Abstract
Introduction
The US FDA is interested in a tool that would enable pharmacovigilance safety evaluators to automate the identification of adverse drug events (ADEs) mentioned in FDA prescribing information. The MITRE Corporation (MITRE) and the FDA organized a shared task—Adverse Drug Event Evaluation (ADE Eval)—to determine whether the performance of algorithms currently used for natural language processing (NLP) might be good enough for real-world use.
Objective
ADE Eval was conducted to evaluate a range of NLP techniques for identifying ADEs mentioned in publicly available FDA-approved drug labels (package inserts). It was designed specifically to reflect pharmacovigilance practices within the FDA and model possible pharmacovigilance use cases.
Methods
Pharmacovigilance-specific annotation guidelines and annotated corpora were created. Two metrics modeled the experiences of FDA safety evaluators: one measured the ability of an algorithm to identify correct Medical Dictionary for Regulatory Activities (MedDRA®) terms for the text from the annotated corpora, and the other assessed the quality of evidence extracted from the corpora to support the selected MedDRA® term by measuring the portion of annotated text an algorithm correctly identified. A third metric assessed the cost of correcting system output for subsequent training (averaged, weighted F1-measure for mention finding).
Results
In total, 13 teams submitted 23 runs: the top MedDRA® coding F1-measure was 0.79, the top quality score was 0.96, and the top mention-finding F1-measure was 0.89.
Conclusion
While NLP techniques do not perform at levels that would allow them to be used without intervention, it is now worthwhile exploring making NLP outputs available in human pharmacovigilance workflows.
Electronic supplementary material
The online version of this article (10.1007/s40264-020-00996-3) contains supplementary material, which is available to authorized users.
Key Points
| The MITRE Corporation and the US FDA conducted Adverse Drug Event Evaluation (ADE Eval) to evaluate the ability of software systems to find adverse drug events in package inserts (drug labels) using guidelines and annotated training data for adverse drug event detection customized for the pharmacovigilance needs of FDA safety evaluators. |
| In total, 13 teams submitted 23 system runs, evaluated using metrics to model the experience of FDA safety evaluators, including a novel metric to estimate the cost of correcting system output for subsequent training. |
| Varied approaches achieved high performance, suggesting that the technology is now mature enough to experiment with using natural language processing in human pharmacovigilance workflows. |
Introduction
The US FDA is interested in a tool that would enable pharmacovigilance safety evaluators (SEs) to automate the identification of adverse drug events (ADEs) mentioned in FDA prescribing information, which could facilitate the triage, review, and processing of postmarket ADE reports, also known as individual case safety reports (ICSRs). The FDA continually receives ICSRs describing ADEs observed during the use of marketed drug and therapeutic biologic products from drug manufacturers, healthcare professionals, and consumers. In addition, the FDA continues to approve new drug labels,1 also known as package inserts (PIs),2 and update already approved PIs with newly identified postmarket ADE information. In 2019, over 2 million ICSRs were submitted to the FDA Adverse Event Reporting System (FAERS) database [1], and 48 novel drug products were approved by the FDA [2]. As part of the FDA’s postmarket drug safety surveillance activities, SEs are tasked with reviewing increasing volumes of ICSRs to identify safety concerns associated with drug products to promote and protect public health.
Safety concerns can be new unlabeled ADEs (those that are not yet described in the relevant drug label) or an increase in severity or frequency of a labeled ADE. The large volume of reports means that SEs face challenges in screening and prioritizing ICSRs that implicate a causal association between a drug and an ADE of interest. SEs must frequently decide whether ADEs that are described in the ICSRs are mentioned in the appropriate section (e.g., boxed warning, warnings and precautions, contraindication) of the relevant PI.
However, within the current SE workflow, the process of determining and comparing the labeled status of an ADE in a PI with that of the ADEs described in ICSRs is a manual one. This is because the ADEs reported in each ICSR are standardized to the Medical Dictionary for Regulatory Activities (MedDRA®; https://www.meddra.org) terminology3 but the ADEs mentioned in a PI are not and may appear as unstructured text in forms that do not exactly match any of the alternative terms listed in MedDRA® for the relevant ADE. Because a common terminology is crucial for SEs to readily determine and compare the labeled status of ADEs with that of the MedDRA®-coded ADEs reported in ICSRs, it would be extremely useful for the FDA to have a tool that could summarize, for a set of PIs, the particular ADEs mentioned, using MedDRA® as the reference vocabulary, and could locate, within the particular PI sections, the evidence for the ADEs mentioned.
The adverse drug event (ADE) evaluation (ADE Eval) shared task was sponsored by the FDA to evaluate a range of natural language processing (NLP) techniques for identifying ADEs mentioned in publicly available FDA PIs. The ADE Eval task consisted of identifying mentions of ADEs in specific sections of PIs and mapping those mentions to associated terms in MedDRA®. The aim of the task was to determine whether the performance of current NLP algorithms might be good enough to support real-world pharmacovigilance use in cases such as those described.
Related Work
Evaluation of the ability of NLP systems to extract ADEs from unstructured text is a natural consequence of growing interest in the application of NLP for postmarket pharmacovigilance. Initial evaluation of more general NLP-based information extraction systems has been followed by the development and evaluation of systems designed more specifically to recognize ADEs and related concepts in a variety of textual sources, including biomedical literature, electronic health records (EHRs), social media, and PIs.
US FDA Center for Drug Evaluation and Research, National Institute for Standards and Technology Text Analysis Conference (TAC) Adverse Drug Reactions (ADRs), and the Motivation for ADE Eval
The FDA Center for Drug Evaluation and Research (CDER) has a long-standing interest in the ability to automatically extract ADEs from PIs for the purpose of pharmacovigilance.
Ly et al. [3] evaluated the performance of the following three NLP systems for their ability to extract ADE terms from PI labels and normalize the terms to MedDRA® PTs:
Event-Based Text Mining of Health Electronic Records (ETHER) [4, 5], which was designed to extract clinical terms and time statements from free-text ADE descriptions in postmarket reports;
I2E [6], an NLP-based text-mining application designed to extract information from a variety of textual sources, including scientific literature, EHRs, patents, news feeds, clinical trials data, and proprietary content; and
MetaMap [7], an NLP-based system developed by the National Library of Medicine and designed to process biomedical literature and map concepts to the Unified Medical Language System (UMLS) Metathesaurus.
