Abstract
Natural killer cells constitute a phenotypically diverse population of innate lymphoid cells with a broad functional spectrum. Classically defined as cytotoxic lymphocytes with the capacity to eliminate cells lacking self‐MHC or expressing markers of stress or neoplastic transformation, critical roles for NK cells in immunity to infection in the regulation of immune responses and as vaccine‐induced effector cells have also emerged. A crucial feature of NK cell biology is their capacity to integrate signals from pathogen‐, tumor‐ or stress‐induced innate pathways and from antigen‐specific immune responses. The extent to which innate and acquired immune mediators influence NK cell effector function is influenced by the maturation and differentiation state of the NK cell compartment; moreover, NK cell differentiation is driven in part by exposure to infection. Pathogens can thus mould the NK cell response to maximise their own success and/or minimise the damage they cause. Here, we review recent evidence that pathogen‐ and vaccine‐derived signals influence the differentiation, adaptation and subsequent effector function of human NK cells.
Keywords: differentiation, malaria, NK cells, vaccines, viruses
Natural killer (NK) cells integrate signals from both innate and adaptive immune components to act as regulatory and effector cells in the immune response to pathogens. Here, we review how distinct viruses, malaria parasites and emerging infections interact with NK cells and influence the differentiation of NK cells and the consequences for their phenotypic and functional characteristics.

Human NK cell differentiation – straight from innate to adaptive?
NK cells differentiate from haematopoietic bone marrow precursors under the influence of innate cytokines and stromal cell‐derived factors and, in humans, are defined by the expression of CD56, whilst lacking CD3epsilon. It has been estimated that there are more than 100 000 distinct NK cell phenotypes in human peripheral blood alone 1 ; lymphoid and tissue‐resident and migratory NK cells add to this diversity. 2 The distribution of these different NK cell subsets in the circulation, their tissue residence or their capacity to home to different tissues undoubtedly influences their capacity to respond to damaged, cancerous or infected cells.
Initial observations pointed to a potential differentiation pathway for peripheral blood NK cells in which cells expressing high levels of CD56 were considered as less differentiated precursors of cells with lower CD56 expression. 3 Several important pieces of evidence support this broad phenotypic characterisation including the higher intrinsic proliferative capacity and greater telomere length of CD56bright NK cells compared to CD56dim NK cells. 4 , 5 , 6 However, any relationship between CD56bright and CD56dim NK cells is unlikely to be direct and the exact relationship remains uncertain. A recent single‐cell transcriptomic analysis indicated the presence of eight major differentiation phenotypes of peripheral blood of NK cells; some of these have the potential to interconvert in response to cytokine signals whilst others may represent more terminally differentiated lineages. 7 More recently, cord blood ILC‐1‐like precursors were shown to differentiate into CD56dimKIR+NKG2A−Peforin+GzB+ NK cells on OP9 stromal cells in the presence of interleukin (IL)‐2, IL‐7 and IL‐15. 8 However, NK cells derived under these conditions contained relatively low frequencies of CD16+ NK cells. 8
CD56bright NK cells express high levels of IL‐12R, IL‐15R and IL‐18R enabling them to respond very efficiently to these cytokines. CD56dim NK cells express lower levels of these receptors, instead expressing markers associated with more advanced differentiation (CD57), functional education in the context of HLA‐ligands [Killer cell immunoglobulin‐like receptors (KIR)], antibody‐dependent activation (CD16 and CD32) and cytotoxicity (granzyme B and perforin). 9 However, these different differentiation states do not necessarily correlate with a cell's ability to integrate innate or adaptive signals per se. A subset of CD56bright NK cells, for example, expresses the high‐affinity IL‐2 receptor heterodimer (CD25/CD122), responds to picogram concentrations of IL‐2 and interacts with CD4+ T cells in secondary lymphoid tissues, suggesting that these cells amplify the responses of antigen‐specific memory T cells. 10 However, CD56dim NK cells that express high levels of FcγRIII (CD16) could be viewed as adapted for integration of antibody‐dependent signals but also acquire the high‐affinity IL‐2R on activation. 11 Furthermore, detailed examination of transcription factor and transmembrane signalling adaptor expression of human blood NK cells reveals a polarisation of CD56dim NK cell functional phenotypes from ‘canonical’ cells (expressing the proteomyelocytic zinc finger (PLZF) molecule) to ‘adaptive’ NK cells (lacking the expression of this transcription factor). 12 , 13 Reduced expression of PLZF is associated with loss of FcεR1γ and associated signalling components including Syk and EAT‐2 and alternative signalling via the CD3zeta chain. 12 , 13
Paradoxically, in vitro cross‐linking of NKG2C combined with long‐term IL‐15 stimulation not only induces expansion of highly differentiated ‘adaptive’ CD56dimCD57+NKG2C+ NK cells but also promotes their co‐expression of NKG2A and the checkpoint inhibitors PD1 and LAG3 and induces trans‐differentiation from CD45RA to CD45RO isoform expression. 14 Hence, chronic stimulation may promote further differentiation of NK cells. Interestingly, CD56bright NK cells and a subset of CD56dim NK cells also lack PLZF and FcεR1γ suggesting that there is significant plasticity in signalling pathways across distinct NK cell differentiation stages. 13 In many ways, this plasticity of NK cell differentiation and adaptation should not be surprising considering the role of epigenetic changes in the diversification process. ‘Adaptive’ NK cells undergo gene silencing via methylation of loci associated with expression and responsiveness to innate cytokines whereas demethylation promotes a lower threshold for activation via alternative pathways including at the IFN‐γ locus. 13
NK cell differentiation is driven, in part, by HCMV
The spectrum of NK cell differentiation varies between individuals reflecting, in part, their prior infection history. 7 Pathogens express immune‐activating molecules (pathogen‐associated molecular patterns) or induce tissue damage‐associated ‘danger’ signals that contribute to NK cell activation either directly or via cytokines produced by intermediary ‘accessory’ cells such as monocytes and macrophages. The phenotypic and functional repertoire of NK cells responding to a distinct pathogen, and the magnitude of that response, will therefore vary according to the strength and duration of these pathogen‐derived signals.
