Abstract
Next-generation sequencing (NGS) technologies have changed the process of genetic diagnosis from a gene-by-gene approach to syndrome-based diagnostic gene panel sequencing (DPS), diagnostic exome sequencing (DES), and diagnostic genome sequencing (DGS). A priori information on the causative genes that might underlie a genetic condition is a prerequisite for genetic diagnosis before conducting clinical NGS tests. Theoretically, DPS, DES, and DGS do not require any information on specific candidate genes. Therefore, clinical NGS tests sometimes detect disease-related pathogenic variants in genes underlying different conditions from the initial diagnosis. These clinical NGS tests are expensive, but they can be a cost-effective approach for the rapid diagnosis of rare disorders with genetic heterogeneity, such as the glycogen storage disease, familial intrahepatic cholestasis, lysosomal storage disease, and primary immunodeficiency. In addition, DES or DGS may find novel genes that that were previously not linked to human diseases.
Keywords: Next-generation sequencing, High-throughput nucleotide sequencing, Diagnostic exome sequencing, Diagnostic genome sequencing
INTRODUCTION
A pediatrician requested a genetic test for a patient suspected of having congenital chloride diarrhea (CLD). A PubMed search confirmed that SLC26A3 is the only causative gene of CLD. After extracting the DNA from the blood sample of the patient, Sanger sequencing on all coding exons of SLC26A3 was performed, which revealed two disease-related variants. Next, the two variants were examined in the parents, and it was confirmed that each variant was inherited from one parent; therefore, the genetic diagnosis has been made successfully [1].
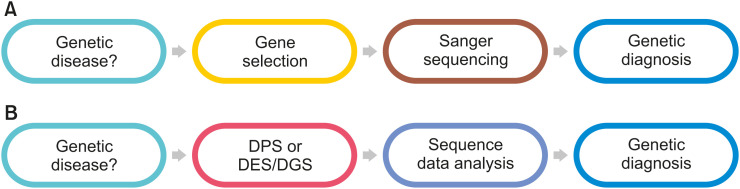
This example illustrates the traditional genetic diagnosis process. In other words, in order to perform a genetic diagnosis in a patient suspected of having a genetic disease, the gene that causes the disease must be first identified. If the causative gene is identified, a genetic test is performed using Sanger sequencing; if a disease-related variants are found, a genetic diagnosis is reached (Fig. 1A).
Fig. 1. Paradigm change of genetic diagnosis. (A) Traditional gene-by-gene approach using Sanger sequencing. (B) Syndrome-based genetic diagnosis process by diagnostic gene panel sequencing (DPS), diagnostic exome sequencing (DES), or diagnostic genome sequencing (DGS) using next-generation sequencing.
In Turkey 2009, a 5-month-old boy suffered from failure to thrive and dehydration, and various diseases such as the Batter syndrome were suspected, considering that the base was wet despite the presence of dehydration. Since the diagnosis was unclear, he was referred for whole exome sequencing (WES), which was emerging as a new technology at the time. WES is a test method that analyzes the coding exons of more than 20,000 human genes at once by next-generation sequencing (NGS). In this child, a disease-related variant was unexpectedly found in the SLC26A3 gene, and thus, the patient was diagnosed with CLD [2].
This case is very important from several perspectives. First, it shows that despite the unclear clinical diagnosis, genetic testing can lead to an accurate diagnosis. Second, it proves that WES can be used for clinical diagnosis of rare diseases. Third, it has been suggested that genes may be identified using WES in diseases where the causative gene has not yet been identified.
NGS has changed the paradigm of genetic diagnosis. In the past, the gene that caused the disease had to be identified for genetic diagnosis. Specific genes were selected and analyzed one after the other through the Sanger sequencing method. However, when a genetic disease is suspected as in the aforementioned case, it is possible to analyze dozens, hundreds, or 20,000 genes at once, find disease-related variants, and then confirm the clinical diagnosis (Fig. 1B).
In this review, the latest findings on the clinical application of NGS, which has changed the paradigm of genetic diagnosis is presented.