Ly et al. [3] compared each system’s output to MedDRA® PT ADE lists that had been manually mapped by FDA pharmacovigilance experts. I2E had the highest precision (0.77), recall (0.83), and F measure (0.79). The goal of the study was to demonstrate the feasibility of using NLP tools to discover these ADEs without human intervention, and the authors concluded that existing tools were insufficient to meet their needs but that their performance was sufficient to consider further development.
In further support of their interest in automated extraction of ADE terms, several offices within the FDA cosponsored a track of the 2017 National Institute for Standards and Technology (NIST) Text Analysis Conference (TAC) [8]. The NIST TAC adverse drug reaction (ADR) track [9] addressed identification of ADRs in structured PIs. The top performing system achieved an F1 score of 0.852 on extraction of ADRs and 0.853 macro-averaged F1 on MedDRA® term mapping.4
The TAC ADR evaluation was designed for a generic use case that did not align specifically to the FDA FAERS review use case. For instance, mentions of death qualify as ADRs for the TAC ADR evaluation; however, for the pharmacovigilance use case, death represents an outcome.5 Where ADRs represent medical conditions and are accompanied by associated symptoms, TAC ADR track guidelines instruct participants to annotate the reaction and the symptoms, whereas pharmacovigilance guidelines stipulate annotation of only the ADR and not of their symptoms or outcomes.
This led to the design of ADE Eval, built on pharmacovigilance-specific definitions of ADEs and providing diagnostic insight into how well existing systems could support the pharmacovigilance use cases. Details of how and why these two evaluations differ can be found in Sect. A.1 in the electronic supplementary material (ESM).
Other Adverse Drug Event (ADE)-Related Shared Tasks
A number of other ADE-related shared tasks have been conducted to support the development and evaluation of systems designed specifically to extract ADEs and related concepts from unstructured text. The scope of these tasks is broader than TAC ADR or ADE Eval; because they do not focus on PI documents (and are thus not focused specifically on a single drug), they must include both a medication recognition task and a medication–ADE relation extraction task.
The Medication and Adverse Drug Events from Electronic Health Records (MADE) 1.0 challenge for extracting medication, indication, and ADEs from EHR notes was held in 2018 [10]. It consisted of three tasks: (1) identifying medications and their attributes (dosage, route, duration, and frequency), indications, ADEs, and severity; (2) identifying relations between the entities (with named entities provided in input): medication–indication, medication–ADE, and attribute relations; and (3) performing end-to-end entity and relation identification on unlabeled input.
National NLP Clinical Challenges (n2c2) held a shared task on ADEs and medication extraction in EHRs in 2018 [11]. Track 2 of the shared task included (1) identifying medications, their signature information, and ADEs; (2) identifying relationships between medications and their attributes and between medications and ADEs; and (3) building an end-to-end system that extracts concepts and finds relations of those concepts to their medications.
Three Social Media Mining for Health (SMM4H) shared tasks included extraction of ADEs from Twitter tweets. All three shared tasks included classification of tweets according to whether or not they mentioned an ADR. SMM4H 2017 [12, 13] and SMM4H 2018 [14] also included classification of posts based on medication mention and medication intake status. SMM4H 2019 [15] added extraction of ADR mentions from tweets and normalization of extracted ADRs to MedDRA® PT identifiers. For further details, see Tables A-1, A-2, and A-3 in the ESM. Additional pharmacovigilance evaluations involving social media include work by Caster and colleagues [16, 17] and Pierce et al. [18].
All of these shared tasks focused on ADE mentions, whereas TAC and ADE Eval were oriented towards MedDRA® codes.
Methods
The ADE Eval consisted of two tasks: (1) finding ADE mentions and (2) coding ADE mentions to MedDRA®. The specificity of the pharmacovigilance use case provided a concrete opportunity to evaluate NLP technology for ADE detection by coordinating the design of annotation guidelines, corpus definition, and metrics.
Training and Test Corpora
The training data for ADE Eval consisted of 100 annotated documents, 50 of which were also included in the 2017 NIST TAC ADR test set. The test data for the evaluation consisted of 2000 unannotated test documents, 100 of which were annotated for evaluation. The identity of this 100-document test subset was not revealed to performers. Each document consisted of a subset of the sections found in a single PI label, accessed from DailyMed [19] (https://dailymed.nlm.nih.gov/dailymed/). The sections of interest were adverse reactions, boxed warnings, and either one or two sections devoted to warnings and precautions. All documents contained an ADR section; the other sections might or might not appear in a given document. For the purposes of the evaluation, the raw text to the relevant sections was extracted from the PIs.6
The completed annotated corpus is complex and extensive: more than 60,000 mention annotations (each of them MedDRA® coded) over approximately 690,000 words. These test mention annotations amount to almost 14,000 MedDRA® code occurrences across the specified sections of the test corpus documents. Detailed annotation statistics for the full corpus can be found in Sect. B.1 of the ESM.
Guidelines and Annotation Schema
The annotation guidelines reflected pharmacovigilance SEs’ interpretations of PIs as well as application of that expertise to ICSR screening; it followed the FDA labeling guidance, in which adverse experience is defined as “any adverse event associated with the use of a drug in humans, whether or not considered drug related, including the following: an adverse event occurring in the course of the use of a drug product in professional practice; an adverse event occurring from drug overdose whether accidental or intentional; an adverse event occurring from drug abuse; an adverse event occurring from drug withdrawal; and any failure of expected pharmacological action”. See Sect. A.1 in the ESM for more details.
To explore potentially confusable phrase types, the FDA created separate annotation categories for phrases meeting the use-case-specific ADE definitions as well as phrases that might be confusable with this definition, along with the reason for classifying each phrase. These latter categories were labeled in the training and test data but used only for diagnostics and not for scoring.
SEs from the Office of Surveillance and Epidemiology (OSE)—the FDA CDER office responsible for monitoring ICSRs reported to FAERS—annotated the training and test corpus, and the categories were named accordingly. The category names OSE_Labeled_AE, NonOSE_AE, and Not_AE_Candidate represent OSE’s workflow and do not have regulatory implications. The category names were chosen to distinguish between the present evaluation and the previous annotations made by the FDA for the 2017 NIST TAC ADR test set. The ADE Eval annotation schema consisted of three top-level annotation categories:7
OSE_Labeled_AE Primary ADEs listed in a drug product label and associated with that particular drug exposure. This category was the only category scored.
