Abstract
Pseudokinases are members of the protein kinase superfamily but signal primarily through noncatalytic mechanisms. Many pseudokinases contribute to the pathologies of human diseases, yet they remain largely unexplored as drug targets owing to challenges associated with modulation of their biological functions. Our understanding of the structure and physiological roles of pseudokinases has improved substantially over the past decade, revealing intriguing similarities between pseudokinases and their catalytically active counterparts. Pseudokinases often adopt conformations that are analogous to those seen in catalytically active kinases and, in some cases, can also bind metal cations and/or nucleotides. Several clinically approved kinase inhibitors have been shown to influence the noncatalytic functions of active kinases, providing hope that similar properties in pseudokinases could be pharmacologically regulated. In this Review, we discuss known roles of pseudokinases in disease, their unique structural features and the progress that has been made towards developing pseudokinase-directed therapeutics.
Introduction
Protein kinases constitute essential components of almost every signalling pathway. Most kinases are characterized by a highly conserved kinase domain fold that enables catalysis of phosphorylation, with specificity for serine, threonine or tyrosine residues in metazoans. Dysregulation of kinase function has been linked to numerous pathologies, most notably cancer, as well as neurological, immunological and metabolic diseases. In fact, the kinase domain is the most frequently observed domain in known oncogenes1, and a multitude of genetic alterations in kinases, including changes in expression levels and mutations, have been described as driver events in malignant transformation. The therapeutic potential of kinases was evident shortly after their discovery, but the high degree of structural similarity in their active sites was initially thought to be prohibitive to the development of highly selective kinase inhibitors. Now, more than 30 small-molecule kinase inhibitors have been approved by the US Food and Drug Administration (FDA). These inhibitors largely consist of reversible ATP-competitive compounds and therefore directly target the ability of kinases to catalyse phosphorylation and counteract their hyperactivation in diseases2.
Unbiased screens for somatic kinase mutations in cancer have identified a surprising number of mutations that are expected to impair kinase activity, rather than potentiate it, by replacing conserved residues in the active site3,4. Although some of these inactivating mutations occur in kinases that act as tumour suppressors, such as LKB1 (also known as serine/threonine-protein kinase STK11), many others map to the kinase domains of bona fide oncogenic kinases that are typically hyperactivated in cancer, including BRAF, epidermal growth factor receptor (EGFR) and KIT. Indeed, kinase-inactivating mutations are the most common type of BRAF mutation in non-small-cell lung cancer (NSCLC)5, and recently, catalytically impaired BRAF mutants were shown to induce lung adenocarcinoma6. These mutations enhance a nonenzymatic function of BRAF, resulting in increased affinity for BRAF heterodimerization with CRAF, enhanced allosteric activation of CRAF and elevated mitogen-activated protein kinase (MAPK) pathway signalling7–9.
The ability of BRAF kinase-inactivating mutations to drive disease underscores the importance of the noncatalytic functions of kinases in signalling. In recent years, there has been a growing body of evidence demonstrating that many, if not all, kinases possess such functions in addition to their canonical role10,11,12. Kinases can act as scaffolds, as observed for receptor-interacting serine/threonine-protein kinase 1 (RIPK1) and RIPK3, which are involved in assembling apoptotic signalling complexes13–15, or function as allosteric regulators of other enzymes, such as ERK2 (also known as MAPK1), which activates dual specificity protein phosphatase 6 (DUSP6; also known as MKP3)16,17. Systematic analysis of the human genome has revealed that one-tenth of all protein kinases are predicted to be catalytically inactive and to therefore primarily signal through alternative mechanisms18. Members of this subset of the protein kinase superfamily are called pseudokinases and have inactivating mutations in critical catalytic motifs.
Pseudokinases, like their catalytically active counterparts, play pivotal roles in cellular signalling and are often dysregulated in a variety of diseases19–21. Compared with active kinases, however, pseudokinases pose a much greater challenge for drug design. The nucleotide-binding site is, by far, the most ‘druggable’ pocket in protein kinases owing to its evolved ability to interact with a small molecule. However, as discussed below, for many pseudokinases, it is unclear how molecules that compete with nucleotide binding could disrupt nonenzymatic functions. Moreover, a substantial number of pseudokinases do not bind a nucleotide, and several do not even seem to have a structurally defined pocket that could be accessible to ATP-competitive small molecules.
Although there have yet to be any clinically approved therapeutics that target pseudokinases, we are better equipped to meet this challenge than might be apparent at first glance. Structural and biochemical studies over the past decade have revealed that the regulation of noncatalytic kinase signalling carries substantial parallels to that of active kinases11,12. The on and off states of catalytically competent kinases are characterized by defined architectures of the nucleotide-binding site and other structural elements within the kinase domain. Catalysis of phosphorylation relies on a regulated transition between these states. Although it is unclear whether such transitions also occur in pseudokinases, several pseudokinases can adopt conformations that recapitulate features of either the on or off state of catalytically active protein kinases, and in many cases, these conformations are critical for their physiological roles. Therefore, molecules that could force pseudokinases to adopt another, non-functional conformation could be used for therapeutic targeting. Encouraged by examples of conformationally selective small molecules that modulate the noncatalytic functions of active kinases, which we discuss below, there is now growing interest in applying the principles of conformational selection to develop pharmacological agents that target pseudokinases. Here, we discuss these possibilities in light of our increasing knowledge of the structural mechanisms that underlie pseudokinase signalling.
Pseudokinases as disease targets
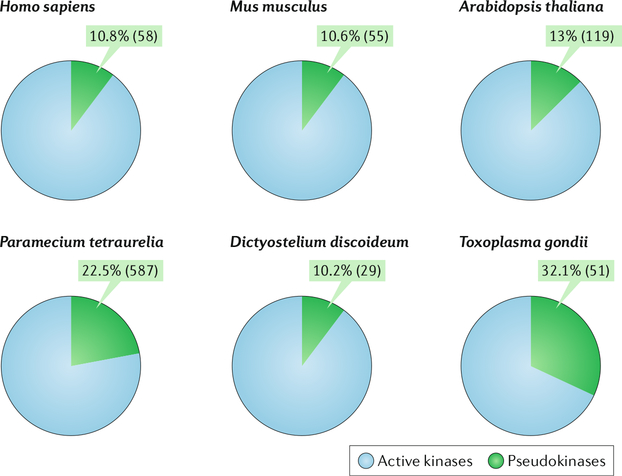
Pseudokinases are well conserved in evolution and comprise a notable proportion of the kinomes of not only humans and mice but also diverse species, including Arabidopsis thaliana, Paramecium tetraurelia, Dictyostelium discoideum and Toxoplasma gondii18,22–28 (Fig. 1). Although the functions of the vast majority of these pseudokinases remain poorly understood, studies conducted thus far highlight crucial roles for many of them in development, the immune response and metabolism. Consequently, mutations in pseudokinases and dysregulation of their expression have been linked to several developmental and morphological disorders, as well as a wide range of diseases, such as cancer, neurological disorders, metabolic disorders and autoimmune diseases19–21,29 (Table 1; see Supplementary Table 1 for a complete table with references).
Fig. 1: Prevalence of pseudokinases in the kinomes of diverse species.
Pie charts showing the percentages of active kinases (blue) and pseudokinases (green) in the kinomes of the indicated organisms. Pseudokinases are defined as kinases that carry mutations in one or more conserved catalytic motifs (the β3 lysine within the VAIK (Val-Ala-Ile-Lys) motif, the aspartate in the DFG (Asp-Phe-Gly) motif and the aspartate in the HRD (His-Arg-Asp) motif).
Table 1.
Known roles of pseudokinases in disease
| Pseudokinase | Links to disease | Relevant pseudokinase domain mutations |
|---|---|---|
| ADCK3 | Mutations associated with cerebellar ataxia result in defects in coenzyme Q10 synthesis | Y514C |
| ANP-A (encoded byNPR1) | Polymorphisms associated with increased left ventricular mass index in essential hypertension; deletion in 5΄ flanking region associated with increased susceptibility to essential hypertension or left ventricular hypertrophy | NA |
| ANP-B (encoded byNPR2) | Loss-of-function mutations associated with acromesomelic dysplasia, Maroteaux type | Y708C, R776W |
| BUBR1 | Mutations associated with mosaic variegated aneuploidy; overexpression associated with tumour cell proliferation in gastric cancer and chromosomal instability in bladder cancer | R727C, R814H, I909T, L1012P |
| CASK | Mutations associated with X-linked mental retardation and FG syndrome | R28L, Y268H |
| DIA1R (encoded byCXORF36) | Mutations associated with X-linked mental retardation; deletions withinCXORF36associated with autism spectrum disorders | R128K |
| EPHA10 | Overexpression associated with prostate cancer and correlated with lymph node metastasis and stage progression in breast cancer | NA |
| EPHB6 | Downregulated in metastatic melanoma; expression level associated with tumour stage and survival in neuroblastoma and ovarian serous carcinoma; deletion mutation enhances cell migration and promotes a pro-metastatic phenotype in NSCLC cells | NA |
| FAM20A | Mutations associated with amelogenesis imperfecta prevent allosteric activation of FAM20C and exhibit reduced secretion | L173R, G331D, D403N |
| GCN2 | Promotes survival and proliferation in fibrosarcoma cells; genetic deletion improved synaptic plasticity and spatial memory in a mouse model of Alzheimer disease; promotes efficacy of yellow fever vaccine; mutations associated with pulmonary veno-occlusive disease | NA |
| HER3 (also known asERBB3) | Increased expression and persistent phosphorylation of HER3 contribute to drug resistance in cancer types such as breast and lung cancer; mutations promote oncogenesis and are found in several tumour types including breast, lung and gastrointestinal cancer; downregulated in dorsolateral prefrontal cortex in schizophrenia; overexpressed in Charcot–Marie–Tooth type1 disease | V695M, Q790R, S827I, E909G |
| HSER (encoded byGUCY2C) | Loss-of-function mutations linked to meconium ileum; activating mutation linked to familial diarrhoea syndrome; mRNA expression in CRC linked to time to recurrence and disease-free survival; suppresses intestinal tumour formation by inhibiting AKT | NA |
| ILK | Overexpression promotes cell migration and invasion in lung cancer and CRC | NA |
| IRAK2 | Hypofunctional mutations associated with reduced survival in CRC and reduced hepatitis C clearance | R214G, L392V |
| IRAK3 | Overexpression associated with deactivation of inflammatory response in monocytes upon exposure to tumour cells; mutations associated with early-onset, persistent asthma | L400V, R429Q |
| JAK1 JH2 | Mutations associated with acute leukaemias | A634D, H647Y, L653F, V658F, R724H, T782M, L783F |
| JAK2 JH2 | Mutations associated with myeloproliferative neoplasms, such as polycythaemia vera, and acute leukaemias | K607N, L611S, V617F, V717F |
| JAK3 JH2 | Inhibitory mutations associated with severe combined immunodeficiency; activating mutations associated with acute leukaemias | A572V, A573V, M576L, A593T, V715I, V722I |
| KSR1 | KSR1-null mice exhibit impaired tumorigenesis in RAS-driven cancers | NA |
| KSR2 | Mutations associated with obesity and insulin resistance; overexpression promotes anchorage-independent growth and transformation of oncogenic RAS-mutant cells | P662L, E667V, R684C, I801L, G816D, R818Q, R823H, D843N, S904L |
| MLKL | Required for necroptosis; overexpressed in autoimmune hepatitis | NA |
| NRBP1 | Downregulated in multiple cancers, including lung and colon adenocarcinomas; low expression levels in lung tumours correlate with reduced survival | NA |
| NRBP2 | Downregulated in hepatocellular carcinoma; high expression levels correlated with greater chemosensitivity and better prognosis | NA |
| PEAK1 (also known as SGK269) | Overexpressed in pancreatic cancer and a subset of breast cancers; required for KRAS-induced cell expansion in pancreatic cancer | NA |
| POMK (also known asSGK196) | Required for Lassa virus entry; loss-of-function mutations associated with Walker–Warburg syndrome, neuromuscular disorders and congenital muscular dystrophy | L137R, Q258R |
| Pragmin (also known as SGK223 or PRAG1) | Overexpressed in pancreatic cancer; required for SRC-mediated invasion in CRC | NA |
| PTK7 | Mutations lead to idiopathic scoliosis and neural tube defects; overexpression associated with lung and breast cancer; downregulated in metastatic melanoma and ovarian cancer | NA |
| PXK | Genetic variants that reduce B cell receptor internalization associated with systemic lupus erythematosus | NA |
| RETGC1 (encoded byGUCY2D) | Mutations associated with Leber congenital amaurosis and autosomal dominant cone–rod retinal dystrophy | F589S |
| RETGC2 (encoded byGUCY2F) | Mutations associated with pancreatic, lung and breast cancers | K672T |
| RNase L | Mediates innate immunity against viral and bacterial infections; overexpression observed in chronic fatigue syndrome; mutations associated with elevated risk of prostate cancer | R462Q |
| ROR1 | Overexpressed in several types of cancer, including CLL, ALL, gastric carcinoma and NSCLC | NA |
| ROR2 | Germline deletions associated with autosomal dominant brachydactyly type B; loss-of function mutations associated with autosomal recessive Robinow syndrome; overexpression associated with osteosarcoma and renal cell carcinoma | N620K |
| RPS6KL1 | Knockdown reduces cell survival of cervical carcinoma cells | NA |
| RYK | Inhibition of axon regeneration following spinal cord injury; overexpressed in epithelial ovarian cancer and associated with decreased overall survival | NA |
| S6KΔ1 (encoded byRPS6KC1) | Fusion of PX domain with AKT3 results in constitutively activated oncogenic fusion protein that promotes tumour growth of HR+breast cancer cells | NA |
| SCYL1 | Frameshift mutations associated with recessive form of spinocerebellar neurodegeneration and a hepatocerebellar neuropathy syndrome | NA |
| SCYL2 | Reduces release of HIV-1 by promoting Vpu dephosphorylation | NA |
| SCYL3 | Oncogenic fusion of TRKA kinase domain with SCYL3 pseudokinase domain and HEAT repeats is associated with CRC and promotes cell proliferation | NA |
| STK31 (also known asSGK396) | Overexpressed in CRC; inhibits differentiation; promotes cell migration and invasion | NA |
| STK40 (also known asSGK495) | Knockdown reduces cell viability in cervical carcinoma cells | NA |
| STRADα | Allosterically activates tumour suppressor LKB1; deletion of exons 9–13 associated with PMSE | NA |
| STRADβ | Allosterically activates tumour suppressor LKB1 | NA |
| STYK1 | Overexpressed in castration-resistant prostate cancer; low expression correlates with improved survival in hepatocellular carcinoma and decreased cell growth, migration and invasion | NA |
| TBCK | Mutations associated with infantile syndromic encephalopathy, intellectual disability and hypotonia | NA |
| Titin | Mutations associated with cardiomyopathy | NA |
| TRIB1 | Overexpression associated with AML and prostate cancer; gain-of-function mutation associated with acute megakaryocytic leukaemia; polymorphisms associated with altered lipid metabolism and cardiovascular disease | R107L |
| TRIB2 | Overexpressed in AML, lung cancer and liver cancer; promotes drug resistance in several cancer types, including melanoma and breast and renal cancers | NA |
| TRIB3 | Gain-of-function mutation associated with insulin resistance; overexpression associated with lung, colon and breast cancers; overexpression associated with Parkinson disease | Q84R |
| TRRAP | Necessary for MYC and E1A-mediated transformation | NA |
| TYK2 JH2 | Inhibitory mutations associated with autoimmune diseases, including systemic lupus erythematosus, Crohn’s disease and psoriasis; required for amyloid-β-induced neuronal apoptosis | I684S |
| ULK4 | Deletions of exons 21–34 and 33–34 in theULK4gene associated with schizophrenia | NA |
| VRK3 | Phosphorylation of VRK3 by CDK5 protects neuronal cells from oxidative stress-induced cell death by inhibiting ERK | NA |
A complete table including references can be found in the supplementary information. ALL, acute lymphocytic leukaemia; AML, acute myeloid leukaemia; ANP-A, atrial natriuretic peptide receptor type A; CDK5, cyclin-dependent kinase 5; CLL, chronic lymphocytic leukaemia; CML, chronic myeloid leukaemia; CRC, colorectal cancer; DIA1R, deleted in autism-related protein 1; EPHA10, pseudokinase ephrin receptor; HER, human epidermal growth factor receptor; HR, hormone receptor; HSER, heat-stable enterotoxin receptor; ILK, integrin-linked protein kinase; IRAK2, interleukin-1 receptor-associated kinase-like 2; JAK, Janus kinase; JH2, JAK homology 2; KSR, kinase suppressor of RAS; MLKL, mixed lineage kinase domain-like protein; NA, not applicable; NRBP1, nuclear receptor-binding protein 1; NSCLC, non-small-cell lung cancer; PMSE, polyhydramnios, megalencephaly, symptomatic epilepsy; POMK, protein O-mannose kinase; PTK7, protein tyrosine kinase 7; RETGC1, retinal guanylyl cyclase 1; RPS6KL1, ribosomal protein S6 kinase-like 1; RYK, receptor-like tyrosine kinase; SCYL1, SCY-like protein 1; STK, serine/threonine-protein kinase; STRADα, STE20-related adaptor-α; TRRAP, transformation/transcription domain-associated protein; TRIB1, Tribbles homologue 1.
HER3
One of the pseudokinases that emerged early as a potential therapeutic target is human epidermal growth factor receptor 3 (HER3; also known as ERBB3), a member of the EGFR (also known as HER or ERBB) family of receptor tyrosine kinases (RTKs). The known physiological functions of HER3 depend on its role as a co-receptor of catalytically active HER family members, including HER2 and EGFR, with which HER3 forms complexes upon growth factor binding. In these complexes, the HER3 pseudokinase domain allosterically activates its partner HER kinase by stabilizing it in the active conformation through the formation of an asymmetric kinase dimer30–32. This allosteric functionality is encoded in all receptors in the EGFR family; HER3 has only this allosteric role and no catalytic role33,34.
HER3 overexpression alone is insufficient to drive transformation35, but its presence is required for the transformation of NIH3T3 cells by HER236. In HER2-overexpressing breast cancers and in EGFR-mutant-driven lung cancer and head and neck squamous cell carcinoma (SCCHN), HER3 promotes resistance to HER2 and EGFR inhibitors, respectively37–39. HER3 is also associated with resistance to anti-oestrogen therapies in oestrogen receptor-positive breast cancer and with resistance to insulin-like growth factor 1 receptor (IGF1R) inhibitors in hepatocellular carcinomas40,41. Drug-induced elevation of HER3 expression and phosphorylation are the primary mechanisms that contribute to this resistance (Fig. 2a). Somatic mutations that map to extracellular or intracellular regions of HER3, including the pseudokinase domain, have also been identified in various types of cancer, including NSCLC, gastric cancer and colon cancer3,42–48. The mutations in the extracellular domain might destabilize its autoinhibited conformation, thereby favouring HER3 dimerization with other members of the HER family. Several of these mutations promote ligand-independent cell survival and tumour growth both in vitro and in vivo in a HER2-dependent manner48,49 (Fig. 2a). The pseudokinase domain mutations map most frequently to the asymmetric kinase dimer interface and increase the affinity of the HER3–EGFR kinase heterodimer, which enhances the HER3-dependent allosteric activation of EGFR32,48.