A role for infection in driving the differentiation and functional adaptation of human NK cells is particularly well documented for human cytomegalovirus (HCMV). Expansions of NK cells expressing CD57 and NKG2C (a receptor recognising cognate HLA‐E stabilised on HCMV‐infected cells by peptides from the UL40 viral protein) and lacking PLZF and FcεR1γ are frequently found in bone marrow transplant patients experiencing HCMV infection or reactivation and are elevated in frequency in HCMV seropositive compared to seronegative individuals. 12 However, expansions of NK cells lacking FcεR1γ, EAT‐2 and Syk1 but with intermediate levels of CD57 expression are also observed in HCMV seronegative individuals, highlighting the potential for peripheral NK cell differentiation to be driven by other mechanisms. 15 The potential for a non‐linear, HCMV‐independent pathway for functional NK cell diversification is further supported by the observation of dynamic, intra‐individual fluctuations in the frequencies of these cells over a protracted period of time in HCMV seronegative individuals. 15
HCMV‐infected myeloid cells and fibroblasts are in many ways the archetypic example of pathogen‐induced NK cell activation and differentiation, both directly via binding of HCMV‐induced ligands on infected host cells to NK cell surface receptors including NKG2C, LILRB1 and activating KIR and indirectly through the action of pro‐inflammatory cytokines, interleukin‐2 and antibodies emanating from other immune cells. 16 , 17
Human ‘adaptive’ NK cells (defined by the expression of NKG2C or loss of PLZF/Fcεr1γ associated pathways and induced by cognate interaction between NKG2C and HLA‐E on HCMV‐infected cells) share many features of murine ‘memory’ NK cells induced by binding of the murine cytomegalovirus (MCMV) m157 protein to NK Ly49h receptors. 18 In both cases, NK cell expansion and differentiation are supported by accessory cell secretion of IL‐12 and leads to generation of effector cells with superior killing activity and control of CMV infection. 18 , 19 Additionally, ‘cytokine‐induced‐memory‐like’ (CIML) NK cells can be generated in vitro in both humans and mice with combinations of accessory cytokines (IL‐12, IL‐18 and IL‐15); these cells respond to subsequent in vitro stimulation by enhanced cytokine secretion, and this enhanced activity is retained after adoptive transfer. 20 , 21 CIML NK cells are distinguishable from human adaptive NK cells and murine memory NK cells in their lack of specificity for individual pathogens and their independence from cognate CMV‐NKG2C/Ly49h interactions (reviewed in Pahl et al. 22 ).
In the ‘real world’ of complex individual infection histories, these categorisations may be less clear with adaptive/memory cells acquiring additional CIML‐like properties after in vivo exposure to acute infection, inflammation or vaccination. CIML NK cells are likely to be further expanded and differentiated by subsequent CMV infection. Nevertheless, the dominant role of HCMV in promoting the differentiation and functional adaptation of human NK cells (especially after infection in early life) has the potential to directly influence responses to third party infections. HCMV‐associated ‘adaptive’ NK cells in humans have enhanced capacity for antibody‐mediated activation and killing of infected target cells (antibody‐dependent cellular cytotoxicity, ADCC) 12 , 13 , 18 although their long‐term fate during HCMV infection or reactivation may ultimately depend on the presence or absence of co‐stimulatory signals.
Endemicity of HCMV infection is greatly influenced by environment, for which geographical location and ethnicity have been used as proxies. Generally speaking, HCMV seroprevalence increases with equatorial proximity, with the highest prevalence being reported in sub‐Saharan Africa. 23 Rates of HCMV infection in infants and children are also highest in these settings with between 83% and 98% of individuals becoming infected by the age of 14 years. 23 In a UK population, higher seroprevalence of HCMV (and, incidentally, herpes simplex virus, HSV) was observed in children of Asian heritage than amongst children of white British heritage raising the possibility that immune differentiation, and thus disease susceptibility or presentation, may vary amongst communities of differing ethnicity within the same geographical location. 24 NK cell differentiation occurs noticeably more rapidly (i.e. in younger age groups) in settings of high HCMV endemicity with, for example, maximal expansion of CD57+NKG2C+ and CD57+NKG2A+ NK cells being reached by 10 years of age in an African population with near universal HCMV infection in the first year of life. 25 This is accompanied by loss of NK cell responsiveness to innate cytokines but maintenance of robust antibody‐dependent activation 25 and emergence of high frequencies of ‘adaptive’ FcεR1γ−CD56dim NK cells in childhood and early adulthood. 26 , 27 Similar effects are observed in temperate areas of the northern hemisphere, but over a more protracted period, with advanced NK cell differentiation mostly occurring later in life in these lower HCMV seroprevalence settings. 28 , 29 , 30
The role of other pathogens in NK cell differentiation
Whilst HCMV infection is associated with robust NK cell differentiation and adaptation, perhaps setting the stage for subsequent responses, other pathogens also influence the expansion and differentiation of particular NK cell subsets. In experimental settings, several examples have emerged of direct activation of NK cells by binding of pathogen‐encoded ligands to either invariant or polymorphic NK cell receptors. One recent, well‐characterised example is that of a conserved peptide epitope from bacterial recombinase A (rec A) that is presented by HLA‐C*0501 and recognised by the human NK cell receptor KIR2DS4 enabling NK cell activation by diverse bacteria including Brucella, Campylobacter, Chlamydia and Helicobacter species. 31 Similarly, a highly conserved epitope in the helicase of flaviviruses is presented by HLA‐C*0102 and binds to KIR2DS2, inducing NK cell degranulation and cytotoxicity 32 and several HIV‐derived peptides have been identified that either provoke or inhibit NK cell responses in the context of KIR‐HLA interactions. 33 , 34 By contrast, the fungal pathogen Aspergillus fumigatus is reported to bind directly to NK cell CD56, resulting in NK cell activation, beta chemokine production and a reduction in CD56 expression. 35 Whilst A. fumigatus appears to interact with both CD56bright and CD56dim NK cell subsets, fungal activation reportedly affects expression of other receptors which may be unevenly distributed across the NK cell differentiation spectrum, including NKG2D and NKp46. 36 Furthermore, A. fumigatus‐derived signals synergise with indirect, dendritic cell‐derived signals to activate, and prevent exhaustion of, less differentiated NK cell subsets. 37 Direct interaction of influenza A virus haemagglutinin with NKp46, NTB‐A and 2B4 has also been implicated in the direct activation of NK cells. 38
Certain viruses have the capacity to directly infect human NK cells, with evidence of tropism for and impact on NK cell differentiation phenotype. Varicella‐Zoster virus (VZV), for example, targets CD56dim NK cells, resulting in acquisition of CD57 but, paradoxically, reducing surface CD16 expression despite the absence of VZV‐specific antibodies in the culture system. 39 Less differentiated NK cells can be infected during chronic active Epstein–Barr virus infection 40 : EBV‐infected CD56dim NK cells (with CD2+CCR7−CD11a+CD11b−NKG2A+NKG2C−NKG2D–CD57− phenotype) exhibit increased STAT1 phosphorylation and Akt activation with consequences for viral latency. 40
Evidence is also accumulating of indirect changes in NK cell phenotype and function in people actively infected with a variety of, mostly viral, infections (summarised in Table 1); potential points of interaction between distinct pathogens and different NK cell differentiation subsets are summarised in Figure 1. In many cases, these infections seem to augment the effects of underlying HCMV infection, although data on HCMV infection are lacking in some studies (Table 1). For example, many of the effects of chronic HIV‐1 infection on NK cells are indistinguishable from those associated with recurrent infection/reactivation of HCMV including loss of CD56bright cells and emergence of CD57+NKG2C+FcεR1γ− cells. 41 , 42 , 43 Indeed, reversal of NKG2A/NKG2C frequencies over a 24‐month period of antiretroviral therapy likely reflects enhanced immune control of HCMV infection. 44 Accelerated acquisition of highly differentiated, ‘adaptive’ NK cell phenotypes is observed in HIV‐1‐infected individuals in Europe but these effects are less marked in HIV‐1+ Africans amongst whom NK cell differentiation is typically already well advanced due to HCMV infection early in life. 45 However, persistent untreated HIV‐1 infection further extends NK cell differentiation with accumulation of CD56− NKG2C+ cells. 46 Proteomic analysis suggests that these cells emerge from a CD56dim precursor population 47 but it as yet unclear whether their differentiation is driven by HIV‐1 infection itself (with or without HCMV co‐infection), whether opportunistic infections (including with fungal pathogens such as A. fumigatus) also drive this NK cell differentiation, 35 or whether the activating signals are direct (pathogen ligands binding to NK cell receptors) or indirect (e.g. mediated by inflammatory cytokines).