NonOSE_AE Adverse events (AEs) other than OSE_Labeled_AE that are potentially confusable with OSE_Labeled _AE, such as ADEs identified in an unapproved use of the drug, ADEs occurring in the context of animal exposure, ADEs representing a sign/symptom/manifestation of an OSE_Labeled_AE, and ADEs resulting from a drug interaction. ADEs that result from drug interactions are not associated with either drug alone but are associated with exposure to the drug combination. This is why pharmacovigilance reviewers consider ADEs resulting from drug–drug interactions as different from OSE_Labeled_AEs and, for the purpose of the study, we categorized them as NonOSE_AEs. A label may state an ADE (categorized as an OSE_Labeled_AE) and include its typical manifestations (NonOSE_AEs). Manifestations are categorized as NonOSE_AEs because they could potentially be associated with multiple primary AEs (OSE_Labeled_AEs) or could present as a stand-alone ADE with a distinct mechanism, thus warranting further exploration/characterization. (See Table B-1 in the ESM for the different subtypes of NonOSE_AE). This category was not scored.
Not_AE_Candidate Terms that describe a condition unrelated to AEs such as the drug’s indication, contraindication, and patient’s medical history. Like NonOSE_AE terms, the terms in this class are potentially confusable with AEs but occur in a different context. This category was not scored.
Each annotated mention bore a number of additional attributes, which fell into three distinct groups:
Discontinuities Attributes that help determine the exact span of the mention in the case of so-called discontinuous mentions (i.e., mentions whose text is not an uninterrupted phrase). An example of this sort of discontinuity is found in the phrase “suicidal thoughts and behaviors,” where the phrase “suicidal … behaviors” is a candidate ADE that is discontinuous. The discontinuity attributes were used in the scoring of discontinuous mentions.
MedDRA® Attributes that capture the MedDRA® information (PT and code, LLT and code) associated with the mention. MedDRA® PTs/codes were used in scoring.
Reasons Attributes that indicate the reason for the choice of top-level category. Each of the three annotation categories is associated with a different set of reasons (e.g., the AE_animal reason is associated with the NonOSE_AE category because ADRs observed in animal data, although informative, do not necessarily translate to AEs observed in humans). The reason attributes were recorded for the purposes of information and data analysis only and were not scored. The specific values for the reason attributes are listed and defined in Table B-1 in the ESM.
Corpus Preparation
Before human annotators examined the documents, each document was pre-tagged for possible ADEs using MITRE’s jCarafe conditional random field mention-finding tool [20], trained on the NIST TAC ADR data set. A team of 17 pharmacovigilance SEs produced the annotations by correcting and reviewing this pre-tagging using a customized version of the MITRE Annotation Toolkit [21]. All documents were double-annotated during this phase, and the annotations were reconciled by a team of two pharmacovigilance adjudicators. Subsequently, a team of two MedDRA® experts, working in consultation, jointly annotated the mentions for MedDRA® LLTs and PTs. Once annotation was complete, MITRE and the FDA jointly conducted a detailed quality control review to ensure the consistency of the annotated corpus.
After an initial tranche of mention annotation, MITRE computed a pairwise inter-annotator agreement rate [22] of approximately 0.65 on mention annotation (where exact agreement of annotation category, annotation extent, and annotation reason was required), and the FDA revised and clarified the guidelines. At the end of the annotation process, MITRE once again computed pairwise inter-annotator agreement for the initial tranche of annotation and for the remainder of the annotation. For this second review, MITRE focused specifically on the inter-annotator agreement rate for the OSE_Labeled_AE category, since it was the category scored in the evaluation, and the other two categories were not intended to have been completely annotated. MITRE also used a more generous comparison requiring category match and extent overlap (not match) and did not require the reason attribute to match. MITRE discovered that, under this comparison metric, the pairwise inter-annotator agreement rate for OSE_Labeled_AE was 0.81 after the initial tranche of mention annotation and 0.87 for the remainder of the annotation, for a reduction in error of almost 30% after additional clarification of the guidelines. The overall pairwise inter-annotator agreement rate for OSE_Labeled_AE under this latter comparison metric was 0.86.
Comparison with the TAC ADR Corpus
The ADE Eval annotation schema included the NonOSE_AE and Not_AE_Candidate categories, and the reasons for assigning mentions to these various categories, to enable better diagnostics in the ADE Eval and to analyze and quantify differences in the FDA and TAC ADR corpora. The inventory of reasons, and their distribution in the ADE Eval corpus, are shown in Tables B-2 and B-3 in the ESM.
As noted in Sect. 3.1, 50 of the drug labels in the ADE Eval training corpus were chosen for overlap with the NIST TAC ADR evaluation. The ADE Eval annotation scheme made it possible to see the effect of differences in the two evaluations. For further details, see Table B-4 in the ESM.
Evaluation Metrics
The ADE Eval envisioned two types of use cases, described in Sects. 3.2.1 and 3.2.2, referred to as “front office” and “back office”. The front office use case is supported in the ADE Eval by two section- and label-level metrics intended to measure the submission’s ability to discover MedDRA® codes and their supporting evidence (namely, at least one corresponding mention within that section or label). The back office use case is supported by more traditional mention-level precision and recall metrics, weighted in a way that attempts to model the effort involved in correcting any mention annotation errors, with a view to creating completely annotated training data for machine-learning-based NLP systems.
The evaluation metrics assume that the gold and submission mentions are paired with each other and divided into exact match mention pairs (which match in span and MedDRA® PT code), inexact match mention pairs (which overlap in span but do not count as exact matches), missing gold mentions, and spurious submission mentions, which have no gold counterpart. The process that produces this pairing is described in Sect. B.1 in the ESM.
Front Office Use Case
In the front office use case, pharmacovigilance SEs need to know whether a MedDRA® code-associated ADE is present in a given section of the PI label and may want to see evidence of the presence of this ADE. The section-level analysis is crucial because the SE needs to know what level of severity the PI reflects for the given ADE, and the different severity levels are associated with specific sections.