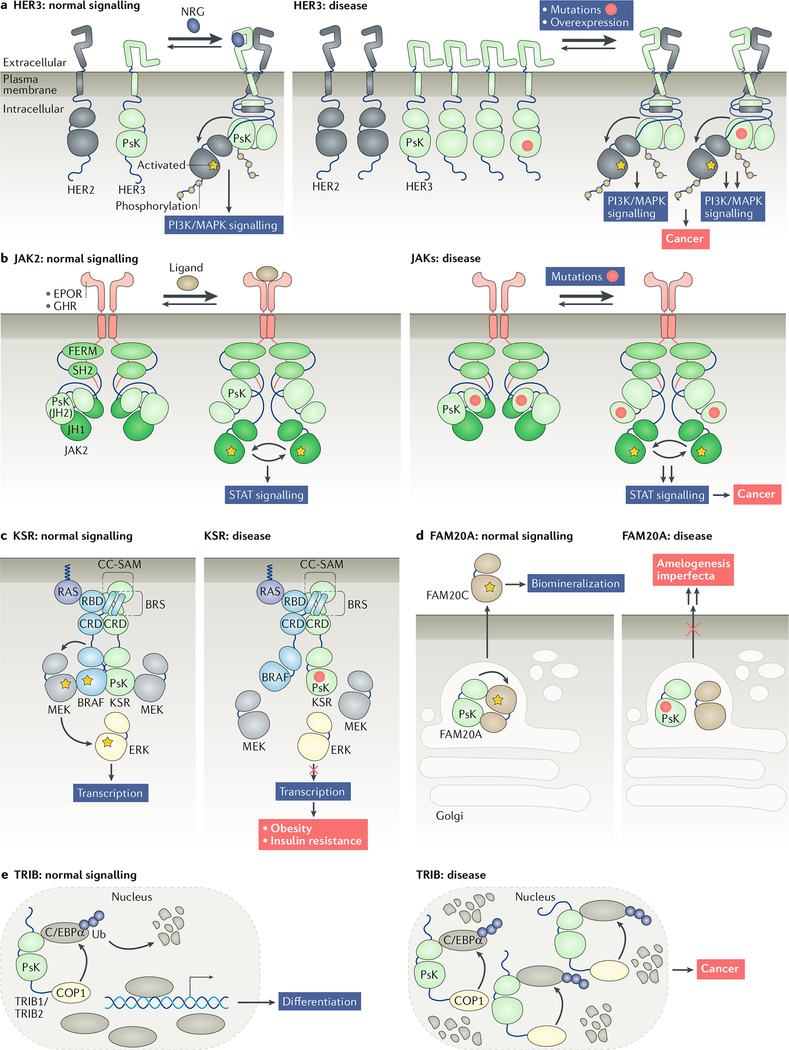
Fig. 2: Dysregulation of pseudokinase signalling in disease.
a | In normal cells, human epidermal growth factor receptor 3 (HER3) allosterically activates other HER kinases through heterodimerization in a ligand-dependent manner (left panel). Overexpression or mutation of HER3 drives ligand-independent association with its dimerization partners, such as HER2, and activation of signalling pathways that promote tumorigenesis (right panel). b | In normal cells, Janus kinase 2 (JAK2) associates with its cognate receptor at the plasma membrane, and the JAK homology 2 (JH2) pseudokinase domain of JAK2 allosterically inhibits the JH1 kinase domain. This inhibition is relieved upon ligand binding to the receptor, allowing transphosphorylation and activation of the kinase domain (left panel). Mutations in the pseudokinase domain associated with cancers, such as myeloproliferative neoplasms, are believed to disrupt the pseudokinase–kinase domain interaction, causing constitutive activation of the kinase domain (right panel). c | Under normal conditions, kinase suppressor of RAS (KSR) allosterically activates BRAF to promote phosphorylation of MEK and activate mitogen-activated protein kinase (MAPK) signalling. KSR also interacts directly with MEK, and this interaction enhances the allosteric activator function of KSR (left panel). Mutations associated with obesity and insulin resistance impair the ability of KSR to activate the MAPK pathway by disrupting its interactions with MEK and BRAF (right panel). d | Under normal conditions, FAM20A and FAM20C localize to the Golgi apparatus. FAM20A allosterically activates FAM20C, which becomes secreted and phosphorylates extracellular proteins that are important for biomineralization and proper enamel development (left panel). Mutations in FAM20A linked to amelogenesis imperfecta fail to activate FAM20C or promote FAM20C secretion (right panel). e | The transcription factor CCAAT enhancer-binding protein-α (C/EBPα) promotes myeloid cell differentiation. Tribbles homologue 1 (TRIB1) and TRIB2 interact with C/EBPα and the E3 ubiquitin ligase COP1 to mediate ubiquitylation and proteasomal degradation of C/EBPα (left panel). Overexpression of TRIB1 or TRIB2 induces leukaemogenesis by causing excessive degradation of C/EBPα in a COP1-dependent manner (right panel). CRD, cysteine-rich domain; NRG, neuregulin; PI3K, phosphoinositide 3-kinase; STAT, signal transducer and activator of transcription.
In addition to its oncogenic role in cancer, HER3 is also a potential target in neurological disorders. HER3 is essential for the growth and development of Schwann cells by functioning as a receptor for the growth factor neuregulin 1 (NRG1)50. Mice homozygous for deletion of HER3 manifest severe defects in myelination, which lead to impaired motor and sensory neuron development; most animals die during early embryonic stages50. Likely owing to the importance of HER3 for myelination, aberrant HER3 expression in humans is associated with a range of neurological diseases. Expression of HER3 is downregulated in the dorsolateral prefrontal cortex of patients with schizophrenia51–53, and HER3 overexpression is observed in the Schwann cells of patients with Charcot–Marie–Tooth type 1 disease, one of the most frequently inherited neuropathies51. While the value of modulating the NRG1–HER3 axis in neurological disorders continues to be debated52,53, its utility will ultimately be tested through the development of therapeutics that specifically and efficiently target HER3.
ROR1, ROR2, PTK7 and RYK
An unrelated group of RTKs, ROR1, ROR2, protein tyrosine kinase 7 (PTK7; also known as colon carcinoma kinase (CCK4)) and receptor-like tyrosine kinase (RYK), which are all categorized as pseudokinases, are co-receptors in WNT signalling pathways. Collectively, these pseudokinase receptors have been implicated in the pathology of several cancers and developmental disorders. ROR1 and ROR2 were originally identified as orphan RTKs that are highly expressed during embryonic development and have critical roles in skeletal and neural organogenesis54. ROR1 and ROR2 have since been found to interact with the ligand WNT5A to mediate noncanonical WNT signalling through the planar cell polarity (PCP) pathway55,56. Although ROR1 and ROR2 are not expressed in most normal adult tissues, the re-emergence of their expression through transcriptional upregulation is associated with various types of cancer. This makes RORs very attractive cancer targets, as they would potentially have few adverse effects in normal tissues. ROR1 is often overexpressed in haematological malignancies, including chronic lymphocytic leukaemia, in which WNT5A induces hetero-oligomerization of ROR1 and ROR2 to promote chemotaxis and proliferation57. Upregulation of ROR1 is also observed in cancers that overexpress MET (also known as hepatocyte growth factor receptor), such as gastric carcinoma and NSCLC58. Phosphorylation of the proline-rich domain and pseudokinase domain of ROR1 in these cells by MET and SRC, respectively, is important for MET-driven cell proliferation and invasion59. Knockdown of ROR1 was found to reduce cell proliferation in multiple cancer cell lines and inhibited tumorigenesis in vivo in xenograft models58,60.
ROR2 overexpression also promotes cell proliferation and migration in several types of cancer, such as osteosarcoma and renal cell carcinoma, and knockdown of ROR2 inhibits tumour growth both in vitro and in vivo61,62. In some cancers, such as colon cancer and hepatocellular carcinoma, elevated ROR2 expression has the opposite effect and suppresses tumour growth63,64. These contrasting roles of ROR2 may reflect its ability to both inhibit canonical β-catenin-dependent WNT signalling and activate noncanonical WNT signalling through the PCP pathway56,65. Germline mutations in ROR2 are associated with autosomal dominant brachydactyly type B (BDB), a condition that is characterized by terminal deficiency of the fingers and toes, as well as an autosomal recessive form of Robinow syndrome (RRS), manifested by severe skeletal malformations and abnormal craniofacial features66–69. RRS can be modelled using Ror2-deficient mice, suggesting that RRS-associated mutations lead to loss of function68–70. BDB-associated mutations result in truncations of ROR2 before and after the pseudokinase domain, which are likely dominant-negative forms of the protein66.
PTK7 interacts with WNT receptors, such as Frizzled and ROR2, as well as several WNT ligands, including WNT3A, WNT8 and WNT5A, to modulate both canonical and noncanonical WNT signalling71,72. The PTK7 pseudokinase domain is essential for associating with β-catenin and interacts with the scaffold protein receptor of activated protein C kinase 1 (RACK1) to recruit Dishevelled to the plasma membrane73,74. Although the mechanisms underlying its signalling remain poorly understood, PTK7 regulates several developmental and physiological processes that rely on cell migration, cell polarity and epidermal wound repair. Germline loss-of-function mutations in PTK7 are associated with idiopathic scoliosis and neural tube defects75,76. The role of PTK7 in cancer remains somewhat mysterious. PTK7 is upregulated in some cancers, such as colon cancer, lung adenocarcinoma and breast cancer, but downregulation of PTK7 has been observed in ovarian carcinoma and metastatic melanoma77–81. Experimental evidence suggests that inhibition of PTK7 signalling would likely be beneficial in cancers that overexpress PTK7. Knockdown of PTK7 reduces cell viability and increases apoptosis in lung cancer cells, inhibits tumour growth in lung adenocarcinoma xenografts and decreases motility and invasiveness in breast cancer cells78,79.
RYK also contributes to both canonical and noncanonical WNT signalling, including the PCP and WNT–Ca2+ pathways. Like PTK7, RYK interacts with Frizzled and Dishevelled, as well as multiple WNT ligands, including WNT1, WNT3A and WNT5A82. At present, little is known regarding the mechanisms that underlie signalling by this pseudokinase receptor. RYK plays a pivotal role in the central nervous system by controlling axon guidance and neuorogenesis83,84. RYK also inhibits axon regeneration following spinal cord injuries in rats; therefore, blocking RYK-mediated signalling might improve recovery from such injuries85. Overexpression of RYK has been associated with decreased overall survival and shorter progression-free survival in epithelial ovarian cancer86, but the value of targeting RYK in cancer has yet to be explored.
JAKs
The Janus kinase (JAK) family of non-receptor tyrosine kinases, consisting of JAK1, JAK2, JAK3 and TYK2, contain a tandem kinase domain module in which the catalytically active tyrosine kinase domain (JAK homology 1, JH1) is preceded by a pseudokinase domain (JH2). The JH2 pseudokinase domain binds to the active JH1 kinase domain and stabilizes it in an inactive conformation87,88. JAKs associate with the cytoplasmic portions of cytokine receptors and become activated by ligand binding to the receptors, resulting in the release of the autoinhibition imposed on the JH1 domain by the JH2 pseudokinase domain. Several mutations in the JH2 domains of JAK1, JAK2 or JAK3 result in constitutive activation of the JH1 kinase domain and are associated with myeloproliferative neoplasms, such as polycythaemia vera, and with acute myeloid leukaemia (AML)89–92. Many of these mutations, such as V617F in the JH2 domain of JAK2, map near the JH1–JH2 interface, which likely disrupts the autoinhibitory lock that controls the activity of the active JH1 kinase domain93,94 (Fig. 2b). By contrast, some disease mutations in the JAK3 and TYK2 JH2 domains reduce activity of their JH1 domains95,96, possibly by enhancing the autoinhibitory interaction between the pseudokinase and kinase domains. These JAK3 mutations have been linked to severe combined immunodeficiency95, while the TYK2 mutations are associated with increased susceptibility to autoimmune disease96. Thus, modulation of the extent of pseudokinase–kinase interactions in either direction in the JAKs could be beneficial in disease.
MLKL
As occurs in JAK proteins, the pseudokinase domain of mixed lineage kinase domain-like protein (MLKL) regulates the activity of an adjacent domain, denoted as the four-helix bundle (4HB) domain. MLKL is a key effector in necroptosis owing to its role in inducing plasma membrane permeabilization — a prerequisite for necroptotic cell death97–99. Under normal conditions, the pseudokinase domain of MLKL stabilizes the 4HB domain in an inhibited conformation. Phosphorylation of the pseudokinase domain by RIPK3 triggers release of the 4HB domain, MLKL oligomerization and localization of MLKL to the cell membrane, where it induces permeabilization97–99. The resulting loss of membrane integrity causes leakage of cellular contents, resulting in necroptotic cell death and, frequently, an inflammatory response that contributes to diseases, such as neurodegeneration, myocardial infarction and stroke, atherosclerosis, ischaemic–reperfusion injury and inflammatory bowel disease100. Targeting necroptosis in these diseases is an attractive concept, and as the key effector of necroptosis, MLKL could serve as a novel target for the development of specific inhibitors that could avoid some of the undesirable effects of inhibitors developed to target RIPKs. Selective inhibitors of RIPK3 have been developed, but high concentrations of these compounds trigger apoptosis, and therefore they have limited therapeutic potential15. In addition to necroptosis, RIPK1 is involved in other processes, including nuclear factor-κB signalling and apoptosis101, which will inadvertently be affected by RIPK1 inhibitors.
KSR
Kinase suppressor of RAS 1 (KSR1) and KSR2 are pseudokinases in the RAF kinase family that function as allosteric regulators of active kinases, similar to HER3 and the JH2 domains of JAKs. Both KSR1 and KSR2 promote activation of the MAPK pathway through interactions with RAF, MEK and ERK. An important step in RAF kinase activation is formation of a ‘side-to-side’ dimer in which one kinase allosterically stabilizes the active conformation in another102. This allosteric activator function is not dependent on catalytic activity, and hence KSR can activate RAF through heterodimerization, which results in MEK phosphorylation102,103. KSR also binds directly to MEK, and recently, structural and biochemical studies revealed that this interaction further enhances the association between KSR and BRAF104.
Owing to its positive effect on MAPK signalling, KSR has become a therapeutic target of interest in the treatment of RAS-driven cancers. KSR1 is necessary for tumorigenesis in a mouse model of skin cancer driven by the oncogenic RAS G12V mutation, and deletion of KSR1 in mouse embryonic fibroblasts prevents RAS G12V-induced transformation105,106. KSR1-null mice develop normally, which indicates that inhibition of KSR1 in adults with cancer might have low toxicity105,107. By contrast, KSR2-null mice develop diabetes and hypertension108,109, and mutations in KSR2, including several in the pseudokinase domain, are associated with insulin resistance and obesity110. These mutations impair the ability of KSR2 to stimulate MAPK signalling and are predicted to disrupt interactions with RAF and MEK (Fig. 2c). Hence, inhibition of KSR might disrupt glucose homeostasis108,109, unless inhibitors specific to KSR1 are developed.
FAM20A
FAM20A is another pseudokinase that modulates the activity of a closely related active kinase, FAM20C. FAM20A and FAM20C are members of a recently characterized family of atypical Golgi-localized kinases that undergo secretion to the extracellular space111,112. Both of these proteins have critical roles in biomineralization and enamel development. FAM20A allosterically activates FAM20C and promotes its secretion112,113. These functions of FAM20A are abrogated by mutations associated with the biomineralization disorder amelogenesis imperfecta112,113,114 (Fig. 2d). Mutations in FAM20C result in Raine syndrome, a rare disorder characterized by severe and often lethal osteosclerotic bone dysplasia115. FAM20C was recently found to be responsible for phosphorylating the majority of the secreted phosphoproteome — a function disrupted in Raine syndrome by mutations that impair FAM20C catalytic activity112,116. Strikingly, FAM20A potentiates the activity of FAM20C Raine syndrome mutants in vitro112, suggesting that enhancing the interaction between FAM20A and FAM20C could be beneficial in the treatment of Raine syndrome.
Tribbles
The Tribbles family of pseudokinases (TRIB1, TRIB2 and TRIB3) stands out among pseudokinases because these proteins have been described as scaffolds for diverse signalling proteins in seemingly unrelated signalling pathways, such ubiquitylation of transcription factors or regulation of kinases in the MAPK and AKT signalling pathways. Consequently, TRIBs have been implicated in a variety of diseases, including several types of cancer and metabolic disease.
One of the best understood functions of TRIBs is their role in regulation of ubiquitylation by the E3 ubiquitin ligase COP1. The C-terminal tails of TRIBs contain a highly conserved motif that recruits COP1, which then ubiquitylates substrates that interact with TRIB, such as the transcription factor CCAAT enhancer-binding protein-α (C/EBPα) and the metabolic enzyme acetyl-CoA carboxylase117–120. Evolutionary analysis of TRIB homologues, compared with the rest of the kinome, revealed that the C-terminal COP1-binding motif is a defining feature of this family of kinases121. Overexpression of TRIB1 or TRIB2 induces leukaemogenesis in mice through depletion of C/EBPα in a COP1-dependent manner, implicating upregulation of COP1 function by TRIB1 and TRIB2 in AML117,119,122 (Fig. 2e). In addition, likely owing to the role of C/EBPα in lipogenesis, genome-wide association studies have identified variants at the TRIB1 locus that significantly associate with cardiovascular disease and plasma levels of triglycerides, cholesterol, low-density lipoprotein and high-density lipoprotein123.
The mechanisms by which TRIB family pseudokinases modulate kinases within the MAPK and AKT pathways are much less understood. TRIB1, TRIB2 and TRIB3 interact with MEK through a conserved motif in the C-terminal lobes (C-lobes) of their pseudokinase domains, and this interaction increases ERK phosphorylation124. TRIB-mediated activation of the MAPK pathway has been implicated in both leukaemia and breast cancer124,125. In the AKT pathway, different TRIBs seem to play opposing roles. TRIB3 promotes insulin resistance by binding to and inhibiting AKT, which leads to increased apoptosis of pancreatic β-cells and reduced insulin secretion126,127. Elevated levels of TRIB3 and the resulting inhibition of AKT are also associated with neuronal cell death in Parkinson disease through the promotion of apoptosis128,129. By contrast, TRIB2 enhances AKT signalling, and overexpression of TRIB2 promotes drug resistance in cancer cells130. Collectively, these findings indicate that there is still much to learn about the mechanisms of TRIB signalling and that inhibition of TRIB function would potentially be therapeutically beneficial in a variety of diseases.
TRRAP
Another pseudokinase that functions as a scaffold is transformation/transcription domain-associated protein (TRRAP), a member of the phosphatidylinositol 3-kinase-related kinase (PIKK) family. TRRAP plays a critical role in tumorigenesis, as it is essential for transformation driven by the oncogenic transcription factors, MYC and E2F, as well as by the adenovirus E1A oncoprotein131,132. TRRAP serves as a component of several histone acetyltransferase (HAT) complexes, including the STAGA, TFTC and TIP60 complexes133–135. Through its interactions with oncoproteins, such as MYC and E1A, TRRAP recruits HAT complexes to chromatin to promote histone acetylation and stimulate transcription136–138. The pseudokinase domain of TRRAP is required for assembly of functional HAT complexes and for MYC-driven oncogenic transformation139. Although the TRRAP pseudokinase domain does not bind directly to MYC, it does interact with HATs139. Consequently, small molecules targeting TRRAP could potentially be used therapeutically to target MYC-driven cancers by preventing HAT recruitment to sites of MYC-dependent transcription.
Structural features of pseudokinases
The crystal structures of pseudokinase domains that have been resolved to date show that pseudokinases have the same overall kinase domain fold as catalytically active kinases11,12. This highly conserved fold is composed of two lobes, the amino-terminal lobe (N-lobe) and the C-lobe, which are connected via a flexible hinge region (Fig. 3). In active kinases, the relative motions of these lobes during catalysis are associated with conformational transitions in the active site, located between the N-lobes and C-lobes and composed from structural elements provided by both lobes. These motions serve to accommodate nucleotide and substrate binding, ATP hydrolysis, phosphotransfer and the eventual release of ADP and the phosphorylated substrate140,141. The conformational states that correspond to the different steps of catalysis have been captured in crystal structures of active kinases and, interestingly, are emulated by some pseudokinases in their crystal structures.