Table 1.
Impacts of active infections on human NK cell differentiation
| Pathogen | Impact on NK cell differentiation | References |
|---|---|---|
| Dengue virus | Robust proliferation across the differentiation spectrum although CD56bright and less differentiated CD56 NK cells dominate, emergence of skin homing CLA+ NK cell phenotype | 54 |
| Ebola virus | Early acute reduction in CD56bright NK cells precedes proliferation of both CD56bright and CD56dim subsets. Emergence of CD56−CD16+ NK cells persisting after EVD recovery | 69, 71 |
| Hantavirus | IL‐15 and HLA‐E dependent expansions of CD57+NKG2C+ NK cells. High proportion of HCMV co‐infected individuals | 52 |
| Hepatitis C virus | Redistribution of CD56bright and CD56dim cell subsets in both acute infection and self‐resolving infections. Reduced CD56dim NK cell frequencies and expansion of CD56−CD16+ NK cells. Expansion of CD57+PD‐1+ NK cells. HCMV co‐infection not reported | 55, 56, 58 |
| HIV‐1 | Reduced frequencies of CD56bright and expansion of CD56dimCD57+NKG2C+ NK cells and adaptive NK cells in chronic infection. Increased frequencies of CD56−CD16+ NK cells. Partial resolution after treatment. Likely role of HCMV co‐infection | 41, 42, 43, 44, 45, 46 |
| Influenza virus | Acute infection is associated with reduced CD56bright NK cell frequencies. Activation and proliferation across the NK cell differentiation spectrum. Emergence of CD49α+CD16−CXCR3+ with lung homing capacity | 48, 49 |
| Malaria | Increased frequencies of activated NKp30+ cells across differentiation spectrum in CHIM with CD56dim subsets dominating the responses. Emergence of CD38dimHLA‐DR+CD45RO+ NK cells in sickle cell trait with low parasitaemia. Role of HCMV co‐infection not reported | 60, 61 |
| Mycobacterium | Enrichment of CD45RO+ NK cells in pleural effusions of pulmonary tuberculosis patients | 64, 65 |
| SARS‐CoV‐2 | Activation of CD56bright NK cells early in infection. High frequency and increased proliferation of CD56dimNKG2C+Ksp37+ associated with disease severity. Adaptive expansions present in a higher proportion of HCMV+ patients with severe COVID‐19 compared to control or mild disease. Enrichment of CD56dimCD57+ NK cells in ARDS | 74, 78 |
Figure 1.
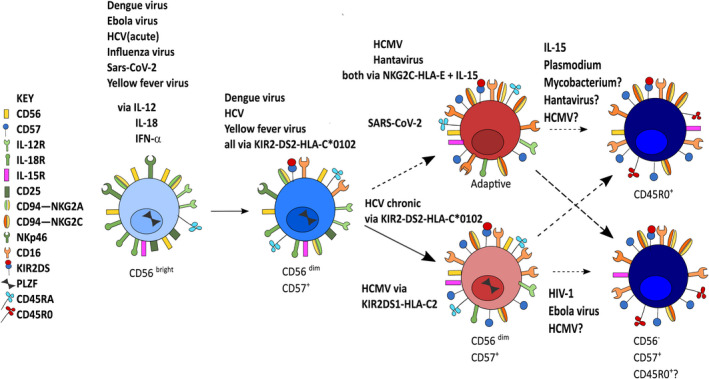
Schematic summary of the natural killer cell differentiation pathway with potential points of interaction with different pathogens. Many viral pathogens induce innate cytokines and type 1 interferons which activate and expand less differentiated CD56bright and CD56dimCD57− NK cells during early/acute infection. Viral pathogens also utilise direct mechanisms including via activating KIR‐HLA interactions, in some cases synergising with IL‐15. HCMV promotes expansion of adaptive NK cells, forming a potential template for the role of persistent viral infections and chronic or recurrent infections (e.g. by malaria parasites) to promote further expansion of adaptive NK cells and/or their terminal differentiation, as defined by the loss of CD56 (CD56−) and/or the expression of CD45RO.
Influenza infection is associated with reduced frequencies of CD56bright NK cells, coincident with NK cell production of antiviral and inflammatory cytokines, 48 as well as upregulation of activation/functional markers (CD69, CD38 and granzyme B) and a tendency for increased proliferation of both CD56bright and CD56dimCD16+ NK cells. 49 Crucially, a small population of CD49α+CD16−CXCR3+ NK cells with potential lung homing capacity has been identified during acute influenza infection and equivalent cells isolated from lung tissue have been shown to have antiviral activity in vitro. 49 Importantly, many of these influenza‐induced phenotypic changes are short‐lived, with reversal to pre‐infection characteristics during convalescence, compatible with redistribution and regulation of the NK cell response. 49
The importance of infection in promoting activation of relatively undifferentiated, tissue‐resident NK cells and migration to distinct anatomical sites is also evident in recall responses to VZV and hepatitis B viruses. 50 , 51 NK cells with characteristics of liver‐resident NK cells (CD56highCXCR6+CD16+NKG2D+CD69+CD62L+) were enriched in skin blisters after challenge with VZV skin test antigen, 50 and CD49α+CD16− human liver NK cells acquire epigenetic modifications and migrate to the skin after hepatitis B vaccination. 51
Hantavirus infection leads to rapid and persistent in vivo expansion of NK cells across the differentiation spectrum including CD57+NKG2C+ NK cells. 52 Such expansions are associated with elevated levels of IL‐15 and with the Hantavirus‐driven upregulation of HLA‐E on infected cells. Furthermore, over 80% of infected individuals were HCMV seropositive in the study, consistent with the possibility that Hantavirus further expands cells initially expanded by HCMV infection. 52 In this context, the activation of more differentiated NK cells is supported by hantavirus‐induced upregulation of IL‐15‐IL‐15R on epithelial cells. 53
Acute dengue virus infection is associated with robust proliferation of all NK cell subsets, irrespective of their differentiation status, although CD56bright and CD56dimCD57− NK cells dominate the response; this is associated with increased IL‐18 concentrations in plasma (and in experimentally induced skin blister fluid) and activation of IL‐18‐induced signalling pathways. 54 In this case, the cells preferentially homing to the skin were CD56bright and expressed a unique combination of chemokine receptors, including CLA‐1 which is typically associated with skin homing. 54
Acute hepatitis C virus (HCV) infection leads to increased blood frequencies of CD56bright cells, and correspondingly decreased frequencies of CD56dim NK cells, although both subsets of NK cells appear activated. 55 However, NK cell frequencies subsequently normalised in patients who went on to clear their infections and in those who developed chronic infection and no long‐term impacts on NK cells were reported. 55 Similarly, another study reported increased frequencies of CD56− NK cells and concomitantly reduced frequencies of CD56dim, NKG2D+, NKp30+ and NKp46+ NK cells during HCV infections. 56 Importantly, frequencies of NKG2A+ and CD94+ NK cells were increased in acute and chronic infection but not in those whose infections resolved, whereas frequencies of NKp30+, NKp46+ and NKG2D+ cells were lower in those who resolved their infections than in those who became chronically infected. 56 Consistent with these observations, a recent study of HCMV seropositive patients reported that high frequencies of adaptive CD57+FcεR1γ− NK cells and elevated expression of PD‐1 were associated with high viral load in chronic HCV infection and were reduced after direct‐acting antiviral therapy (DAA). 57 In a separate study, chronic HCV infection was similarly associated with higher frequencies of CD57+ and PD‐1+ NK cells compared to uninfected control individuals. 58 However, although PD1 expression frequency reduced after DAA, proportions of CD57+ and KLRG1+ cells increased after treatment, suggesting ongoing expansion of highly differentiated NK cells. 58 The slightly different NK cell outcomes in these two studies may be explained by differences in HCMV status, HCV viral genotype, viral load and treatment regimen. 57 , 58
Impacts of chronic, persistent or recurrent infection on NK cell differentiation – evidence from malaria and tuberculosis
Human malaria infections provide insights into the impacts of chronic and repeated infection on the NK cell compartment. Blood stage malaria parasites can induce high concentrations of inflammatory and NK cell‐activating cytokines with associated NK cell activation seen in animal models and human co‐culture systems in vitro. 59 Experimental, controlled human malaria infections (CHMI) have given valuable insights into the activation of NK cells during primary infection in malaria‐naïve individuals. Surprisingly, despite the known role for inflammatory cytokines (IL‐12, IL‐18) in activating less differentiated NK cells, the primary acute response during CHMI is characterised by activation (CD69 expression) of NKp30+ NK cells distributed across the differentiation spectrum but with CD56dim cells dominating over CD56bright cells 60 ; interestingly, these expansions were closely linked to the course of parasitaemia and resolved after treatment. 60 It is likely that both expansion and homing of activated CD56bright NK cells to – and their retention in – secondary lymphoid tissues contribute to redistribution of NK cell subsets during active malaria infection.
