Abstract
Background
Long non-coding RNA (lncRNA) has been recognized as the new regulator and biomarker for cancers. However, in clear cell renal cell carcinoma (ccRCC), the functions of lncRNAs are not well characterized. This research aimed to probe the function of lncRNA PCED1B-AS1 in the progression of ccRCC.
Materials and Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) was conducted to detect the expression levels of PCED1B-AS1, microRNA-484 (miR-484), and zinc finger E-box binding homeobox 1 (ZEB1) in 40 pairs of human ccRCC tissues and corresponding adjacent kidney tissue samples. Chi-square test was employed to evaluate the association between PCED1B-AS1 expression level and clinicopathological characteristics. The effects of PCED1B-AS1, miR-484 and ZEB1 on the cell proliferation, migration and epithelial-mesenchymal transition (EMT) process of ccRCC cells were studied by CCK-8 assay, EdU cell proliferation assay, wound healing test and Western blotting. The regulatory relationships among PCED1B-AS1, miR-484, ZEB1 were examined by luciferase reporter gene assay and RNA immunoprecipitation assay.
Results
PCED1B-AS1 was remarkably up-regulated in ccRCC tissues and cell lines. High expression of PCED1B-AS1 was associated with poor prognosis of the patients. Loss-of-function experiments showed that PCED1B-AS1 could regulate the proliferation, migration and EMT of ccRCC cells. PCED1B-AS1 sponged miR-484 to suppress its expression, and miR-484 targeted the 3ʹ-UTR of ZEB1 to repress the expression of ZEB1. MiR-484 counteracted the functions of PCED1B-AS1 in promoting the proliferation, migration and EMT of ccRCC cells, and PCED1B-AS1 promotes the expression of ZEB1 via repressing miR-484.
Conclusion
PCED1B-AS1/miR-484/ZEB1 axis is involved in regulating the progression of ccRCC.
Keywords: ccRCC, PCED1B-AS1, miR-484, ZEB1
Introduction
Renal cell carcinoma (RCC) is the second most common cause of death in patients with urinary tumors, which has a high mortality worldwide.1 In 2015, there are about 66.8 thousands of new RCC cases in China, and clear cell renal cell carcinoma (ccRCC) is the most common subtype of RCC, taking up about 75% of all cases.2,3 Although the treatment of ccRCC has been improved, the prognosis is still very poor due to its high rate of metastasis and recurrence, and the median survival time of the patients is only about 1.5 years.4 In addition, ccRCC cells are less sensitive to conventional chemotherapy and radiotherapy. Hence, an in-depth understanding of the molecular regulatory mechanisms in the occurrence and development of ccRCC will help develop novel treatment strategy for ccRCC.
Accumulating studies have shown that long non-coding RNA (lncRNA) is implicated in a variety of biological processes, such as cell proliferation, cell cycle, cell differentiation, and apoptosis.5–7 LncRNA dysregulation has been found to be associated with the carcinogenesis and development of ccRCC.8–11 For example, high expression of lncRNA PANDAR is related to high TNM stage, lymph node metastasis and distant metastasis of ccRCC patients, and silencing of PANDAR induces cell cycle arrest and apoptosis of ccRCC cells.10 LncRNA TP73-AS1 promotes viability and inhibits apoptosis of ccRCC cells via interacting with EZH2 and repressing the PI3K/Akt/mTOR pathway to suppress the expression of KISS1.11 PC-esterase domain containing 1B antisense RNA 1 (PCED1B-AS1) is a rarely studied lncRNA, and its role and mechanism in ccRCC are unclear.
MicroRNA (miRNA) can down-regulate target gene expression by binding to the 3ʹ -untranslated region (3ʹ-UTR), playing a prominent role in regulating diverse biological processes.12 Aberrantly expressed miRNAs participate in the growth and metastasis of ccRCC.13–16 For example, miR-663 is down-regulated in ccRCC tissues and cell lines, and miR-663 can directly target PAK4 to impede the proliferation and invasion of ccRCC cells.15 Additionally, in ccRCC, miR-124 and miR-203 participate in the activation of the epithelial-mesenchymal transition (EMT) pathway by regulating ZEB2.16 Accumulating studies show that lncRNA can function as a competitive endogenous RNA (ceRNA) to regulate downstream gene expression at the post-transcriptional level by sponging miRNAs.17 For example, lncRNA SNHG12 regulates MDM4 expression and the p53 pathway during the development of ccRCC by acting as a ceRNA for miR-125-5p.18
In the research, we demonstrated that PCED1B-AS1 was up-regulated in ccRCC tissues, and PCED1B-AS1 expression was related to clinical stage and Fuhrman grade of ccRCC. It was further found that silencing of PCED1B-AS1 suppressed the proliferation, migration and epithelial-mesenchymal transition (EMT) of ccRCC cells. Additionally, it was revealed that, functioning as a ceRNA, PCED1B-AS1 directly targeted miR-484 to increase the expression of zinc finger E-box binding homeobox 1(ZEB1) to promote ccRCC progression.
Materials and Methods
Sample Collection
The study was approved by the Ethics Committee of the Affiliated Hospital of Southwest Medical University (Approval Number: 201,708,042). All samples were available from patients undergoing radical or partial nephrectomy in the Affiliated Hospital of Southwest Medical University. A total of 40 pairs of ccRCC samples and matching normal kidney tissue samples were obtained. All samples were confirmed by postoperative pathological examination. All tissues were frozen and stored in liquid nitrogen until RNA extraction. The clinicopathological characteristics of these patients were recorded, and each patient provided written informed consent.