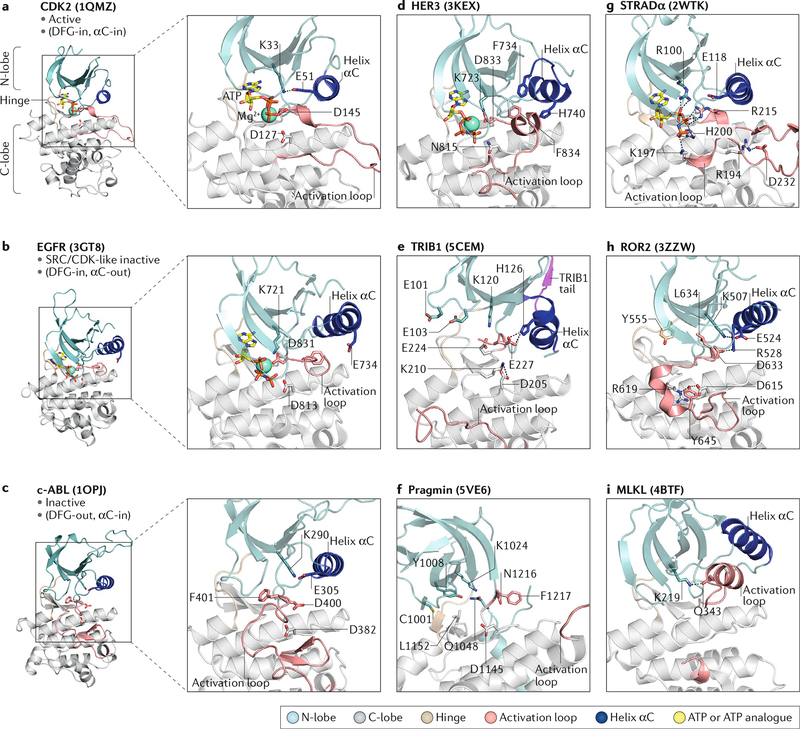
Fig. 3: Structural features of pseudokinases.
Left panels show crystal structures of the CDK2, epidermal growth factor receptor (EGFR) and ABL kinase domains in the active, SRC/CDK-like inactive and inactive conformations, respectively. Insets show zoomed views of the active sites. Right panels show zoomed views of the pseudoactive sites in the crystal structures of the indicated pseudokinases. a | In the active conformation (CDK2 is shown), the β3 lysine (K33) forms a salt bridge with E51 in helix αC, and the DFG (Asp-Phe-Gly) aspartate (D145) coordinates Mg2+ in the active site. b | In the SRC/CDK-like inactive conformation (EGFR is shown), helix αC is shifted away from the active site, preventing a salt bridge between the β3 lysine (K721) and E734. c | In the inactive conformation (ABL is shown), the DFG motif is flipped so that phenylalanine (F401) blocks nucleotide binding. d | Helix αC in human epidermal growth factor receptor 3 (HER3) is partially unwound and adopts a SRC/CDK-like inactive conformation. e | Helix αC is unusually short in TRIB1 and adopts a kink, resulting in a shallow nucleotide-binding pocket. f | Pragmin has a disordered helix αC and occluded nucleotide-binding pocket. g | STE20-related adaptor-α (STRADα) coordinates ATP using positively charged residues. h | The ROR2 DLG motif is shifted ~4 Å compared with canonical DFG motifs and interacts with helix αC, pushing it away from the pseudoactive site. i | Mixed lineage kinase domain-like protein (MLKL) has an unusual activation loop helix that interacts with the β3 lysine (K219). The corresponding Protein Data Bank codes for each structure shown are indicated in parentheses. C-lobe, carboxyl-terminal lobe; N-lobe, amino-terminal lobe.
By definition, pseudokinases lack one or more key conserved catalytic residues: the lysine within the VAIK (Val-Ala-Ile-Lys) motif, the aspartate within the HRD (His-Arg-Asp) motif and the aspartate within the DFG (Asp-Phe-Gly) motif18. The VAIK motif is located in the β3 strand of the N-lobe, which positions the catalytic lysine to coordinate the α and β phosphates of ATP. Proper coordination of ATP is dependent on salt bridge formation between the catalytic lysine and a conserved glutamate residue in helix αC, which is rotated towards the active site in the kinase active state (Fig. 3a). This salt bridge is disrupted in an inactive conformation of the kinase, referred to as the SRC or cyclin-dependent kinase (SRC/CDK)-like inactive conformation, in which helix αC is rotated away from the active site142 (Fig. 3b). Mutation of the β3 lysine is often used as an inactivating mutation to generate a ‘kinase-dead’ mutant. Alteration of this lysine is observed in some pseudokinases, including KSR and STE20-related adaptor-α (STRADα). Other pseudokinases compromise the salt bridge through mutation of the helix αC glutamate, which is sometimes accompanied by even more pronounced structural changes in helix αC. In HER3, the canonical glutamate is replaced by a histidine, and helix αC is partially unwound30,143. A central residue in the unstructured region, Phe734, which is absent in active HER kinases, forms hydrophobic interactions that stabilize the activation loop and helix αC in the SRC/CDK-like inactive conformation (Fig. 3d). Similar to HER3, helix αC in TRIB1 lacks the conserved glutamate and is shorter than it is in most protein kinases. In addition, it adopts an atypical kink that interferes with the canonical structure of the nucleotide-binding pocket and likely contributes to the inability of TRIB1 to bind nucleotides143 (Fig. 3e). Pragmin (also known as SGK223 or PRAG1) and PEAK1 (also known as SGK269) also lack the conserved glutamate residue, and their structures hint at either disorder or a high degree of conformational dynamics within the helix αC region. The N-terminal region of helix αC in PEAK1 has only two helical turns visible in the structure144, while in structures of Pragmin, helix αC is entirely disordered145,146 (Fig. 3f).
The HRD motif located in the catalytic loop is another key motif required for catalysis. The aspartate residue in this motif serves as the catalytic base during ATP hydrolysis, and thus its mutation renders kinases catalytically inactive. Several pseudokinases carry HRD mutations or are missing this motif entirely, including HER3, integrin-linked protein kinase (ILK) and MLKL (Table 2). Other sequence alterations in pseudokinases include substitutions in the glycine-rich loop located in the N-lobe. The glycine-rich loop usually conforms to the consensus sequence GXGXXG in active kinases. The absence of side chains in the glycine residues allows for close contact of the glycine-rich loop with the adenosine ring of ATP, which enables nucleotide binding and proper positioning of ATP for catalysis. In pseudokinases, the glycines are often replaced by larger amino acids, frequently negatively charged, that interfere with ATP binding. This is exemplified by VRK3, in which Asp175 and Gln177 within the glycine-rich loop protrude into the putative ATP-binding pocket147. In TRIB1, Glu101 and Glu103 in the glycine-rich loop are predicted to interfere with ATP binding, and the loop is also shorter than usual, which makes the ATP-binding pocket relatively shallow143 (Fig. 3e).
Table 2.
Classes of pseudokinases
| Pseudokinase | Degraded glycine-rich loop | VAIK motif | HRD motif | DFG motif | Crystal structures | Refs |
|---|---|---|---|---|---|---|
| Class 1 — pseudokinases that do not bind nucleotides or cations | ||||||
| BIR2 (A. thaliana) | Y | Y | N | Y | 4L68 | 259 |
| BUBR1 | Y | Y | Y | Y | NA | 159 |
| GCN2 | Y | N | N | Y | NA | 159 |
| IRAK3 | N | Y | N | Y | NA | 159 |
| MviN (M. tuberculosis) | Y | N | N | N | 3OTV | 159, 160 |
| NRBP1 | Y | N | N | N | NA | 159 |
| Pragmin | Y | Y | Y | N | 5VE6, 6EWX | 145, 146, 159 |
| PTK7 | N | Y | Y | N | NA | 159 |
| ROP8 (T. gondii) | Y | N | N | N | 3BYV | 166 |
| ROR1 | N | Y | Y | Y | NA | 159 |
| ROR2 | N | Y | Y | Y | 3ZZW, 4GT4 | 154 |
| RYK | N | Y | Y | Y | NA | 159 |
| SCYL1 | Y | N | N | N | NA | 159 |
| STK40 | Y | Y | Y | N | 5L2Q | 159, 260 |
| Titin | N | Y | Y | N | 4JNW, 1TKI | 261, 262 |
| TRIB1 | Y | Y | Y | N | 5CEK, 5CEM, 6DC0 | 120, 143 |
| VRK3 | Y | Y | N | N | 2JII | 147 |
| Class 2 — pseudokinases that bind nucleotides in the absence of cations | ||||||
| CASK | N | Y | Y | N | 3C0G, 3C0H, 3C0I, 3MFR (4 M), 3MFS (4 M), 3MFU (4 M), 3TAC | 163, 263, 264 |
| EPHB6 | N | N | N | N | NA | 159 |
| FAM20A | Y | Y | Y | Y | 5WRR, 5WRS, 5YH2, 5YH3 | 112, 164, 215 |
| MLKL | Y | Y | N | N | 4BTF, 4M67, 4M68, 4M69, 4MWI, 5KNJ, 5KO1 | 97, 155,156,– 157 |
| STRADα | N | N | N | N | 2WTK, 3GNI | 152, 153 |
| TRIB2 | Y | Y | Y | N | NA | 162 |
| TRIB3 | Y | Y | Y | N | NA | 162 |
| ULK4 | N | N | Y | N | NA | 159 |
| Class 3 — pseudokinases that bind cations but not nucleotides | ||||||
| PEAK1 | Y | Y | Y | N | 6BHC | 144, 159 |
| ROP2 (T. gondii) | Y | N | Y | N | 2W1Z, 3DZO | 159, 165, 166 |
| Class 4 — pseudokinases that bind to nucleotides and cations | ||||||
| ADCK3 | Y | Y | Y | Y | 4PED | 265 |
| ANP-A | N | Y | N | Y | NA | 266 |
| BSK8 (A. thaliana) | Y | Y | N | N | 4I92, 4I93, 4I94 | 267 |
| HER3 | N | Y | N | Y | 3KEX, 3LMG, 4OTW, 4RIW, 4RIX (Q790R), 4RIY (E909G) | 30, 32, 151 |
| HSER | Y | Y | N | Y | NA | 268 |
| ILK | Y | Y | N | Y | 3KMU, 3KMW, 3REP, 6MIB (L207W) | 209, 211 |
| IRAK2 | Y | Y | N | N | NA | 269 |
| JAK1 JH2 | N | Y | N | Y | 4L00, 4L01 (V685F) | 270 |
| JAK2 JH2 | N | Y | N | Y | 4FVP, 4FVQ, 4FVR (V617F), 5I4N (E596A or V617F), 5USZ, 5UTO, 5UT1, 5UT2, 5UT3, 5UT4, 5UT5, 5UT6 | 169, 171, 203, 204, 271 |
| KSR1 | N | N | Y | Y | NA | 103 |
| KSR2 | N | N | Y | Y | 2Y4I, 5KKR | 172, 199 |
| PAN3 | Y | Y | N | N | 4BWK, 4BWX, 4BWP, 4CYI, 4CYJ, 4CZY, 4XR7 | 272,273,274,– 275 |
| POMK | Y | N | Y | Y | 5GZA, 5GZ8, 5GZ9 | 276,277,– 278 |
| RNase L | Y | Y | Y | Y | 4OAV, 4OAU, 4O1P, 4O1O | 149, 150, 205 |
| ROP5B (T. gondii) | N | Y | N | Y | 3Q5Z, 3Q60, 4LV5 | 213, 214 |
| STKLD1 | N | Y | N | Y | NA | 159 |
| TYK2 JH2 | N | Y | N | Y | 3ZON, 4OLI, 4WOV, 5TKD | 93, 200, 201 |
All protein structures are as per the Protein Data Bank. ANP-A, atrial natriuretic peptide receptor type A; A. thaliana, Arabidopsis thaliana; DFG, Asp-Phe-Gly; HER, human epidermal growth factor receptor; HRD, His-Arg-Asp; HSER, heat-stable enterotoxin receptor; ILK, integrin-linked protein kinase; IRAK2, interleukin-1 receptor-associated kinase-like 3; JAK, Janus kinase; JH2, JAK homology 2; KSR, kinase suppressor of RAS; M. tuberculosis, Mycobacterium tuberculosis; MLKL, mixed lineage kinase domain-like protein; NA, not available; NRBP1, nuclear receptor-binding protein 1; POMK, protein O-mannose kinase; PTK7, protein tyrosine kinase 7; ROP8, rhoptry protein 8; RYK, receptor-like tyrosine kinase; SCYL1, SCY-like protein 1; STK, serine/threonine-protein kinase; STRADα, STE20-related adaptor-α; T. gondii, Toxoplasma gondii; TRIB1, Tribbles homologue 1; VAIK, Val-Ala-Ile-Lys.
Conformational transitions of the activation loop, which adopts a tethered conformation in the inactive state and extends during activation, are necessary for catalysis and substrate binding in active kinases (Fig. 3a–c). This extended conformation is typically promoted by phosphorylation within the activation loop and is sometimes stabilized by an interaction with a binding partner140. Conformational changes within the activation loop are sometimes accompanied by the motions of the N-terminally located DFG motif. In the active ‘DFG-in’ conformation, the aspartate points into the active site and coordinates a Mg2+ ion that interacts with the β-phosphate and γ-phosphate of ATP. In the inactive, ‘DFG-out’ state, the aspartate rotates ~180° away from the active site, and its position in the ATP-binding site is occupied by the neighbouring phenylalanine (Fig. 3c). The DFG-in versus DFG-out states of kinases often control the interactions of active kinases with their binding partners. For example, ERK2 loses the ability to serve as an allosteric activator of the phosphatase MKP3 when stabilized in the DFG-out conformation by a type II inhibitor. By contrast, locking ERK2 in the DFG-in state using a type I inhibitor enhances MKP3 activation148.
Many pseudokinases carry mutations in their DFG motifs or lack this motif entirely (Fig. 2). Those that retain an intact DFG motif often have other alterations that likely prevent the canonical conformational changes observed in active kinases, such as the unusually short activation loops in RNase L and TYK293,149,150. Nevertheless, the emerging theme from structural studies on pseudokinases is that, even when they do not retain canonical motifs like the DFG motif, pseudokinases seem to adopt conformations that resemble the DFG-in, DFG-out or SRC/CDK-like inactive states. These states are achieved in pseudokinases through novel interactions that accommodate their divergent sequences. HER3, for example, has been repeatedly crystallized in the SRC/CDK-like inactive conformation in which it is stabilized through hydrophobic interactions that are missing in other HER kinases30,32,151. STRADα adopts a conformation that resembles the active conformation seen in active kinases152,153, which is stabilized in a unique fashion by a salt bridge between Asp232 in the activation loop of STRADα and Arg194 in the catalytic loop (Fig. 3g). ROR2, however, seems to adopt a conformation that is a hybrid of the DFG-out and SRC/CDK-like inactive states. In ROR2, the phenylalanine in the DFG motif is replaced by a leucine residue that points into the pseudoactive site, mimicking a DFG-out-like state154. This DLG motif in ROR2 is positioned 4.0–4.5 Å away from the usual location of the DFG motif in active kinases, making room for the activation loop to fold into the active site and subsequently push helix αC away, in a manner reminiscent of the SRC/CDK-like inactive state154 (Fig. 3h). Tyr555 in the hinge region of ROR2 directly inserts into the nucleotide-binding pocket and structurally occludes ATP binding in this unique conformational state of the ROR2 pseudokinase (Fig. 3h). Thus, structural features of pseudokinase domains, while mirroring conformational states of active kinases, also seem to be unique and are likely intimately coupled to pseudokinase functions.
Conformational transitions in pseudokinases
Although we know much less about the ability of pseudokinases to toggle between different conformational states and how such changes regulate their functions, structural studies point towards the potential for such mechanisms. Human and murine MLKL adopt strikingly different conformations in crystal structures, suggesting the potential regulation of MLKL function through conformational changes in the activation loop and helix αC. The human MLKL pseudokinase domain was crystallized in a canonical ‘active’ conformation in which the β3 lysine (Lys230) forms a salt bridge with Glu250 in helix αC155–157. In structures of murine MLKL, however, the β3 lysine (Lys219) interacts with Gln343 in the activation loop, which forms an unusual helix that takes the position in the kinase domain typically occupied by helix αC97,155 (Fig. 3i). Mutations that disrupt the noncanonical interaction between the β3 lysine and the activation loop helix result in constitutive activation of MLKL-mediated necroptosis97. Thus, the MLKL pseudokinase domain could potentially toggle between these two different conformational states to control necroptosis.
More direct evidence that conformational transitions underlie pseudokinase signalling has been recently provided by structures of TRIB1. A crystal structure of the TRIB1 pseudokinase domain with its C-terminal tail revealed that the tail binds to a pocket formed by helix αC in the N-lobe of the pseudokinase domain in an interaction reminiscent of the autoinhibitory binding of the C-terminal tails of AGC family kinases to the 3-phosphoinositide-dependent protein kinase 1 (PDK1)-interacting fragment (PIF) pocket143,158 (Fig. 3f). This intramolecular interaction effectively buries the COP1-binding motif, located in the TRIB1 C-terminal tail, such that, in this conformation, TRIB1 would be unable to bind COP1. More recently, binding of the COP1 substrate C/EBPα to the C-lobe of TRIB1 was found to induce long-range conformational changes in the TRIB1 pseudokinase domain, including ordering of the activation loop into a DFG-in-like state and displacement of the C-terminal tail from the N-lobe120. Hence, binding of C/EBPα to TRIB1 has an allosteric effect and exerts positive feedback on COP1-dependent C/EBPα ubiquitylation by increasing the accessibility of the TRIB1 C-terminal tail for binding to COP1. These studies present a compelling example of how the conformational dynamics of a pseudokinase domain can be coupled to its function.
Classes of pseudokinases
Pseudokinases diverge from the canonical structural features of catalytically active kinases to different extents. Numerous pseudokinases retain the capacity to interact with nucleotides and divalent cations, and within this group, a few pseudokinases even have measurable kinase activity in vitro. A comprehensive analysis of these features across a broad spectrum of pseudokinases led to their categorization into four distinct classes. Class 1 pseudokinases do not bind nucleotides or cations; class 2 pseudokinases bind nucleotides in the absence of cations; class 3 pseudokinases bind cations but not nucleotides; and class 4 pseudokinases bind nucleotides and cations159 (Fig. 4; Table 2).
Fig. 4: Accessibility of the putative nucleotide-binding pocket in different classes of pseudokinases.
Surface representations of crystal structures of active kinases in the active DFG-in (CDK2), SRC/CDK-like inactive (EGFR) and inactive DFG-out conformations (c-ABL) (part a), pseudokinases that do not bind nucleotides or cations (part b), pseudokinases that bind nucleotides in the absence of cations (part c), pseudokinases that bind cations but not nucleotides (part d) and pseudokinases that bind nucleotides and cations (part e). For each structure, bound ATP or ATP analogues are shown as sticks, while Mg2+ ions are shown as spheres. Insets show zoomed views of the putative nucleotide-binding pocket for each kinase or pseudokinase. The corresponding Protein Data Bank codes for each crystal structure shown are indicated in parentheses. αC, helix αC; AMP-PNP, nonhydrolysable ATP analogue; C-lobe, carboxyl-terminal lobe; EGFR, epidermal growth factor receptor; HER3, human EGFR; N-lobe, amino-terminal lobe; JAK2, Janus kinase 2; JH2, JAK homology 2; KSR2, kinase suppressor of RAS 2; ROP2, rhoptry protein 2; STRADα, STE20-related adaptor-α.