The potential for chronic or repeated malaria infections to influence NK cell phenotype and function is illustrated by the identification of a novel, activated CD38+HLA−DR+CD45RO+ NK cell population in individuals with sickle cell trait who have persistent low‐density parasitaemia. 61 A possible explanation for this is that chronic in vitro stimulation of NK cells via NKG2C in combination with IL‐15 induces CD45 isoform switching on CD57+NKG2C+ cells, resulting in a CD45RO+ population with high proliferative potential, characteristic of central memory T cells. 14 Increased expression of checkpoint inhibitors including TIM3 and PD‐1 has also been observed after repeated malaria exposure with increasing age in endemic populations, 62 again with parallels to the emergence of LAG3 and PD1+ adaptive NK cells after activating receptor (NKp30, NKG2D, NKG2C) or HCMV‐mediated activation in vitro. 14 Interestingly, a CD25+CD45RO+ NK cell population has been identified in patients with metastatic melanoma treated with anti‐PD‐1 62 , 63 further supporting a potential for progressive differentiation of NK cells in a manner analogous to that seen in T cells. There is also evidence that persistent Mycobacterium tuberculosis infections, and the associated chronic inflammatory response, lead to the emergence of CD45RO+ NK cells and enrichment of these cells in the pleural effusion fluid of pulmonary tuberculosis patients. 64 These cells are potent producers of both IL‐22 and IFN‐γ that are implicated, respectively, in tissue repair and inflammation. 65 Whether these NK cell expansions rely on prior infection or co‐infection with other pathogens is unknown although there is some evidence that the course of tuberculosis can be modulated by HCMV co‐infection. 66 , 67
Emerging pathogens and NK cells
As described above, an abundance of experimental and observational studies suggests a role for infections, especially repeated or persistent parasite or viral infections, in gradual and extensive NK cell differentiation. Furthermore, studies of emerging pathogens are providing additional insights into the relative impacts of innate and adaptive immune pathways in inducing NK cell differentiation. Notably, severe systemic inflammation is a hallmark of severe Ebola and SARS‐CoV‐2 (COVID‐19) disease; this inflammation may in turn induce expansion and differentiation of NK cells with potential long‐term consequences.
Ebola virus is an example of a novel pathogen with only very limited adaptation to a human host. Ebola virus‐induced overproduction of the NK cell‐activating inflammatory cytokines IFN‐α2 and IL‐18, insufficiently counterbalanced by anti‐inflammatory cytokines (IL‐10), is associated with poor outcomes in Ebola virus disease (EVD). 68 During EVD, there is a rapid reduction in the frequency of CD56bright NK cells, within days of infection, and a subsequent increase in the frequencies of proliferating CD56bright and CD56dim cells. 69 In vitro, Ebola virus glycoprotein directly induces the secretion of both pro‐inflammatory cytokines (including IL‐18 which contributes significantly to NK cell activation, degranulation and IFN‐γ production) and anti‐inflammatory IL‐10 (which restricts these responses). 70 In this scenario, NK cell responses are dominated by CD56bright and CD56dimCD57− cells, raising the possibility that these less differentiated, cytokine‐producing cells may act to further amplify the inflammatory cascade (and thus disease severity) in vivo. Interestingly, increases in the frequency of CD56−CD16+ NK cells are observed up to 2 years after recovery from severe EVD, suggestive of persistent activation and/or terminal differentiation from CD56dimCD16+ NK cells. 47 , 71 However, in experimental studies in mice, adoptive transfer of dendritic cells exposed to virus‐like particles expressing the Ebola virus glycoprotein can prime NK cells to protect against subsequent lethal Ebola virus infection, raising the possibility that NK cell activation by direct binding of a viral ligand together with cytokine‐mediated co‐stimulation could induce protective NK cell effector mechanisms. 72 Indeed, human NK cells expressing NKp30 have been implicated in the cytotoxic response to Ebola virus after interaction with infected dendritic cells. 73
More recently, NK cells have been implicated in both protection against and pathogenesis of SARS‐CoV‐2 infection (COVID‐19). As in other viral infections, and irrespective of outcome in terms of disease, innate inflammatory pathways are triggered early in SARS‐CoV‐2 infection. CD56bright NK cells are activated, and their increased expression of perforin and granzyme B is associated with inflammatory markers of disease severity, in particular with high concentrations of IL‐6. 74 In addition, reduced expression of CD16 on CD56dim NK cells in people with COVID‐19 75 is consistent with ongoing activation of this subset by SARS‐CoV‐2 antigen/antibody immune complexes. 76 Importantly, HCMV infection and the associated expansion of the subset of adaptive CD56dimNKG2C+ NK cells were associated with severe COVID‐19. Over 90% of severe COVID‐19 cases in this cohort were HCMV+ (compared with ~60% of controls and 80% of mild COVID‐19 cases). 74 More importantly, ~65% of people with severe COVID‐19 had expansion of adaptive NK cells compared with only 10–20% of controls and those with moderate disease. 74 Whether these expansions of adaptive NK cells in HCMV+ people were driven by SARS‐CoV‐2, or whether they predated SARS‐CoV‐2 infection, is not known but these data do raise the possibility that adaptive NK cells may contribute to tissue damage in COVID‐19. In support of this hypothesis, in COVID‐19 patients these adaptive cells also express Ksp37, a marker of cytotoxic lymphocytes associated with lung disease, 77 and, despite their lower intrinsic proliferative capacity, also express markers of recent proliferation. 74 This is reminiscent of the increased proliferative capacity of adaptive CD45RO+ NK cells in chronic malaria infection as discussed above 61 , 74 and is also consistent with reports of enrichment of highly differentiated CD56dimCD57+ NK cells with high proliferative capacity in the peripheral blood in a patient with SARS‐CoV‐2 infection and acute respiratory distress syndrome (ARDS). 78
A cluster of genes enriched in terminally differentiated CD8+ T cells and CD56dimCD57+ NK cells was downregulated in the blood of children with multisystem inflammatory syndrome after SARS‐CoV‐2 infection 79 suggesting that there is no straightforward relationship between NK cell subset activation and disease severity. However, a causal relationship between activation of highly differentiated NK cells and severe COVID‐19 is, as yet, unproven and homing of less differentiated CD56dim NK cells to the tissues during severe disease may also contribute to alterations in cell frequencies in peripheral blood.
