Abstract
Bats carry diverse severe acute respiratory syndrome-related coronaviruses (SARSr-CoVs). The suspected interspecies transmission of SARSr-CoVs from bats to humans has caused two severe CoV pandemics, the SARS pandemic in 2003 and the recent COVID-19 pandemic. The receptor utilization of SARSr-CoV plays the key role in determining the host range and the interspecies transmission ability of the virus. Both SARS-CoV and SARS-CoV-2 use angiotensin-converting enzyme 2 (ACE2) as their receptor. Previous studies showed that WIV1 strain, the first living coronavirus isolated from bat using ACE2 as its receptor, is the prototype of SARS-CoV. The receptor-binding domain (RBD) in the spike protein (S) of SARS-CoV and WIV1 is responsible for ACE2 binding and medicates the viral entry. Comparing to SARS-CoV, WIV1 has three distinct amino acid residues (442, 472, and 487) in its RBD. This study aimed at exploring whether these three residues could alter the receptor utilization of SARSr-CoVs. We replaced the three residues in SARS-CoV (BJ01 strain) S with their counterparts in WIV1 S, and then evaluated the change of their utilization of bat, civet, and human ACE2s using a lentivirus-based pseudovirus infection system. To further validate the S-ACE2 interactions, the binding affinity between the RBDs of these S proteins and the three ACE2s were verified by flow cytometry. The results showed that the single amino acid substitution Y442S in the RBD of BJ01 S enhanced its utilization of bat ACE2 and its binding affinity to bat ACE2. On the contrary, the reverse substitution in WIV1 S (S442Y) significantly attenuated the pseudovirus utilization of bat, civet and human ACE2s for cell entry, and reduced its binding affinity with the three ACE2s. These results suggest that the S442 is critical for WIV1 adapting to bats as its natural hosts. These findings will enhance our understanding of host adaptations and cross-species infections of coronaviruses, contributing to the prediction and prevention of coronavirus epidemics.
Keywords: SARS-CoV, SARSr-CoV, ACE2, Receptor utilization, Spike, Receptor binding domain
1. Introduction
Coronaviruses (CoVs) are enveloped, single-stranded and positive-sense RNA viruses with a genome of 27−32 kb, which can cause diseases in respiratory, gastrointestinal, hepatic and central nervous systems of humans, livestock and wild animals (Cui et al., 2019). CoVs are classified in the subfamily Orthocoronavirinae, family Coronaviridae, suborder Cornidovirineae, order Nidovirales (Lu et al., 2020). According to the genetic features, CoVs are grouped into four genera, Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. Betacoronavirus is further classified into 5 subgenera, Embecovirus (previous lineage A), Hibecovirus, Sarbecovirus (previous lineage B), Merbecovirus (previous lineage C) and Nobecovirus (previous lineage D) (https://talk.ictvonline.org/taxonomy/).
Severe acute respiratory syndrome-related coronaviruses (SARSr-CoVs) belong to the subgenus Sarbecovirus, containing various bat SARSr-CoVs and two pandemic human viruses, SARS-CoV and SARS-CoV-2 (Gorbalenya et al., 2020). SARS-CoV and SARS-CoV-2 have caused the 2002–2003 SARS pandemic and the ongoing COVID-19 pandemic, respectively (Wu et al., 2020; Zhou et al., 2020). The outbreaks of SARS brought over 8000 cases of infection with a fatality rate of 10 %, resulting in tremendous social unrest and economic loss. The COVID-19 pandemic is even worse. As of November 12, 2020, SARS-CoV-2 has infected 51,848,261 people globally (https://www.who.int/emergencies/diseases/novel-coronavirus-2019), causing 1,280,868 deaths, which is still increasing rapidly. These pandemics remind us that investigations on the diversity and interspecific transmission of coronavirus remain insufficient.
Previous studies on molecular epidemiology and genetic evolution of viruses have confirmed that bats carry a variety of SARSr-CoVs, including Rp3, Rs672, HKU3 and others (Drexler et al., 2010; Lau et al., 2005; Li et al., 2005a; Tong et al., 2009). Recent studies based on virus isolation, receptor utilization and viral infection test have revealed that bat CoVs WIV1 and RsSHC014 from Rhinolophus sinicus are closely related to SARS-CoV (Ge et al., 2013). The WIV1, RsSHC014, WIV16 and some other strains of bat SARSr-CoVs use ACE2 as their receptor, which may be the direct ancestors of SARS-CoV and the potential pathogens for future outbreaks of CoV epidemics (Ge et al., 2013; Menachery et al., 2016; Yang et al., 2016). Especially, the CoVs from Rhinolophus sinicus are the most homologous to SARS-CoV, which is worth more attention.