Class 1 — pseudokinases that do not bind nucleotides or cations
A substantial number of pseudokinases, including TRIB1, VRK3, ROR2, Pragmin and the bacterial pseudokinase MviN, lack the ability to bind traditional ligands, such as nucleotides and metal cations, in their pseudoactive sites159. Crystal structures of several of these pseudokinases offer an explanation for this deficiency by revealing highly distorted nucleotide-binding sites, often occluded by hydrophobic residues143,145–147,154,160 (Figs 3,4). Effectively, such pseudokinases do not have the canonical druggable pocket present in active kinases, which constitutes a major challenge for targeting using small-molecule therapeutics (Fig. 4b). However, these divergent pseudoactive sites could potentially accommodate nonconventional ligands, either physiological or experimentally developed. As discussed in more detail below, close homologues of several class 1 pseudokinases, such as TRIB2 and PEAK1, retain the ability to bind canonical kinase ligands even though their pseudoactive sites are similarly distorted.
Class 2 — pseudokinases that bind nucleotides in the absence of cations
Divalent metal binding, most typically of Mg2+, neutralizes the net negative charge of the nucleotide and enables nucleotide binding to the kinase active site; divalent metals are also essential for ATP coordination and the chemistry of phosphotransfer161. However, several pseudokinases, such as STRADα, MLKL, FAM20A, CASK and TRIB2, retain the ability to bind to ATP even in the absence of cations97,152,162–164. In fact, the presence of Mg2+ inhibits nucleotide binding to some of these pseudokinases97,162,164. With the exception of FAM20A, most of the pseudokinases within this class lack the canonical metal-coordinating aspartate residue found in the DFG motifs of active kinases (Table 2). In the remarkable case of STRADα, the pseudoactive site lacks the β3 lysine, DFG motif and the catalytic aspartate in the HRD motif but still binds ATP in an orientation similar to that observed for active kinases. The phosphates of ATP are coordinated by basic residues that occupy the space in the STRADα pseudoactive site where Mg2+ is coordinated by the DFG aspartate in active kinases152 (Fig. 3g). In FAM20A, although present, the DFG motif is not involved in ATP binding, perhaps owing to the lack of metal coordination. In a manner somewhat similar to STRADα, ATP is coordinated by basic residues in FAM20A, but ATP binds to FAM20A in a unique orientation in which it is inverted relative to the position the nucleotide adopts in canonical kinases164.
Unexpectedly, CASK and TRIB2 have been reported to catalyse phosphorylation in vitro, but only in the presence of a metal chelator, such as EDTA162,163. Although there is no crystal structure of TRIB2 to date, the crystal structure of the CASK pseudokinase domain bound to a nonhydrolysable ATP analogue, AMP-PNP, does not reveal an obvious mechanism for ATP hydrolysis. The β-phosphate and γ-phosphate of the nucleotide, which are typically coordinated by Mg2+ in structures of canonical kinases, are disordered in the CASK structure163. Owing to the high cellular concentrations of Mg2+, it also remains unclear how CASK and TRIB2 would be able to efficiently catalyse phosphorylation in cells. As CASK plays a key role in synaptic function, it has been proposed that its catalytic activity could be regulated by the changes in cellular Mg2+ and Ca2+ concentrations associated with synaptic activity163.
Class 3 — pseudokinases that bind cations but not nucleotides
One surprising outcome of the analysis performed by Murphy and colleagues was the identification of a distinct class of pseudokinases that bind divalent cations but not nucleotides or ATP-competitive small molecules159. This small class of pseudokinases is currently composed of the Toxoplasma protein rhoptry protein 2 (ROP2) and PEAK1, a close homologue of Pragmin. Crystal structures of the pseudokinase domains of PEAK1 and ROP2 reveal that both possess highly occluded pseudoactive sites that seem structurally incompatible with nucleotide binding144,165 (Fig. 4d). Thus far, divalent cations have not been resolved in any of the crystal structures of either PEAK1 or ROP2, which leaves their binding site undefined144,165,166. As neither of these pseudokinases possesses an intact DFG motif (Table 2), it is likely that these binding sites will not resemble those of canonical kinases. It remains unclear whether cations bind to PEAK1 and ROP2 in a physiological setting and what role metal ion binding plays in the function of these pseudokinases. A documented example of MAPK kinase signalling regulation by copper binding sets an intriguing precedent for the unconventional allosteric role of metal ligands in the regulation of protein kinases167.
Class 4 — pseudokinases that bind nucleotides and cations
Class 4 pseudokinases retain the ability to bind both nucleotides and divalent cations and include HER3, KSR, RNase L and the JH2 pseudokinase domains of the JAKs (Table 2). Typically, these pseudokinases have an intact DFG aspartate, but there are exceptions, such as the interleukin-1 receptor-associated kinase-like 2 (IRAK2) and the plant pseudokinase BSK8 (Table 2). Many of the pseudokinases in this class, including HER3, the JH2 domain of JAK2 and KSR, have measurable kinase activity, typically a few orders of magnitude lower than that of catalytically active kinases. For example, HER3 does not have measurable activity towards a generic tyrosine substrate peptide in vitro30, but its autophosphorylation activity towards its C-terminal tail can be detected in vitro, albeit the activity is ~1,000-fold lower than that of EGFR towards its respective C-terminal tail151. HER3 pseudokinase domain mutations that further reduce this activity do not have a measurable effect on HER3-mediated signalling in cells168.
The JH2 pseudokinase domain of JAK2 was demonstrated to have weak autophosphorylation activity in vitro169. Engineered mutations that disrupt ATP binding to the JH2 domain have only minor effects on wild-type JAK2 signalling, but they prevent the hyperactivation that occurs in the pathogenic JAK2-V617F mutant170. Mutation of the putative JH2 autophosphorylation sites has no effect on the hyperactivity of the JAK2-V617F mutant170, indicating that the ability of the JH2 domain to bind ATP — not JH2 catalytic activity — contributes to the pathogenic signalling by JAK2-V617F. As suggested by molecular dynamics simulations, ATP binding might play a structural role in stabilizing helix αC in the JH2 domain, which is intrinsically more disordered in disease-associated JAK2 mutants170,171. Although no activity has been associated with the highly homologous JH2 domain of JAK1, hyperactivating disease-associated mutations in JAK1 JH2 that are analogous to those found in JAK2 JH2, such as JAK1-V658F, also seem to rely on ATP binding to the JH2 domain for pathological signalling170.
KSR1 and KSR2 were reported to become catalytically activated through heterodimerization with BRAF or CRAF, resulting in phosphorylation of MEK103,172. This phenomenon was detected upon treatment with the RAF inhibitors PLX4720 or GDC-0879 and led to the hypothesis that KSR kinase activity might be involved in paradoxical activation of the MAPK pathway103,172. More recently, however, mutations in the KSR1 active site that were expected to ablate KSR activity had no impact on MEK phosphorylation104. By contrast, inactivating mutations in BRAF reduced MEK phosphorylation, indicating that the kinase activity of BRAF, and not that of KSR, is required for MEK phosphorylation by the BRAF–KSR complex104. Consequently, the allosteric function of KSR, rather than its potential catalytic activity, appears to be more important for its role in MAPK signalling. Thus, as with the class 2 pseudokinases, CASK and TRIB2, the physiological relevance of the catalytic activity associated with class 4 pseudokinases remains unclear.
Targeting the nucleotide-binding site
Lessons from paradoxical effects of inhibitors targeting active kinases
Occupancy of the nucleotide-binding pocket in catalytically active protein kinases can be coupled to conformational changes in distal parts of the kinase domain and, in this way, allosterically regulate interactions of kinases with their binding partners11,173–175. The importance of these long-range interactions is underscored by numerous studies that revealed paradoxical effects of ATP-competitive inhibitors on their target kinases. A prototypical example is provided by BRAF inhibitors, including both type I and type II inhibitors, such as GDC-0879 and AZ-628, respectively, that counterintuitively cause activation of the MAPK pathway at subsaturating concentrations176. These inhibitors stabilize helix αC of BRAF in the ‘in’ position and promote dimerization with CRAF, resulting in its allosteric activation and enhanced binding of BRAF to RAS-GTP177–181. RAF dimerization also undermines the efficiency of another class of RAF inhibitors, including vemurafenib and dabrafenib, which stabilize helix αC in the ‘out’ position. The RAF dimerization interface is not sterically compatible with both protomers adopting an αC-out conformation. Therefore, only the inhibitor-bound protomer is in an αC-out state, and the other protomer is forced to adopt an αC-in conformation that is competent to bind ATP and activate MAPK signalling180. Understanding the structural basis for the activating effect of these inhibitors enabled design of next-generation BRAF inhibitors that stabilize the kinase in a conformation that is not compatible with BRAF–CRAF dimerization or interaction with RAS-GTP180,182,183. Another strategy to avoid paradoxical activation utilizes pan-RAF inhibitors that target both BRAF and CRAF to prevent the CRAF-mediated phosphorylation of MEK that is induced by BRAF-selective inhibitors184.
A somewhat similar mechanism appears to regulate PKR-like ER kinase (PERK; also known as EIF2αK3), a transmembrane kinase sensor involved in the unfolded protein response. Subsaturating concentrations of an ATP-competitive inhibitor of the PERK kinase domain induce dimerization and activation of PERK kinase activity185. This is thought to occur through a mechanism reminiscent of the paradoxical activation of BRAF in which the inhibitor stabilizes the kinase domain of PERK in a dimer where only the active site of one protomer is occupied by the compound. This inhibitor-bound protomer could then allosterically activate an apo protomer that is still capable of binding ATP.
Another example of successful pharmacological modulation of noncatalytic kinase signalling is that of IRE1, another transmembrane regulator of the unfolded protein response. IRE1 oligomerization induced by binding of misfolded proteins on the lumenal side of the endoplasmic reticulum results in activation of the kinase on the cytosolic side. Activation and autophosphorylation of the kinase domain then promote RNase domain activation. The same effect can be achieved by binding of a type I inhibitor to the IRE1 kinase domain186,187. By stabilizing the kinase domain in an active DFG-in conformation, type I inhibitors promote IRE1 oligomerization, which is the critical determinant of RNase domain activation186–188. By contrast, compounds that lock the IRE1 kinase domain in an inactive DFG-out conformation prevent both kinase oligomerization and activation of the RNase domain188–190.
Conformation-selective inhibitors of Aurora kinase A were also found to modulate the extent of its protein–protein interactions. Aurora kinase A is often overexpressed in cancer, and one of its therapeutically important functions is to stabilize the oncogenic transcription factor N-MYC through a direct interaction191. This function of Aurora kinase A is independent of its kinase activity. Type I kinase inhibitors, such as hesperadin, do not interfere with N-MYC binding to Aurora kinase A. By contrast, another class of Aurora kinase A inhibitors, including MLN8054, MLN8037 and CD532, successfully disrupts binding of N-MYC by stabilizing an unusual conformation of the DFG motif that shifts helix αC away from the active site of Aurora kinase A192,193.
Most recently, the ATP-competitive HER2 inhibitor, lapatinib, was found to cooperate at low concentrations with the growth factor NRG1, a HER3 ligand, to promote the proliferation of HER2+ breast cancer cells194. Lapatinib stabilizes a SRC/CDK-like inactive conformation of the HER2 kinase domain that is incompatible with heterodimerization with HER3 in the canonical asymmetric dimer mode in which HER2 is allosterically activated by HER332,33,195,196. Instead, lapatinib-bound HER2 was shown to engage in different HER2–HER3 heterodimers driven by symmetrical ‘head-to-head’ N-lobe interactions, which were previously observed in crystal structures of the HER3 pseudokinase domain30,194 and the inactive HER4 kinase domain196. These symmetrical heterodimers composed of inhibited HER2 and the pseudokinase HER3 both have their allosteric activator interfaces available for the formation of canonical asymmetric dimers with other HER receptors. Indeed, lapatinib was shown to promote higher-order HER2 and HER3 receptor oligomers that are thought to serve as nucleation points for the recruitment of other HER receptors and their activation through asymmetric kinase dimerization194. These larger clusters of receptors could explain how lapatinib promotes downstream HER2–HER3 signalling, much as EGFR clustering has been functionally linked to EGFR signalling197,198.
Known ATP-competitive modulators of pseudokinases
Collectively, what we have learned from these studies of active kinases is that ATP-competitive small molecules can be used to manipulate the noncatalytic functions of kinases by stabilizing conformations that disrupt existing interactions or by promoting novel interactions that result in a modified signalling output. Multiple examples support this approach as a viable strategy for therapeutic targeting of pseudokinases that have an accessible ATP-binding pocket29 (Table 3).
Table 3.
Known modulators of pseudokinase function
| Molecule or antibody name | Type | Mechanism of action | Refs |
|---|---|---|---|
| EPHA10 | |||
| BsAb | mAb | Bispecific antibody that targets EPHA10 and CD3; induces redirected T cell-mediated lysis of EPHA10-overexpressing cells | 257 |
| Db-1 and Db-2 | Diabody | Heterodimeric (Db-1) and homodimeric (Db-2) diabodies that target EPHA10 and CD3; induce redirected T cell-mediated lysis of EPHA10-overexpressing cells | 279 |
| HER3 | |||
| Elgemtumab (LJM716) | mAb | Binds to inactive conformation of ECD but does not prevent ligand binding; inhibits ligand-dependent and ligand-independent HER3 signalling | 280 |
| Duligotuzumab (MEHD-7945A) | mAb | Dual specificity antibody that recognizes the inactive conformations of the HER3 and EGFR ECDs; blocks ligand binding | 248, 249 |
| Seribantumab (MM-121) | mAb | Inhibits ligand-induced phosphorylation of HER3 | 242 |
| Patritumab (U3–1287 or AMG-888) | mAb | Promotes receptor internalization; inhibits ligand-induced HER3 signalling | 245, 246, 281 |
| U3–1402 | ADC | ADC consisting of patritumab conjugated to the topoisomerase I inhibitor DX-8951 | 282 |
| Lumretuzumab (RG7116) | mAb | Binds to inactive conformation of ECD and blocks ligand binding; inhibits ligand-induced HER3 signalling; exhibits enhanced ADCC due to glycoengineering of Fc moiety | 243 |
| FL518 | mAb | Four-in-one antibody made by combining duligotuzumab and bH1–44 that recognizes EGFR, HER2, HER3 and VEGF; inhibits ligand-dependent and ligand-independent signalling | 252 |
| AV-203 | mAb | Blocks ligand binding and prevents dimerization with HER2; inhibits ligand-dependent and ligand-independent HER3 signalling | 283 |
| GSK2849330 | mAb | Disrupts ligand-dependent HER3 signalling; engineered to exhibit enhanced ADCC and cell division cycle | 284 |
| KTN3379 | mAb | Prevents HER2–HER3 heterodimerization; inhibits both ligand-dependent and ligand-independent HER3 signalling | 285 |
| REGN1400 | mAb | Blocks ligand binding | 286 |
| MCLA-128 | mAb | Bispecific antibody targeting both HER2 and HER3 that exhibits enhanced ADCC | 287 |
| TX2–121-1 | Small molecule | Irreversible ATP-competitive inhibitor that forms covalent adduct with Cys721 in HER3; induces degradation of HER3 and interferes with heterodimerization with HER2 and MET | 232 |
| Bosutinib | Small molecule | ATP-competitive SRC and ABL inhibitor that promotes HER3 heterodimerization with HER2 and promotes proliferation of HER2+ breast cancer cells | 194 |
| CT8 | Small molecule | Binds Sec61 translocon and interferes with cotranslational insertion of HER3 into the ER, resulting in HER3 degradation | 235 |
| JAK2 JH2 | |||
| JNJ-7706621 | Small molecule | ATP-competitive pan-CDK and Aurora A and Aurora B inhibitor that binds to both JAK2 JH1 and JH2 | 203, 204 |
| AT-9283 | Small molecule | ATP-competitive Aurora A, Aurora B, JAK2 and JAK3 inhibitor that binds to both JAK2 JH1 and JH2 | 203 |
| KSR2 | |||
| APS-2–79 | Small molecule | ATP-competitive inhibitor that stabilizes an inactive conformation of the pseudokinase domain; prevents heterodimerization with RAF kinases and stabilizes inactive complex with MEK | 199 |
| ASC-24 | Small molecule | ATP-competitive inhibitor that blocks KSR kinase activity in vitro but does not prevent heterodimerization with RAF kinases | 172 |
| MLKL | |||
| Compound 1 | Small molecule | ATP-competitive VEGFR and RIPK1 inhibitor that stabilizes DFG-out conformation of MLKL pseudokinase domain; inhibits necroptosis | 99, 157 |
| Compound 4 | Small molecule | ATP-competitive small molecule that stabilizes DFG-in conformation of MLKL pseudokinase domain; more selective for MLKL than compound 1 but fails to inhibit necroptosis | 157 |
| PTK7 | |||
| PF-06647020 | ADC | ADC consisting of the anti-PTK7 mAb h6M24 conjugated to the auristatin microtubule inhibitor Aur0101 | 258 |
| RNase L | |||
| Sunitinib | Small molecule | ATP-competitive inhibitor of PKR, PDGFR and VEGFR; inhibits RNase L activity and dimerization | 206 |
| Flavonols | Small molecule | ATP-competitive activators of yeast IRE1 RNase activity; inhibit RNase L activation | 206 |
| ROR1 | |||
| 3B8, 1C11, 1D8, 4C10, 4A7 | mAb | mAbs raised against immunoglobulin (3B8), CRD (1C11, 1D8) and KNG (4A7, 4C10) domains of ROR1 ECD; induce apoptosis of patient-derived CLL cells | 253 |
| Cirmtuzumab (UC-961) | mAb | Inhibits WNT5a-induced activation of RAC1 | 288 |
| ROR1-CAR | CAR | ROR1-specific CAR generated based on a mAb that binds to the ECD of ROR1; T cells transduced with ROR1–CAR selectively lyse ROR1-overexpressing cancer cells | 289 |
| RYK | |||
| RWD1 | mAb | Binds to WIF domain of ECD; inhibits WNT5A-induced signalling and neurite outgrowth | 290 |
| TRIB2 | |||
| Afatinib | Small molecule | Irreversible ATP-competitive inhibitor of EGFR and HER2; forms covalent adduct with Cys96 in TRIB2; destabilizes TRIB2 in vitro and induces its degradation in cells | 233 |
| Neratinib, osimertinib | Small molecule | Irreversible ATP-competitive inhibitors of EGFR and HER2; destabilize TRIB2 in vitro | 233 |
| Lapatinib, TAK-285, GW693881A | Small molecule | Reversible ATP-competitive inhibitors of EGFR and HER2; stabilize TRIB2 in vitro | 233 |
| GW804482X | Small molecule | Reversible ATP-competitive inhibitor of PLK; destabilizes TRIB2 in vitro | 233 |
| TYK2 JH2 | |||
| Compound 1, BMS-066 | Small molecule | ATP-competitive inhibitor that blocks TYK2-mediated signalling through IL-23R in T cells | 200 |
| Compound 35 | Small molecule | ATP-competitive inhibitor that blocks TYK2-mediated signalling through IL-23R in T cells with greater selectivity and metabolic stability than compound 1 | 201 |
| BMS-986165 | Small molecule | ATP-competitive inhibitor that blocks TYK2-mediated signalling through IL-12, IL-23 and type I interferon signalling | 202 |
ADC, antibody–drug conjugate; ADCC, antibody-dependent cell-mediated cytotoxicity; CAR, chimeric antigen receptor; CDK, cyclin-dependent kinase; CLL, chronic lymphocytic leukaemia; CRD, cysteine-rich domain; DFG, Asp-Phe-Gly; ECD, extracellular domain; EGFR, epidermal growth factor receptor; ER, endoplasmic reticulum; Fc, crystallizable fragment of an antibody; HER, human epidermal growth factor receptor; IL-23R, interleukin-23 receptor; JAK, Janus kinase; JH, JAK homology; KNG, kringle; KSR, kinase suppressor of RAS; mAb, monoclonal antibody; MET, hepatocyte growth factor; MLKL, mixed lineage kinase domain-like protein; PTK7, protein tyrosine kinase 7; RIPK1, serine/threonine-protein kinase 1; RYK, receptor-like tyrosine kinase; TRIB2, Tribbles homologue 2; VEGF, vascular endothelial growth factor; VEGFR, VEGF receptor; WIF, WNT-inhibitory factor.