Cell Culture and Transfection
Human ccRCC cell lines (786-O, A498, ACHN, and Caki-1) and immortalized normal human renal tubular epithelial cell line (HK-2) were procured from the Institute of Cell Research, Chinese Academy of Sciences (Shanghai, China). A498, 786-O, and HK-2 cells were cultured in RPMI-460 medium (Hyclone, Logan, UT, USA). ACHN cells were cultured in DMEM (Invitrogen, Waltham, MA, USA), and Caki-1 cells were cultured in McCoy’s 5A medium (Gibco, Los Angeles, CA, USA). All medium contained 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA), and the cells were placed in an incubator at 37 °C and in 5% CO2.
siRNA specifically targeting PCED1B-AS1 (si-PCED1BAS1) and siRNA negative control (si-NC), as well as miR-484 mimics, NC mimics, miR-484 inhibitors and NC inhibitors were provided by GenePharma (Shanghai, China). In accordance with the manufacturer’s instruction, they were transfected into A498 and Caki-1 cells using LipofectamineTM 3000 Reagent (Invitrogen, Carlsbad, CA, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from the tissue samples and cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The total RNA was reverse transcribed into cDNA using PrimeScript RT reagent kit with gDNA Eraser (Takara, Dalian, China). For miR-484, reverse transcription was performed using the One Step PrimeScript miRNA cDNA Synthesis Kit (Takara, Dalian, China). qRT-PCR was carried out on an ABI 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) using SYBR Premix Ex Taq (Takara, Dalian, China). The primers are as follows: PCED1B-AS1: 5ʹ-TTTTGATGTTGGCCAATGCCG-3ʹ and 5ʹ-GGGCAGGGAGTCTTCATAGC-3ʹ; ZEB1: 5ʹ-GCACCTGAAGAGGACCAGAG-3ʹ and 5ʹ-TGCATCTGGTGTTCCATTTT-3ʹ; glyceraldehyde-3-phosphate dehydrogenase (GAPDH): 5ʹ-AGAGGCAGGGATGATGTTCTG-3ʹ and 5ʹ-GACTCATGACCACAGTCCATGC-3ʹ; miR-484: 5ʹ-TGCAGTCAGGCTCAGTCCCC-3ʹ; U6: 5ʹ-TGCGGGTGCTCGCTTCGGCAGC-3ʹ and 5ʹ-CCAGTGCAGGGTCCGAGGT-3ʹ.
Cell Proliferation Experiment
After transfection, the ccRCC cells were inoculated in a 96-well plate (2×103 cells/well). At 24, 48, 72 and 96 h, 10 μL of cell counting kit-8 (CCK-8) solution (Dojindo, Kumamoto, Japan) was supplemented to each well, respectively, and incubated at 37 °C for 2 h. Then, the optical density of the cells was measured at 450nm using a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
The EdU proliferation detection kit (Beyotime, Shanghai, China) was also employed to monitor the proliferation of ccRCC cells. Briefly, after 2 h of incubation with EdU kit, the cells were fixed with 4% paraformaldehyde for 30 min. After performing Apollo staining at room temperature for 30 min, DAPI staining solution was added and the cells were incubated at room temperature for 20 min in the dark. After the cells were washed with PBS, EdU-positive cells were observed, photographed and counted under a fluorescence microscope (Olympus, Tokyo, Japan).
Wound Healing Assay
Cell migration was assessed by wound healing experiment. In short, when the cells covered the bottom of the well, the cells were scratched with a sterile 200 μL pipette tip. After washing, the cells were cultured in a serum-free medium. After 0 and 24 h of culture, the images of the scratches were collected under a microscope (Olympus, Tokyo, Japan), respectively, and the width of the wound was measured.
Dual-Luciferase Reporter Gene Assay
HEK-293 cells were inoculated into 24-well plates, and miR-484 mimics or NC mimics and pmirGLO plasmids containing wild-type or mutant sequence of PCED1B-AS1 or 3ʹ-UTR of ZEB1 were co-transfected into HEK-293 cells by LipofectamineTM 3000 reagent (Invitrogen, Carlsbad, CA, USA). Forty-eight hours later, the luciferase activity of the cells in each group was determined by Dual-Luciferase® Reporter Assay System (Promega, Beijing, China). The firefly luciferase activity was normalized by renilla luciferase activity.
RNA Immunoprecipitation (RIP)
The binding relationship between PCED1B-AS1 and miR-484 was detected using a Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Bedford, MA, USA). The ccRCC cells were lysed with RIP buffer. Cell extracts were then incubated with magnetic beads, which were coupled with anti-Ago2 antibody or control IgG (Millipore, Billerica, MA, USA). Subsequently, the immunoprecipitate was incubated with proteinase K (Sigma-Aldrich, Louis, MO, USA), and total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and ultimately, the enrichment of PCED1B-AS1 and miR-484 was detected by qRT-PCR.
Western Blotting
Protein was extracted from cells with RIPA buffer (Beyotime, Shanghai, China), and the total protein was separated on 10% SDS-PAGE and transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membrane was then blocked with 5% skimmed milk, and the membrane was incubated with the primary antibody at 4 °C overnight. Thereafter, the membrane was incubated with the secondary antibody for 1 h at room temperature. β-Actin was used as the endogenous control, and the protein bands were exposed with enhanced chemiluminescence kit (Millipore, Billerica, MA, USA). The information of primary antibody is as follows: anti-ZEB1 (1:1000, Abcam, ab203829), anti-N-cadherin (1:1000, Abcam, ab202030), anti-E-cadherin (1:1000, Abcam, ab40772), anti-β-actin (1:2000, Abcam, Ab8226). All of the primary antibodies were obtained from Abcam (Cambridge, UK). The secondary antibodies were obtained from Beyotime (Shanghai, China).