Spike (S) proteins of CoVs are responsible for cell recognition and host adaptation (Gui et al., 2017). The S protein can be cleaved by proteases into an N-terminal S1 subunit for receptor recognition and binding, and a C-terminal S2 region mediating fusion of the virus and host cell membranes (Belouzard et al., 2009; Bertram et al., 2011). CoVs are RNA viruses with insufficient proofreading capability, which leads to high mutation rate, driving viral evolution and genome variability. Especially, mutations in the S protein of CoVs impose the major effect on viral infectivity and host range. For example, recent research reported that the furin protease cleavage site and the mutation D614 G of SARS-CoV-2 enhances viral infectivity and increases transduction of multiple human cell types, raising the concern about the existence of more highly transmissible or more virulent SARS-CoV-2 forms in the future (Daniloski et al., 2020; Plante et al., 2020; Wang et al., 2020b; Zhang et al., 2020). The defined receptor-binding domain (RBD) includes a 193-residue fragment (amino acids 318–510) in the SARS-CoV S1 domain, which shows more sufficient binding to ACE2 than the full S1 domain (Babcock et al., 2004; Wong et al., 2004; Xiao et al., 2003). In essence, RBD determines cellular receptor recognition, cell tropism, and host specificity. For instance, bat SARSr-CoV Rp3 S protein cannot use ACE2 for cell entry regardless of the origin of the ACE2 molecule (Ren et al., 2008). However, when Rp3 RBD is replaced with SARS-CoV S proteins, bat SARSr-CoV Rp3 can utilize AEC2 receptor for viral entry (Becker et al., 2008; Ren et al., 2008). In addition, genetic variance of the RBD can cause viral virulence alteration and cross-species infections. Even minor changes in amino acid residues of the RBD of SARS-CoV S protein are able to alter the host range of several coronaviruses. For instance, there are four prototypic SARS-CoV strains: hTor02 (a human strain isolated in 2002–2003), hGd03 (a human strain isolated in 2003–2004), cSz02 (a civet strain isolated in 2002–2003), and cGd05 (a civet strain isolated in 2005–2006). The affinity of their RBDs to human ACE2 varies, but all of them show a high affinity to the civet ACE2. As a result, humans are highly susceptible to hTor02, moderately susceptible to hGd03, and poorly susceptible to cSz02 and cGd05, while civets are susceptible to all four viral strains (Liu et al., 2007). Especially, there were two key residues (amino acids 479 and 487) alteration in S proteins of SARS-CoV isolated during minor 2003–2004 outbreak, probably explaining why the virus had failed to transmit from human to human (Li, 2008; Li et al., 2005b). Collectively, CoV spike protein variance significantly influences the host range and cross-species transmission of the viurs.
Interactions between five residues (amino acids 442, 472, 479, 487 and 491) in RBD and four residues in ACE2 (residues 31, 35, 38, and 353) are important for SARS progression and tropism (Li, 2008, 2013; Wu et al., 2012). In the present study, three residues (amino acids 442, 472, and 487) of these RBDs were investigated. In order to explore whether these three residues could alter the host range of SARS-CoV and SARSr-CoV, we substituted these three residues in SARS-CoV (BJ01 strain) RBD with the corresponding residues of WIV1 RBD, and then evaluating the change of their capability mediating cell entry of pseudovirus via bat, civet, and human ACE2s. Meanwhile, the abilities of wild-type and mutant SARS-CoV and SARSr-CoV RBDs to bind to different ACE2 were verified by protein binding assay. This study identified that 442th residue of the spike protein of SARSr-CoV is critical for host adaptation and interspecies transmission. These results suggest that mutation of S protein is probably affects the host rang of SARSr-CoV and knowing about these residues may be helpful for assessing the risk of new SARSr-CoV outbreak in the future.
2. Materials and methods
2.1. Cell culture
HEK293T, GP2-293, HeLa and Vero-E6 cells were cultured in DMEM (Gibco Grand Island, USA) supplemented with 10 % fetal bovine serum (FBS) (Gibco). All cells were maintained at 37 °C in a humidified incubator containing 5% CO2.
2.2. Generation of cells stably expressing the bat, civet and human ACE2
Firstly, the full length ACE2 genes of human, civet, and R. sinicus bat (GenBank number KC881004.1) were cloned into pQCXIH vector with the C-terminal HA tag. Secondly, 3 μg retroviral expression vectors (VSV-G) and 3 μg expression plasmids (pQCXIH-h/b/c-ACE2-HA) were cotransfected into GP2-293 cells seeded in 6 cm cell dish using Lipofectamine 2000 (Invitrogen, USA) respectively. After 6 h transfection, media was removed and replaced with fresh DMEM with 10 % FBS. After 48 h, viral supernatants were harvested and then cleared cell debris by centrifugation. Following filtrated through a 0.45 μM sterilized membrane (Millipore, USA), and finally aliquots were stored at −80 °C.
HeLa cells were seeded in 6-well plates and grown for 18 h to reach ∼70 % confluence, discarding the cell culture supernatant, infected by 2 mL virus for 7 h, then replaced the virus with the growth medium containing 250 μg/mL hygromycin to eliminate the non ACE2-expressing cells. Hygromycin-resistant clones were validated via western blot (WB) and immunofluorescence assay (IFA).
