Introduction
Desmin is an important intermediate filament protein involved in extrasarcomeric cytoskeleton and cellular function. By interacting with its binding partners, desmin as well as other cytoskeletal proteins form a complex intracellular network that connects cell membrane, nuclear envelope, and organelles.1 The 3-dimensional structure provides an important basis for cellular mechanics, organelle transportation, and intracellular signaling.2 The DES gene, which is located in chromosome 2q35 and mainly expresses in muscle tissues, plays vital roles in myocyte development, degeneration, and cellular function.3
Various DES mutations were found to be related with myopathies. DES mutation carriers can present neurological signs, cardiologic signs, myopathy, or a combination of these manifestations.3,4 According to a meta-analysis enrolling 159 DES mutation carriers with 40 different mutations,5 74% of carriers presented neurological signs and cardiologic signs; 70% of carriers had myopathy and one-third of them had normal serum creatine kinase level; 50% of carriers had cardiomyopathy; and 60% had cardiac conduction disease or arrhythmia, among which atrioventricular block (AVB) was quite common. Isolated left bundle branch block progressing to complete heart block and asystole has been found in pediatric patients with DES mutation,6 but alternating bundle branch block (ABBB) remained scarcely reported.
Here we report a 25-year-old male patient diagnosed with mild left ventricular hypertrophy and ABBB that finally progressed to AVB, caused by a de novo DES gene mutation.
Key Teaching Points.
-
•
We uncover an unpublished desmin variation in patients with hypertrophic cardiomyopathy and rare alternating bundle branch block.
-
•
Whole exome sequencing was an effective tool in clinical work to uncover potential disease.
-
•
Transcriptome sequencing should be valued because it helps to identify molecular mechanisms, especially in genetic diseases.
Case report
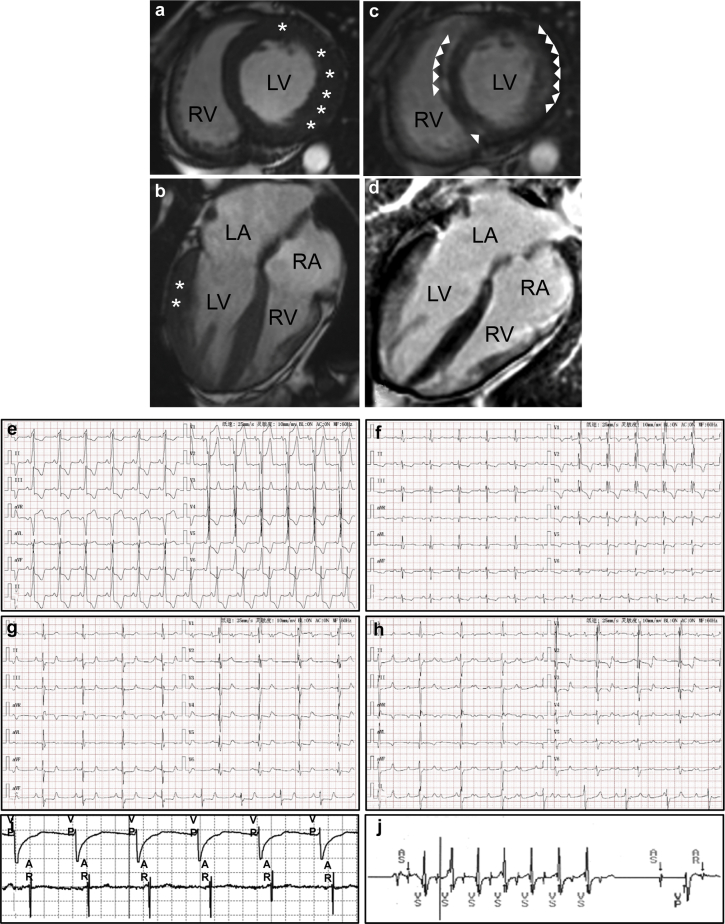
A 25-year-old male patient was admitted because of an episode of syncope. His serum creatine kinase and creatine kinase-MB were normal, but high-sensitive troponin I was elevated (Supplemental Figure 1). Transthoracic echocardiography indicated normal left ventricular systolic function with ejection fraction at 65%. Cardiac magnetic resonance imaging showed hypertrophic anterior and lateral walls of the left ventricle, with maximum thickness reaching 13 mm (Figure 1a and b). Late gadolinium enhancement showed abnormal signal and partial delayed myocardial enhancement in most parts of the interventricular septum and left ventricular walls (Figure 1c and d). ABBB was noted on electrocardiogram. On admission, complete left bundle branch block was documented with sinus rhythm (heart rate of 84 beats per minute) and a PR interval of 0.145 seconds (Figure 1e). The subsequent electrocardiogram in the next 4 weeks revealed combination of complete right bundle branch block and left anterior fascicle block with first-degree (Figure 1f) or second-degree AVB (Figure 1g). Incomplete trifascicular block lasted for 3 days and progressed to a complete AVB with bradycardia (Figure 1h). The patient underwent temporary transvenous pacemaker placement owing to hemodynamic instability. After 3 weeks observation, complete AVB persisted and a dual-chamber permanent pacemaker (Model Sensia SED01; Medtronic, Minneapolis, MN) was implanted.