For the front office use case, the scorer treated as legal matches both exact match mention pairs and inexact match mention pairs matching in MedDRA® code, where any degree of overlap was acceptable. The intuition behind this decision is that, in this use case, the SEs are looking for evidence, and as long as the mention draws the SE’s attention to the evidence, it is acceptable.
For this use case, we computed two primary metrics. The first metric is macro-averaged precision/recall/F1-measure (P/R/F1) on MedDRA® codes. The scope of the macro-averaging was the section; P/R/F1 were computed per section and the values averaged together. In this computation, a correct MedDRA® code was one that was realized by at least one legal match. (We refer to these codes as properly grounded.) All other MedDRA® codes were judged to be either missing (i.e., it was the MedDRA® code for at least one mention in the gold standard, but none of those mentions were paired with a similarly coded submission mention) or spurious (i.e., it was the MedDRA® code for at least one mention in the submission, but none of those mentions were paired with a similarly coded gold standard mention).
We also introduced a second, new metric that attempts to assess the quality of the evidence for the properly grounded MedDRA® codes. The metric is designed in such a way that the higher the score, the more likely it is that a mention for any properly grounded code is valid evidence for that code. This metric was a macro-averaged precision measure on submission mentions associated with each correct code. Each correct MedDRA® code was assigned a score based on the fraction of the linked submission mentions that were paired with an identically coded gold mention. Each mention score was scaled by the overlap of the mention with its gold pair. The reason for scaling the score by the overlap was to give more credit to more precise evidence. The score for the MedDRA® codes within a section were averaged to create the section-level score, and these scores were macro-averaged across the corpus.
Back Office Use Case
The back office use case tests the scenario where pharmacovigilance SEs use an automated system to identify ADEs, and some of the PI labels annotated by the system are hand corrected by human annotators to create a completely annotated reference. In this use case, every mention is important and some corrections are more time consuming than others.
The primary metric for this use case was weighted, micro-averaged, corpus-level P/R/F1 measure on mentions. A perfect score was awarded to each exact match mention pair, and all other elements were weighted in an attempt to model the time cost of correcting the errors. Given a count of M exact match mention pairs, C inexact match mention pairs (differing in span or MedDRA® code), N missing mentions, and S spurious mentions,
N′ = N (missing mentions are weighted 1, because adding a mention is hard).
S′ = 0.25 × S (spurious mentions are weighted 0.25, since deleting a mention is easy).
C′ = 0.5 × C (errors are weighted 0.5, since correcting a mention is hard but likely not as hard as adding one).
M′ = M + (0.5 × C) (matches accrue the correct share of the clash).
P/R/F1 measure are now computed normally:
P = M′/(M′ + C′ + S′).
R = M′/(M′ + C′ + N′).
F = (2 × P × R)/(P + R).
Results
Summary of Results
In total, 13 teams collectively returned 23 system submissions for ADE Eval. The submission scores are listed in Table 1.
Table 1.
ADE Eval submission scores
| Team | Run | Mentions—exact match, weighted (P/R/F1) | MedDRA® coding—macro-averaged by section (P/R/F1) | Quality |
|---|---|---|---|---|
| BetaResearch | Submission1 | 0.90/0.70/0.79 | 0.75/0.57/0.61 | 0.91 |
| BetaResearch | Submission2 | 0.87/0.78/0.82 | 0.64/0.62/0.60 | 0.90 |
| CONDL | CONDL_E19 | 0.97/0.68/0.80 | 0.89/0.55/0.63 | 0.96 |
| CONDL | CONDL_E46 | 0.97/0.69/0.80 | 0.87/0.56/0.63 | 0.95 |
| GMU-VCU-VASaltLake | Ensemble | 0.89/0.73/0.80 | 0.67/0.59/0.50 | 0.85 |
| GMU_VASALTLAKE | Submission_run1 | 0.89/0.71/0.79 | 0.70/0.57/0.48 | 0.84 |
| GMU_VASALTLAKE | Submission_run2_resubmission | 0.87/0.75/0.80 | 0.68/0.59/0.47 | 0.81 |
| JHU | JHU_System_Submission_1st_Run | 0.86/0.73/0.79 | 0.66/0.62/0.58 | 0.84 |
| Linguamatics | AEs | 0.85/0.83/0.84 | 0.66/0.82/0.70 | 0.79 |
| Linguamatics | Baseline | 0.80/0.82/0.81 | 0.57/0.83/0.64 | 0.76 |
| MC-UC3M | Run1 MC-UC3M fixed | 0.82/0.74/0.78 | 0.58/0.53/0.53 | 0.92 |
| MC-UC3M | Run2 MC-UC3M fixed | 0.83/0.72/0.77 | 0.58/0.50/0.51 | 0.93 |
| MayoNLPTest | Test_sub_r1 | 0.82/0.75/0.79 | 0.61/0.64/0.59 | 0.85 |
| MayoNLPTest | Test_sub_r2 | 0.81/0.68/0.74 | 0.57/0.55/0.52 | 0.86 |
| MelaxNLP | Run1_submission | 0.93/0.85/0.89 | 0.83/0.79/0.79 | 0.93 |
| MelaxNLP | Run2_submission | 0.92/0.86/0.89 | 0.80/0.81/0.79 | 0.92 |