2.3. Immunofluorescence confocal microscopy
HeLa cells stably expressing human, civet and bat ACE2 were seeded on coverslips in 24-well plates (Corning, USA). The following day, the cells were washed with PBS twice and fixed with 4% formaldehyde for 15 min, then permeabilized with 0.2 % triton-100 for 5 min at room temperature, after being blocked with 3% BSA for 1 h, the cells were incubated with mouse anti-HA antibody (CST, USA) at 4 °C overnight followed by incubation with Alexa 555-conjugated goat anti-mouse IgG (Invitrogen, USA) at room temperature for 30 min. Finally, cells were incubated with DAPI (Sigma, USA) for 10 min at room temperature for nuclear staining. Each incubation step was followed by washing with PBS three times. After the final wash, the slides were fixed with 3 μL fluorescent quencher and observed under a confocal microscope (Nikon, Japan).
2.4. Preparation and detection of HIV-based pseudotyped viruses
By comparing the S sequences of BJ01 (GenBank number AY278488) and WIVI (GenBank number KC881007), the sequence of the BJ01 strain was chosen to generate a series of BJ01 variants using site-directed mutagenesis method. The mutated amino acid codon for each mutant is presented as follows: Y442S, L472 F, T487 N, Y442S/L472 F, Y442S/T487 N, L472 F/T487 N, Y442S/L472 F/T487 N. In addition, the WIV1 strain was chosen to generate the variant S442Y, the Rp3 strain (GenBank number DQ071615) serves as the negative control.
HIV-based pseudoviruses carrying the S protein were prepared as described previously (Nie et al., 2004). Briefly, 15 μg plasmids containing the respective S gene or mutants (in vector pcDNA3.1) and 10 μg HIV backbone plasmid (pNL4−3.luc.RE) were cotransfected into HEK 293 T cells using Polyethylenimine/PEI (Polysciences, USA). After 6 h transfection, the medium was replaced with fresh media. Next, the cell culture supernatant was collected after 48 h transfection, and the cell debris was cleared by centrifugation and further removed by passing through a 0.45 μm filter (Millipore). HIV-1-pseudotyped virus were aliquoted and stored at −80 °C. Pseudoviruses was separated on a 10 % SDS-PAGE gel and transferred to a PVDF membrane (Millipore). After blocking with 5% skimmed milk in PBS and incubated with antibody against the SARS-CoV S protein polyclonal antibody (Invitrogen, USA) and anti-p24 antibodies (Abnova, USA) at 4 °C for 12 h. The membrane was washed three times with PBS containing Tween-20 (0.1 %) followed by incubation with horseradish peroxidase-conjugated secondary antibody diluted 1:5000. Then, the membrane was washed three times and proteins were visualized with the SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific).
2.5. Infection assay with pseudovirus
A human immunodeficiency virus (HIV)-based pseudovirus system was employed to compare difference in receptor usage between SARSr-CoV WIV1, SARS-CoV BJ01 and their mutants. The luciferase activity was measured using the luciferase assay system (Promega, USA), which was expressed from the reporter gene carried by the pseudovirus. Briefly, cells were lysed after infection 48 h by adding 60 μL of lysis buffer provided with the kit, and then 20 μL of the resulting lysates were added to 20 μL of luciferase substrate, mixed thoroughly and measured for luciferase activity. Each infection experiment was conducted in triplicate.
2.6. RBD expression and purification
First of all, the RBD sequences (residues 317–569) of the BJ01 strain were cloned into pCAGGS vector, introducing a secretion signal peptide and an S-tag in order to subsequent experiment. Similarly, a series of RBD mutants in the same site were generated by the same method. Secondly, a series of RBD variants were conducted and then transfected into human embryonic kidney HEK293 T cells cultured in 10 cm dishes using PEI, further cultured with 293 SFM II-based expression medium (Life Technologies) for 48 h. The RBD protein in supernatants and cell lysis were detected by western blot using anti-S antibodies, respectively. Next, Large-scale supernatants containing the soluble RBD proteins were harvested, cleared the cell debris by centrifugation, and filtrated through a 0.45 μm sterilized membrane, then added S-protein Agarose (Millipore, USA) and incubated for 1 h at room temperature. The Agarose was added into the column with filter cartridge and eluted with 3 M MgCl2. The purified protein was concentrated by ultrafiltration using an ultrafiltration tube (Millipore, USA) and then dissolved in PBS. Finally, the concentration of purified RBDs was quantified by BCA assay, and then the purified RBDs were aliquoted.
2.7. Flow cytometric analysis of RBD/ACE2 interactions
To measure the binding affinity between various RBD protein and bACE2, cACE2 and hACE2. HeLa cells stably expressing bACE2, cACE2 and hACE2 were seeded in 10 cm dishes. Discarding the cell culture supernatant and washing twice with PBS after 18 h, the cells were digested from culture dishes using 5 mM EDTA solution, and incubated with purified various S-tagged RBD proteins (5 μg) for 1 h at room temperature, following blocked with PBS supplemented by 3% BSA for 30 min. Subsequently, the cells were resuspended in 200 μL of PBS with 2% FBS containing DyLight® 488-labeled anti-S antibodies (Abcam, UK) and incubated for 1 h at room temperature in dark. The fluorescence intensity was performed using a Gallios cytometer (Beckman Coulter) by counting 20 000 events.
