2020.109041. 109041-109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.W.H. Organization . 2020. Use of Chest Imaging in COVID-19; pp. 1–56. [Google Scholar]
- 231.Yusuf G.T., Wong A., Rao D., Tee A., Fang C., Sidhu P.S. The use of contrast-enhanced ultrasound in COVID-19 lung imaging. J Ultrasound. 2020:1–5. doi: 10.1007/s40477-020-00517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ni Q., Sun Z.Y., Qi L., Chen W., Yang Y., Wang L., Zhang X., Yang L., Fang Y., Xing Z.J.E.r. vol. 30. 2020. pp. 6517–6527. (A Deep Learning Approach to Characterize 2019 Coronavirus Disease (COVID-19) Pneumonia in Chest CT Images). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wang S., Zha Y., Li W., Wu Q., Li X., Niu M., Wang M., Qiu X., Li H., Yu H.J.E.R.J. 2020. A Fully Automatic Deep Learning System for COVID-19 Diagnostic and Prognostic Analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Narayanan R., Werahera P.N., Barqawi A., Crawford E.D., Shinohara K., Simoneau A.R., Suri J.S. Adaptation of a 3D prostate cancer atlas for transrectal ultrasound guided target-specific biopsy. Phys. Med. Biol. 2008;53:N397–N406. doi: 10.1088/0031-9155/53/20/N03. [DOI] [PubMed] [Google Scholar]
- 235.Shen F., Narayanan R., Suri J.S. Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual International Conference. 2008. Rapid motion compensation for prostate biopsy using GPU; pp. 3257–3260. 2008. [DOI] [PubMed] [Google Scholar]
- 236.State of the Art in Neural Networks and Their Applications - 1st Edition.
- 237.Acharya U.R., Vinitha Sree S., Mookiah M.R., Yantri R., Molinari F., Zieleźnik W., Małyszek-Tumidajewicz J., Stępień B., Bardales R.H., Witkowska A., Suri J.S. Diagnosis of Hashimoto's thyroiditis in ultrasound using tissue characterization and pixel classification. Proc. IME H J. Eng. Med. 2013;227:788–798. doi: 10.1177/0954411913483637. [DOI] [PubMed] [Google Scholar]
- 238.Kandemirli S.G., Altundag A., Yildirim D., Tekcan Sanli D.E., Saatci O. Olfactory bulb MRI and paranasal sinus CT findings in persistent COVID-19 anosmia. Acad. Radiol. 2020;28(1):28–35. doi: 10.1016/j.acra.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Cuadrado-Godia E., Dwivedi P., Sharma S., Santiago A.O., Gonzalez J.R., Balcells M., Laird J., Turk M., Suri H.S., Nicolaides A.J.J.o.s. vol. 20. 2018. p. 302. (Cerebral Small Vessel Disease: a Review Focusing on Pathophysiology, Biomarkers, and Machine Learning Strategies). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Maniruzzaman M., Kumar N., Abedin M.M., Islam M.S., Suri H.S., El-Baz A.S., Suri J.S.J.C.m., biomedicine p.i. vol. 152. 2017. pp. 23–34. (Comparative Approaches for Classification of Diabetes Mellitus Data: Machine Learning Paradigm). [DOI] [PubMed] [Google Scholar]
- 241.Acharya R., Ng Y.E., Suri J.S. Artech House; 2008. Image Modeling of the Human Eye. [Google Scholar]
- 242.El-Baz A., Suri J. CRC Press; 2019. Lung Imaging and CADx. [Google Scholar]
- 243.El-Baz A., Suri J.S. CRC Press; 2011. Lung Imaging and Computer Aided Diagnosis. [Google Scholar]
- 244.Online COVID-19 Diagnosis with Chest CT Images: Lesion-Attention Deep Neural Networks, Rescognito, Inc.