| NLP@VCU | Test_submission | 0.94/0.19/0.31 | 0.88/0.17/0.22 | 0.96a |
| NaCTeM | Run1_HYPHEN | 0.93/0.79/0.86 | 0.79/0.68/0.70 | 0.96 |
| NaCTeM | Run2_Neural | 0.75/0.64/0.69 | 0.56/0.48/0.50 | 0.94 |
| UMLBioNLP | Submission1 | 0.92/0.83/0.87 | 0.83/0.74/0.75 | 0.93 |
| UMLBioNLP | Submission2 | 0.92/0.86/0.89 | 0.81/0.77/0.76 | 0.93 |
| UPennHLP | Run1_unsupervised | 0.76/0.79/0.77 | 0.50/0.66/0.53 | 0.79 |
| UPennHLP | Run2_supervised | 0.84/0.83/0.84 | 0.61/0.75/0.65 | 0.86 |
BetaResearch Uppsala Monitoring Centre, CONDL University of North Dakota, School of Medicine and Health Sciences, GMU George Mason University, JHU Johns Hopkins School of Medicine, MayoNLPTest Mayo Clinic, MC MeaningCloud, MedDRA® Medical Dictionary for Regulatory Activities, MelaxNLP Melax Technologies, Inc., NaCTeM National Centre for Text Mining, University of Manchester, NLP@VCU Natural Language Processing at Virginia Commonwealth University, P/R/F1 precision/recall/F1-measure, UC3M Universidad Carlos III de Madrid, UMLBioNLP University of Massachusetts at Lowell, UPennHLP University of Pennsylvania, VASaltLake Veterans Administration, Salt Lake City, VCU Virginia Commonwealth University
aNote that the Quality metric should be interpreted with care when the recall in the middle column is very low
Mention Finding
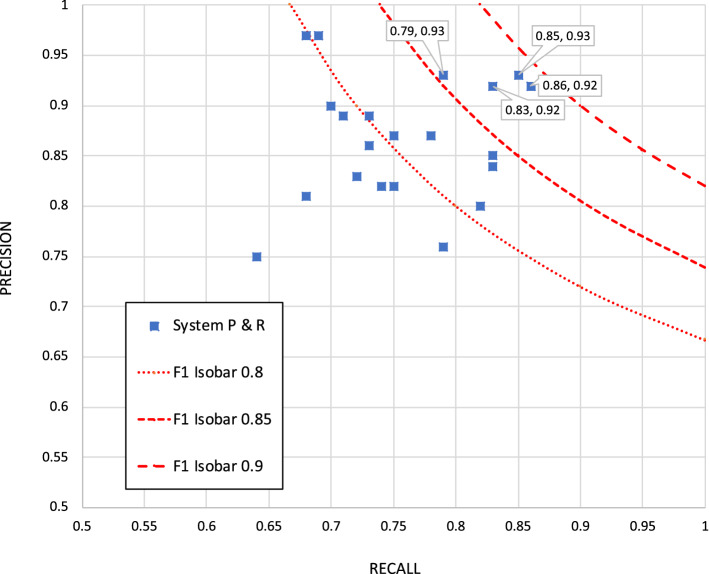
The primary mention-finding metric for the back office use case (see Table 1, column 3) is based on a match of both extent and MedDRA® code, weighted as described in Sect. 3.2.2.8 The highest F1 score here was 0.89, achieved by both submissions of the MelaxNLP system (using technology from University of Texas – Houston, a top performer in TAC ADR) and one of the UMLBioNLP submissions. About half of the submissions achieved an F1 of ≥ 0.8, including submissions from NaCTeM, Linguamatics, UPenn, BetaResearch, CONDL, and GMU. Figure 1 shows a precision versus recall graph.
Fig. 1.
Precision vs. recall for exact mention match (weighted)
The distribution of mention-finding methods among the responding sites reflects the NLP community’s current preference for statistical approaches: only two of 13 sites (JHU, Linguamatics) used a nonstatistical approach. The distribution of statistical approaches also reflected the current direction of NLP work. Using standard sequence tagging approaches such as conditional random fields (CRFs) [23, 24] alone is falling out of favor as the community moves toward neural-network-oriented approaches; only one site (NLP@VCU) used a CRF alone. In many cases, CRFs are used as a layer on top of a complex neural network architecture known as bidirectional long short-term memory (Bi-LSTM) [25, 26]. At least five of the 13 responding sites used Bi-LSTM + CRF for at least one of their runs, and a sixth (GMU-VCU-VASaltLake) used such an approach as a component of its ensemble. Four others that did not explicitly use Bi-LSTM + CRF used neural-based deep-learning approaches as some component of their mention-finding approach.
While all the Bi-LSTM + CRF submissions achieved an F1 of ≥ 0.8, using this technique is not a fundamental requirement for good results; for example, Linguamatics did relatively well with a nonstatistical approach. See Table C.1 in the ESM for further details. Further analysis of the mention-finding results, including score breakdowns by error type and significance testing, can be found in Sect. C.1 of the ESM.
MedDRA® Coding
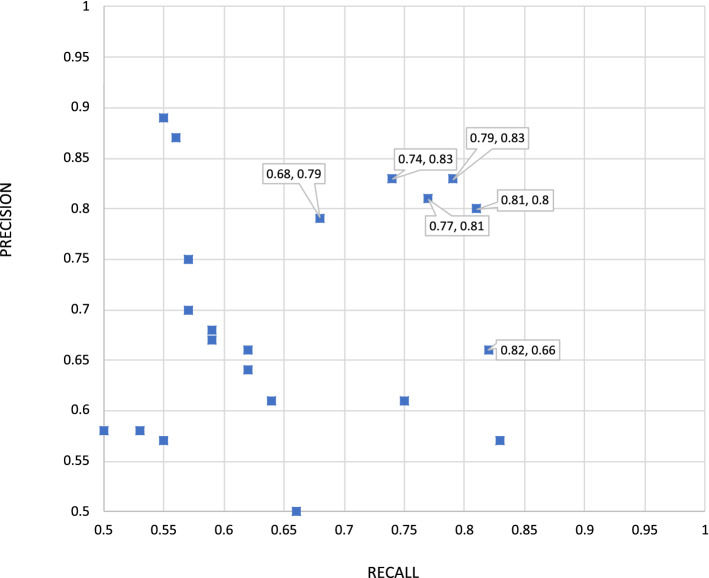
The first primary front office metric is MedDRA® code retrieval, macro-averaged by section. The top performer here, again, was MelaxNLP, with an F1 of 0.79 for both runs, followed by UMLBioNLP, with scores of 0.76 and 0.75. Overall, four teams had runs scoring ≥ 0.7 F1, including Linguamatics and NaCTeM with scores of 0.70. This metric is more demanding than the equivalent TAC ADR metric because the MedDRA® codes must be properly grounded in a correctly matching gold mention in order to count as a match, and the scope of the macro-averaging is the individual PI section, rather than the union of PI sections as in TAC ADR.
Figure 2 shows a precision versus recall graph to illustrate the relative strengths of each system in MedDRA® coding.
Fig. 2.
Precision vs. recall for Medical Dictionary for Regulatory Activities (MedDRA®) retrieval (macro-averaged by section)
The correlation between the mention-finding submission score order and the MedDRA® code retrieval submission score order, at least among the higher performing systems, was striking; once we created groupings of F1 scores based on statistical significance thresholds, the members of the top two equivalence classes (taken together) across the two metrics were identical (see Tables C-3 and C-4 in the ESM). This is reminiscent of TAC ADR; clearly, the quality of the mention finding is a dominant factor.