3. Results
3.1. Establishment of stable cell lines expressing ACE2s of different species
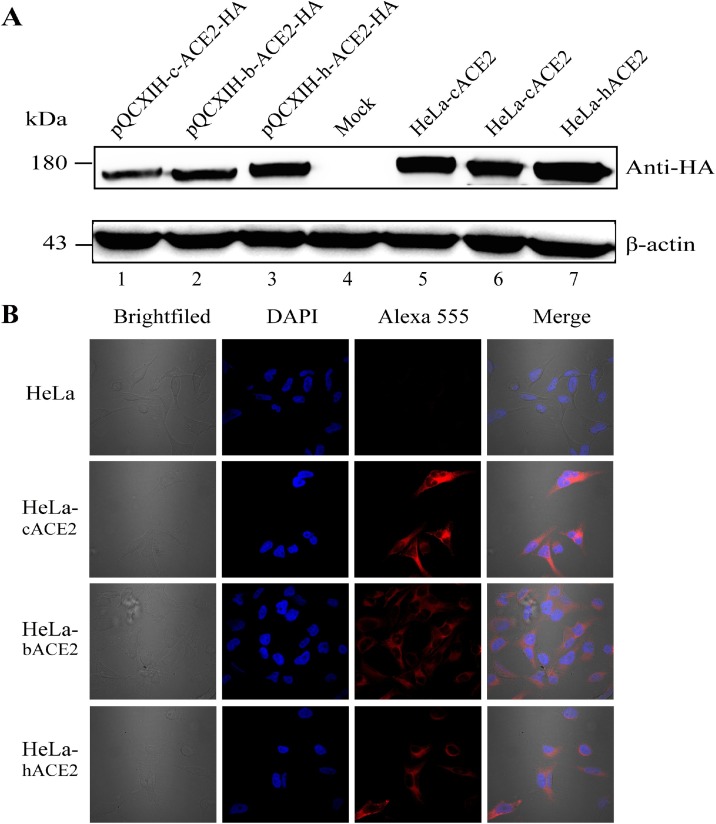
In order to investigate the host receptor adaptations and cross-species infections, HeLa cells, a cell line not expressing ACE2 natively, were used to establish stable cell lines expressing human, civet or bat ACE2 (hACE2, cACE2, and bACE2). Western blot analysis indicated that all the three stable cell lines showed similar levels of ACE2 expression which were higher than transiently transfected cells (Fig. 1 A). Immunofluorescence assay showed that stable expression of hACE2, cACE2, and bACE2s were localized to the cell membrane (Fig. 1B). Taken all together, the stable cell lines expressing hACE2, cACE2, and bACE2 were established successfully.
Fig. 1.
Generation of cells stably expressing civet, bat and human ACE2. (A) Detection of ACE2 expression in HeLa cells by western-blot using anti-HA tag antibodies. Lanes 1 to 3: transiently transfectedcells expressing civet (c), bat (b) and human (h) ACE2; Lane 4: HeLa cell acts as a negative control; Lanes 5 to 7: cell lines stably expressing civet, bat and human ACE2 cell. (B) HeLa cells stably expressing civet, bat and human ACE2 were verified by immunofluorescence. ACE2 expression was detected with mouse anti-HA antibody followed by Alexa 555-conjugated goat anti-mouse IgG. Nuclei were stained with DAPI (6-diamidino-2-phenylindole). The top row shows HeLa cells as a negative control. Rows 2 to 4 show HeLa cells stably expressing hACE2, bACE2, and cACE2. The columns (from left to right) show cells of brightfield, staining of cell nuclei (blue), staining of expressed ACE2-HA (red fluorescence of Alexa 555), and the merged double-stained image.