The expansion of CD56dimNKG2C+ NK cells in COVID‐19 patients also raises the possibility that increased expression of HLA‐E in conjunction with inflammatory mediators may mediate this adaptive NK cell expansion. 74 Indeed, a recent study suggested that a peptide derived from the SARS‐CoV‐2 spike 1 protein can stabilise HLA‐E on lung epithelial cells leading to activation of NKG2C+ NK cells. 80 Interestingly, an in silico study also identified SARS‐CoV‐2 peptides predicted to bind HLA‐C alleles recognising activating KIR. 81
Vaccination and NK cells
An indirect role for NK cells in regulating virus‐ or vaccine‐induced responses has been highlighted in several experimental systems. For example, NK cells have been shown to regulate CD4+ T‐cell responses and follicular T helper cell (Tfh) activity, thereby influencing the breadth and depth of the antibody response and effector CD8+ T‐cell response to vaccinia virus and LCMV infections in murine models. 82 , 83 , 84 Moreover, adaptive/highly differentiated NK cells, which express higher levels of the endosomal effector protein RAB111FIP5, impact on the generation of broadly neutralising antibodies in HIV‐1‐infected individuals. 85
However, mounting evidence also suggests that NK cell differentiation is also affected by vaccination (summarised in Table 2). Less differentiated CD56bright and CD56dimCD57− NK cells are activated after influenza virus vaccination 48 , 86 , 87 with evidence suggesting that common γ chain cytokines (IL‐2, IL‐12 and IL‐15) prime myeloid dendritic cells to secrete IL‐12 to support NK cell responses. 88 , 89 Post‐vaccination increases in the frequencies of these less differentiated CD56dimCD57− NK cells in HCMV+ individuals are consistent with IL‐2 family cytokines also supporting maintenance and expansion of less differentiated, cytokine‐responsive subsets. However, more differentiated CD56dimCD57+ cells are also enhanced post‐vaccination in the presence of influenza‐specific antibody rather than relying on cytokines for their activation. 76 In line with this, increased frequencies of CD56dimCD16+NKG2C+ (CD57− and CD57+) NK cells are observed up to 14 days after seasonal influenza vaccination in individuals with high titres of haemagglutination inhibiting antibodies. 90
Table 2.
Impacts of vaccination on NK cell differentiation
| Vaccine | Impact on NK cell differentiation subset | References |
|---|---|---|
| Ebola | Increased absolute numbers of CD56bright and CD56dim NK subsets. Increased Ki67 and CD25 expression in CD56bright NK cell | 70, 93 |
| HBsAg | Priming of CD56dimCD57+KLRG1+ NK cell subset revealed on in vitro restimulation | 91 |
| Influenza | Activation, proliferation or expansion of CD56bright and CD56dimCD57− NK cells. Priming of CD56bright and CD56dimCD57− NK cells for enhanced response to cytokines. Increased frequencies of CD56dimCD16+NKG2C+ (both CD57− and CD57+) NK cells in individuals with high HAI titres after vaccination | 48, 86, 90 |
| Yellow Fever | Proliferation and expansion of CD56bright and CD56dimCD57− subsets | 92 |
A recent study demonstrated increased cytotoxic and proliferative responses to antigen‐pulsed monocyte‐derived dendritic cells by highly differentiated CD56dimCD57+KLRG1+ NK cells after hepatitis B subunit vaccination. 91 Interestingly, these responses did not require the presence of immune serum. Yellow fever vaccine 17D induces proliferation and expansion of less differentiated CD56bright and CD56dimCD57− NK cells; these cells also demonstrate enhanced responsiveness to in vitro restimulation with innate cytokines. 92 Similarly, increased absolute numbers of CD56bright and CD56dim NK cells are observed within 3 days of immunisation with the rVSV‐ZEBOV Ebola virus vaccine; increased NK cell expression of CXCR6 is an independent correlate of rVSV‐ZEBOV vaccine responsiveness whilst reduced frequencies of NKG2D+ and increased frequencies of NKp30 and KIR+ NK cells are inversely correlated with plasma cytokine and chemokine concentrations, indicating likely recirculation of NK cell subsets within 72 h of vaccination. 93 The Ad26.ZEBOV Ebola virus vaccine also activates CD56bright NK cells as assessed by the induction of CD25 and Ki67 expression up to 14 days after the second dose. 70 The extent to which different vaccine vectors, delivery platforms and adjuvating systems impact on NK cell differentiation will likely depend on their primary cellular tropism, interactions with pattern recognition receptors and downstream production of NK cell‐activating cytokines; this aspect of vaccination has as yet received very little attention.
There are also many examples of potent, Fc receptor‐mediated degranulation and cytokine production by more highly differentiated NK cells in response to vaccination‐induced antibody. 76 , 94 , 95 , 96 Moreover, subunit, virally vectored and whole viral vaccines, with or without chemical adjuvants, will provide distinct cytokine signatures for co‐stimulation of NK cell responses with type 1 interferons, IL‐15 and IL‐18, all described to support antibody‐dependent responses of adaptive NK cells. 27 , 95 , 97 , 98 However, it is not known whether these cytokine‐ and antibody‐mediated signals contribute to further differentiation of these particular NK cells.
Conclusions
Exposure to an infection, be it in early life, with a novel pathogen or after vaccination, is accompanied by bystander, accessory‐cell‐dependent proliferation and expansion of the ‘less differentiated’ pool of NK cells. Cumulative exposure to diverse pathogens, exacerbated by chronic or recurrent infection, can lead to progressive activation and expansion of ‘more differentiated’ NK cells working in tandem with the adaptive immune response. Cytokines from antigen‐specific T cells and pathogen‐specific antibodies amplify these NK cell responses, thereby co‐opting NK cells into the adaptive immune response. Important questions remain, however, as to how pathogens or vaccines influence NK cell differentiation and functional diversification and what this means for the ability of NK cells to then contribute to protection from, or susceptibility to, both infectious and non‐communicable diseases. Amongst these are the potential for maternal antibodies to protect the infant from infection by non‐neutralising mechanisms and to induce NK cell activation and differentiation in early life. Differentiation of CD3−CD16+ cells from cord blood ILC1‐like precursors provides an opportunity for antibody‐dependent effector cells to contribute to immune responses, perhaps earlier in life than hitherto appreciated. The extent to which NK cell subsets re‐equilibrate after a pathogen is controlled or eliminated, and the mechanisms whereby uncontrolled infections (e.g. Hantavirus, severe SARS‐CoV‐2) or chronic or repeated infections (e.g. hepatitis C and malaria) induce lasting effects on the NK cell repertoire, are other key issues that need further investigation (Figure 1). The role, and the longer‐term functional consequences, of IL‐15 in the expansion and terminal differentiation of adaptive NK cells also merits further investigation in the context of different pathogens. In summary, individual experiences of infection and vaccination over the life course will shape not only the adaptive immune cell population but also the NK cell population, with consequences for susceptibility to subsequent infections and, potentially, non‐communicable disease such as cancers as inflammatory conditions.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
Martin R Goodier: Conceptualization; Writing‐original draft; Writing‐review & editing. Eleanor M Riley: Conceptualization; Writing‐review & editing.