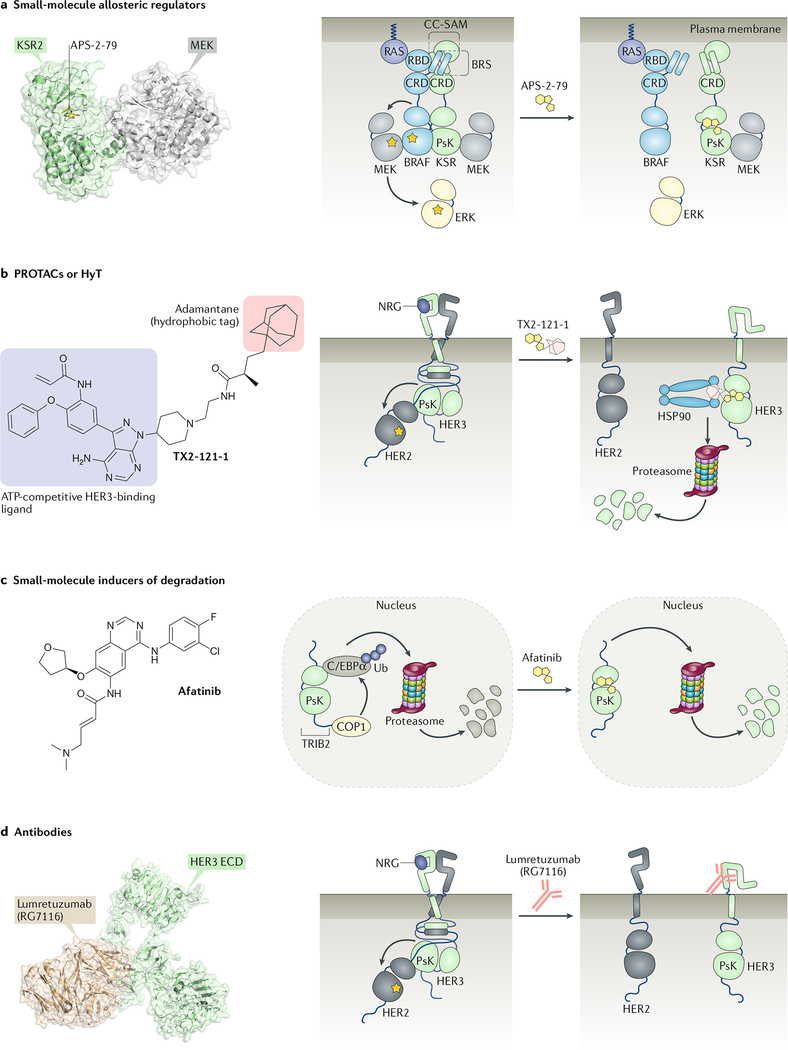
The allosteric function of KSR2 can be modulated by an ATP-competitive small molecule, APS-2–79, which stabilizes a conformation of KSR2 that is incompatible with its interaction with RAF. This compound reduces RAF-mediated phosphorylation of MEK by promoting the interaction of KSR2 with MEK, which results in occlusion of the Ser218 and Ser222 phosphorylation sites on MEK199 (Fig. 5a). By contrast, the type I inhibitor ASC-24, which stabilizes a more ‘active-like’ conformation of the KSR2 pseudokinase domain, does not interfere with the ability of KSR2 to heterodimerize with RAF and RAF-mediated phosphorylation of MEK172,199.
Fig. 5: Strategies for pharmacological targeting of pseudokinases.
a | Small molecules can be used to allosterically modulate the conformation and interactions of pseudokinases, as illustrated by APS-2–79, an ATP-competitive inhibitor of kinase suppressor of RAS 2 (KSR2), that stabilizes an inactive conformation of KSR2 that is incompatible with heterodimerization with RAF and that blocks RAF-dependent phosphorylation sites on MEK. A crystal structure of KSR2 bound to APS-2–79 and MEK is shown in the left panel. b | Proteolysis-targeting chimaeras (PROTACs) or hydrophobic tagging (HyT) can be used to induce degradation of a protein of interest. The human epidermal growth factor receptor 3 (HER3) inhibitor TX2–121-1 consists of a portion that binds covalently to the HER3 nucleotide-binding pocket (blue) connected to adamantane (red), a hydrophobic moiety that mimics the presence of an unfolded protein. Binding of TX2–121-1 leads to proteasomal degradation of HER3, a process that is aided by chaperones, including HSP70 and HSP90. c | Small molecules other than the bifunctional ligands employed for PROTAC-mediated or HyT-mediated protein degradation can be used to destabilize pseudokinases and induce their degradation. Covalent inhibitors of epidermal growth factor receptor (EGFR) and HER2, such as afatinib, destabilize the pseudokinase TRIB2 and promote its proteasomal degradation. d | Antibodies can be used to target the extracellular domains (ECDs) of receptor tyrosine kinases that possess pseudokinase domains. The monoclonal antibody lumretuzumab (RG7116) locks the HER3 ECD in an inactive conformation that prevents ligand binding. A crystal structure of an antigen-binding fragment, derived from lumretuzumab, bound to the HER3 ECD is shown in the left panel. C/EBPα, CCAAT enhancer-binding protein-α; CRD, cysteine-rich domain; NRG, neuregulin.
In HER3, occupancy of the nucleotide-binding pocket seems to play a structural role that enables it to heterodimerize with HER2 through both asymmetric and symmetric dimers194. Mutations in HER3 that prevent ATP binding abrogate NRG-induced phosphorylation of HER3 by HER2, as well as lapatinib-induced symmetric heterodimerization with HER2. By contrast, the ATP-competitive inhibitor bosutinib, which binds to HER3 with nanomolar affinity and stabilizes the same conformation of the HER3 pseudokinase domain as ATP does, promotes HER2–HER3 heterodimerization and ligand-independent proliferation of HER2+ breast cancer cells194. These findings suggest that molecules that occupy the nucleotide-binding pocket of HER3 in a similar fashion to ATP promote its heterodimerization with other HER kinases and downstream signalling.
Compounds that bind to the JH2 pseudokinase domain of TYK2, but not the JH1 kinase domain, were identified in a chemogenomic screen for kinase inhibitors that antagonize interleukin-23 receptor (IL-23R) signalling. These molecules inhibited TYK2 autophosphorylation, as well as TYK2-mediated phosphorylation of signal transducer and activator of transcription 3 (STAT3) in cellular assays, but had no effect on the enzymatic activity of the purified JH1 kinase domain in vitro200. This effect could be achieved if the compounds prevent activation of the JH1 domain indirectly through binding to the JH2 pseudokinase domain200. In the crystal structure of the JH2 pseudokinase domain bound to one of these compounds, BMS-066, the JH2 pseudokinase domain adopts a SRC/CDK-like inactive conformation, similar to that observed in the structure of the autoinhibited form of the pseudokinase–kinase module of TYK293,200. Thus, the inhibitory effect of compounds, such as BMS-066, was proposed to result from stabilization of an autoinhibited conformation of TYK2. Further optimization of the top hits from the screen yielded inhibitors with even greater potency, metabolic stability and selectivity for the TYK2 JH2 domain201. In a phase II clinical trial, BMS-986165, which binds selectively to the TYK2 JH2 domain and potently inhibits activation of the JH1 kinase domain, was found to substantially enhance skin clearance in patients with moderate-to-severe psoriasis202. Phase III studies evaluating BMS-986165 in psoriasis are currently enrolling (NCT03611751 and NCT03624127), while phase II studies of this compound for treatment of Crohn’s disease and systemic lupus erythematosus are ongoing (NCT03599622 and NCT03252587).
Several ATP-competitive small molecules, including the pan-CDK and Aurora inhibitor JNJ-7706621 and the Aurora, JAK2 and JAK3 inhibitor AT-9283, were found to bind to the JH2 domain of JAK2203,204. In co-crystal structures of the JAK2 JH2 pseudokinase domain in complex with these compounds, the JH2 domain adopts the same conformation as TYK2 JH2 bound to BMS-066, suggesting that these compounds could potentially inhibit JAK2 signalling by stabilizing an autoinhibitory interaction between the JH1 and JH2 domains200,203. Further studies are needed to increase the selectivity of these compounds and to determine whether they can indeed modulate JAK2 signalling.
Following the mode of its active counterpart, IRE1, in which nucleotide binding to the kinase domain activates the adjacent RNase domain through oligomerization, the pseudokinase domain of RNase L requires nucleotide binding to allosterically activate its RNase domain150,205. ATP, ADP and, to a slightly lesser extent, AMP-PNP exert this activating effect through binding to the RNase L pseudokinase domain150. Structural studies demonstrate that these nucleotides stabilize the pseudokinase domain in a conformation reminiscent of the active DFG-in state of catalytically active kinases, indicating that, as observed for IRE1, this conformation of the pseudokinase domain is important for RNase activation in RNase L149,150. Several compounds, including sunitinib, an ATP-competitive inhibitor of multiple RTKs, as well as flavonols, such as quercetin, were found to inhibit RNase L activity and dimerization206. Addition of an excess of ADP overcomes the inhibitory effect of these compounds, indicating that both sunitinib and the flavonols act through the ATP-binding pocket of the RNase L pseudokinase domain, likely by stabilizing a conformation that is incompatible with activation of the RNase domain206.
ATP-binding pseudokinases with unclear relevance for modulation by ATP-competitive small molecules
For the majority of pseudokinases with demonstrated nucleotide-binding ability, the feasibility of targeting their functions using ATP-competitive compounds remains to be determined. Recently, a type II kinase inhibitor, referred to as compound 1, that binds the nucleotide-binding pocket of the MLKL pseudokinase domain was identified. Upon binding, compound 1 inhibited the translocation of MLKL to the plasma membrane and the induction of necroptosis in response to tumour necrosis factor (TNF), SMAC-mimetic and the pan-caspase inhibitor Q-VD-OPh. These findings led to the hypothesis that the compound interferes with the conformational change induced by RIPK3-mediated phosphorylation of the MLKL activation loop that is necessary for MLKL translocation and oligomerization99. Although apo MLKL adopts a DFG-in conformation in crystal structures, it crystallizes in the DFG-out conformation when bound to compound 1, supporting the idea that MLKL signalling could be regulated by conformationally selective ATP-competitive small molecules155–157. Regrettably, compound 1 is not very specific and was found to inhibit several other kinases, including RIPK1157. Whether a highly selective type II inhibitor of MLKL would inhibit necroptosis, and thus whether ATP-competitive small molecules can inhibit necroptosis through binding to MLKL, remains unclear.
The pseudokinase ILK is overexpressed in multiple types of cancer, including colorectal cancer and NSCLC, in which ILK promotes cell migration and invasion by triggering F-actin filament bundling207–209. The ILK pseudokinase domain was crystallized in complex with the focal adhesion protein α-parvin both with and without Mg2+-ATP bound in the pseudoactive site210. In both structures, ILK and α-parvin interact in a fashion that resembles a canonical kinase–substrate interaction. In this binding mode, the ILK pseudoactive site, including a portion of the activation loop, forms a major part of the interface with α-parvin. The binding interface is nearly identical in the apo and ATP-bound structures, and mutations that disrupt ATP binding have no effect on the interaction with α-parvin or the localization of ILK to focal adhesions211. Interestingly, another mutation (L207W) in the ILK pseudoactive site that prevents ATP binding markedly impairs the ability of ILK to promote F-actin bundling209. This observation hints at the potential for regulation of ILK function in cell migration using ATP-competitive small molecules.
Another pseudokinase in which disruption of ATP binding does not seem to have a functional consequence is the T. gondii pseudokinase ROP5, which contributes to virulence by allosterically inhibiting the immunity-related interferon-inducible GTPase 1 (IIGP1; also known as IRGA6)212. There are no noticeable differences in the conformation of ROP5 in the crystal structures of the apo and ATP-bound states of the pseudokinase domain213. The same conformation of the ROP5 pseudokinase domain was also observed in a co-crystal structure of ROP5 in complex with ADP and IIGP1214. Thus, the role of nucleotide binding in ROP5-dependent virulence remains poorly understood.
Likewise, the secretory pathway pseudokinase FAM20A, which binds to ATP and, to a lesser extent, to ADP, GTP, CTP and UTP112,164, adopts the same conformation in crystal structures in the apo and ATP-bound states164. Whether ATP binding plays a role in regulating FAM20A function has yet to be determined. Recent structural studies suggest that ATP-binding stabilizes heterotetramerization of the FAM20A–FAM20C kinase complex (in 2:2 stoichiometry), but the importance of this oligomeric state in signalling remains unknown215.
Alternative approaches
For pseudokinases in which nucleotide-binding seems to not affect conformational state or function and for those whose divergent putative nucleotide-binding pockets preclude ligand binding, alternative strategies need to be considered. These approaches could rely on allosteric modulators that use binding sites outside of the canonical nucleotide-binding pocket, molecules that recruit degradation machinery to target pseudokinases and therapeutic antibodies.
Allosteric inhibitors
At present, there are no examples of non-ATP-competitive compounds that modulate pseudokinase function. A growing number of such molecules have been reported for active kinases, demonstrating the feasibility of this strategy for targeting kinase functions. In 2013, the type III inhibitor of MEK1 and MEK2, trametinib, became the first allosteric kinase inhibitor to be approved by the FDA, and since then, several other allosteric kinase inhibitors have entered clinical trials. Type III MEK inhibitors, such as trametinib, bind in a small pocket adjacent to the nucleotide-binding site and stabilize a SRC/CDK-like inactive conformation of the kinase domain216. Thus, trametinib provides an example of conformational selection that is incompatible with MEK1 and MEK2 signalling even when ATP binding is preserved.
The allosteric EGFR inhibitor EAI045 binds selectively to the drug-resistant EGFR-T790M mutant. Like trametinib, EAI045 does not preclude ATP binding because it occupies an adjacent pocket within the active site that only becomes available in the SRC/CDK-like inactive conformation owing to displacement of helix αC away from the active site217. Because EAI045 cannot bind to the kinase domain in the active state, its efficacy is greatly improved when used in combination with the anti-EGFR antibody cetuximab. Cetuximab, which inhibits EGFR dimerization, promotes the SRC/CDK-like inactive state of the EGFR kinase and hence improves EAI045 binding217, providing an elegant example of a potentially very effective combination therapy.
Another class of allosteric inhibitors explores functionally important sites on kinases that are located distally from the nucleotide-binding pocket. A series of AKT inhibitors, including MK-2206 and AKT inhibitor VIII, bind to the interface between the N-lobe of the kinase domain and the adjacent Pleckstrin homology (PH) domain and thereby lock the AKT kinase domain in an inactive conformation that prevents ATP binding, membrane association and activation loop phosphorylation218,219. Similarly, small molecules that bind to the myristate-binding pocket in the C-lobe of the kinase domain of ABL modulate association of the SRC homology 2 (SH2) and SH3 domains with the kinase domain to control ABL activity. These type IV inhibitors can lead to opposing changes in ABL activity depending on the nature of their effect on inhibition of the kinase domain mediated by the SH2 or SH3 domain. The small molecule GNF-2 locks ABL in an autoinhibited state in which the SH2 and SH3 domains interact tightly with the kinase domain, whereas bulkier ligands, such as the small molecule DPH, activate ABL kinase activity by preventing the SH2 and SH3 domains from adopting the autoinhibited conformation220,221. In addition to its effect on stabilizing an inactive conformation of ABL, GNF-2 also sensitizes ABL to ATP-competitive inhibitors, thereby capitalizing on the allosteric connection between the kinase C-lobe and the active site222.
For multidomain targets in which the kinase domain is functionally controlled by other domains, effective allosteric inhibitors could exert their effect by binding to sites outside the kinase domain. A precedent for such regulation is set by the small molecule SSR128129E, which binds to the extracellular domain of the fibroblast growth factor receptor (FGFR) and induces a conformational change that biases signalling223,224. Recent studies of RTK and cytokine receptor signalling have further demonstrated the remarkable communication between the conformation of receptor extracellular domains and downstream signalling kinetics and pathway bias198,225,226. While the extracellular domains of RTKs are typically targeted using antibody-based therapeutics, the example of SSR128129E illustrates how small molecules could be used to modulate downstream signalling through binding to the extracellular domain. This approach could therefore be applied to regulate signalling by pseudokinase RTKs.
Allosteric inhibitors have also been successfully used to disrupt intermolecular protein–protein interactions that involve protein kinases. Small molecules that bind to the PIF pocket, located in the N-lobe of the PDK1 kinase domain, disrupt the interaction of PDK1 with substrates that specifically bind this pocket through a conserved hydrophobic motif227,228. A site analogous to the PIF pocket is involved in the receiver interface of the asymmetric dimer formed by HER kinases33. Small molecules targeting this interface would be expected to prevent asymmetric dimer formation and thereby inhibit HER signalling.
Induced protein degradation
A promising approach for pharmacological targeting of pseudokinases relies on induced protein degradation through proteolysis-targeting chimaeras (PROTACs) or hydrophobic tagging (HyT)229. PROTACs are bifunctional small molecules that bind to a target protein and recruit a ubiquitin ligase, such as von Hippel–Lindau disease tumour suppressor (VHL) or cereblon, to promote proteasomal degradation of the target. In HyT, a hydrophobic moiety, such as adamantane or tert-butyl carbamate-protected arginine (Boc3Arg), is appended to the target protein to induce degradation through the unfolded protein response. Using ATP-competitive small molecules, PROTACs have been developed for several active protein kinases, such as RIPK2, CDK9 and TBK1, as well as the RTKs EGFR, HER2 and MET230,231. HyT has been used to induce degradation of the pseudokinase HER3232. In this approach, an adamantane moiety was added to a covalent ATP-competitive HER3 binder, TX1–185-1, to generate TX2–121-1, which induced proteasomal degradation of HER3 and reduced its heterodimerization with HER2 and MET232 (Fig. 5b). Analogous molecules could be particularly useful for targeting other pseudokinases that are capable of nucleotide binding but whose signalling is not sensitive to conformational changes induced by the binding of ATP analogues.
Small molecules other than PROTAC and HyT bifunctional ligands can also induce protein degradation. For example, ATP-competitive covalent inhibitors of EGFR and HER2, including afatinib and neratinib, destabilize the pseudokinase TRIB2 in vitro and induce its ubiquitylation and degradation in cells (Fig. 5c; Table 3). The inhibitors form a covalent adduct with a highly conserved cysteine residue located in helix αC of TRIB2 that is absent in other TRIB pseudokinases233. Although the mechanism for the destabilizing effect is not entirely clear, inhibitor binding to TRIB2 was shown to result in displacement of the C-terminal tail from its binding site in the N-lobe of the pseudokinase domain and, in doing so, perhaps interferes with interactions with unknown stabilizing factors233. This specific C-terminal-tail-mediated interaction with the N-lobe is a unique feature of TRIB pseudokinases and was first observed in the crystal structure of the closely related pseudokinase TRIB1143,233.