3.2. Y442 is key for the utilization of bat ACE2 by SARSr-CoVs
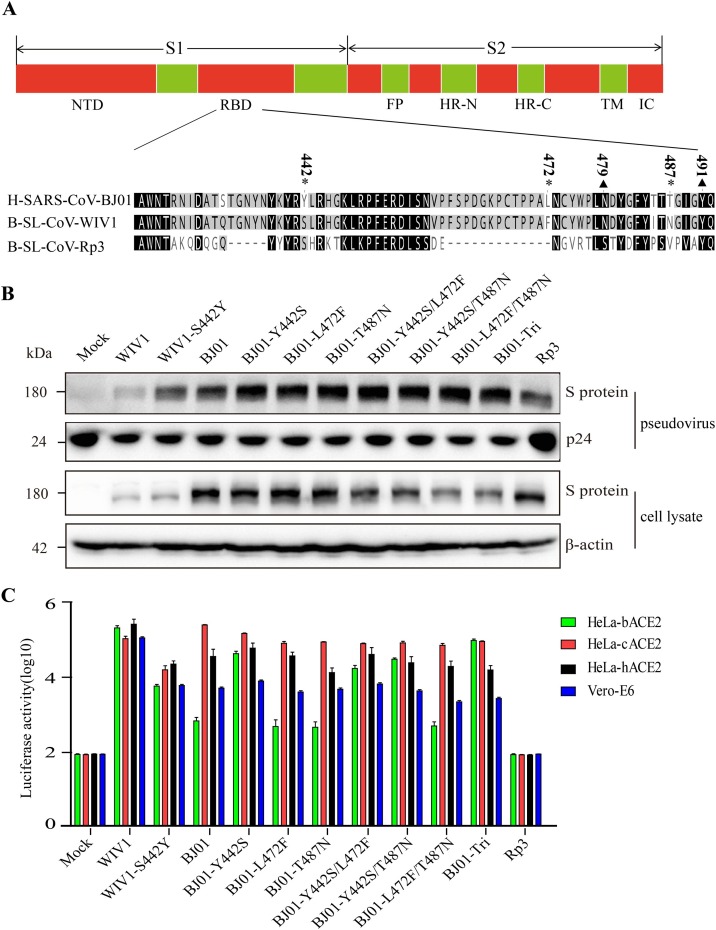
Previous structure and genetic analyses reported that five key residues (amino acids 442, 472, 479, 487 and 491) in the RBD of SARS-CoV S protein play an essential role in receptor binding (Wu et al., 2012). The S of SARS-CoV BJ01 is different from that of SARSr-CoV WIV1 at amino acids residues 442, 472, and 487 (Fig. 2 A). In order to explore the role of these three residues in receptor binding, the 442th (Tyr, Y), 472th (Leu, L) and 487th (Thr, T) residues in the BJ01 spike was replaced by the WIV1 counterpart residues 442 (Ser, S), 472 (Phe, F) and 487 (Asn, N), respectively, and the double and triple substitutions were also generated. As shown in Fig. 2B, these substitutions had no effect on the expression of S proteins.
Fig. 2.
Construction and functional analysis of pseudoviruses. (A) Sequence homology of the RBD of the spike gene of human SARS-CoV BJ01 (H-SARS-CoV-BJ01), bat SARSr-CoV WIV1 (B-SL-CoV-WIV1) and bat SARSr-CoV Rp3 (B-SL-CoV-RP3). The * represents different amino acid sites, and the▲represents the same sites among the key residues between H-SARS-CoV-BJ01 and B-SL-CoV-WIV1. (B) Detection of pseudoviruses bearing different S proteins by western-blot. Pseudoviruses were probed with C-oV S protein polyclonal pntibody (top) and with P24-specific mAbs (bottom) as a control. (C) Measurement of pseudovirus infectivity by determining luciferase activity. HIV-1-luciferase pseudotyped with S protein of the H-SARS-CoV-BJ01, B-SL-CoV-WIV1 and their variants, was incubated with HeLa-bACE2, HeLa-cACE2, HeLa-hACE2 and Vero-E6 cells. After 48 h infection, cell lysates were measured as luciferase activity. The results were presented as the means and standard deviations of data from three independent experiments. The error bars indicate standard deviations.
Subsequently, we prepared pseudotyped viruses harboring these S proteins and verified the packaging efficiency of pseudoviruses by western-blot detecting SARS-CoV S (Fig. 2B). The pseudovirus infectivity was tested in Vero-E6 cells, a cell line expressing ACE2 natively. As shown in Fig. 2C, pseudoviruses expressing BJ01-S and WIV1-S entered Vero-E6 efficiently, while Rp3 pseudoviruses could not infect Vero E6 cells, as reported previously (Ren et al., 2008). Then we carried out the pseudovirus entry assay using HeLa cells stably expressing hACE2, cACE2, and bACE2. The results showed that WIV1 pseudovirus could efficiently infect all the HeLa cell lines expressing hACE2, cACE2, and bACE2. In contrast, BJ01 pseudovirus efficiently utilized hACE2 and cACE2 but used bACE2 less efficiently, which is consistent with previous reports (Ren et al., 2008). In addition, the BJ01 mutants had no significant change in entering cells expressing cACE2 and hACE2. As for bACE2, only Y442S substitution elevated BJ01 capability of invading bACE2-expressing cells while other substitutions showed no influence on bACE2 utilization. In addition, the double or triple substitutions including Y442S (Y442S/L472F, Y442S/T487N and Y442S/L472 F/T487 N) could enhance pseudovirus infection to cells expressing bACE2. To further determine the roles of Y442 in SARS-CoV infection and transmission, a reverse substitution of S442Y on WIV1 S protein was made and its utilization efficiency of bACE2, cACE2 and hACE2 was tested. The result showed that S442Y substitution on WIV1 S proteins greatly attenuated its infection to HeLa cells expressing any one of the three ACE2s (Fig. 2C). Collectively, Y442 is critical for the utilization of bat ACE2 by SARS-CoV BJ01 and SARSr-CoV WIV1.
3.3. Successful purification of RBD mutants
In order to further confirm the binding ability of ACE2s to different RBD mutants, a series of RBD mutants were purified and used for binding assay. As shown in Fig. 3 A, the extopically expressed RBDs were detected not only in the cell lysate, but also in the cell supernatants, suggesting that the cells could secrete soluble RBD protein into the cell supernatant successfully. Subsequently, the RBDs were purified from the supernatants. The purified RBDs were then characterized by western blot and coomassie blue staining, which showed the correct size of the 37 kDa. Interestingly, we observed two bands for Rp3 RBD, the lower band is about 35 kDa which is smaller than the other RBDs as expected since Rp3 RBD is 19 amino-acid shorter (Fig. 3A and B). These soluble RBDs were used in the following flow cytometric experiment.