Acknowledgment
We thank Asia‐Sophia Wolf for assistance with preparing the figure.
Contributor Information
Martin R Goodier, Email: martin.goodier@lshtm.ac.uk.
Eleanor M Riley, Email: eleanor.riley@ed.ac.uk.
References
- 1. Horowitz A, Strauss‐Albee DM, Leipold M et al Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 2013; 5: 208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dogra P, Rancan C, Ma W et al Tissue determinants of human NK cell development, function, and residence. Cell 2020; 180: 749–763 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer‐cell subsets. Trends Immunol 2001; 22: 633–640. [DOI] [PubMed] [Google Scholar]
- 4. Chan A, Hong DL, Atzberger A et al CD56bright human NK cells differentiate into CD56dim cells: role of contact with peripheral fibroblasts. J Immunol 2007; 179: 89–94. [DOI] [PubMed] [Google Scholar]
- 5. Ouyang Q, Baerlocher G, Vulto I, Lansdorp PM. Telomere length in human natural killer cell subsets. Ann NY Acad Sci 2007; 1106: 240–252. [DOI] [PubMed] [Google Scholar]
- 6. Romagnani C, Juelke K, Falco M et al CD56brightCD16– killer Ig‐like receptor‐ NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol 2007; 178: 4947–4955. [DOI] [PubMed] [Google Scholar]
- 7. Smith SL, Kennedy PR, Stacey KB et al Diversity of peripheral blood human NK cells identified by single‐cell RNA sequencing. Blood Adv 2020; 4: 1388–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bennstein SB, Weinhold S, Manser AR et al Umbilical cord blood‐derived ILC1‐like cells constitute a novel precursor for mature KIR+NKG2A– NK cells. Elife 2020; 9: e55232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Freud AG, Mundy‐Bosse BL, Yu J, Caligiuri MA. The broad spectrum of human natural killer cell diversity. Immunity 2017; 47: 820–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fehniger TA, Cooper MA, Nuovo GJ et al CD56bright natural killer cells are present in human lymph nodes and are activated by T cell‐derived IL‐2: a potential new link between adaptive and innate immunity. Blood 2003; 101: 3052–3057. [DOI] [PubMed] [Google Scholar]
- 11. White MJ, Nielsen CM, McGregor RH, Riley EH, Goodier MR. Differential activation of CD57‐defined natural killer cell subsets during recall responses to vaccine antigens. Immunol 2014; 142: 140–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee J, Zhang T, Hwang I et al Epigenetic modification and antibody‐dependent expansion of memory‐like NK cells in human cytomegalovirus‐infected individuals. Immunity 2015; 42: 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schlums H, Cichocki F, Tesi B et al Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 2015; 42: 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Merino A, Zhang B, Dougherty P et al Chronic stimulation drives human NK cell dysfunction and epigenetic reprograming. J Clin Invest 2019; 129: 3770–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gyurova IE, Schlums H, Sucharew H et al Dynamic changes in natural killer cell subset frequencies in the absence of cytomegalovirus infection. Front Immunol 2019; 10: 2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goodier MR, Jonjic S, Riley EM, Juranic LV. CMV and natural killer cells: shaping the response to vaccination. Eur J Immunol 2018; 48: 50–65. [DOI] [PubMed] [Google Scholar]
- 17. van der Ploeg K, Chang C, Ivarsson MA, Moffett A, Wills MR, Trowsdale J. Modulation of human leukocyte antigen‐C by human cytomegalovirus stimulates KIR2DS1 recognition by natural killer cells. Front Immunol 2017; 8: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Min‐Oo G, Lanier LL. Cytomegalovirus generates long‐lived antigen‐specific NK cells with diminished bystander activation to heterologous infection. J Exp Med 2014; 211: 2669–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rölle A, Pollmann J, Ewen E‐M et al IL‐12‐producing monocytes and HLA‐E control HCMV‐driven NKG2C+ NK cell expansion. J Clin Invest 2014; 124: 5305–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. Cytokine‐induced memory‐like natural killer cells. Proc Natl Acad Sci USA 2009; 106: 1915–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Romee R, Schneider SE, Leong JW et al Cytokine activation induces human memory‐like NK cells. Blood 2012; 120: 4751–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pahl JHW, Cerwenka A, Ni J. Memory‐like NK cells: remembering a previous activation by cytokines and NK cell receptors. Front Immunol 2018; 9: 2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bates M, Brantsaeter AB. Human cytomegalovirus (CMV) in Africa: a neglected but important pathogen. J Virus Erad 2016; 2: 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pembrey L, Waiblinger D, Griffiths P, Patel M, Azad R, Wright J. Cytomegalovirus, Epstein‐Barr virus and varicella zoster virus infection in the first two years of life: a cohort study in Bradford, UK. BMC Infect Dis 2017; 17: 2319–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goodier MR, White MJ, Darboe A et al Rapid NK cell differentiation in a population with near‐universal human cytomegalovirus infection is attenuated by NKG2C deletions. Blood 2014; 124: 2213–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hart GT, Tran TM, Theorell J et al Adaptive NK cells in people exposed to Plasmodium falciparum correlate with protection from malaria. J Exp Med 2019; 216: 1280–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sherratt S, Patel A, Baker DA, Riley EM, Goodier MR. Differential IL‐18 dependence of canonical and adaptive NK cells for antibody dependent responses to P. falciparum . Front Immunol 2020; 11: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Juelke K, Killig M, Luetke‐Eversloh M et al CD62L expression identifies a unique subset of polyfunctional CD56dim NK cells. Blood 2010; 116: 1299–1307. [DOI] [PubMed] [Google Scholar]
- 29. Björkström NK, Riese P, Heuts F et al Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK‐cell differentiation uncoupled from NK‐cell education. Blood 2010; 116: 3853–3864. [DOI] [PubMed] [Google Scholar]
- 30. Nielsen CM, White MJ, Bottomley C et al Impaired NK cell responses to pertussis and H1N1 influenza vaccine antigens in human cytomegalovirus‐infected individuals. J Immunol 2015; 194: 4657–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sim MJW, Rajagopalan S, Altmann DM, Boyton RJ, Sun PD, Long EO. Human NK cell receptor KIR2DS4 detects a conserved bacterial epitope presented by HLA‐C. Proc Natl Acad Sci USA 2019; 116: 12964–12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Naiyer MM, Cassidy SA, Magri A et al KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA‐C. Sci Immunol 2017; 2: eaal5296. [DOI] [PubMed] [Google Scholar]