An indirect strategy to induce degradation of a target protein involves the selective inhibition of cellular processes necessary for protein maturation and trafficking. This concept could be especially relevant for pseudokinase RTKs, which, as membrane proteins, must undergo a series of maturation steps as they proceed through the secretory pathway. One of these steps is the insertion of their transmembrane domains into the endoplasmic reticulum membrane, a process mediated by the Sec61 translocon. A class of cyclic peptides known as cotransins are Sec61 inhibitors that selectively disrupt the insertion of proteins that contain specific signal peptide sequences234. One of the cotransins, CT8, prevents Sec61-mediated cotranslational membrane insertion of HER3, leading to HER3 degradation and disruption of HER3 signalling in cancer cells235. Remarkably, CT8 acts selectively on HER3, but not other HER receptors, owing to unique determinants within the HER3 signal peptide sequence. Future structure–function studies to elucidate the mechanisms that underlie signal peptide recognition by Sec61 and inhibition by cotransins could lead to the development of molecules that selectively block cotranslational insertion and could be used to target other pseudokinase receptors.
Antibodies against pseudokinase RTKs
Pseudokinase RTKs offer a unique opportunity for targeting using antibody-based approaches. Owing to its role in drug resistance in EGFR-driven and HER2-driven cancers, there has been much interest in developing HER3 antibodies for clinical use236. Several HER3 antibodies were well tolerated in phase I clinical trials and have been tested in combination with other established therapeutic agents for treatment of patients with advanced or metastatic solid tumours in phase II studies237–241 (Table 3). These antibodies, such as seribantumab (MM-121) and lumretuzumab (RG7116), inhibit HER3 by preventing ligand binding and/or its dimerization with other HER receptors242,243 (Fig. 5d). Patritumab (U3–1287) is the only HER3 antibody so far to advance to a phase III clinical trial244. Although its mechanism of action remains poorly understood, patritumab promotes HER3 internalization and has been shown to be effective in reducing tumour growth in several tumour cell lines, including cellular models of NSCLC, breast cancer and pancreatic cancer245,246. Despite promising results in a phase II study, however, treatment with a combination of patritumab and the EGFR inhibitor erlotinib failed to increase progression-free survival or overall survival compared with erlotinib alone in patients with advanced NSCLC in a phase III clinical trial244,247.
Another strategy to inhibit HER3 signalling involved efforts to develop bifunctional antibodies that, in addition to HER3, also target its co-receptor EGFR. The dual specificity antibody duligotuzumab (MEHD-7945A) was able to overcome acquired resistance to the EGFR antibody cetuximab or the EGFR kinase inhibitor erlotinib in NSCLC and SCCHN cells. This antibody binds to the inactive ‘tethered’ conformation of both HER3 and EGFR and is believed to block ligand binding248,249. Phase II trials for duligotuzumab, however, failed to improve progression-free survival or the objective response rate when compared with cetuximab treatment in patients with SCCHN or RAS wild-type metastatic colorectal cancer250,251. Subsequently, duligotuzumab was combined with another two-in-one antibody — bH1–44 — that binds to HER2 and vascular endothelial growth factor (VEGF) to create the ‘four-in-one’ antibody FL518, which recognizes EGFR, HER2, HER3 and VEGF. This antibody was more effective than either parental bispecific antibody at inhibiting tumour growth of various cancer cell lines, as well as xenografts252.
Several other antibodies that target pseudokinase RTKs have been reported (Table 3). ROR1, which is expressed in chronic lymphocytic leukaemia (CLL) cells and only rarely in normal adult tissues, is potentially a very selective therapeutic target for the treatment of CLL. Antibodies against ROR1 have been shown to induce apoptosis of CLL cells but not normal cells253, and the anti-ROR1 antibody cirmtuzumab (UC-961) is currently being evaluated in several phase I clinical trials254. The pseudokinase ephrin receptor, EPHA10, is also rarely expressed in healthy tissues but is overexpressed in breast and prostate cancers255,256. Treatment with an anti-EPHA10 antibody results in complement-dependent cytotoxicity in prostate cancer cells in an EPHA10-dependent manner256. A bispecific antibody that targets both EPHA10 and the T cell co-receptor CD3 has also been reported to induce T cell-dependent cytotoxicity in cell lines overexpressing EPHA10, as well as in xenograft models257. Finally, the RTK pseudokinase PTK7 was found to be enriched in tumour-initiating cells in patient-derived xenografts (PDXs) of triple-negative breast cancer, ovarian cancer and NSCLC258. Treatment of tumour cells with PF-06647020, an antibody–drug conjugate consisting of an anti-PTK7 antibody and the auristatin microtubule inhibitor Aur0101, resulted in cytotoxicity in vitro and sustained tumour regression in PDX mice258. PF-06647020 is currently being evaluated in phase I trials for patients with advanced solid tumours and metastatic triple-negative breast cancer.
Conclusions
As the functions of pseudokinases continue to be uncovered, the importance of these unusual kinases for normal physiological processes and in disease is becoming increasingly apparent. A landmark example is the EGFR family of receptors, in which signalling by the pseudokinase HER3 underlies resistance to inhibitors that target the active receptors EGFR and HER2, suggesting that only combination therapy directed towards multiple EGFR family members will ultimately be effective. Although several HER3-targeted antibodies have already been tested in clinical trials in combination with small molecules or antibodies that target EGFR or HER2 for the treatment of advanced solid tumours, so far, these combinations have not demonstrated greater efficacy than existing treatment regimens. More careful selection of study participants or alternative treatment modalities aimed at HER3 might be necessary to achieve clinical benefits.
In this Review, we discuss several examples in which modulation of pseudokinase signalling can be achieved through a pocket that, in active kinases and some pseudokinases, binds nucleotides. The remarkable structural diversity of these pockets in pseudokinases provides an opportunity to achieve a high degree of specificity in pseudokinase-directed small molecules. For pseudokinases that do not bind nucleotides, the structural features of some of the ‘pseudo’-nucleotide-binding pockets might still offer this possibility. The underlying goal of these pseudokinase-binding molecules is to disrupt a functional conformation of the pseudokinase domain that may mediate protein–protein interactions that are necessary for signal propagation. Alternatively, a small molecule can be used to mediate target degradation. This strategy is particularly appealing for targeting pseudokinases. The degradation-inducing molecules discussed in this article target nucleotide-binding pockets in pseudokinases and active kinases, but small molecules that bind to distal allosteric sites in the pseudokinase domain or adjacent domains could also be used to develop molecules that induce degradation and inhibit pseudokinase signalling. Finally, interfering with the function of cell surface-exposed pseudokinases has been successful in preclinical and early clinical studies and could be further used to develop antibody-based, pseudokinase-directed therapeutics.
Recent reports of the unexpected effects of small-molecule inhibitors on the noncatalytic functions of active kinases emphasize the need to carefully consider the physiological consequences of kinase inhibition in cells. Although the emerging paradoxical effects of active kinase inhibitors are a cautionary tale, they should also guide the design of molecules that can modulate pseudokinase signalling. Structural and mechanistic studies of pseudokinases show striking parallels between pseudokinase regulation and that of their active counterparts. This finding indicates that much of what we have learned from drugging active kinases can be applied to targeting pseudokinases and vice versa. Encouragingly, the chemical space for pseudokinase drug design is likely much larger than that of active kinases owing to the uniqueness of the architectures of pseudoactive sites, a consideration to keep in mind when screening for novel small molecules that bind pseudokinases.
Supplementary Material
Acknowledgements
The authors sincerely apologize to all colleagues whose work was omitted in this Review owing to space constraints. N.J. and J.E.K. are supported by a grant from the National Institute of General Medical Sciences to N.J. (R01 GM109176), Susan G. Komen Foundation Training Grant to N.J. (CCR14299947), National Science Foundation Graduate Research Fellowship to J.E.K. and University of California at San Francisco (UCSF) Discovery Fellowship to J.E.K.
Glossary
- Salt bridge
A non-covalent interaction between oppositely charged residues.
- Type II inhibitor
An ATP-competitive small molecule that binds to the inactive DFG (Asp-Phe-Gly)-out conformation of a kinase.
- Type I inhibitor
An ATP-competitive small molecule that binds to the active DFG (Asp-Phe-Gly)-in conformation of a kinase.
- Protomers
Individual subunits of a multimeric protein complex. In the context of a BRAF homodimer, a protomer would be a single BRAF monomer.
- Apo
A state of a protein when it has no ligands bound.
- Type III inhibitor
A small-molecule kinase inhibitor that binds to an allosteric site adjacent to the ATP-binding pocket and does not interfere with ATP binding.
- Type IV inhibitors
Small-molecule kinase inhibitors that bind to allosteric sites distal from the ATP-binding site.
- Sec61 translocon
A protein complex that mediates insertion of the transmembrane domains of membrane proteins into the endoplasmic reticulum membrane.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Futreal PA et al. A census of human cancer genes. Nat Rev Cancer 4, 177–183 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu P, Nielsen TE & Clausen MH FDA-approved small-molecule kinase inhibitors. Trends Pharmacol. Sci. 36, 422–439 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Greenman C et al. Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Creixell P et al. Kinome-wide decoding of network-attacking mutations rewiring cancer signaling. Cell 163, 202–217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang MT et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nature Biotechnology 34, 155–163 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nieto P et al. A Braf kinase-inactive mutant induces lung adenocarcinoma. Nature (2017). doi: 10.1038/nature23297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan PTC et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855–867 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Garnett MJ et al. Wild-Type and Mutant B-RAF Activate C-RAF through Distinct Mechanisms Involving Heterodimerization. Molecular Cell 20, 963–969 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Yao Z et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 548, 234–238 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobsen AV & Murphy JM The secret life of kinases: insights into non-catalytic signalling functions from pseudokinases. Biochem. Soc. Trans. 45, 665–681 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Kung JE & Jura N Structural Basis for the Non-catalytic Functions of Protein Kinases. Structure 24, 7–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw AS, Kornev AP, Hu J, Ahuja LG & Taylor SS Kinases and pseudokinases: lessons from RAF. Molecular and Cellular Biology 34, 1538–1546 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger SB et al. Cutting Edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J. Immunol. 192, 5476–5480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton K et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science 343, 1357–1360 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Mandal P et al. RIP3 induces apoptosis independent of pronecrotic kinase activity. Molecular Cell 56, 481–495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camps M et al. Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science 280, 1262–1265 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Zhou B et al. Mapping ERK2-MKP3 Binding Interfaces by Hydrogen/Deuterium Exchange Mass Spectrometry. Journal of Biological Chemistry 281, 38834–38844 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Manning G, Whyte DB, Martinez R, Hunter T & Sudarsanam S The protein kinase complement of the human genome. Science 298, 1912–1934 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Boudeau J, Miranda-Saavedra D, Barton GJ & Alessi DR Emerging roles of pseudokinases. Trends in Cell Biology 16, 443–452 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Reiterer V, Eyers PA & Farhan H Day of the dead: pseudokinases and pseudophosphatases in physiology and disease. Trends in Cell Biology 24, 489–505 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Bailey FP, Byrne DP, McSkimming D, Kannan N & Eyers PA Going for broke: targeting the human cancer pseudokinome. Biochem. J. 465, 195–211 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Caenepeel S, Charydczak G, Sudarsanam S, Hunter T & Manning G The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proceedings of the National Academy of Sciences 101, 11707–11712 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castells E & Casacuberta JM Signalling through kinase-defective domains: the prevalence of atypical receptor-like kinases in plants. J. Exp. Bot. 58, 3503–3511 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Bemm F, Schwarz R, Förster F & Schultz J A kinome of 2600 in the ciliate Paramecium tetraurelia. FEBS Letters 583, 3589–3592 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Goldberg JM et al. The dictyostelium kinome--analysis of the protein kinases from a simple model organism. PLoS Genet. 2, e38 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peixoto L et al. Integrative genomic approaches highlight a family of parasite-specific kinases that regulate host responses. Cell Host & Microbe 8, 208–218 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter T & Manning G in Receptor Tyrosine Kinases: Structure, Functions and Role in Human Disease 1–15 (Springer, New York, NY, 2015). doi: 10.1007/978-1-4939-2053-2_1 [DOI] [Google Scholar]
- 28.Wilson LJ et al. New Perspectives, Opportunities, and Challenges in Exploring the Human Protein Kinome. Cancer Research 78, 15–29 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Byrne DP, Foulkes DM & Eyers PA Pseudokinases: update on their functions and evaluation as new drug targets. Future Med Chem 9, 245–265 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Jura N, Shan Y, Cao X, Shaw DE & Kuriyan J Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. U.S.A. 106, 21608–21613 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monsey J, Shen W, Schlesinger P & Bose R Her4 and Her2/neu Tyrosine Kinase Domains Dimerize and Activate in a Reconstituted in Vitro System. Journal of Biological Chemistry 285, 7035–7044 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Littlefield P et al. Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations. Science Signaling 7, ra114–ra114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 33.Zhang X, Gureasko J, Shen K, Cole PA & Kuriyan J An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Liu L et al. Regulation of Kinase Activity in the Caenorhabditis elegans EGF Receptor, LET-23. Structure 26, 270–281.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang K et al. Transformation of NIH 3T3 cells by HER3 or HER4 receptors requires the presence of HER1 or HER2. J. Biol. Chem. 271, 3884–3890 (1996). [PubMed] [Google Scholar]
- 36.Hellyer NJ, Kim MS & Koland JG Heregulin-dependent activation of phosphoinositide 3-kinase and Akt via the ErbB2/ErbB3 co-receptor. J. Biol. Chem. 276, 42153–42161 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Erjala K et al. Signaling via ErbB2 and ErbB3 Associates with Resistance and Epidermal Growth Factor Receptor (EGFR) Amplification with Sensitivity to EGFR Inhibitor Gefitinib in Head and Neck Squamous Cell Carcinoma Cells. Clin. Cancer Res. 12, 4103–4111 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Engelman JA et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Sergina NV et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature 445, 437–441 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller TW et al. Loss of Phosphatase and Tensin Homologue Deleted on Chromosome 10 Engages ErbB3 and Insulin-Like Growth Factor-I Receptor Signaling to Promote Antiestrogen Resistance in Breast Cancer. Cancer Research 69, 4192–4201 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desbois-Mouthon C et al. Insulin-Like Growth Factor-1 Receptor Inhibition Induces a Resistance Mechanism via the Epidermal Growth Factor Receptor/HER3/AKT Signaling Pathway: Rational Basis for Cotargeting Insulin-Like Growth Factor-1 Receptor and Epidermal Growth Factor Receptor in Hepatocellular Carcinoma. Clin. Cancer Res. 15, 5445–5456 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Jeong EG et al. ERBB3 kinase domain mutations are rare in lung, breast and colon carcinomas. Int. J. Cancer 119, 2986–2987 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Ding L et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kan Z et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Stransky N et al. The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaiswal BS et al. Oncogenic ERBB3 mutations in human cancers. Cancer Cell 23, 603–617 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Mishra R et al. Activating HER3 mutations in breast cancer. Oncotarget 9, 27773–27788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riethmacher D et al. Severe neuropathies in mice with targeted mutations in the ErbB3 receptor. Nature 389, 725–730 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Massa R et al. Overexpression of ErbB2 and ErbB3 receptors in Schwann cells of patients with Charcot-Marie-tooth disease type 1A. Muscle Nerve 33, 342–349 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Kanazawa T et al. Schizophrenia is not associated with the functional candidate gene ERBB3: results from a case-control study. Am. J. Med. Genet. B Neuropsychiatr. Genet. 144B, 113–116 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Watanabe Y et al. No association between the ERBB3 gene and schizophrenia in a Japanese population. Neurosci. Res. 57, 574–578 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Al-Shawi R, Ashton SV, Underwood C & Simons JP Expression of the Ror1 and Ror2 receptor tyrosine kinase genes during mouse development. Dev. Genes Evol. 211, 161–171 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Fukuda T et al. Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc. Natl. Acad. Sci. U.S.A. 105, 3047–3052 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oishi I et al. The receptor tyrosine kinase Ror2 is involved in non-canonical Wnt5a/JNK signalling pathway. Genes to Cells 8, 645–654 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Yu J et al. Wnt5a induces ROR1/ROR2 heterooligomerization to enhance leukemia chemotaxis and proliferation. J. Clin. Invest. 126, 585–598 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gentile A, Lazzari L, Benvenuti S, Trusolino L & Comoglio PM Ror1 is a pseudokinase that is crucial for Met-driven tumorigenesis. Cancer Research 71, 3132–3141 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Gentile A, Lazzari L, Benvenuti S, Trusolino L & Comoglio PM The ROR1 pseudokinase diversifies signaling outputs in MET-addicted cancer cells. Int. J. Cancer 135, 2305–2316 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi T et al. NKX2–1/TITF1/TTF-1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell 21, 348–361 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Morioka K et al. Orphan receptor tyrosine kinase ROR2 as a potential therapeutic target for osteosarcoma. Cancer Science 100, 1227–1233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wright TM et al. Ror2, a developmentally regulated kinase, promotes tumor growth potential in renal cell carcinoma. Oncogene 28, 2513–2523 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lara E et al. Epigenetic repression of ROR2 has a Wnt-mediated, pro-tumourigenic role in colon cancer. Mol. Cancer 9, 170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Geng M et al. Loss of Wnt5a and Ror2 protein in hepatocellular carcinoma associated with poor prognosis. World J. Gastroenterol. 18, 1328–1338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mikels AJ & Nusse R Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 4, e115 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oldridge M et al. Dominant mutations in ROR2, encoding an orphan receptor tyrosine kinase, cause brachydactyly type B. Nat Genet 24, 275–278 (2000). [DOI] [PubMed] [Google Scholar]
- 67.Schwabe GC et al. Distinct mutations in the receptor tyrosine kinase gene ROR2 cause brachydactyly type B. Am. J. Hum. Genet. 67, 822–831 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Afzal AR et al. Recessive Robinow syndrome, allelic to dominant brachydactyly type B, is caused by mutation of ROR2. Nat Genet 25, 419–422 (2000). [DOI] [PubMed] [Google Scholar]
- 69.van Bokhoven H et al. Mutation of the gene encoding the ROR2 tyrosine kinase causes autosomal recessive Robinow syndrome. Nat Genet 25, 423–426 (2000). [DOI] [PubMed] [Google Scholar]
- 70.Schwabe GC et al. Ror2 knockout mouse as a model for the developmental pathology of autosomal recessive Robinow syndrome. Dev. Dyn. 229, 400–410 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Peradziryi H et al. PTK7/Otk interacts with Wnts and inhibits canonical Wnt signalling. EMBO J. 30, 3729–3740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martinez S et al. The PTK7 and ROR2 Protein Receptors Interact in the Vertebrate WNT/Planar Cell Polarity (PCP) Pathway. Journal of Biological Chemistry 290, 30562–30572 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wehner P et al. RACK1 is a novel interaction partner of PTK7 that is required for neural tube closure. Development 138, 1321–1327 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Puppo F et al. Protein tyrosine kinase 7 has a conserved role in Wnt/β-catenin canonical signalling. EMBO Rep. 12, 43–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hayes M et al. ptk7 mutant zebrafish models of congenital and idiopathic scoliosis implicate dysregulated Wnt signalling in disease. Nat Comms 5, 4777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang M et al. Role of the planar cell polarity gene Protein tyrosine kinase 7 in neural tube defects in humans. Birth Defects Res. Part A Clin. Mol. Teratol. 103, 1021–1027 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Mossie K et al. Colon carcinoma kinase-4 defines a new subclass of the receptor tyrosine kinase family. Oncogene 11, 2179–2184 (1995). [PubMed] [Google Scholar]
- 78.Chen R et al. A meta-analysis of lung cancer gene expression identifies PTK7 as a survival gene in lung adenocarcinoma. Cancer Research 74, 2892–2902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gärtner S et al. PTK 7 is a transforming gene and prognostic marker for breast cancer and nodal metastasis involvement. PLoS ONE 9, e84472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang H et al. PTK7 protein is decreased in epithelial ovarian carcinomas with poor prognosis. Int J Clin Exp Pathol 7, 7881–7889 (2014). [PMC free article] [PubMed] [Google Scholar]
- 81.Easty DJ et al. Loss of expression of receptor tyrosine kinase family genes PTK7 and SEK in metastatic melanoma. Int. J. Cancer 71, 1061–1065 (1997). [DOI] [PubMed] [Google Scholar]
- 82.Lu W, Yamamoto V, Ortega B & Baltimore D Mammalian Ryk Is a Wnt Coreceptor Required for Stimulation of Neurite Outgrowth. Cell 119, 97–108 (2004). [DOI] [PubMed] [Google Scholar]
- 83.Keeble TR et al. The Wnt receptor Ryk is required for Wnt5a-mediated axon guidance on the contralateral side of the corpus callosum. J. Neurosci. 26, 5840–5848 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Y et al. Ryk-mediated Wnt repulsion regulates posterior-directed growth of corticospinal tract. Nat. Neurosci. 8, 1151–1159 (2005). [DOI] [PubMed] [Google Scholar]
- 85.Liu Y et al. Repulsive Wnt signaling inhibits axon regeneration after CNS injury. J. Neurosci. 28, 8376–8382 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katso RM et al. Overexpression of H-Ryk in epithelial ovarian cancer: prognostic significance of receptor expression. Clin. Cancer Res. 6, 3271–3281 (2000). [PubMed] [Google Scholar]
- 87.Saharinen P, Takaluoma K & Silvennoinen O Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Molecular and Cellular Biology 20, 3387–3395 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saharinen P & Silvennoinen O The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J. Biol. Chem. 277, 47954–47963 (2002). [DOI] [PubMed] [Google Scholar]
- 89.Kralovics R et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 352, 1779–1790 (2005). [DOI] [PubMed] [Google Scholar]
- 90.Walters DK et al. Activating alleles of JAK3 in acute megakaryoblastic leukemia. Cancer Cell 10, 65–75 (2006). [DOI] [PubMed] [Google Scholar]
- 91.Flex E et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. Journal of Experimental Medicine 205, 751–758 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jeong EG et al. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clin. Cancer Res. 14, 3716–3721 (2008). [DOI] [PubMed] [Google Scholar]
- 93.Lupardus PJ et al. Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc. Natl. Acad. Sci. U.S.A. 111, 8025–8030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shan Y et al. Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nature Structural & Molecular Biology 21, 579–584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen M et al. Complex effects of naturally occurring mutations in the JAK3 pseudokinase domain: evidence for interactions between the kinase and pseudokinase domains. Molecular and Cellular Biology 20, 947–956 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Z et al. Two rare disease-associated Tyk2 variants are catalytically impaired but signaling competent. J. Immunol. 190, 2335–2344 (2013). [DOI] [PubMed] [Google Scholar]
- 97.Murphy JM et al. The Pseudokinase MLKL Mediates Necroptosis via a Molecular Switch Mechanism. Immunity 39, 443–453 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Wang H et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Molecular Cell 54, 133–146 (2014). [DOI] [PubMed] [Google Scholar]
- 99.Hildebrand JM et al. Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc. Natl. Acad. Sci. U.S.A. 111, 15072–15077 (2014).This study identifies an ATP-competitive compound that binds to the pseudoactive site of MLKL and inhibits necroptosis.