TYPES OF CLINICAL NGS TESTS
While NGS is inadequate for analyzing a single gene due to the nature of the test, it is useful to analyze tens to hundreds or thousands of genes simultaneously. If the Sanger sequencing and NGS methods are compared to means of transportation, Sanger sequencing is expensive, such as a taxi or a private plane, but can selectively and conveniently move a small number of passengers, whereas NGS is a method used to move a large number of passengers, such as a large commercial aircraft.
Clinical diagnosis methods using NGS can be divided into gene panel, exome, and genome according to the composition of the gene and the range to be analyzed [3]. Considering the purpose of genetic diagnosis, these methods are named diagnostic gene panel sequencing (DPS), diagnostic exome sequencing (DES), or diagnostic genome sequencing (DGS).
The difference between DPS, DES, and DGS for clinical NGS testing may be expressed in terms of how many of the three billion nucleotide sequences of the human genome are analyzed (genome coverage) and the number of times a sequence is read and analyzed (sequencing depth). In the case of DGS, the genome coverage is 90–95% and the sequencing depth is approximately 30–60×; for DES, the genome coverage is approximately 1–2% and the sequencing depth is approximately 100–200×; and for DPS, the genome coverage is 0.01–0.1% and the sequencing depth is approximately 200–500× (Table 1).
Table 1. Comparison of clinical next-generation sequencing test.
| Charateristics | DPS | DES | DGS | |
|---|---|---|---|---|
| Genome coverage | Low | Intermediate | High | |
| Sequencing depth | High | Intermediate | Low | |
| Number of genes | Up to thousands | More than 20,000 | More than 20,000 | |
| Capture bias | Yes | Yes | No | |
| Diagnostic yield | ||||
| SNV/INDEL | High | High to intermediate | High | |
| Intron variant | Low | Low | High | |
| CNV | Intermediate | Intermediate | High | |
| Gene rearrangement | Low | Low | High | |
| Re-analysis potential | Low | Intermediate to high | High | |
| Cost | Low | Intermediate | High | |
DPS: diagnostic gene panel sequencing, DES: diagnostic exome sequencing, DGS: diagnostic genome sequencing, SNV: single nucleotide variant, INDEL: insertion and deletion, CNV: copy number variation.
DIAGNOSTIC GENE PANEL SEQUENCING
DPS is called targeted gene panel, targeted exome, and focused NGS panel. This test selects tens to hundreds of genes related to a specific disease or syndrome, capturing the coding exon and adjacent intron sites of the corresponding genes, and performing NGS analysis. DPS is the most widely used for clinical diagnosis.
DPS may show a wide variety of diagnostic yields depending on the disease group, the method of selecting a study subject, and the gene panel composition. In Korean patients, the diagnostic yields of DPS have been reported in disorders of sex development (29.5%) [4], syndromic growth disorder (46.2%) [5], inherited peripheral neuropathies (27.0%) [6], developmental and epileptic encephalopathy (EIEE) (37.1%) [7], intractable early onset epilepsy (37.8%) [8], developmental delay and/or intellectual disability (29%) [9], infantile nystagmus syndrome (58.3%) [10], autism spectrum disorder, and comorbid epilepsy (17.5%) [11].
DIAGNOSTIC EXOME SEQUENCING
DES is a method of NGS analysis that captures coding exons and adjacent intron sites of all disease-related genes (Mendeliome) or all human genes (Whole Exome). DES has been clinically used before DPS. DES may include genes that are not related to genetic diseases; therefore, even if a gene causing disease is not identified in the current analysis, a reanalysis of data may be conducted to identify new causative genes [12].
When DES was initially used for genetic diagnosis, it was expected that the diagnosis rate of genetic diseases would be very high because all disease-causing genes or all human genes could be analyzed at once. However, when DES was conducted in 2013 in 250 patients with genetic diseases, the diagnostic yield was only 24.8% (62/250) [13]. In a follow-up study, the researchers reported a 25.2% (504/2,000) diagnostic yield through DES in 2,000 patients with genetic disorders [14]. However, it was reported that the diagnostic yield was as high as 36.1% in the specific neurological disease group, while it was as low as 20.1% in the non-neuronal disease group. Another research team reported that 26% of patients with suspected genetic diseases were diagnosed through DES [15]. The Trio DES, which was performed simultaneously with parents and patients, resulted in a diagnostic yield of 31%. Since then, DES diagnostic yields ranging from less than 10% to more than 50% have been reported in various disease groups, showing a wide variety of diagnosis rates according to the disease group [16].