Fig. 3.
Detection of expression and purification of RBD protein. (A) Detection of RBD protein in supernatant and lysate by western-blot using anti-S tag mAbs respectively. (B) Detection of the purified protein by western-blot using anti-S tag mAbs and coomassie blue staining.
3.4. Y442 is key for RBD/ACE2 interactions
To further assess the host adaption of genetic variations in the RBD domain, the binding ability between a series of equivalent amount of RBDs and stable cell lines expressing ACE2 was analyzed by flow cytometry. As shown in Fig. 4 , both RBDs of SARS-CoV BJ01 and SARSr-CoV WIV1 could bind to hACE2 and cACE2 efficiently. In contrast, the binding affinity of BJ01-RBD and WIV1-RBD to bACE2 was lower than hACE2 and cACE2. Interestingly, we found that the mutants of Y442S, Y442S/L472F, Y442S/T487N and Y442S/L472F/T487N could increase the binding affinity between BJ01-RBD and bACE2, but the other three mutants, L472F, T487N and L472F /T487N, had no effect. Compared to the wild-type WIV1, the single amino acid substitution S442Y could significantly reduce the interaction of WIV1 RBD with bACE2, cACE2 and hACE2, which was consistent with pseudovirus assay results. Compared to wild-type BJ01 RBD, the single-amino-acid substitutions Y442S and L472F slightly increased their affinity to hACE2, but reduced their affinity to cACE2, while T487N decreased the RBD/hACE2 and the RBD/cACE2 affinity obviously. In addition, introduction of WIV1 residues at 487 in BJ01 RBD (Y442S/T487N, L472F/T487N and Y442S/L472F/T487N) decreased the RBD/hACE2 and the RBD/cACE2 affinity to various levels. As the negative control, the RBD of SARSr-CoV Rp3 showed almost no binding ability to any ACE2s.
Fig. 4.
Solution RBD binding assays of bat, civet, and human ACE2. HeLa cells stably expressing bat (A), civet (B), and human ACE2 (C) were resuspended with BJ01-RBD, WIV1-RBD and Rp3-RBD proteins and their mutants (50 μg/mL) and then performed flow cytometry. The gray shaded area represents the control of HeLa cells without ACE2 expression.
4. Discussion
The S protein of coronavirus is responsible for receptor recognition, binding and facilitating viral cell entry, which are key steps for infection, host range and cross-species transmission of the virus (Li et al., 2005a). During the transmission of SARS-CoV from its natural reservoir (bats) to the intermediate host (civets), and then the final host (humans), the S protein is challenged by strong positive selection to adapt to different viral hosts, and the key nonsynonymous variations in the S gene are mainly located within the RBD (Qu et al., 2005). This study was focused on 3 residues (amino acids 442, 472, and 487) of human SARS-CoV BJ01 RBD which are distinct from bat SARSr-CoV WIV1 RBD and may be responsible for the host adaption.
In this study, the effects of the spike variations on SARS-CoV BJ01 infectivity in hACE2, cACE2, and bACE2 cells were investigated by the pseudovirus system. Compared with wild-type BJ01 pseudotyped virus, all mutants containing Y442S dramatically increased the infectivity. Introduction of the 442th residue of WIV1 to BJ01 S (BJ01 S mutants Y442S/L472F, Y442S/T487N and Y442S/L472F/T487N) substantially enhanced its utilization of bACE2, whereas any other substitutions had no obvious effects (Fig. 2B). In addition, the protein binding affinity of the RBD variations to bACE2, cACE2 and hACE2 cells was determined through flow cytometry. Similarly, the BJ01 RBD mutants Y442, Y442S/L472F, Y442S/T487N and Y442S/L472F/T487N notably enhanced binding affinity to bat ACE2. To further determine the roles of the 442th residue in SARS-CoV BJ01 infection and transmission, we substituted this amino acid of SARSr-CoV WIV1 S protein with the 442th residues of SARS-CoV BJ01 S protein and then determined its’ effects on infection and binding affinity to bat, civet and human ACE2 expressing cells. The single amino acid substitution S442Y almost diminished WIV1 S protein-pseudotyped virus infection to HeLa cells expressing all the ACE2s. Correspondingly, the binding affinity between WIV1-RBD and the ACE2s was also greatly decreased. Taken together, the substitutions at positions 442 is essential for receptor binding and utilization.