Statistical Analysis
SPSS 20.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 8 software (GraphPad Software Inc., San Diego, CA, USA) were used for statistical analysis and graphing. Each experiment was repeated for at least three times, and all data were expressed as mean ± standard deviation. Data were analyzed using one-way ANOVA or Student’s t-test. Pearson’s correlation coefficient was employed to analyze the correlations among PCED1B-AS1, miR-484 and ZEB1. P < 0.05 indicated statistical significance.
Results
PCED1B-AS1 Was Up-Regulated in ccRCC Tissues and Cell Lines and Was Associated with Adverse Prognosis
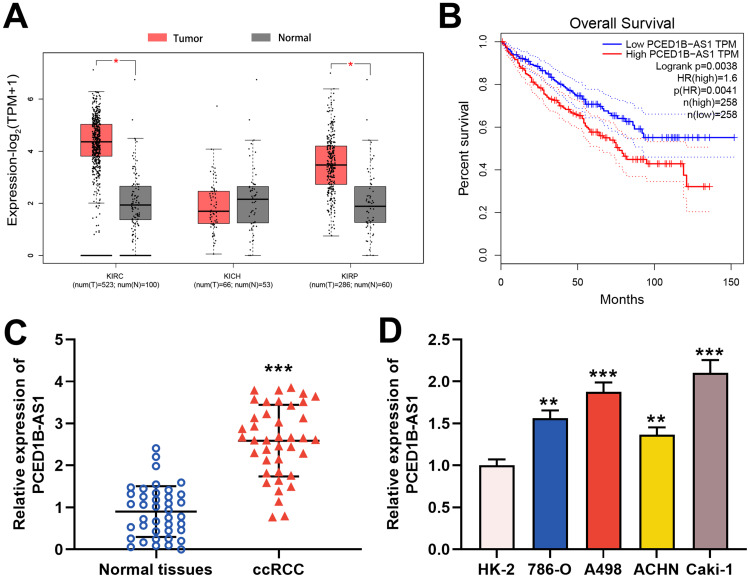
To explore the expression characteristics and prognostic value of PCED1B-AS1 in ccRCC, we first analyzed the data in the GEPIA database. The results showed that compared with normal tissues, PCED1B-AS1 was highly expressed in ccRCC (KIRC) and renal papillary cell carcinoma (KIRP), but not changed in kidney chromophobe (KICH) (Figure 1A). The overall survival time (OS) of ccRCC patients with high expression of PCED1B-AS1 was significantly shorter than those with low expression of PCED1B-AS1 (Figure 1B). We also measured the expression level of PCED1B-AS1 in ccRCC tissues and cell lines using qRT-PCR, and the results revealed that the expression level of PCED1B-AS1 in ccRCC tissues was remarkably higher than normal kidney tissue (Figure 1C). Additionally, the expression levels of PCED1B-AS1 in 786-O, A498, ACHN, and Caki-1 cell lines were also remarkably up-regulated compared with HK-2 cells (Figure 1D). What is more, we further analyzed the relationship between PCED1B-AS1 expression and the clinicopathological characteristics of ccRCC patients (Table 1). ccRCC patients were divided into low expression group (n=20) and high expression group (n=20). The results showed that high expression of PCED1B-AS1 was associated with high tumor stage and high Fuhrman grade, but not related to age, gender, tumor diameter, and lymph node metastasis. The data indicated that PCED1B-AS1 may play a tumor-promoting role in ccRCC.
Figure 1.
PCED1B-AS1 was up-regulated in ccRCC tissues and cell lines and was associated with adverse prognosis of the patients. (A) In the GEPIA database, PCED1B-AS1 was highly expressed in ccRCC tissues (red: cancerous tissues; gray: normal tissues). (B) GEPIA database was used to analyze the relationship between the expression of PCED1B-AS1 in ccRCC patients and the overall survival time of patients. (C) qRT-PCR was employed to detect the expression level of PCED1B-AS1 in ccRCC tissue and normal kidney tissue. (D) qRT-PCR was used to detect the expression of PCED1BAS1 in normal renal tubular epithelial cell line HK-2 and ccRCC cell lines (786-O, A498, ACHN and Caki-1). *P<0.05, **P <0.01, and ***P < 0.001.
Table 1.
Correlation Between PCED1B-AS1 Expression and Clinicopathologic Features of ccRCC Patients
| Factors | Total (n=40) | Expression of PCED1B-AS1 | P-value | |
|---|---|---|---|---|
| Low | High | |||
| Diagnosis age (years) | 0.337 | |||
| ≤60 | 17 | 10 | 7 | |
| >60 | 23 | 10 | 13 | |
| Gender | 0.204 | |||
| Male | 18 | 7 | 11 | |
| Female | 22 | 13 | 9 | |
| Tumor stage | 0.022* | |||
| pT1 | 25 | 16 | 9 | |
| pT2/T3 | 15 | 4 | 11 | |
| Fuhrman grade | 0.011* | |||
| I+II | 22 | 7 | 15 | |
| III+IV | 18 | 13 | 5 | |
| Tumor diameter (cm) | 0.113 | |||
| ≤7 cm | 21 | 8 | 13 | |
| >7 cm | 19 | 12 | 7 | |
| Lymph node metastasis | 0.185 | |||
| Negative | 26 | 15 | 11 | |
| Positive | 14 | 5 | 9 | |
Note: *P<0.05 (chi-square test).