Even though DES is a very efficient method for genetic diagnosis, it has some limitations. DES may miss regions such as GC-rich regions, and copy number variation (CNV) such as large deletions or duplications difficult to detect [17].
DIAGNOSTIC GENOME SEQUENCING
DGS is the most comprehensive method to analyze entire human genomes, including not only genes, but also the intergenic region between genes and the intron region between exons. However, the data size is too big to handle, and the sequencing cost is too high. Recently, the cost of NGS analyses decreased, and the advantages of DGS are emphasized because the genome data analyzed through DGS have higher uniformity than DPS and DES, and most of the GC-rich regions can be analyzed. In addition, even with DES or DPS, a significant number of patients do not receive a genetic diagnosis. Therefore, DGS has been proposed as a first-tier genetic test [18].
DGS was performed as a first-tier test in 103 pediatric patients with suspected genetic diseases. The diagnostic yield was 41%, which was significantly higher than the 24% of the conventional method [19]. In another study, DGS was performed on 14 early infantile EIEE patients, and disease-related variants were found in all the patients [20]. Two of these patients were not diagnosed with DES and another three were previously tested through DPS with negative results.
DGS has several advantages over DPS and DES. First, the CNV, such as large deletions and duplications as well as genomic rearrangements can be detected. DPS and DES can also detect part of the CNV, but DGS can detect most types of CNV and genomic rearrangements [21]. According to the results of DGS as a first-tier test in 60 rare, undiagnosed, or genetic diseases, clinically important genomic findings were observed in 41 patients (68.3%), of which 20 were CNV or gross chromosome abnormalities [22]. Second, it is possible to detect a deep intronic variant. In some cases where disease-related pathogenic variants are not found in DPS or DES, the mRNA sequence may be affected by deep intronic variants [23]. For example, deep intron variants that were detected by DGS in patients with retinal dystrophy and nephronophthisis have been reported [24,25].
CONCLUSION
NGS-based DPS, DES, and DGS changed the paradigm of genetic diagnosis. DPS is most commonly used, and DES is used less frequently, but serves a prominent role in clinical settings. DGS is rapidly approaching financial feasibility for use in everyday practice. However, even when these NGS-based genetic tests are used, approximately 50% of the patients do not receive a definite genetic diagnosis. There are many genetic reasons for this, including repeat expansion variants, somatic variants, and deep intron variants with indeterminate splicing effects, yet inadequate clinical information and the consequent inappropriate application of clinical NGS tests remain the most important cause of negative results. Therefore, the provision of detailed and accurate clinical information is a prerequisite for achieving the optimal diagnostic yield.
Clinicians and laboratory medicine doctors need to communicate closely and effectively in the process of selecting appropriate genetic tests and interpreting their results.
Footnotes
Conflict of Interest: The author have no financial conflicts of interest.
References
- 1.Lee ES, Cho AR, Ki CS. Identification of SLC26A3 mutations in a Korean patient with congenital chloride diarrhea. Ann Lab Med. 2012;32:312–315. doi: 10.3343/alm.2012.32.4.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams DR, Eng CM. Next-generation sequencing to diagnose suspected genetic disorders. N Engl J Med. 2018;379:1353–1362. doi: 10.1056/NEJMra1711801. [DOI] [PubMed] [Google Scholar]
- 4.Kim JH, Kang E, Heo SH, Kim GH, Jang JH, Cho EH, et al. Diagnostic yield of targeted gene panel sequencing to identify the genetic etiology of disorders of sex development. Mol Cell Endocrinol. 2017;444:19–25. doi: 10.1016/j.mce.2017.01.037. [DOI] [PubMed] [Google Scholar]
- 5.Kim YM, Lee YJ, Park JH, Lee HD, Cheon CK, Kim SY, et al. High diagnostic yield of clinically unidentifiable syndromic growth disorders by targeted exome sequencing. Clin Genet. 2017;92:594–605. doi: 10.1111/cge.13038. [DOI] [PubMed] [Google Scholar]