Furthermore, the single amino acid substitutions Y442S and L472F in SARS-CoV BJ01 RBD enhanced their affinity to hACE2, possibly owing to the affinities of RBD/hACE2 interfaces. Based on the structural interactions, the Y442 of SARS-CoV has partial steric clash with alkyl side chain of hACE2 L31, while the substitution Y442S partially relieves this steric clash and strengthens the structure of hotspot-31, in addition, alcohol hydroxyl group of serine has a higher affinity with the amino of lysine than the phenolic hydroxyl group of tyrosine (Li, 2013). In addition, L472F may make stronger van der Waals contact with M82 and thus further slightly strengthens RBD/hACE2 interactions. As for RBD/cACE2 interfaces, RBD residue Y442 forms a hydrogen bond with T31 and thereby binds to cACE2 efficiently. F472 would have partial steric clash with the hydroxyl group of chimeric ACE2 T82 (Wu et al., 2012). Therefore, the single amino acid mutation Y442S or L472F slightly decreases the RBD/cACE2 binding affinity. Furthermore, mutation T487N of the human SARS-CoV BJ01 which mimics WIV1 RBD significantly decreases the RBD/hACE2 and RBD/cACE2 binding affinity. The methyl group of T487 is capable of forming salt bridge with D38 and K353 of hACE2 in a hydrophobic environment, while T487N removes an energetically favorable hydrophobic interaction with hACE2 K353 and becomes difficult for formation of the salt bridge (Li, 2008). Similarly, the methyl group of T487 also supports the cACE2 K353-E38 salt bridge, and thus, mutation T487N blocks the formation of the salt bridge (Wu et al., 2012). Introduction of WIV1 residues at 487, the BJ01 RBD mutants Y442S/T487 N, L472F/T487N and Y442S/L472F/T487N lower the RBD/hACE2 and RBD/cACE2 binding affinity to various levels. Therefore, T487 is essential for high affinity association with hACE2 and cACE2, which is critical or sufficient for the adaptation of SARS-CoV to human cells (Li et al., 2005b). However, there are some shortcomings in this study, some protein binding data were not consistent with pseudovirus-entry results, which may be due to the different mechanisms of the two assays. Briefly, peusodvirus entry depends on not only RBD-ACE2 binding but also other steps such as membrane fusion and particle internalization. In addition, the purified monomeric or oligomeric soluble RBD proteins may not completely maintain their natural conformation. Living virus infection could be conducted to verify the results in the future. Interestingly, there are two bands for Rp3 RBD, which may be due to post-translational modification, protien cleaving or other unknown reasons,which may potentially lead to distinct receptor utilization of Rp3.
Bats are highly diverse with the second largest number of species in mammals, carrying a wide variety of coronaviruses (Burgin et al., 2018; Wong et al., 2019). Bats are the only group of mammals capable of flying, which allows bats to disseminate the viruses widely leading to high chance of interspecies transmission (Woo et al., 2012). Coronaviruses are capable of adapting to new hosts through amino acid residue changes in RBD domain with relative shortcuts which could facilitate cross-species infections and human-to-human transmission (Drexler et al., 2014; Li et al., 2020b; Luk et al., 2019). SARS-CoV, SARS-CoV-2 and some other SARSr-CoVs can use ACE2 of many mammal species as their functional receptors, which indicates broad host range of these viruses (Qiu et al., 2020; Wang et al., 2020a). Therefore, the threats of coronaviruses to public health are constant and long-term (Li et al., 2020a). Research on the RBD/receptor interactions will help to clarify the viral infectivity and provide theoretical basis for vaccine development and others.
CRediT authorship contribution statement
Jin-Yan Li: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing - original draft. Qiong Wang: Investigation, Methodology, Formal analysis, Validation. Ce-Heng Liao: Methodology, Investigation. Ye Qiu: Funding acquisition, Supervision, Validation, Writing - review & editing. Xing-Yi Ge: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing - review & editing.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
This work was jointly funded by the National Natural Science Foundation of China [grant number 32041001, 31470260, and 81902070], and Provincial Natural Science Foundation of Hunan Province [grant number 2019JJ20004, and 2019JJ50035].