PCED1B-AS1 Promoted the Proliferation and Migration of ccRCC Cells
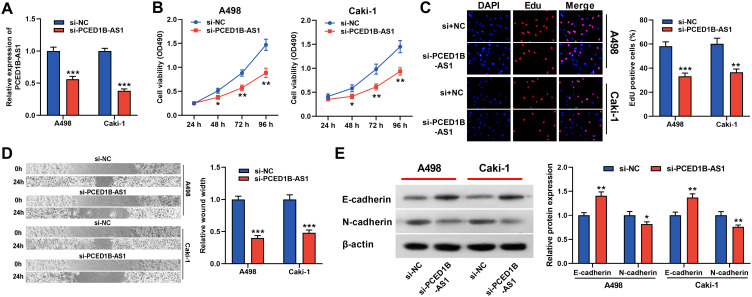
To probe the function of PCED1B-AS1 in ccRCC, we investigated its biological function in A498 and Caki-1 cells by knocking down PCED1B-AS1 expression using siRNA (Figure 2A). We measured cell proliferation using CCK-8 and EdU assays. The results showed that PCED1B-AS1 knockdown markedly suppressed the proliferation of A498 and Caki-1 cells (Figure 2B and C). Wound healing experiments were used to examine the effect of PCED1B-AS1 on cell migration, the results of which demonstrated that the migration ccRCC cells with PCED1B-AS1 silenced was significantly inhibited (Figure 2D). Western blotting was used to detect the expression of EMT-related proteins, and the data demonstrated that silencing of PCED1B-AS1 significantly increased E-cadherin expression and decreased N-cadherin expression in ccRCC cells (Figure 2E).
Figure 2.
PCED1B-AS1 promoted the proliferation and migration of ccRCC cells. (A) qRT-PCR was used to detect the transfection efficiency of si-PCED1B-AS1. (B, C) The effect of silencing PCED1B-AS1 on the proliferation of ccRCC cells was determined by CCK-8 and EdU experiments. (D) Wound healing experiment was employed to detect the effect of silencing PCED1B-AS1 on the migration of ccRCC cells. (E) Western blotting was used to detect the expression levels of EMT-related proteins. *P < 0.05; **P < 0.01; ***P < 0.001.
PCED1B-AS1 Acted as ceRNA of miR-484 in ccRCC
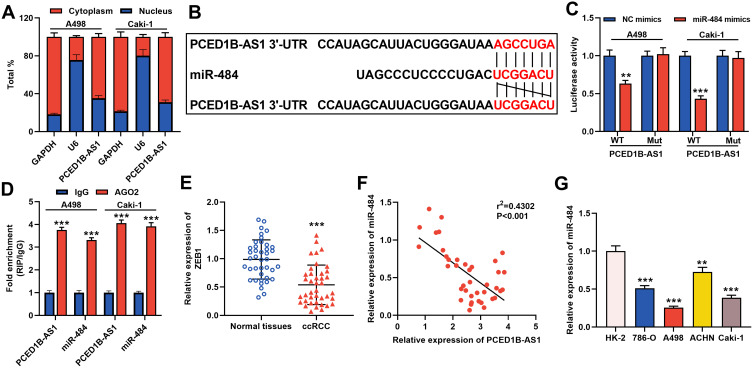
The results of cytoplasmic/nuclear separation experiments showed that PCED1B-AS1 was mainly expressed in the cytoplasm of A498 and Caki-1 cells, suggesting it could probably function as a ceRNA (Figure 3A). Next, we performed bioinformatics predictions on the DIANA database, and it was predicted that PCED1B-AS1 sequence that could potentially bind with miR-484 (Figure 3B, Supplementary Table 1). To verify the targeting relationship between them, we co-transfected PCED1B-AS1-WT luciferase reporter vector or PCED1B-AS1-MUT luciferase reporter vector and miR-484 mimics or NC mimics into A498 and Caki-1 cells. The results revealed that the luciferase activity in PCED1B-AS1-WT group was remarkably inhibited after transfection with miR-484 mimics, while the luciferase activity in PCED1B-AS1-MUT-transfected cells remained unchanged (Figure 3C). Additionally, RIP analysis showed that anti-Ago2 antibodies were able to immunoprecipitate PCED1B-AS1 and miR-484 in both A498 and Caki-1 cell lysates (Figure 3D). Moreover, qRT-PCR found that miR-484 was markedly down-regulated in ccRCC tissues compared with normal kidney tissues (Figure 3E), and Pearson’s correlation analysis indicated that there was a negative correlation between PCED1B-AS1 expression and miR-484 expression in ccRCC tissues (Figure 3F). Additionally, compared with HK-2 cells, miR-484 expression in ccRCC cells was down-regulated (Figure 3G). These data suggested that PCED1B-AS1 could sponge miR-484 and inhibit its expression in ccRCC.
Figure 3.