- 6.Nam SH, Hong YB, Hyun YS, Nam E, Kwak G, Hwang SH, et al. Identification of genetic causes of inherited peripheral neuropathies by targeted gene panel sequencing. Mol Cells. 2016;39:382–388. doi: 10.14348/molcells.2016.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko A, Youn SE, Kim SH, Lee JS, Kim S, Choi JR, et al. Targeted gene panel and genotype-phenotype correlation in children with developmental and epileptic encephalopathy. Epilepsy Res. 2018;141:48–55. doi: 10.1016/j.eplepsyres.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Rim JH, Kim SH, Hwang IS, Kwon SS, Kim J, Kim HW, et al. Efficient strategy for the molecular diagnosis of intractable early-onset epilepsy using targeted gene sequencing. BMC Med Genomics. 2018;11:6. doi: 10.1186/s12920-018-0320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han JY, Jang JH, Park J, Lee IG. Targeted next-generation sequencing of Korean patients with developmental delay and/or intellectual disability. Front Pediatr. 2018;6:391. doi: 10.3389/fped.2018.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rim JH, Lee ST, Gee HY, Lee BJ, Choi JR, Park HW, et al. Accuracy of next-generation sequencing for molecular diagnosis in patients with infantile nystagmus syndrome. JAMA Ophthalmol. 2017;135:1376–1385. doi: 10.1001/jamaophthalmol.2017.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J, Ha S, Lee ST, Park SG, Shin S, Choi JR, et al. Next-generation sequencing in Korean children with autism spectrum disorder and comorbid epilepsy. Front Pharmacol. 2020;11:585. doi: 10.3389/fphar.2020.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewans LJ, Schofield D, Shrestha R, Zhu Y, Gayevskiy V, Ying K, et al. Whole-exome sequencing reanalysis at 12 months boosts diagnosis and is cost-effective when applied early in Mendelian disorders. Genet Med. 2018;20:1564–1574. doi: 10.1038/gim.2018.39. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 17.Meienberg J, Bruggmann R, Oexle K, Matyas G. Clinical sequencing: is WGS the better WES? Hum Genet. 2016;135:359–362. doi: 10.1007/s00439-015-1631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall CR, Bick D, Belmont JW, Taylor SL, Ashley E, Dimmock D, et al. The Medical Genome Initiative: moving whole-genome sequencing for rare disease diagnosis to the clinic. Genome Med. 2020;12:48. doi: 10.1186/s13073-020-00748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lionel AC, Costain G, Monfared N, Walker S, Reuter MS, Hosseini SM, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med. 2018;20:435–443. doi: 10.1038/gim.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostrander BEP, Butterfield RJ, Pedersen BS, Farrell AJ, Layer RM, Ward A, et al. Whole-genome analysis for effective clinical diagnosis and gene discovery in early infantile epileptic encephalopathy. NPJ Genom Med. 2018;3:22. doi: 10.1038/s41525-018-0061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brewer MH, Chaudhry R, Qi J, Kidambi A, Drew AP, Menezes MP, et al. Whole genome sequencing identifies a 78 kb insertion from chromosome 8 as the cause of Charcot-Marie-Tooth neuropathy CMTX3. PLoS Genet. 2016;12:e1006177. doi: 10.1371/journal.pgen.1006177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scocchia A, Wigby KM, Masser-Frye D, Del Campo M, Galarreta CI, Thorpe E, et al. Clinical whole genome sequencing as a first-tier test at a resource-limited dysmorphology clinic in Mexico. NPJ Genom Med. 2019;4:5. doi: 10.1038/s41525-018-0076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaz-Drago R, Custódio N, Carmo-Fonseca M. Deep intronic mutations and human disease. Hum Genet. 2017;136:1093–1111. doi: 10.1007/s00439-017-1809-4. [DOI] [PubMed] [Google Scholar]
- 24.Di Scipio M, Tavares E, Deshmukh S, Audo I, Green-Sanderson K, Zubak Y, et al. Phenotype driven analysis of whole genome sequencing identifies deep intronic variants that cause retinal dystrophies by aberrant exonization. Invest Ophthalmol Vis Sci. 2020;61:36. doi: 10.1167/iovs.61.10.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larrue R, Chamley P, Bardyn T, Lionet A, Gnemmi V, Cauffiez C, et al. Diagnostic utility of whole-genome sequencing for nephronophthisis. NPJ Genom Med. 2020;5:38. doi: 10.1038/s41525-020-00147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]