References
- Babcock G.J., Esshaki D.J., Thomas W.D., Jr., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78(9):4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M.M., Graham R.L., Donaldson E.F., Rockx B., Sims A.C., Sheahan T.P., Pickles R.J., Davide C., Johnston R.E., Baric R.S., Denison M.R. Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice. Proc. Natl. Acad. Sci. U. S. A. 2008;105(50):19944–19949. doi: 10.1073/pnas.0808116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V., Whittaker G. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Muller M.A., Lavender H., Gnirss K., Nehlmeier I., Niemeyer D., He Y., Simmons G., Drosten C., Soilleux E.J., Jahn O., Steffen I., Pohlmann S. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011;85(24):13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin C.J., Colella J.P., Kahn P.L., Upham N.S. How many species of mammals are there? J. Mammal. 2018;99(1):1–14. [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Guo X., Sanjana N.E. 2020. The D614G Mutation in SARS-CoV-2 Spike Increases Transduction of Multiple Human Cell Types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J., Florian G., Jorg G., Victor M., Doreen M., Matthias Gea. Genomic characterization of severe acute respiratory syndrome-related coronavirus in european bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J. Virol. 2010;84(21):11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J., Luo C.M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.Y., Wang L.F., Daszak P., Shi Z.L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. Gorbalenya Alexander, E, Baker Susan, C, Baric Ralph, S, de Groot Raoul, J, Drosten Christian, Gulyaeva Anastasia, A, Haagmans Bart, L, Lauber Chris, Leontovich Andrey, M, Neuman Benjamin, W, Penzar Dmitry, Perlman Stanley, Poon Leo L M, Samborskiy Dmitry, V, Sidorov Igor, A, Sola Isabel, Ziebuhr John. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27(1):119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.W., Wong B.H.Lea. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Natl Acad. Sci. U. S. A. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J. Virol. 2008;82(14):6984–6991. doi: 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res. 2013;100(1):246–254. doi: 10.1016/j.antiviral.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Y., You Z., Wang Q., Zhou Z.J., Qiu Y., Luo R., Ge X.Y. The epidemic of 2019-novel-coronavirus (2019-nCoV) pneumonia and insights for emerging infectious diseases in the future. Microbes Infect. 2020;22(2):80–85. doi: 10.1016/j.micinf.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.Q., Wu J.J., Nie J.H., Zhang L., Hao H., Liu S., Zhao C.Y., Zhang Q., Liu H., Nie L.L., Qin H.Y., Wang M., Lu Q., Li X.Y., Sun Q.Y., Liu J.K., Zhang L.Q., Li X.G., Huang W.J., Wang Y.C. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Fang Q., Deng F., Wang H., Yi C.E., Ba L., Yu W., Lin R.D., Li T., Hu Z., Ho D.D., Zhang L., Chen Z. Natural mutations in the receptor binding domain of spike glycoprotein determine the reactivity of cross-neutralization between palm civet coronavirus and severe acute respiratory syndrome coronavirus. J. Virol. 2007;81(9):4694–4700. doi: 10.1128/JVI.02389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk H.K.H., Li X., Fung J., Lau S.K.P., Woo P.C.Y. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect. Genet. Evol. 2019;71:21–30. doi: 10.1016/j.meegid.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., Swanstrom J., Sheahan T.P., Pickles R.J., Corti D., Randell S.H., Lanzavecchia A., Marasco W.A., Baric R.S. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U. S. A. 2016;113(11):3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y., Wang P., Shi X., Wang G., Chen J., Zheng A., Wang W., Wang Z., Qu X., Luo M., Tan L., Song X., Yin X., Chen J., Ding M., Deng H. Highly infectious SARS-CoV pseudotyped virus reveals the cell tropism and its correlation with receptor expression. Biochem. Biophys. Res. Commun. 2004;321(4):994–1000. doi: 10.1016/j.bbrc.2004.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Zhao Y.B., Wang Q., Li J.Y., Zhou Z.J., Liao C.H., Ge X.Y. Predicting the angiotensin converting enzyme 2 (ACE2) utilizing as the of SARS-CoV-2. Microbes Infect. 2020;22(4–5):221–225. doi: 10.1016/j.micinf.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X.X., Hao P., Song X.J., Jiang S.M., Liu Y.X., Wang P.G., Rao X., Song H.D., Wang S.Y., Zuo Y., Zheng A.H., Luo M., Wang H.L., Deng F., Wang H.Z., Hu Z.H., Ding M.X., Zhao G.P., Deng H.K. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 2005;280(33):29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Qu X., Li W., Han Z., Yu M., Zhou P., Zhang S.Y., Wang L.F., Deng H., Shi Z. Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and SARS-like coronavirus of bat origin. J. Virol. 2008;82(4):1899–1907. doi: 10.1128/JVI.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Conrardy C., Ruone S., Kuzmin I.V., Guo X., Tao Y., Niezgoda M., Haynes L., Agwanda B., Breiman R.F., Anderson L.J., Rupprecht C.E. Detection of novel SARS-like and other coronaviruses in bats from Kenya. Emerg. Infect. Dis. 2009;15(3):482–485. doi: 10.3201/eid1503.081013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Qiu Y., Li J.Y., Liao C.H., Zhou Z.J., Ge X.Y. Receptor utilization of angiotensin-converting enzyme 2 (ACE2) indicates a narrower host range of SARS-CoV-2 than that of SARS-CoV. Transbound. Emerg. Dis. 2020 doi: 10.1111/tbed.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Qiu Y., Li J.Y., Zhou Z.J., Liao C.H., Ge X.Y. A unique protease cleavage site predicted in the spike protein of the novel pneumonia coronavirus (2019-nCoV) potentially related to viral transmissibility. Virol. Sin. 2020;35(3):337–339. doi: 10.1007/s12250-020-00212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.C.P., Li X., Lau S.K.P., Woo P.C.Y. Global epidemiology of bat coronaviruses. Viruses. 2019;11(2):174. doi: 10.3390/v11020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Peng G., Wilken M., Geraghty R., Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012;287(12):8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. The sars-cov s glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312(4):1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.L., Hu B., Wang B., Wang M.N., Zhang Q., Zhang W., Wu L.J., Ge X.Y., Zhang Y.Z., Daszak P., Wang L.F., Shi Z.L. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J. Virol. 2016;90(6):3253–3256. doi: 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. 2020. The D614G Mutation in the SARS-CoV-2 Spike Protein Reduces S1 Shedding and Increases Infectivity. [DOI] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]