PCED1B-AS1 acted as a ceRNA of miR-484 in ccRCC. (A) qRT-PCR was used to detect the subcellular location of PCED1B-AS1 in A498 and Caki-1 cells. (B) Bioinformatics was used to predict the binding site for miR-484 on PCED1B-AS1 sequence. (C) MiR-484 mimics or NC mimics and PCED1B-AS1-WT luciferase reporter or PCED1B-AS1-MUT luciferase reporter were co-transfected into A498 and Caki-1 cells, and the relative luciferases activity in each group was detected. (D) The direct interaction between PCED1B-AS1 and miR-484 was detected by RIP method. (E) qRT-PCR was used to detect the relative expression level of miR-484 in ccRCC tissue and normal kidney tissue. (F) Pearson’s correlation analysis was used to evaluate the correlation between PCED1B-AS1 and miR-484 expression levels in ccRCC tissues. (G) qRT-PCR was used to detect the expression of miR-484 in normal renal tubular epithelial cell line HK-2 and ccRCC cell lines (786-O, A498, ACHN and Caki-1). **P < 0.01, and ***P < 0.001.
miR-484 Played a Tumor-Suppressive Role in ccRCC
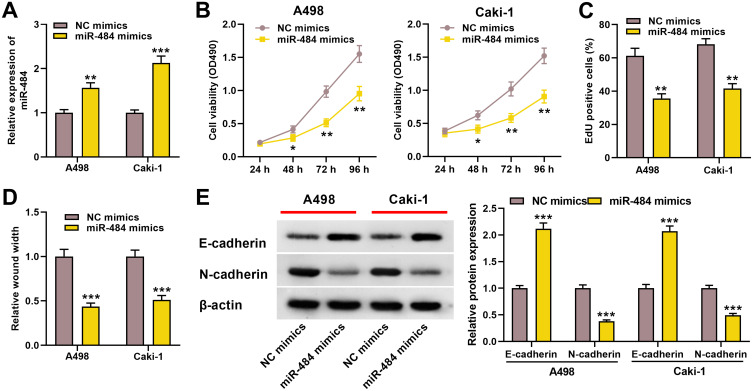
We further explore the role of miR-484 in ccRCC. We transfected miR-484 mimics or NC mimics into A498 and Caki-1 cells, and as shown, miR-484 expression in the cells transfected with miR-484 mimics was remarkably increased (Figure 4A). Moreover, CCK-8 and EdU assays showed miR-484 overexpression repressed the proliferation of ccRCC cells (Figure 4B and C). Wound healing assay indicated that miR-484 overexpression blocked the migration of ccRCC cells (Figure 4D). Western blotting revealed that compared with the control group, E‐cadherin was increased and N‐cadherin was decreased in the ccRCC cells in the miR-484 overexpression group (Figure 4E). These results indicated that miR-484 had an inhibitory effect on the progression of ccRCC.
Figure 4.
miR-484 played a tumor-suppressive role in the progress of ccRCC. (A) qRT-PCR was used to detect miR-484 expression level of A498 and Caki-1 cells after miR-484 mimics or NC mimics were transfected. (B, C) CCK-8 and EdU assays were used to detect the effect of miR-484 on the proliferation of A498 and Caki-1 cells. (D) The effect of miR-484 on the migration of A498 and Caki-1 cells was detected by the wound healing experiment. (E) Western blotting was used to detect the expression level of EMT-related proteins. *P < 0.05, **P < 0.01, and ***P < 0.001.
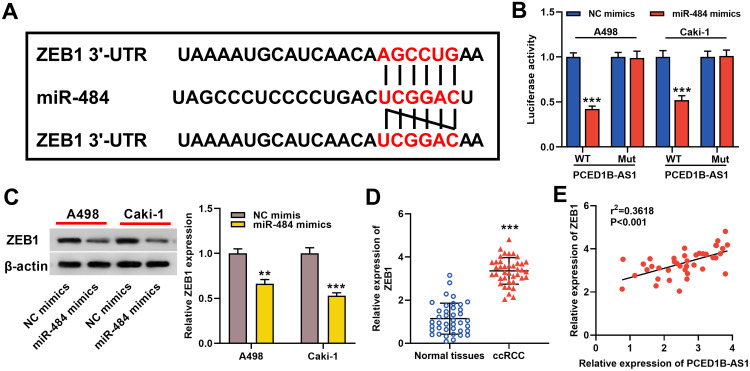
ZEB1 Was a Direct Target Gene of miR-484
Furthermore, TargetScan database predicted that ZEB1 was a candidate target of miR-484 (Figure 5A). The dual-luciferase reporter experiment revealed that transfection of miR-484 mimics reduced luciferase activity of ZEB1-WT reporter, but had no significant effect on the luciferase activity in ZEB1-MUT group (Figure 5B). Western blotting indicated that ZEB1 protein expression was markedly inhibited in miR-484 mimics-transfected cells (Figure 5C). In addition, we found that ZEB1 expression was remarkably higher in ccRCC tissues than in normal kidney tissues (Figure 5D), and correlation analysis showed that ZEB1 was positively correlated with PCED1B-AS1 expression (Figure 5E). The data indicated that ZEB1 was a direct target of miR-484 in ccRCC cells, and could probably be regulated by PCED1B-AS1.
Figure 5.