- 33. van Teijlingen NH, Hölzemer A, Körner C et al Sequence variations in HIV‐1 p24 Gag‐derived epitopes can alter binding of KIR2DL2 to HLA‐C*03:04 and modulate primary natural killer cell function. AIDS 2014; 28: 1399–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ziegler MC, Nelde A, Weber JK et al HIV‐1 induced changes in HLA‐C*03: 04‐presented peptide repertoires lead to reduced engagement of inhibitory natural killer cell receptors. AIDS 2020; 34: 1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ziegler S, Weiss E, Schmitt A‐L et al CD56 is a pathogen recognition receptor on human natural killer cells. Sci Rep 2017; 7: 6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Santiago V, Rezvani K, Sekine T, Stebbing J, Kelleher P, Armstrong‐James D. Human NK cells develop an exhaustion phenotype during polar degranulation at the Aspergillus fumigatus hyphal synapse. Front Immunol 2018; 9: 2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weiss E, Ziegler S, Fliesser M et al First insights in NK‐DC cross‐talk and the importance of soluble factors during infection with Aspergillus fumigatus . Front Cell Infect Microbiol 2018; 8: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duev‐Cohen A, Bar‐On Y, Glasner A et al The human 2B4 and NTB‐A receptors bind the influenza viral hemagglutinin and co‐stimulate NK cell cytotoxicity. Oncotarget 2016; 7: 13093–13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Campbell TM, McSharry BP, Steain M, Ashhurst TM, Slobedman B, Abendroth A. Varicella zoster virus productively infects human natural killer cells and manipulates phenotype. PLoS Pathog 2018; 14: e1006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Howe MK, Dowdell K, Kuehn HS et al Patients with natural killer (NK) cell chronic active Epstein‐Barr virus have immature NK cells and hyperactivation of PI3K/Akt/mTOR and STAT1 pathways. J Infect Dis 2020; 222: 1170–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mela CM, Burton CT, Imami N et al Switch from inhibitory to activating NKG2 receptor expression in HIV‐1 infection: lack of reversion with highly active antiretroviral therapy. AIDS 2005; 19: 1761–1769. [DOI] [PubMed] [Google Scholar]
- 42. Gumá M, Cabrera C, Erkizia I et al Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV‐1‐positive patients. J Infect Dis 2006; 194: 38–41. [DOI] [PubMed] [Google Scholar]
- 43. Peppa D, Pedroza‐Pacheco I, Pellegrino P, Williams I, Maini MK, Borrow P. Adaptive reconfiguration of natural killer cells in HIV‐1 infection. Front Immunol 2018; 9: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brunetta E, Fogli M, Varchetta S et al Chronic HIV‐1 viremia reverses NKG2A/NKG2C ratio on natural killer cells in patients with human cytomegalovirus co‐infection. AIDS 2010; 24: 27–34. [DOI] [PubMed] [Google Scholar]
- 45. Cubero EM, Ogbe A, Pedroza‐Pacheco I et al Subordinate effect of ‐21M HLA‐B dimorphism on NK cell repertoire diversity and function in HIV‐1 infected individuals of African origin. Front Immunol 2020; 11: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gregson JN, Kuri‐Cervantes L, Mela CM, Gazzard BG, Bower M, Goodier MR. Short communication: NKG2C+ NK cells contribute to increases in CD16+CD56– cells in HIV type 1+ individuals with high plasma viral load. AIDS Res Hum Retroviruses 2013; 29: 84–88. [DOI] [PubMed] [Google Scholar]
- 47. Voigt J, Malone DFG, Dias J et al Proteome analysis of human CD56– NK cells reveals a homogeneous phenotype surprisingly similar to CD56dim NK cells. Eur J Immunol 2018; 48: 1456–1469. [DOI] [PubMed] [Google Scholar]
- 48. Jost S, Quillay H, Reardon J et al Changes in cytokine levels and NK cell activation associated with influenza. PLoS One 2011; 6: e25060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scharenberg M, Vangeti S, Kekäläinen E et al Influenza A virus infection induces hyperresponsiveness in human lung tissue‐resident and peripheral blood NK cells. Front Immunol 2019; 10: 1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nikzad R, Angelo LS, Aviles‐Padilla K et al Human natural killer cells mediate adaptive immunity to viral antigens. Sci Immunol 2019; 4: eaat8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stary V, Pandey RV, Strobl J et al A discrete subset of epigenetically primed human NK cells mediates antigen‐specific immune responses. Sci Immunol 2020; 5: eaba6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Björkström NK, Lindgren T, Stoltz M et al Rapid expansion and long‐term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med 2011; 208: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Braun M, Björkström NK, Gupta S et al NK cell activation in human hantavirus infection explained by virus‐induced IL‐15/IL15Rα expression. PLoS Pathog 2014; 10: e1004521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zimmer CL, Cornillet M, Solà‐Riera C et al NK cells are activated and primed for skin‐homing during acute dengue virus infection in humans. Nat Commun 2019; 10: 3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Amadei B, Urbani S, Cazaly A et al Activation of natural killer cells during acute infection with hepatitis C virus. Gastroenterology 2010; 138: 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Alter G, Jost S, Rihn S et al Reduced frequencies of NKp30+NKp46+, CD161+, and NKG2D+ NK cells in acute HCV infection may predict viral clearance. J Hepatol 2011; 55: 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mele D, Oliviero B, Mantovani S et al Adaptive natural killer cell functional recovery in hepatitis C virus cured patients. Hepatology 2020. 10.1002/hep.31273 [DOI] [PubMed] [Google Scholar]
- 58. Wijaya RS, Read SA, Selvamani SP et al Hepatitis C virus eradication with interferon free, DAA‐based therapy results in KLRG1+, hepatitis C virus‐specific memory natural killer cells. J Infect Dis 2020; jiaa492 10.1093/infdis/jiaa492 [DOI] [PubMed] [Google Scholar]
- 59. Goodier MR, Wolf AS, Riley EM. Differentiation and adaptation of natural killer cells for anti‐malarial immunity. Immunol Rev 2020; 293: 25–37. [DOI] [PubMed] [Google Scholar]
- 60. Walk J, Sauerwein RW. Activatory receptor NKp30 predicts NK cell activation during controlled human malaria infection. Front Immunol 2019; 10: 2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Loiseau C, Doumbo OK, Traore B et al A novel population of memory‐activated natural killer cells associated with low parasitaemia in Plasmodium falciparum‐exposed sickle‐cell trait children. Clin Transl Immunol 2020; 9: e1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Moebius J, Guha R, Peterson M et al PD‐1 expression on NK cells in malaria‐exposed individuals is associated with diminished natural cytotoxicity and enhanced antibody‐dependent cellular cytotoxicity. Infect Immun 2020; 88: e00711‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kasanen H, Hernberg M, Mäkelä S et al Age‐associated changes in the immune system may influence the response to anti‐PD1 therapy in metastatic melanoma patients. Cancer Immunol Immunother 2020; 69: 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fu X, Liu Y, Li LI et al Human natural killer cells expressing the memory‐associated marker CD45RO from tuberculous pleurisy respond more strongly and rapidly than CD45RO– natural killer cells following stimulation with interleukin‐12. Immunol 2011; 134: 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fu X, Yu S, Yang B, Lao S, Li B, Wu C. Memory‐like antigen‐specific human NK cells from TB pleural fluids produced IL‐22 in response to IL‐15 or Mycobacterium tuberculosis antigens. PLoS One 2016; 11: e0151721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Müller