- 100.Linkermann A & Green DR Necroptosis. N Engl J Med 370, 455–465 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ting AT, Pimentel-Muiños FX & Seed B RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J. 15, 6189–6196 (1996). [PMC free article] [PubMed] [Google Scholar]
- 102.Rajakulendran T et al. A dimerization-dependent mechanism drives RAF catalytic activation. Nature 461, 542–545 (2009). [DOI] [PubMed] [Google Scholar]
- 103.Hu J et al. Mutation that blocks ATP binding creates a pseudokinase stabilizing the scaffolding function of kinase suppressor of Ras, CRAF and BRAF. Proceedings of the National Academy of Sciences 108, 6067–6072 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lavoie H et al. MEK drives BRAF activation through allosteric control of KSR proteins. Nature (2018). doi: 10.1038/nature25478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lozano J et al. Deficiency of kinase suppressor of Ras1 prevents oncogenic ras signaling in mice. Cancer Research 63, 4232–4238 (2003). [PubMed] [Google Scholar]
- 106.Kortum RL & Lewis RE The molecular scaffold KSR1 regulates the proliferative and oncogenic potential of cells. Molecular and Cellular Biology 24, 4407–4416 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nguyen A et al. Kinase Suppressor of Ras (KSR) Is a Scaffold Which Facilitates Mitogen-Activated Protein Kinase Activation In Vivo. Molecular and Cellular Biology 22, 3035–3045 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Costanzo-Garvey DL et al. KSR2 is an essential regulator of AMP kinase, energy expenditure, and insulin sensitivity. Cell Metab. 10, 366–378 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Revelli J-P et al. Profound obesity secondary to hyperphagia in mice lacking kinase suppressor of ras 2. Obesity (Silver Spring) 19, 1010–1018 (2011). [DOI] [PubMed] [Google Scholar]
- 110.Pearce LR et al. KSR2 Mutations Are Associated with Obesity, Insulin Resistance, and Impaired Cellular Fuel Oxidation. Cell 155, 765–777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tagliabracci VS et al. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science 336, 1150–1153 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cui J et al. A secretory kinase complex regulates extracellular protein phosphorylation. eLife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ohyama Y et al. FAM20A binds to and regulates FAM20C localization. Sci. Rep. 6, 27784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.O’Sullivan J et al. Whole-Exome sequencing identifies FAM20A mutations as a cause of amelogenesis imperfecta and gingival hyperplasia syndrome. Am. J. Hum. Genet. 88, 616–620 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Simpson MA et al. Mutations in FAM20C are associated with lethal osteosclerotic bone dysplasia (Raine syndrome), highlighting a crucial molecule in bone development. Am. J. Hum. Genet. 81, 906–912 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tagliabracci VS et al. A Single Kinase Generates the Majority of the Secreted Phosphoproteome. Cell 161, 1619–1632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dedhia PH et al. Differential ability of Tribbles family members to promote degradation of C/EBPalpha and induce acute myelogenous leukemia. Blood 116, 1321–1328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qi L et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science 312, 1763–1766 (2006). [DOI] [PubMed] [Google Scholar]
- 119.Keeshan K et al. Transformation by Tribbles homolog 2 (Trib2) requires both the Trib2 kinase domain and COP1 binding. Blood 116, 4948–4957 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jamieson SA et al. Substrate binding allosterically relieves autoinhibition of the pseudokinase TRIB1. Science Signaling 11, eaau0597 (2018).This study demonstrated that substrate binding alters the conformation of the TRIB1 pseudokinase domain, providing evidence that pseudokinases can undergo conformational transitions similar to those observed for active kinases.
- 121.Eyers PA, Keeshan K & Kannan N Tribbles in the 21st Century: The Evolving Roles of Tribbles Pseudokinases in Biology and Disease. Trends in Cell Biology 27, 284–298 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yoshida A, Kato J-Y, Nakamae I & Yoneda-Kato N COP1 targets C/EBPα for degradation and induces acute myeloid leukemia via Trib1. Blood 122, 1750–1760 (2013). [DOI] [PubMed] [Google Scholar]
- 123.Bauer RC, Yenilmez BO & Rader DJ Tribbles-1: a novel regulator of hepatic lipid metabolism in humans. Biochem. Soc. Trans. 43, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yokoyama T et al. Trib1 links the MEK1/ERK pathway in myeloid leukemogenesis. Blood 116, 2768–2775 (2010). [DOI] [PubMed] [Google Scholar]
- 125.Izrailit J, Berman HK, Datti A, Wrana JL & Reedijk M High throughput kinase inhibitor screens reveal TRB3 and MAPK-ERK/TGFβ pathways as fundamental Notch regulators in breast cancer. Proceedings of the National Academy of Sciences 110, 1714–1719 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Du K, Herzig S, Kulkarni RN & Montminy M TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300, 1574–1577 (2003). [DOI] [PubMed] [Google Scholar]
- 127.Liew CW et al. The pseudokinase tribbles homolog 3 interacts with ATF4 to negatively regulate insulin exocytosis in human and mouse β cells. J. Clin. Invest. 120, 2876–2888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zareen N et al. A feed-forward loop involving Trib3, Akt and FoxO mediates death of NGF-deprived neurons. Cell Death Differ 20, 1719–1730 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aimé P et al. Trib3 Is Elevated in Parkinson’s Disease and Mediates Death in Parkinson’s Disease Models. J. Neurosci. 35, 10731–10749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hill R et al. TRIB2 confers resistance to anti-cancer therapy by activating the serine/threonine protein kinase AKT. Nat Comms 8, 14687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 131.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD & Cole MD The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94, 363–374 (1998). [DOI] [PubMed] [Google Scholar]
- 132.Deleu L, Shellard S, Alevizopoulos K, Amati B & Land H Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene 20, 8270–8275 (2001). [DOI] [PubMed] [Google Scholar]
- 133.Martinez E et al. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Molecular and Cellular Biology 21, 6782–6795 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brand M, Yamamoto K, Staub A & Tora L Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 274, 18285–18289 (1999). [DOI] [PubMed] [Google Scholar]
- 135.Ikura T et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102, 463–473 (2000). [DOI] [PubMed] [Google Scholar]
- 136.McMahon SB, Wood MA & Cole MD The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Molecular and Cellular Biology 20, 556–562 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lang SE, McMahon SB, Cole MD & Hearing P E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem. 276, 32627–32634 (2001). [DOI] [PubMed] [Google Scholar]
- 138.Lang SE & Hearing P The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene 22, 2836–2841 (2003). [DOI] [PubMed] [Google Scholar]
- 139.Park J, Kunjibettu S, McMahon SB & Cole MD The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes & Development 15, 1619–1624 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Huse M & Kuriyan J The conformational plasticity of protein kinases. Cell 109, 275–282 (2002). [DOI] [PubMed] [Google Scholar]
- 141.Taylor SS & Kornev AP Protein kinases: evolution of dynamic regulatory proteins. Trends in Biochemical Sciences 36, 65–77 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jura N et al. Catalytic Control in the EGF Receptor and Its Connection to General Kinase Regulatory Mechanisms. Molecular Cell 42, 9–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Murphy JM et al. Molecular Mechanism of CCAAT-Enhancer Binding Protein Recruitment by the TRIB1 Pseudokinase. Structure 23, 2111–2121 (2015). [DOI] [PubMed] [Google Scholar]
- 144.Ha BH & Boggon TJ The crystal structure of pseudokinase PEAK1 (Sugen Kinase 269) reveals an unusual catalytic cleft and a novel mode of kinase fold dimerization. J. Biol. Chem. jbc.RA117.000751 (2017). doi: 10.1074/jbc.RA117.000751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Patel O et al. Structure of SgK223 pseudokinase reveals novel mechanisms of homotypic and heterotypic association. Nat Comms 8, 1157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lecointre C et al. Dimerization of the Pragmin Pseudo-Kinase Regulates Protein Tyrosine Phosphorylation. Structure 26, 545–554.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 147.Scheeff ED, Eswaran J, Bunkoczi G, Knapp S & Manning G Structure of the Pseudokinase VRK3 Reveals a Degraded Catalytic Site, a Highly Conserved Kinase Fold, and a Putative Regulatory Binding Site. Structure 17, 128–138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hari SB, Merritt EA & Maly DJ Conformation-Selective ATP-Competitive Inhibitors Control Regulatory Interactions and Noncatalytic Functions of Mitogen-Activated Protein Kinases. Chemistry & Biology 21, 628–635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Han Y et al. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science 343, 1244–1248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Huang H et al. Dimeric Structure of Pseudokinase RNase L Bound to 2–5A Reveals a Basis for Interferon-Induced Antiviral Activity. Molecular Cell 53, 221–234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shi F et al. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proceedings of the National Academy of Sciences 107, 7692–7697 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zeqiraj E et al. ATP and MO25α Regulate the Conformational State of the STRADα Pseudokinase and Activation of the LKB1 Tumour Suppressor. PLoS Biol. 7, e1000126 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zeqiraj E, Filippi BM, Deak M, Alessi DR & van Aalten DMF Structure of the LKB1-STRAD-MO25 Complex Reveals an Allosteric Mechanism of Kinase Activation. Science 326, 1707–1711 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Artim SC, Mendrola JM & Lemmon MA Assessing the range of kinase autoinhibition mechanisms in the insulin receptor family. Biochem. J. 448, 213–220 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xie T et al. Structural Insights into RIP3-Mediated Necroptotic Signaling. Cell Reports 5, 70–78 (2013). [DOI] [PubMed] [Google Scholar]
- 156.Murphy JM et al. Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL. Biochem. J. 457, 369–377 (2014). [DOI] [PubMed] [Google Scholar]
- 157.Ma B et al. ATP-Competitive MLKL Binders Have No Functional Impact on Necroptosis. PLoS ONE 11, e0165983 (2016).This paper revealed that a previously identified ATP-competitive inhibitor of MLKL stabilizes the pseudokinase domain in the DFG-out state. This compound was also found to inhibit RIPK1, while ATP-competitive molecules that bind to MLKL with greater specificity were unable to inhibit necroptosis.
- 158.Pearce LR, Komander D & Alessi DR The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol 11, 9–22 (2010). [DOI] [PubMed] [Google Scholar]
- 159.Murphy JM et al. A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J. 457, 323–334 (2013).In this study, a wide range of pseudokinases were classified into different classes based on their ability to bind nucleotides and/or cations.
- 160.Gee CL et al. A Phosphorylated Pseudokinase Complex Controls Cell Wall Synthesis in Mycobacteria. Science Signaling 5, ra7–ra7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zheng J et al. 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr Sect D Biol Crystallogr 49, 362–365 (1993). [DOI] [PubMed] [Google Scholar]
- 162.Bailey FP et al. The Tribbles 2 (TRB2) pseudokinase binds to ATP and autophosphorylates in a metal-independent manner. Biochem. J. (2015). doi: 10.1042/BJ20141441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mukherjee K et al. CASK Functions as a Mg2+-independent neurexin kinase. Cell 133, 328–339 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cui J et al. Structure of Fam20A reveals a pseudokinase featuring unique disulfide pattern and inverted ATP-binding. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Labesse G et al. ROP2 from Toxoplasma gondii: A Virulence Factor with a Protein-Kinase Foldand No Enzymatic Activity. Structure/Folding and Design 17, 139–146 (2009). [DOI] [PubMed] [Google Scholar]
- 166.Qiu W et al. Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii. EMBO J. 28, 969–979 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Brady DC et al. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature 509, 492–496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mendrola JM, Shi F, Park JH & Lemmon MA Receptor tyrosine kinases with intracellular pseudokinase domains. Biochem. Soc. Trans. 41, 1029–1036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ungureanu D et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nature Structural & Molecular Biology 18, 971–976 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hammarén HM et al. ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc. Natl. Acad. Sci. U.S.A. (2015). doi: 10.1073/pnas.1423201112This paper demonstrates that ATP binding to the JH2 pseudokinase domain of JAK2 is essential for hyperactivation of pathogenic JAK2 mutants.
- 171.Bandaranayake RM et al. Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nature Structural & Molecular Biology 19, 754–759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Brennan DF et al. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature 472, 366–369 (2011). [DOI] [PubMed] [Google Scholar]
- 173.Cameron AJM & Cameron AJM Occupational hazards: allosteric regulation of protein kinases through the nucleotide-binding pocket. Biochem. Soc. Trans. 39, 472–476 (2011). [DOI] [PubMed] [Google Scholar]
- 174.Dar AC & Shokat KM The Evolution of Protein Kinase Inhibitors from Antagonists to Agonists of Cellular Signaling. Annu. Rev. Biochem. 80, 769–795 (2011). [DOI] [PubMed] [Google Scholar]
- 175.Claus J, Cameron AJM & Parker PJ Pseudokinase drug intervention: a potentially poisoned chalice. Biochem. Soc. Trans. 41, 1083–1088 (2013). [DOI] [PubMed] [Google Scholar]
- 176.Karoulia Z, Gavathiotis E & Poulikakos PI New perspectives for targeting RAF kinase in human cancer. Nat Rev Cancer 17, 676–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hatzivassiliou G et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464, 431–435 (2010). [DOI] [PubMed] [Google Scholar]
- 178.Poulikakos PI, Zhang C, Bollag G, Shokat KM & Rosen N RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464, 427–430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Heidorn SJ et al. Kinase-Dead BRAF and Oncogenic RAS Cooperate to Drive Tumor Progression through CRAF. Cell 140, 209–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Karoulia Z et al. An Integrated Model of RAF Inhibitor Action Predicts Inhibitor Activity against Oncogenic BRAF Signaling. Cancer Cell 30, 485–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jin T et al. RAF inhibitors promote RAS-RAF interaction by allosterically disrupting RAF autoinhibition. Nat Comms 8, 1211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhang C et al. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature 526, 583–586 (2015). [DOI] [PubMed] [Google Scholar]
- 183.Okimoto RA et al. Preclinical efficacy of a RAF inhibitor that evades paradoxical MAPK pathway activation in protein kinase BRAF-mutant lung cancer. Proc. Natl. Acad. Sci. U.S.A. 113, 13456–13461 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Girotti MR et al. Paradox-Breaking RAF Inhibitors that Also Target SRC Are Effective in Drug-Resistant BRAF Mutant Melanoma. Cancer Cell (2014). doi: 10.1016/j.ccell.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Mendez AS et al. Endoplasmic reticulum stress-independent activation of unfolded protein response kinases by a small molecule ATP-mimic. eLife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Papa FR, Zhang C, Shokat K & Walter P Bypassing a kinase activity with an ATP-competitive drug. Science 302, 1533–1537 (2003). [DOI] [PubMed] [Google Scholar]
- 187.Korennykh AV et al. The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687–693 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang L et al. Divergent allosteric control of the IRE1α endoribonuclease using kinase inhibitors. Nature Chemical Biology 8, 982–989 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ghosh R et al. Allosteric Inhibition of the IRE1α RNase Preserves Cell Viability and Function during Endoplasmic Reticulum Stress. Cell 158, 534–548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Waller DD et al. A Covalent Cysteine-Targeting Kinase Inhibitor of Ire1 Permits Allosteric Control of Endoribonuclease Activity. Chembiochem 17, 843–851 (2016). [DOI] [PubMed] [Google Scholar]
- 191.Otto T et al. Stabilization of N-Myc Is a Critical Function of Aurora A in Human Neuroblastoma. Cancer Cell 15, 67–78 (2009). [DOI] [PubMed] [Google Scholar]
- 192.Brockmann M et al. Small Molecule Inhibitors of Aurora-A Induce Proteasomal Degradation of N-Myc in Childhood Neuroblastoma. Cancer Cell 24, 75–89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gustafson WC et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell 26, 414–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Claus J et al. Inhibitor-induced HER2-HER3 heterodimerisation promotes proliferation through a novel dimer interface. eLife 7, 16ra7–23 (2018).This study reveals that the HER2 inhibitor lapatinib promotes heterodimerization of HER2 and HER3 through their N-lobes. This study also demonstrates that ATP binding is essential for the allosteric activator function of HER3 and that bosutinib enhances this function.
- 195.Wood ER et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Research 64, 6652–6659 (2004). [DOI] [PubMed] [Google Scholar]
- 196.Qiu C et al. Mechanism of Activation and Inhibition of the HER4/ErbB4 Kinase. Structure 16, 460–467 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Huang Y et al. Molecular basis for multimerization in the activation of the epidermal growth factor receptor. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Liang SI et al. Phosphorylated EGFR Dimers Are Not Sufficient to Activate Ras. Cell Reports 22, 2593–2600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Dhawan NS, Scopton AP & Dar AC Small molecule stabilization of the KSR inactive state antagonizes oncogenic Ras signalling. Nature 537, 112–116 (2016).This paper identifies an ATP-competitive small molecule that stabilizes an inactive conformation of KSR2 that is incompatible with heterodimerization with B-RAF.