For the 11 sites that reported their MedDRA® coding strategy, the following strategies were reported:
mention finding and MedDRA® coding occurred simultaneously, using retrieval or pattern matching on known MedDRA® terms (four sites);
lookup tables or dictionaries (two sites);
information retrieval-based indexing (three sites);
rules (three sites);
neural approaches (four sites).
Multiple sites used a combination of these approaches, and multiple sites used different MedDRA® coding approaches in different submissions.
Quality
The front office quality metric is intended to determine the user’s experience when encountering system output. It judges the quality of the evidence for each code that has at least one properly grounded mention. These scores were very high, with half > 0.9. This score is informative only in conjunction with the MedDRA® retrieval score because the quality metric does not assign a penalty for a truly spurious code (i.e., a code that the system did not associate with any mention). This metric shows that, where the systems properly find a MedDRA® code, the evidence they provide for that code is of high quality.
The highest-ranking systems were NLP@VCU (achieving a score of 0.96), CONDL (0.96 and 0.95), and NaCTeM (0.96 and 0.94). The top two sites according to the previous metrics—MelaxNLP and UMLBioNLP—followed immediately, along with MC-UC3M, with scores of 0.93 and 0.92. In this case, the best correlation with other metrics, as one might expect, was with MedDRA® coding precision; whereas NLP@VCU scored poorly in F1 on MedDRA® coding, it scored second in precision and achieved the top score here. Eight of the top 11 quality submissions were the top eight submissions for MedDRA® coding precision.
Mention Reasons and Confusability of Spurious Annotations
As part of each submission, systems generated mentions that did not match any OSE_Labeled_AE in the gold standard, i.e., they were spurious. Each mention in the gold standard was marked with a reason that the category was selected (e.g., a mention might be marked as a NonOSE_AE because it describes an ADR in animals, which does not necessarily translate to humans). To diagnose these errors, we applied the ADE Eval pairing algorithm again to all submissions, this time targeting the (unscored) NonOSE_AE or Not_AE_Candidate mentions in the gold standard test corpus. Across the entire range of submissions, a total of 97,173 spurious mentions were generated, of which 44,585 (46%) were paired with some unscored gold standard mention using this alignment process (even though the unscored categories were not exhaustively annotated). In other words, almost half the spurious submission mentions were confusable, aligning with a NonOSE_AE or Not_AE_Candidate gold standard mention. Table C-5 in the ESM shows the confusability data for these two additional categories; see also Sect. C.3 of the ESM.
Discussion
The ADE Eval task was conducted to evaluate a range of NLP techniques for identifying ADEs mentioned in publicly available FDA drug labels. The purpose of the task was to determine whether the performance of algorithms currently used for NLP might be good enough for real-world use. The top performing systems used a combination of Bi-LSTMs and CRFs, but high performance was also achieved by systems using neither of these technologies, suggesting that there are many possible technological paths towards high performance.
The MedDRA® coding metric is the metric most relevant to the front office use case described in Sect. 3.2.1, and it is likely that the best MedDRA® coding performance reported in ADE Eval exceeds the performance found in Ly et al. [1] and TAC ADR. As discussed in Sect. 4.3, the ADE Eval MedDRA® coding metric is stricter than that of TAC ADR (and, also, of Ly et al. [3]); it requires linked evidence and for the MedDRA® code to be found in a specific section rather than in any of the relevant sections. The top performing MelaxNLP F1 score of 0.79 likely represents an advance over the top score reported in Ly et al. [3], which is no higher even though the ADE Eval task is more challenging. Similarly, while a direct comparison with TAC ADR is difficult to quantify, Sect. C.7 of the ESM attempts to elucidate this comparison, exploiting the fact that the UTH-CCB system, a predecessor of MelaxNLP, participated in the TAC ADR evaluation. The best available comparison suggests that UTH-CCB would have achieved an ADE-Eval-equivalent MedDRA® coding F1 score of 0.69 rather than the label-level score of 0.853 reported in TAC ADR.
As discussed in Sect. 2.1, Ly et al. [3] concluded that the NLP tools they investigated did not perform at levels that would allow them to be used without human intervention, and the results of the ADE Eval do not change this conclusion. (See Sect. D.1 in the ESM for a discussion of what might make it hard to find a particular ADE in PI text.) However, there are other ways to use these NLP tools, for example, as inputs to a human correction activity. Multiple studies [27–31] have considered this option in NLP, and—while the results are not universally positive—they are promising enough, and the activity is similar enough to the PI preparation activities required for the pharmacovigilance use cases, that we should begin to consider how and where to insert these tools into the pharmacovigilance workflow to maximize benefit for pharmacovigilance applications.
Conclusion
MITRE and the FDA conducted the ADE Eval, an evaluation of tools designed to identify ADEs mentioned in publicly available FDA PIs. The custom design of the ADE Eval enabled the FDA to assess the applicability of current NLP technologies to its specific use cases. The following were valuable outcomes of ADE Eval:
Some participants were previously unknown to MITRE and the FDA; one (UMLBioNLP) was among the top performers.
Confirmation that the additional complexities related to the pharmacovigilance annotation guidelines did not create an obstacle to good performance.
Computation of finer-grained mention-finding scores by reason, quantitative description of the effect of differences between the pharmacovigilance and TAC ADR annotation guidelines, and better error analysis.
Careful alignment of the ADE Eval results with concrete pharmacovigilance tasks.