J, Tanner R, Matsumiya M et al Cytomegalovirus infection is a risk factor for tuberculosis disease in infants. JCI Insight 2019; 4: e130090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Parra J, Zúñiga M, Zamudio A et al Memory of Natural Killer Cells: A New Chance against Mycobacterium tuberculosis?. Front Immunol 2017; 8: 967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reynard S, Journeaux A, Gloaguen E et al Immune parameters and outcomes during Ebola virus disease. JCI Insight 2019; 4: e125106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. McElroy AK, Akondy RS, Mcllwain DR et al Immunologic timeline of Ebola virus disease and recovery in humans. JCI Insight 2020; 5: e137260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wagstaffe HR, Clutterbuck EA, Bockstal V et al Ebola virus glycoprotein stimulates IL‐18‐dependent natural killer cell responses. J Clin Invest 2020; 130: 3936–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wiedemann A, Foucat E, Hocini H et al Long‐lasting severe immune dysfunction in Ebola virus disease survivors. Nat Commun 2020; 11: 3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Warfield KL, Perkins JG, Swenson DL et al Role of natural killer cells in innate protection against lethal Ebola virus infection. J Exp Med 2004; 200: 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fuller CL, Ruthel G, Warfield KL et al NKp30‐dependent cytolysis of filovirus‐infected human dendritic cells. Cell Microbiol 2007; 9: 962–976. [DOI] [PubMed] [Google Scholar]
- 74. Maucourant C, Filipovic I, Ponzetta A et al Natural killer cell immunotypes related to COVID‐19 disease severity. Sci Immunol 2020; 5: eabd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kuri‐Cervantes L, Pampena MB, Meng W et al Comprehensive mapping of immune perturbations associated with severe COVID‐19. Sci Immunol 2020; 5: eabd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Goodier MR, Lusa C, Sherratt S, Rodriguez‐Galan A, Behrens R, Riley EM. Sustained immune complex‐mediated reduction in CD16 expression after vaccination regulates NK cell function. Front Immunol 2016; 7: 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kuepper M, Bratke K, Julius P et al Increase in killer‐specific secretory protein of 37 kDa in bronchoalveolar lavage fluid of allergen‐challenged patients with atopic asthma. Clin Exp Allergy 2005; 35: 643–649. [DOI] [PubMed] [Google Scholar]
- 78. Bouadma L, Wiedemann A, Patrier J et al Immune alterations in a patient with SARS‐CoV‐2‐related acute respiratory distress syndrome. J Clin Immunol 2020; 40: 1082–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Beckmann ND, Comella PH, Cheng E et al Cytotoxic lymphocytes are dysregulated in multisystem inflammatory syndrome in children. medRxiv 2020; 2020.08.29.20182899. 10.1101/2020.08.29.20182899 [DOI] [Google Scholar]
- 80. Bortolotti D, Gentili V, Rizzo S, Rotola A, Rizzo R. SARS‐CoV‐2 spike 1 protein controls natural killer cell activation via the HLA‐E/NKG2A pathway. Cells 2020; 9: 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Marchan J. Conserved HLA binding peptides from five non‐structural proteins of SARS‐CoV‐2‐An in silico glance. Hum Immunol 2020; 81: 588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hatfield SD, Daniels KA, O'Donnell CL, Waggoner SN, Welsh RM. Weak vaccinia virus‐induced NK cell regulation of CD4 T cells is associated with reduced NK cell differentiation and cytolytic activity. Virology 2018; 519: 131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rydyznski C, Daniels KA, Karmele EP et al Generation of cellular immune memory and B‐cell immunity is impaired by natural killer cells. Nat Commun 2015; 6: 6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rydyznski CE, Cranert SA, Zhou JQ et al Affinity maturation is impaired by natural killer cell suppression of germinal centers. Cell Rep 2018; 24: 3367–3373 e3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bradley T, Peppa D, Pedroza‐Pacheco I et al RAB11FIP5 expression and altered natural killer cell function are associated with induction of HIV broadly neutralizing antibody responses. Cell 2018; 175: 387–399 e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Goodier MR, Rodriguez‐Galan A, Lusa C et al Influenza vaccination generates cytokine‐induced memory‐like NK cells: impact of human cytomegalovirus infection. J Immunol 2016; 197: 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Jost S, Reardon J, Peterson E et al Expansion of 2B4+ natural killer (NK) cells and decrease in NKp46+ NK cells in response to influenza. Immunology 2011; 132: 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wagstaffe HR, Nielsen CM, Riley EM, Goodier MR. IL‐15 Promotes polyfunctional NK cell responses to influenza by boosting IL‐12 production. J Immunol 2018; 200: 2738–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wagstaffe HR, Pickering H, Houghton J et al Influenza vaccination primes human myeloid cell cytokine secretion and NK cell function. J Immunol 2019; 203: 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Riese P, Trittel S, Pathirana RD, Klawonn F, Cox RJ, Guzman CA. Responsiveness to influenza vaccination correlates with NKG2C– expression on NK cells. Vaccines (Basel) 2020; 8: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wijaya RS, Read SA, Truong NR et al HBV vaccination and HBV infection induces HBV‐specific natural killer cell memory. Gut 2021; 70: 357–369. [DOI] [PubMed] [Google Scholar]
- 92. Marquardt N, Ivarsson MA, Blom K et al The human NK cell response to yellow fever virus 17D is primarily governed by NK cell differentiation independently of NK cell education. J Immunol 2015; 195: 3262–3272. [DOI] [PubMed] [Google Scholar]
- 93. Pejoski D, de Rham C, Martinez‐Murillo P et al Rapid dose‐dependent natural killer (NK) cell modulation and cytokine responses following human rVSV‐ZEBOV Ebolavirus vaccination. NPJ Vaccines 2020; 5: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wagstaffe HR, Clutterbuck EA, Bockstal V et al Antibody‐dependent natural killer cell activation after Ebola vaccination. J Infect Dis 2019; jiz657 10.1093/infdis/jiz657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Vanderven HA, Barr I, Reynaldi A et al Fc functional antibody responses to adjuvanted versus unadjuvanted seasonal influenza vaccination in community‐dwelling older adults. Vaccine 2020; 38: 2368–2377. [DOI] [PubMed] [Google Scholar]
- 96. Kristensen AB, Kent SJ, Parsons MS. Contribution of NK cell education to both direct and anti‐HIV‐1 antibody‐dependent NK cell functions. J Virol 2018; 92: e02146‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jegaskanda S, Vanderven HA, Tan H‐X et al Influenza virus infection enhances antibody‐mediated NK cell functions via type I interferon‐dependent pathways. J Virol 2019; 93: e02090‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fisher L, Zinter M, Stanfield‐Oakley S et al Vaccine‐induced antibodies mediate higher antibody‐dependent cellular cytotoxicity after interleukin‐15 pretreatment of natural killer effector cells. Front Immunol 2019; 10: 2741. [DOI] [PMC free article] [PubMed] [Google Scholar]