ZEB1 was a direct target gene of miR-484. (A) Bioinformatics analysis was used to predict the binding site of miR-484 on the ZEB1 3ʹ UTR sequence. (B) MiR-484 mimics or NC mimics and ZEB1-WT luciferase reporter or ZEB1-MUT luciferase reporter were co-transfected into A498 and Caki-1 cells, and the relative luciferase activity of the cells in each group was measured. (C) Western blotting was used to detect ZEB1 protein expression in miR-484 mimics-transfected cells. (D) qRT-PCR was used to detect the relative expression level of ZEB1 in ccRCC tissue and normal kidney tissue. (E) Pearson’s correlation analysis was used to evaluate the correlation between PCED1B-AS1 and ZEB1 expression levels in ccRCC tissues. **P < 0.01, and ***P < 0.001.
PCED1B-AS1 Played a Carcinogenic Role in ccRCC Cells Through Regulating miR-484/ZEB1 Axis
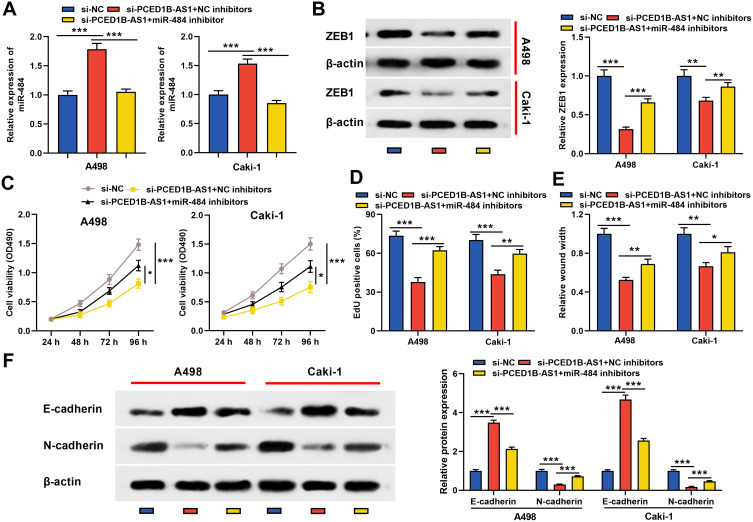
To elucidate whether PCED1B-AS1 modulated cell proliferation, migration, and EMT through miR-484/ZEB1 axis, we co-transfected miR-484 inhibitors into A498 and Caki-1 cells with PCED1B-AS1 silenced, and then “rescue” experiments were performed. qRT-PCR revealed that silencing PCED1B-AS1 increased the expression of miR-484, and the transfection of miR-484 inhibitors remarkably inhibited miR-484 expression (Figure 6A). Western blotting revealed that inhibiting PCED1B-AS1 significantly inhibited the expression of ZEB1, and transfection of miR-484 inhibitors partially restored ZEB1 protein expression (Figure 6B). Functional experiments showed that inhibition of miR-484 counteracted the effects of PCED1B-AS1 knockdown on the proliferation, migration, and EMT of A498 and Caki-1 cells (Figure 6C–F). Briefly, these data indicated that PCED1B-AS1 played a promoting role in the ccRCC progression through regulating the miR-484/ZEB1 axis.
Figure 6.
PCED1B-AS1 played a carcinogenic role in ccRCC cells through the miR-484/ZEB1 axis. miR-484 inhibitors or NC inhibitors were co-transfected into A498 and Caki-1 cells with PCED1B-AS1 silenced. (A) qRT-PCR was employed to detect miR-484 expression. (B) Western blotting was used to detect ZEB1 protein expression. (C, D) CCK-8 and EdU were used to detect the proliferation of A498 and Caki-1 cells. (E) Wound healing test was used to detect the migration of A498 and Caki-1 cells. (F) Western blotting was used to detect the expression level of EMT-related proteins. *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
ccRCC is one of the most common genitourinary cancers around the world. ccRCC patients have limited treatment options, high recurrence rates, and unfavorable prognosis. Therefore, studying the mechanism of ccRCC carcinogenesis and progression and finding potential molecular biomarkers and therapeutic targets are of great significance. Accumulating studies have found that lncRNA plays a key role in the carcinogenesis and development of ccRCC.8,19 For instance, it is reported that lncRNA CRNDE is overexpressed in ccRCC tissues and cell lines, and in vitro functional experiments indicate that CRNDE can enhance the migration and invasion of ccRCC cells.20 Another study reports that lncRNA SNHG5 is up-regulated in ccRCC tissues, and inhibition of SNHG5 reduces the proliferation and invasion of ccRCC cell and promoted apoptosis.21 Nonetheless, the functions and molecular mechanisms of lncRNA in ccRCC progression have not been fully clarified. PCED1B-AS1 is a rarely studied lncRNA. It is reported that lncRNA PCED1B-AS1 is up-regulated in luminal B breast cancer and glioblastoma tissues; PCED1B-AS1 can bind to the 5ʹ-UTR of HIF-1α mRNA and facilitates HIF-1α translation, thereby promoting the Warburg effect and tumorigenesis of glioblastoma.22,23 In addition, PCED1B-AS1 can modulate the miR-194-5p/PCED1B axis to promote glioma cell proliferation and inhibit apoptosis.24 In the current research, for the first time, we investigated the expression and biological function of PCED1B-AS1 in ccRCC. We found that PCED1B-AS1 was up-regulated in ccRCC, and the knockdown of PCED1B-AS1 inhibited cell proliferation, migration, and EMT process, indicating that PCED1B-AS1 functioned as a cancer-promoting factor in ccRCC.