The similarity of the ADE Eval and TAC ADR results suggests that a sufficiently similar evaluation might serve as a valuable proxy in situations where it is not possible, or desirable, to conduct a bespoke evaluation. Finally, while the results of the ADE Eval are not directly comparable with previous evaluations because of the difference in evaluation metrics, the available evidence suggests that NLP tools continue to improve, and it is likely that exploring the benefits of making NLP outputs available in human pharmacovigilance workflows would be worthwhile. One insertion point might be an NLP-enabled curation environment to build a central repository for ADE presence/absence in PIs. SEs currently apply their expert knowledge of PIs, and/or manually consult PIs, to determine ADE labeled status during ICSR evaluation. However, there is currently no such central facility for capturing SE judgments, although individual SEs may create their own tabulations of this information. (See Sect. D.2 and D.3 of the ESM for a discussion of real-world usefulness of NLP to human pharmacovigilance workflows.) A curation environment would allow SEs to record their expert judgments and to validate or correct NLP-based ADEs. Further exploration of these approaches to capture efficiency gains, both quantitative and qualitative, would be informative, in the form of human factors observation and/or human-in-the-loop experimentation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the FDA experts and pharmacovigilance SEs who served as annotators, consensus annotators, and MedDRA® coders, including Carol Pamer, Manish Kalaria, Kelly Cao, Eileen Wu, Cindy Kortepeter, Vicky Chan, Carmen Cheng, Samantha Cotter, Ida-Lina Diak, Charlene Flowers, Neha Gada, Kelly Harbourt, Timothy Jancel, Mihaela Jason, Corrinne Kulick, Monica Munoz, Mohamed Mohamoud, Shital Patel, Kate Phelan, Margee Webster, and Sherry Chang.
Declarations
The views in this manuscript are those of the authors and not necessarily those of the US FDA or The MITRE Corporation.
Funding
This work was funded by the US FDA.
Conflict of interest
Samuel Bayer, Cheryl Clark, Oanh Dang, John Aberdeen, Sonja Brajovic, Kimberley Swank, Lynette Hirschman, and Robert Ball have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval
Not applicable.
Consent to Participate and Consent for Publication
All participants in ADE Eval provided consent to participate, as well as consent for publication of the results.
Availability of data and material
The underlying data for this study are available as structured product labels from https://dailymed.nlm.nih.gov/dailymed/. Access to the annotated training data and guidelines used in this study for research purposes can be requested through the FDA Technology Transfer Program at techtransfer@fda.hhs.gov.
Code availability
Access to the evaluation software used in this study for research purposes can be requested through the FDA Technology Transfer Program at techtransfer@fda.hhs.gov.
Author contributions
All authors contributed to the design and planning of the annotation and the evaluation. OD, SBr and KS contributed to the annotation effort and the MedDRA® coding effort. All authors contributed to the recruitment of participants in the evaluation. SBa, CC, OD, JA, SBr and LH conducted the evaluation. All authors contributed to the manuscript.
Footnotes
In this document, label refers to the structured product label that accompanies a medication. To avoid confusion with this usage of label, descriptions added to text to indicate the semantic category of a text span are always referred to as annotations or categories.
DailyMed is the official provider of FDA label information (PIs). The National Library of Medicine provides this as a public service. https://dailymed.nlm.nih.gov/dailymed/.
MedDRA® terminology is the international medical terminology developed under the auspices of the International Council on Harmonization (ICH) of Technical Requirements for Registration of Pharmaceuticals for Human Use. The MedDRA® trademark is registered by the International Federation of Pharmaceutical Manufacturers and Associations (IFPMA) on behalf of the ICH. MedDRA® is a clinically validated international medical terminology used by regulatory authorities and the regulated biopharmaceutical industry. The terminology is used through the entire regulatory process, from premarketing to postmarketing, and for data entry, retrieval, evaluation, and presentation. MedDRA® formalizes ADEs (medical concepts) as sets of phrases called low-level terms (LLTs); each LLT is associated with a single preferred term (PT). A PT represents a unique AE/medical concept and hierarchically groups the synonymous LLTs for the concept. Every MedDRA® term is associated with a code.
Note that the mapping was performed for already extracted ADR strings.
For the pharmacovigilance use case, an ADE is any undesirable experience associated with the use of a medical product in a patient. Outcome refers to the status of a patient’s health condition, how the patient feels, functions, or survives. An outcome may be attributable to an ADE. Serious outcomes include death, life-threatening experience, hospitalization or prolongation of hospitalization, disability, and birth defects [32].
A production system would require an ability to process the XML in the PIs as well as navigate between the visual presentation and the underlying XML.
For historical reasons, while we refer to the phenomena under discussion as ADEs throughout this paper, the names of the categories and attributes in the ADE Eval use the abbreviation “AE” instead of “ADE.”.
This mention-finding metric is stricter than the one in TAC ADR in that it considers the MedDRA® code when scoring the mention finding but is more generous in granting partial credit for overlapping mentions and discounting the penalty for spurious mentions. The mention-finding task differs from TAC ADR Task 1 in other ways, as described in Sect. A.1 in the ESM, but the MedDRA® coding dependency is the prominent factor here.
Contributor Information
Samuel Bayer, Email: sam@mitre.org.
Cheryl Clark, Email: cclark@mitre.org.
Oanh Dang, Email: oanh.dang@fda.hhs.gov.
John Aberdeen, Email: aberdeen@mitre.org.
Sonja Brajovic, Email: sonja.brajovic@fda.hhs.gov.
Kimberley Swank, Email: kimberley.swank@fda.hhs.gov.
Lynette Hirschman, Email: lynette@mitre.org.
Robert Ball, Email: robert.ball@fda.hhs.gov.
References
- 1.FDA Adverse Event Reporting System (FAERS) Public Dashboard. U.S. Food and Drug Administration. 2020. https://www.fda.gov/drugs/questions-and-answers-fdas-adverse-event-reporting-system-faers/fda-adverse-event-reporting-system-faers-public-dashboard.
- 2.Novel Drug Approvals for 2019. U.S. Food and Drug Administration. 2020. https://www.fda.gov/drugs/new-drugs-fda-cders-new-molecular-entities-and-new-therapeutic-biological-products/novel-drug-approvals-2019.
- 3.Ly T, Pamer C, Dang O, Brajovic S, Haider S, Botsis T, et al. Evaluation of natural language processing (NLP) systems to annotate drug product labeling with MedDRA terminology. J Biomed Inf. 2018;83:73–86. doi: 10.1016/j.jbi.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 4.Botsis T, Jankosky C, Arya D, Kreimeyer K, Foster M, Pandey A, et al. Decision support environment for medical product safety surveillance. J Biomed Inform. 2016;64:354–362. doi: 10.1016/j.jbi.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 5.Pandey A, Kreimeyer K, Foster M, Dang O, Ly T, Wang W, et al. Adverse event extraction from structured product labels using the event-based text-mining of health electronic records (ETHER) system. Health Inf J. 2019;25:1232–1243. doi: 10.1177/1460458217749883. [DOI] [PubMed] [Google Scholar]
- 6.I2E. Full-power, flexible Natural Language Processing. Linguamatics. 2019. https://www.linguamatics.com/products/i2e.