- 200.Tokarski JS et al. Tyrosine Kinase 2-mediated Signal Transduction in T Lymphocytes Is Blocked by Pharmacological Stabilization of Its Pseudokinase Domain. Journal of Biological Chemistry 290, 11061–11074 (2015).This study describes a series of ATP-competitive compounds that inhibit TYK2 signaling by binding to the TYK2 JH2 pseudokinase domain.
- 201.Moslin R et al. Identification of imidazo[1,2- b ]pyridazine TYK2 pseudokinase ligands as potent and selective allosteric inhibitors of TYK2 signalling. MedChemComm (2017). doi: 10.1039/C6MD00560H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Papp Kim, Gordon Kenneth, Diamant Thaçi Akimichi Morita, Gooderham Melinda, Foley Peter, Girgis Ihab G., Kundu Sudeep, Banerjee Subhashis, (2018) Phase 2 Trial of Selective Tyrosine Kinase 2 Inhibition in Psoriasis. New England Journal of Medicine 379 (14):1313–1321 [DOI] [PubMed] [Google Scholar]
- 203.Puleo DE et al. Identification and Characterization of JAK2 Pseudokinase Domain Small Molecule Binders. ACS Med Chem Lett 8, 618–621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Newton AS et al. JAK2 JH2 Fluorescence Polarization Assay and Crystal Structures for Complexes with Three Small Molecules. ACS Med Chem Lett 8, 614–617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dong B & Silverman RH Alternative function of a protein kinase homology domain in 2’, 5’-oligoadenylate dependent RNase L. Nucleic Acids Research 27, 439–445 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jha BK et al. Inhibition of RNase L and RNA-dependent protein kinase (PKR) by sunitinib impairs antiviral innate immunity. Journal of Biological Chemistry 286, 26319–26326 (2011).In this study, activation of RNase L is found to be inhibited by ATP-competitive compounds, including sunitinib and flavonols, such as quercetin.
- 207.Chen D et al. Overexpression of integrin-linked kinase correlates with malignant phenotype in non-small cell lung cancer and promotes lung cancer cell invasion and migration via regulating epithelial-mesenchymal transition (EMT)-related genes. Acta Histochem. 115, 128–136 (2013). [DOI] [PubMed] [Google Scholar]
- 208.Yan Z et al. Overexpression of integrin-linked kinase (ILK) promotes migration and invasion of colorectal cancer cells by inducing epithelial-mesenchymal transition via NF-κB signaling. Acta Histochem. 116, 527–533 (2014). [DOI] [PubMed] [Google Scholar]
- 209.Vaynberg J et al. Non-catalytic signaling by pseudokinase ILK for regulating cell adhesion. Nat Comms 9, 4465 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Fukuda K et al. Biochemical, proteomic, structural, and thermodynamic characterizations of integrin-linked kinase (ILK): cross-validation of the pseudokinase. Journal of Biological Chemistry 286, 21886–21895 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Fukuda K, Gupta S, Chen K, Wu C & Qin J The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions. Molecular Cell 36, 819–830 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Fleckenstein MC et al. A Toxoplasma gondii Pseudokinase Inhibits Host IRG Resistance Proteins. PLoS Biol. 10, e1001358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Reese ML & Boothroyd JC A conserved non-canonical motif in the pseudoactive site of the ROP5 pseudokinase domain mediates its effect on Toxoplasma virulence. Journal of Biological Chemistry 286, 29366–29375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Reese ML, Shah N & Boothroyd JC The Toxoplasma pseudokinase ROP5 is an allosteric inhibitor of the immunity-related GTPases. Journal of Biological Chemistry (2014). doi: 10.1074/jbc.M114.567057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhang H et al. Structure and evolution of the Fam20 kinases. Nat Comms 9, 1218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ohren JF et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nature Structural & Molecular Biology 11, 1192–1197 (2004). [DOI] [PubMed] [Google Scholar]
- 217.Jia Y et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 534, 129–132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Calleja V, Laguerre M, Parker PJ & Larijani B Role of a novel PH-kinase domain interface in PKB/Akt regulation: structural mechanism for allosteric inhibition. PLoS Biol. 7, e17 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wu W-I et al. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLoS ONE 5, e12913 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Adrián FJ et al. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nature Chemical Biology 2, 95–102 (2006). [DOI] [PubMed] [Google Scholar]
- 221.Yang J et al. Discovery and characterization of a cell-permeable, small-molecule c-Abl kinase activator that binds to the myristoyl binding site. Chemistry & Biology 18, 177–186 (2011). [DOI] [PubMed] [Google Scholar]
- 222.Zhang J et al. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature 463, 501–506 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Bono F et al. Inhibition of tumor angiogenesis and growth by a small-molecule multi-FGF receptor blocker with allosteric properties. Cancer Cell 23, 477–488 (2013). [DOI] [PubMed] [Google Scholar]
- 224.Herbert C et al. Molecular mechanism of SSR128129E, an extracellularly acting, small-molecule, allosteric inhibitor of FGF receptor signaling. Cancer Cell 23, 489–501 (2013). [DOI] [PubMed] [Google Scholar]
- 225.Freed DM et al. EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics. Cell (2017). doi: 10.1016/j.cell.2017.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Moraga I et al. Tuning Cytokine Receptor Signaling by Re-orienting Dimer Geometry with Surrogate Ligands. Cell (2015). doi: 10.1016/j.cell.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Engel M et al. Allosteric activation of the protein kinase PDK1 with low molecular weight compounds. EMBO J. 25, 5469–5480 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Busschots K et al. Substrate-selective inhibition of protein kinase PDK1 by small compounds that bind to the PIF-pocket allosteric docking site. Chemistry & Biology 19, 1152–1163 (2012). [DOI] [PubMed] [Google Scholar]
- 229.Lai AC & Crews CM Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov 16, 101–114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jones LH Small-Molecule Kinase Downregulators. Cell Chemical Biology (2017). doi: 10.1016/j.chembiol.2017.10.011 [DOI] [PubMed] [Google Scholar]
- 231.Burslem GM et al. The Advantages of Targeted Protein Degradation Over Inhibition: An RTK Case Study. Cell Chemical Biology 25, 67–77.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Xie T et al. Pharmacological targeting of the pseudokinase Her3. Nature Chemical Biology (2014). doi: 10.1038/nchembio.1658This study describes a covalent ATP-competitive small molecule that induces degradation of HER3.
- 233.Foulkes DM et al. Covalent inhibitors of EGFR family protein kinases induce degradation of human Tribbles 2 (TRIB2) pseudokinase in cancer cells. Science Signaling 11, eaat7951 (2018).This study reveals that compounds previously identified as covalent inhibitors of EGFR and HER2 can also bind to TRIB2 and induce its degradation.
- 234.Garrison JL, Kunkel EJ, Hegde RS & Taunton J A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature 436, 285–289 (2005). [DOI] [PubMed] [Google Scholar]
- 235.Ruiz-Saenz A et al. Targeting HER3 by interfering with its Sec61-mediated cotranslational insertion into the endoplasmic reticulum. Oncogene (2015). doi: 10.1038/onc.2014.455In this paper, an inhibitor of the Sec61 translocon was found to induce HER3 degradation by specifically disrupting cotranslational insertion of HER3 into the ER membrane.
- 236.Jacob W, James I, Hasmann M & Weisser M Clinical development of HER3-targeting monoclonal antibodies: Perils and progress. Cancer Treat. Rev. 68, 111–123 (2018). [DOI] [PubMed] [Google Scholar]
- 237.Im S-A et al. A phase 1 dose-escalation study of anti-HER3 monoclonal antibody LJM716 in combination with trastuzumab in patients with HER2-overexpressing metastatic breast or gastric cancer. Journal of Clinical Oncology (2017). doi: 10.1200/jco.2014.32.15_suppl.2519 [DOI] [Google Scholar]
- 238.Sequist LV et al. Targeting EGFR and ERBB3 in lung cancer patients: Clinical outcomes in a phase I trial of MM-121 in combination with erlotinib. Journal of Clinical Oncology (2017). doi: 10.1200/jco.2012.30.15_suppl.7556 [DOI] [Google Scholar]
- 239.Sarantopoulos J et al. First-in-human phase 1 dose-escalation study of AV-203, a monoclonal antibody against ERBB3, in patients with metastatic or advanced solid tumors. Journal of Clinical Oncology (2017). doi: 10.1200/jco.2014.32.15_suppl.11113 [DOI] [Google Scholar]
- 240.Meulendijks D et al. A first-in-human trial of RG7116, a glycoengineered monoclonal antibody targeting HER3, in patients with advanced/metastatic tumors of epithelial cell origin expressing HER3 protein. Journal of Clinical Oncology (2017). doi: 10.1200/jco.2013.31.15_suppl.2522 [DOI] [Google Scholar]
- 241.Wakui H et al. Phase 1 and dose-finding study of patritumab (U3–1287), a human monoclonal antibody targeting HER3, in Japanese patients with advanced solid tumors. Cancer Chemother Pharmacol 73, 511–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Schoeberl B et al. An ErbB3 antibody, MM-121, is active in cancers with ligand-dependent activation. Cancer Research 70, 2485–2494 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mirschberger C et al. RG7116, a therapeutic antibody that binds the inactive HER3 receptor and is optimized for immune effector activation. Cancer Research 73, 5183–5194 (2013). [DOI] [PubMed] [Google Scholar]
- 244.Paz-Arez L et al. P3.02b-045 Patritumab plus Erlotinib in EGFR Wild-Type Advanced Non–Small Cell Lung Cancer (NSCLC): Part a Results of HER3-Lung Study: Topic: EGFR Clinical. Journal of Thoracic Oncology 12, S1214–S1215 (2017). [Google Scholar]
- 245.Treder M, Hartmann S, Ogbagabriel S & Borges E Fully human Anti-HER3 monoclonal antibodies (mAbs) inhibit oncogenic signaling and tumor cell growth in vitro and in vivo. Proceedings of the American Association for Cancer Research Annual Meeting; Apr (2008). [Google Scholar]
- 246.Treder M et al. Fully human anti-HER3 mAb U3–1287 (AMG 888) demonstrates unique in vitro and in vivo activities versus other HER family inhibitors in NSCLC models (Poster). EJC Suppl. 6, 99 (2008). [Google Scholar]
- 247.Pawel Von, J. et al. Phase 2 HERALD study of patritumab (P) with erlotinib (E) in advanced NSCLC subjects (SBJs). Journal of Clinical Oncology (2014). [Google Scholar]
- 248.Schaefer G et al. A two-in-one antibody against HER3 and EGFR has superior inhibitory activity compared with monospecific antibodies. Cancer Cell 20, 472–486 (2011). [DOI] [PubMed] [Google Scholar]
- 249.Huang S et al. Dual targeting of EGFR and HER3 with MEHD7945A overcomes acquired resistance to EGFR inhibitors and radiation. Cancer Research 73, 824–833 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Fayette J et al. Randomized Phase II Study of Duligotuzumab (MEHD7945A) vs. Cetuximab in Squamous Cell Carcinoma of the Head and Neck (MEHGAN Study). Front Oncol 6, 232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hill AG et al. Phase II study of the dual EGFR/HER3 inhibitor duligotuzumab (MEHD7945A) vs. cetuximab in combination with FOLFIRI in RAS wild-type metastatic colorectal cancer. Clin. Cancer Res. clincanres.0646.2017 (2018). doi: 10.1158/1078-0432.CCR-17-0646 [DOI] [PubMed] [Google Scholar]
- 252.Hu S et al. Four-in-one antibodies have superior cancer inhibitory activity against EGFR, HER2, HER3, and VEGF through disruption of HER/MET crosstalk. Cancer Research 75, 159–170 (2015). [DOI] [PubMed] [Google Scholar]
- 253.Daneshmanesh AH et al. Monoclonal antibodies against ROR1 induce apoptosis of chronic lymphocytic leukemia (CLL) cells. Leukemia 26, 1348–1355 (2012). [DOI] [PubMed] [Google Scholar]
- 254.Choi MY et al. Pre-clinical Specificity and Safety of UC-961, a First-In-Class Monoclonal Antibody Targeting ROR1. Clinical Lymphoma, Myeloma and Leukemia 15, S167–S169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Nagano K et al. Expression of Eph receptor A10 is correlated with lymph node metastasis and stage progression in breast cancer patients. Cancer Med 2, 972–977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nagano K et al. Eph receptor A10 has a potential as a target for a prostate cancer therapy. Biochemical and Biophysical Research Communications 450, 545–549 (2014). [DOI] [PubMed] [Google Scholar]
- 257.Taki S et al. A Novel Bispecific Antibody against Human CD3 and Ephrin Receptor A10 for Breast Cancer Therapy. PLoS ONE 10, e0144712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Damelin M et al. A PTK7-targeted antibody-drug conjugate reduces tumor-initiating cells and induces sustained tumor regressions. Sci Transl Med 9, eaag2611 (2017). [DOI] [PubMed] [Google Scholar]
- 259.Blaum BS et al. Structure of the pseudokinase domain of BIR2, a regulator of BAK1-mediated immune signaling in Arabidopsis. JOURNAL OF STRUCTURAL BIOLOGY 186, 112–121 (2014). [DOI] [PubMed] [Google Scholar]
- 260.Durzynska I et al. STK40 Is a Pseudokinase that Binds the E3 Ubiquitin Ligase COP1. Structure (2016). doi: 10.1016/j.str.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Mayans O et al. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature 395, 863–869 (1998). [DOI] [PubMed] [Google Scholar]
- 262.Bogomolovas J et al. Titin kinase is an inactive pseudokinase scaffold that supports MuRF1 recruitment to the sarcomeric M-line. Open Biol 4, 140041–140041 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Mukherjee K, Sharma M, Jahn R, Wahl MC & Südhof TC Evolution of CASK into a Mg2+-sensitive kinase. Science Signaling 3, ra33–ra33 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wei Z et al. Liprin-mediated large signaling complex organization revealed by the liprin-α/CASK and liprin-α/liprin-β complex structures. Molecular Cell 43, 586–598 (2011). [DOI] [PubMed] [Google Scholar]
- 265.Stefely JA et al. Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. Molecular Cell 57, 83–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Joubert S, Jossart C, McNicoll N & De Léan A Atrial natriuretic peptide-dependent photolabeling of a regulatory ATP-binding site on the natriuretic peptide receptor-A. FEBS J. 272, 5572–5583 (2005). [DOI] [PubMed] [Google Scholar]
- 267.Grütter C et al. Structural Characterization of the RLCK Family Member BSK8: A Pseudokinase with an Unprecedented Architecture. Journal of Molecular Biology 425, 4455–4467 (2013). [DOI] [PubMed] [Google Scholar]
- 268.Jaleel M, Saha S, Shenoy AR & Visweswariah SS The kinase homology domain of receptor guanylyl cyclase C: ATP binding and identification of an adenine nucleotide sensitive site. Biochemistry 45, 1888–1898 (2006). [DOI] [PubMed] [Google Scholar]
- 269.Kawagoe T et al. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat Immunol 9, 684–691 (2008). [DOI] [PubMed] [Google Scholar]
- 270.Toms AV et al. Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases. Nature Structural & Molecular Biology 20, 1221–1223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Leroy E et al. Uncoupling JAK2 V617F activation from cytokine-induced signalling by modulation of JH2 αC helix. Biochem. J. 473, 1579–1591 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wolf J et al. Structural basis for Pan3 binding to Pan2 and its function in mRNA recruitment and deadenylation. EMBO J. 33, 1514–1526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Christie M, Boland A, Huntzinger E, Weichenrieder O & Izaurralde E Structure of the PAN3 Pseudokinase Reveals the Basis for Interactions with the PAN2 Deadenylase and the GW182 Proteins. Molecular Cell 51, 360–373 (2013). [DOI] [PubMed] [Google Scholar]
- 274.Schäfer IB, Rode M, Bonneau F, Schüssler S & Conti E The structure of the Pan2-Pan3 core complex reveals cross-talk between deadenylase and pseudokinase. Nature Structural & Molecular Biology 21, 591–598 (2014). [DOI] [PubMed] [Google Scholar]
- 275.Jonas S et al. An asymmetric PAN3 dimer recruits a single PAN2 exonuclease to mediate mRNA deadenylation and decay. Nature Structural & Molecular Biology 21, 599–608 (2014). [DOI] [PubMed] [Google Scholar]
- 276.Yoshida-Moriguchi T et al. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science 341, 896–899 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zhu Q et al. Structure of protein O-mannose kinase reveals a unique active site architecture. eLife 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Nagae M et al. 3D structural analysis of protein O-mannosyl kinase, POMK, a causative gene product of dystroglycanopathy. Genes to Cells 22, 348–359 (2017). [DOI] [PubMed] [Google Scholar]
- 279.Kamada H et al. Generation and characterization of a bispecific diabody targeting both EPH receptor A10 and CD3. Biochemical and Biophysical Research Communications 456, 908–912 (2015). [DOI] [PubMed] [Google Scholar]
- 280.Garner AP et al. An antibody that locks HER3 in the inactive conformation inhibits tumor growth driven by HER2 or neuregulin. Cancer Research 73, 6024–6035 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Freeman D, Ogbagabriel S, Rothe M & Radinsky R Fully human anti-HER3 monoclonal antibodies (mAbs) have unique in vitro and in vivo functional and antitumor activities versus other HER family …. Proceedings of the American Association for Cancer Research Annual Meeting; Apr (2008). [Google Scholar]
- 282.Yonesaka K, Haratani K, Hirotani K & Nakagawa K Abstract 44: U3–1402, a novel HER3-targeting ADC, and a novel DNA topoisomerase I inhibitor inhibit the growth of non-small cell lung cancer with EGFR mutation. Cancer Research 77, 44–44 (2017). [Google Scholar]
- 283.Vincent S et al. Abstract 2509: AV-203, a humanized ERBB3 inhibitory antibody inhibits ligand-dependent and ligand-independent ERBB3 signaling in vitro and in vivo. Cancer Research 72, 2509–2509 (2012). [Google Scholar]
- 284.Alsaid H et al. Non invasive imaging assessment of the biodistribution of GSK2849330, an ADCC and CDC optimized anti HER3 mAb, and its role in tumor macrophage recruitment in human tumor-bearing mice. PLoS ONE 12, e0176075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Xiao Z et al. A Potent HER3 Monoclonal Antibody That Blocks Both Ligand-Dependent and -Independent Activities: Differential Impacts of PTEN Status on Tumor Response. Mol. Cancer Ther. 15, 689–701 (2016). [DOI] [PubMed] [Google Scholar]
- 286.Zhang L et al. Abstract 2718: REGN1400, a fully-human ERBB3 antibody, potently inhibits tumor growth in preclinical models, both as a monotherapy and in combination with EGFR or HER2 blockers. Cancer Research 72, 2718–2718 (2012). [Google Scholar]
- 287.Geuijen C et al. Abstract LB-261: Mechanism of action of MCLA-128, a humanized bispecific IgG1 antibody targeting the HER2:HER3 heterodimer. Cancer Research 75, LB–261–LB–261 (2015). [Google Scholar]
- 288.Yu J et al. Cirmtuzumab Targets ROR1 to Inhibit Ibrutinib-Resistant, Wnt5a-Induced Rac1 Activation in Chronic Lymphocytic Leukemia. Blood 128, 2034–2034 (2016). [Google Scholar]
- 289.Hudecek M et al. The B-cell tumor-associated antigen ROR1 can be targeted with T cells modified to express a ROR1-specific chimeric antigen receptor. Blood 116, 4532–4541 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Halford MM et al. A Fully Human Inhibitory Monoclonal Antibody to the Wnt Receptor RYK. PLoS ONE 8, e75447 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.