Recent researches show that lncRNAs target miRNAs as ceRNAs to participate in tumorigenesis and cancer progression.25 For example, lncRNA SBF2-AS1 promotes cervical cancer progression by sponging and repressing miR-361-5p to up-regulate FOXM1.26 LncRNA HOTAIR regulates the growth, metastasis, and apoptosis of breast cancer cells through the miR-20a-5p/HMGA2 axis.27 In previous studies, miR-484 has been proved to be a tumor suppressor in several cancers, such as cervical cancer and gastric cancer.28,29 In this study, we found that PCED1B-AS1 can function as ceRNA for miR-484 to inhibit its expression. What is more, miR-484 overexpression inhibited the proliferation and migration of ccRCC cells and EMT process. Our data suggested that miR-484 was a tumor suppressor in ccRCC, and PCED1B-AS1 could negatively regulate its expression.
ZEB1 is a crucial regulatory element of the transcription factor network and mainly controls the EMT process.30,31 ZEB1 is abnormally expressed in a variety of cancers, such as colorectal cancer, lung cancer, breast cancer, and gastric cancer, and it is involved in the proliferation, migration and invasion of cancer cells and is considered to have cancer-promoting functions.32–36 For example, lncRNA TMPO-AS1 facilitates the proliferation, migration, and invasion of cervical cancer cells by modulating the miR-143-3p/ZEB1 axis.37 In this study, we predicted and verified that miR-484 was able to target ZEB1, and that ZEB1 expression in ccRCC patient tissues was positively regulated by PCED1B-AS1. “Rescue” experiments showed that transfection with miR-484 inhibitor not only partially restored ZEB1 expression but also significantly weakened the inhibition of PCED1B-AS1 knockdown on ccRCC cell proliferation, migration, and EMT process. Therefore, these findings suggested that PCED1B-AS1 could regulate the occurrence and development of ccRCC by regulating the miR-484/ZEB1 axis.
In summary, we demonstrate the up-regulation of PCED1B-AS1 in ccRCC and reveal that PCED1B-AS1 enhances ccRCC cell proliferation, migration, and EMT process. Additionally, PCED1B-AS1 positively regulated ZEB1 expression through sponging and repressing miR-484. These findings are helpful to further understand the carcinogenesis and development mechanism of ccRCC, providing a possible direction for the therapy of ccRCC. Notably, PCED1B-AS1 can probably target multiple miRNAs besides miR-484, which may be involved in the regulation of multiple signal pathways. In the future research, a more comprehensive study of the function and mechanism of PCED1B-AS1 in ccRCC is needed.
Funding Statement
This work is supported by the National Natural Science Foundation of China (No. 81800670) and Foundation of Luzhou-Southwest Medical University (No. 2017lzxnyd-j32).
Ethics Statement
The study was approved by the Ethics Committee of the Affiliated Hospital of Southwest Medical University (Number: 201,708,042), and each patient provided informed consent.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. [DOI] [PubMed] [Google Scholar]
- 4.Wu D, Li M, Wang L, et al. microRNA145 inhibits cell proliferation, migration and invasion by targeting matrix metallopeptidase-11 in renal cell carcinoma. Mol Med Rep. 2014;10(1):393–398. doi: 10.3892/mmr.2014.2149 [DOI] [PubMed] [Google Scholar]
- 5.Ernst C, Morton CC. Identification and function of long non-coding RNA. Front Cell Neurosci. 2013;7:168. doi: 10.3389/fncel.2013.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu MH, Tang B, Zeng S, et al. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35(27):3524–3534. doi: 10.1038/onc.2015.413 [DOI] [PubMed] [Google Scholar]
- 8.Li X, Meng X, Wei C, et al. Dissecting LncRNA roles in renal cell carcinoma metastasis and characterizing genomic heterogeneity by single-cell RNA-seq. Mol Cancer Res. 2018;16(12):1879–1888. doi: 10.1158/1541-7786.MCR-17-0776 [DOI] [PubMed] [Google Scholar]
- 9.Sun C, Zhou Z, Shi H, Li F, Zhang G. Identification of long noncoding RNA APOC1P1 as an oncogene in clear cell renal cell carcinoma. Dis Markers. 2019;2019:2814058. doi: 10.1155/2019/2814058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Tong Y, Zhu J, et al. An increase in long non-coding RNA PANDAR is associated with poor prognosis in clear cell renal cell carcinoma. BMC Cancer. 2017;17(1):373. doi: 10.1186/s12885-017-3339-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu G, Zhao X, Zhou J, Cheng X, Ye Z, Ji Z. LncRNA TP73-AS1 promotes cell proliferation and inhibits cell apoptosis in clear cell renal cell carcinoma through repressing KISS1 expression and inactivation of PI3K/Akt/mTOR signaling pathway. Cell Physiol Biochem. 2018;48(1):371–384. doi: 10.1159/000491767 [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lokeshwar SD, Talukder A, Yates TJ, et al. Molecular characterization of renal cell carcinoma: a potential three-microRNA prognostic signature, cancer epidemiology. Biomarkers Prev. 2018;27(4):464–472. doi: 10.1158/1055-9965.EPI-17-0700 [DOI] [PubMed] [Google Scholar]