- 7.Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17:229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.TAC 2017. Adverse Drug Reaction Extraction from Drug Labels [Internet]. U.S National Library of Medicine. 2020. https://bionlp.nlm.nih.gov/tac2017adversereactions/.
- 9.Roberts K, Demner-Fushman D, TAC JT. Overview of the TAC 2017 Adverse Reaction Extraction from Drug Labels Track. 2017. pdfs.semanticscholar.org.
- 10.Jagannatha A, Liu F, Liu W, et al. Overview of the first natural language processing challenge for extracting medication, indication, and adverse drug events from electronic health record notes (MADE 1.0) Drug Saf. 2019;42:99–111. doi: 10.1007/s40264-018-0762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry S, Buchan K, Filannino M, Stubbs A, Uzuner Ö. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J Am Med Inform Assoc. 2019;75:S4–10. doi: 10.1093/jamia/ocz166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarker A, Belousov M, Friedrichs J, Hakala K, Kiritchenko S, Mehryary F, et al. Data and systems for medication-related text classification and concept normalization from Twitter: insights from the Social Media Mining for Health (SMM4H)-2017 shared task. J Am Med Inform Assoc. 2018;25:1274–1283. doi: 10.1093/jamia/ocy114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarker A, Gonzalez-Hernandez G. Overview of the second social media mining for health (SMM4H) shared tasks at AMIA 2017. In: Proceedings of the 2nd social media mining for health research and applications workshop co-located with the American Medical Informatics Association annual symposium (AMIA 2017). 2017. p. 43–8. [DOI] [PMC free article] [PubMed]
- 14.Weissenbacher D, Sarker A, Paul MJ, Gonzalez-Hernandez G. Overview of the third social media mining for health (SMM4H) shared tasks at EMNLP 2018. In: Proceedings of the 2018 EMNLP workshop SMM4H: the 3rd social media mining for health applications workshop and shared task. 2018. p. 13–6.
- 15.Weissenbacher D, Sarker A, Magge A, Daughton A, O’Connor K, Paul M, Gonzalez G. Overview of the fourth social media mining for health (SMM4H) shared tasks at ACL 2019. In: Proceedings of the fourth social media mining for health applications (# SMM4H) workshop & shared task. 2019. p. 21–30.
- 16.Caster O, Juhlin K, Watson S, Norén GN. Improved statistical signal detection in pharmacovigilance by combining multiple strength-of-evidence aspects in vigiRank. Drug Saf. 2014;37:617–628. doi: 10.1007/s40264-014-0204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caster O, Dietrich J, Kürzinger M-L, Lerch M, Maskell S, Norén GN, et al. Assessment of the utility of social media for broad-ranging statistical signal detection in pharmacovigilance: results from the WEB-RADR project. Drug Saf. 2018;41:1355–1369. doi: 10.1007/s40264-018-0699-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierce CE, Bouri K, Pamer C, Proestel S, Rodriguez HW, Van Le H, et al. Evaluation of Facebook and Twitter monitoring to detect safety signals for medical products: an analysis of recent FDA safety alerts. Drug Saf. 2017;40:317–331. doi: 10.1007/s40264-016-0491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DailyMed. U.S. National Library of Medicine. 2020. https://dailymed.nlm.nih.gov/dailymed/.
- 20.Wellner B, Vilain M. Leveraging machine readable dictionaries in discriminative sequence models. In: Language Resources and Evaluation Conference. Genoa: LREC; 2006.
- 21.The MITRE Annotation Toolkit [Internet]. http://mat-annotation.sourceforge.net/. Accessed 11 Apr 2018.
- 22.Fleiss JL. Measuring nominal scale agreement among many raters. Psychol Bull. 1971;76:378–382. doi: 10.1037/h0031619. [DOI] [Google Scholar]
- 23.McCallum A, Li W. Early results for named entity recognition with conditional random fields, feature induction and web-enhanced lexicons. In: Conference of the North American chapter of the Association for Computational Linguistics & Human Language Technologies (NAACL-HLT). 2003. p. 188–91.
- 24.Lafferty J, McCallum A, Pereira F. Conditional random fields: probabilistic models for segmenting and labeling sequence data. In: Proceedings of the international conference on machine learning (ICML). 2001. p. 282–9.
- 25.Graves A, Schmidhuber J. Framewise phoneme classification with bidirectional LSTM and other neural network architectures. Neural Netw. 2005;18:602–610. doi: 10.1016/j.neunet.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 26.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural architectures for named entity recognition. In: Proceedings of the 2016 conference of the North American chapter of the Association for Computational Linguistics: Human Language Technologies. 2016. p. 260–70.
- 27.Kuo TT, Huh J, Kim J et al. The impact of automatic pre-annotation in clinical note data element extraction-the CLEAN Tool. arXiv preprint. 2018. arXiv:1808.03806.
- 28.Greinacher R, Horn F. The DALPHI annotation framework & how its pre-annotations can improve annotator efficiency. arXiv preprint. 2018. arXiv:1808.05558.
- 29.Lingren T, Deleger L, Molnar K, Zhai H, Meinzen-Derr J, Kaiser M, et al. Evaluating the impact of pre-annotation on annotation speed and potential bias—natural language processing gold standard development for clinical named entity recognition in clinical trial announcements. JAMIA. 2014;21:406–413. doi: 10.1136/amiajnl-2013-001837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.South BR, Mowery D, Suo Y, Leng J, Ferrández O, Meystre SM, et al. Evaluating the effects of machine pre-annotation and an interactive annotation interface on manual de-identification of clinical text. J Biomed Inform. 2014;50:162–172. doi: 10.1016/j.jbi.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singhal A, Leaman R, Catlett N, Lemberger T, McEntyre J, Polson S, et al. Pressing needs of biomedical text mining in biocuration and beyond: opportunities and challenges. Database (Oxford). 2016;2016:baw161. [DOI] [PMC free article] [PubMed]
- 32.US Food and Drug Administration. What is a serious adverse event? Silver Spring: US FDA; 2016. https://www.fda.gov/safety/reporting-serious-problems-fda/what-serious-adverse-event.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.