- 14.He YH, Chen C, Shi Z. The biological roles and clinical implications of microRNAs in clear cell renal cell carcinoma. J Cell Physiol. 2018;233(6):4458–4465. doi: 10.1002/jcp.26347 [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Jiao D, Tian Z. MicroRNA663 inhibits the proliferation and invasion of clear cell renal cell carcinoma cells by directly targeting PAK4. Mol Med Rep. 2019;19(1):711–718. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Zhong Y, Li L. miR-124 and miR-203 synergistically inactivate EMT pathway via coregulation of ZEB2 in clear cell renal cell carcinoma (ccRCC). J Transl Med. 2020;18(1):69. doi: 10.1186/s12967-020-02242-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye Y, Li S-L, Wang S-Y, Passos GA. Construction and analysis of mRNA, miRNA, lncRNA, and TF regulatory networks reveal the key genes associated with prostate cancer. PLoS One. 2018;13(8):e0198055. doi: 10.1371/journal.pone.0198055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Z, Chen D, Wang K, Cao C, Xu X. Long non-coding RNA SNHG12 functions as a competing endogenous RNA to regulate MDM4 expression by sponging miR-129-5p in clear cell renal cell carcinoma. Front Oncol. 2019;9:1260. doi: 10.3389/fonc.2019.01260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Chen Y, Hu S, et al. Integrative analysis of protein-coding and non-coding RNAs identifies clinically relevant subtypes of clear cell renal cell carcinoma. Oncotarget. 2016;7(50):82671–82685. doi: 10.18632/oncotarget.12340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding C, Han F, Xiang H, et al. LncRNA CRNDE is a biomarker for clinical progression and poor prognosis in clear cell renal cell carcinoma. J Cell Biochem. 2018;119(12):10406–10414. doi: 10.1002/jcb.27389 [DOI] [PubMed] [Google Scholar]
- 21.Li WZ, Zou Y, Song ZY, et al. Long non-coding RNA SNHG5 affects the invasion and apoptosis of renal cell carcinoma by regulating the miR-363-3p-Twist1 interaction. Am J Transl Res. 2020;12(2):697–707. [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan CL, Jiang XM, Yi Y, et al. Identification of differentially expressed lncRNAs and mRNAs in luminal-B breast cancer by RNA-sequencing. BMC Cancer. 2019;19(1):1171. doi: 10.1186/s12885-019-6395-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Z, Zhang Q, Guo F, et al. Long noncoding RNA PCED1B-AS1 promotes the warburg effect and tumorigenesis by upregulating HIF-1α in glioblastoma. Cell Transplant. 2020;29:963689720906777. doi: 10.1177/0963689720906777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Yu D, Liu X, Changyong E, Yu S. LncRNA PCED1B-AS1 activates the proliferation and restricts the apoptosis of glioma through cooperating with miR-194-5p/PCED1B axis. J Cell Biochem. 2020;121(2):1823–1833. doi: 10.1002/jcb.29417 [DOI] [PubMed] [Google Scholar]
- 25.Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol. 2019;234(7):10080–10100. doi: 10.1002/jcp.27941 [DOI] [PubMed] [Google Scholar]
- 26.Gao F, Feng J, Yao H, Li Y, Xi J, Yang J. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells, Nanomed Biotechnol. 2019;47(1):776–782. doi: 10.1080/21691401.2019.1577883 [DOI] [PubMed] [Google Scholar]
- 27.Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7(3):842–855. doi: 10.1002/cam4.1353 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Hu Y, Xie H, Liu Y, Liu W, Liu M, Tang H. miR-484 suppresses proliferation and epithelial-mesenchymal transition by targeting ZEB1 and SMAD2 in cervical cancer cells. Cancer Cell Int. 2017;17:36. doi: 10.1186/s12935-017-0407-9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Li Y, Liu Y, Yao J, Li R, Fan X. Downregulation of miR-484 is associated with poor prognosis and tumor progression of gastric cancer. Diagn Pathol. 2020;15(1):25. doi: 10.1186/s13000-020-00946-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen JE, Nathan V, Osborne JK, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126(9):3219–3235. doi: 10.1172/JCI76725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caramel J, Ligier M, Puisieux A. Pleiotropic roles for ZEB1 in cancer. Cancer Res. 2018;78(1):30–35. doi: 10.1158/0008-5472.CAN-17-2476 [DOI] [PubMed] [Google Scholar]
- 32.Spaderna S, Schmalhofer O, Hlubek F, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131(3):830–840. doi: 10.1053/j.gastro.2006.06.016 [DOI] [PubMed] [Google Scholar]
- 33.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16(6):488–494. doi: 10.1038/ncb2976 [DOI] [PubMed] [Google Scholar]
- 34.Hou P, Li L, Chen F, et al. PTBP3-mediated regulation of ZEB1 mRNA stability promotes epithelial-mesenchymal transition in breast cancer. Cancer Res. 2018;78(2):387–398. doi: 10.1158/0008-5472.CAN-17-0883 [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, Li J, Ou Y, et al. CDK4/6 inhibition blocks cancer metastasis through a USP51-ZEB1-dependent deubiquitination mechanism. Signal Transduct Target Ther. 2020;5:25. doi: 10.1038/s41392-020-0118-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soleymani Fard S, Sotoudeh M, Saliminejad K, et al. Investigation of the correlation between androgen receptor and ZEB1 and its value in progression of gastric cancer. Avicenna J Med Biotechnol. 2020;12(1):52–60. [PMC free article] [PubMed] [Google Scholar]
- 37.Gang X, Yuan M, Zhang J. Long non-coding RNA TMPO-AS1 promotes cervical cancer cell proliferation, migration, and invasion by regulating miR-143-3p/ZEB1 axis. Cancer Manag Res. 2020;12:1587–1599. doi: 10.2147/CMAR.S226409 [DOI] [PMC free article] [PubMed] [Google Scholar]