Figure 1.
a–d: Cardiac magnetic resonance imaging and electrocardiogram (ECG) of the patient. a: T2-weighted images of short-axis view; b: T2-weighted images of 4-chamber view show mild (11–14 mm) in basal anterior and lateral left ventricular segments; c: delayed contrast enhancement of short-axis view; d: delayed contrast enhancement of 4-chamber view. e: Left bundle branch block (LBBB): Precordial leads V1 through V3 show LBBB pattern (wide QRS complexes of 136 ms and deep S waves with minor r waves). f: First-degree atrioventricular block (AVB), right bundle branch block (RBBB), and left anterior fascicle block (LAFB): A prolonged PR interval (230 ms) is consistent with first-degree AVB; M-shaped QRS complexes in precordial leads V1 through V4 and broad S waves in leads I, aVL, V5, and V6 are consistent with RBBB; small q waves with tall R waves in leads I and aVL and small r waves with deep S waves in in leads II, III, and aVF are consistent with LAFB. g: RBBB, LAFB, and second-degree AVB with 2:1 conduction ratios. h: Complete heart block: the P waves occur every 480 ms and the R-R interval is every 1200 ms, exhibiting an atrioventricular dissociation in impulse rates. i: Atrial (top) and ventricular (bottom) ECGs captured during the pacemaker programming process. Each ventricular paced event (VP) is followed by an atrial event (AR) indicating 1:1 retrograde ventriculoatrial conduction. j: Stored ventricular ECG from the implanted pacemaker with automated ECG capture triggered by a high ventricular rate. AS = atrial sensed event; LA = left atrium; LV = left ventricle; RA = right atrium; RV = right ventricle; VS = ventricular sensed event; star marks myocardial hypertrophy; arrow indicates delayed contrast enhancement.
During 6 months follow-up, the patient was in complete remission. The pacemaker-generated rate histograms showed a high percentage of ventricular pacing (56.8%) with a with programmed AV delay at 280 milliseconds (Supplemental Figure 2a and b). We also observed retrograde ventriculoatrial conduction in the pacemaker programming process (Figure 1i). However, there was no pacemaker-mediated tachyarrhythmia displayed. The 43.2% normal atrioventricular conduction (Supplemental Figure 2c) indicated intermittent AVB, not permanent AVB as we previously considered. Moreover, the rate histograms showed nonsustained ventricular tachycardia of 180 beats per minute, which persisted for 7 beats (Figure 1j).
To elucidate potential causes of the disease, whole exome sequencing of ectopectoralis was performed (Supplemental methods and materials). NM_001927.3:c.735+1G>A, a splicing donor site variant in intron 3 of DES gene, which was likely pathogenic (PVS1,PM2) according to the American College of Medical Genetics and Genomics Standards and Guidelines, was shown to be the most likely candidate variant (Figure 2a and b). Sanger sequencing of the family trio showed the variant was only present in the proband but not in his parents. Further paternity testing by multiplex short tandem repeat typing (Promega, San Luis Obispo, CA) confirmed biological relationship between the proband and his parents (Supplemental Table 1), confirming the de novo nature of the variant.
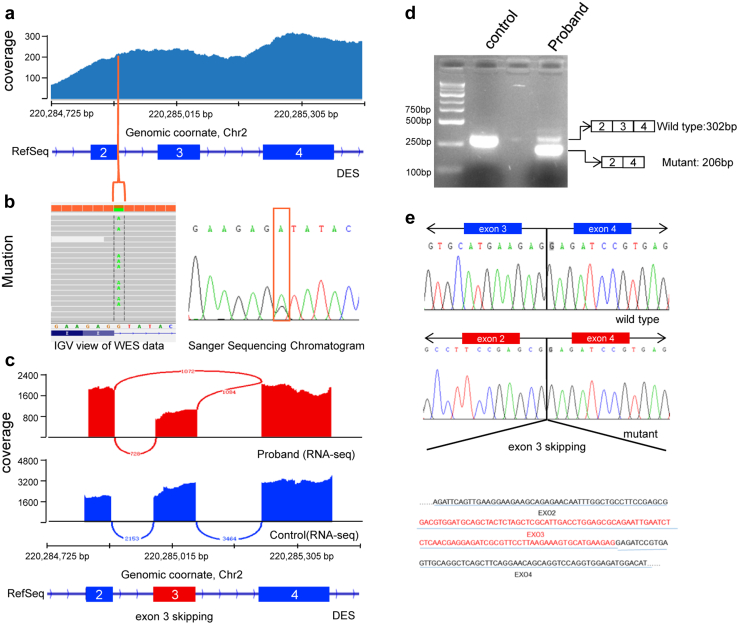
Figure 2.
Identification of the splice site mutation and expression analysis of exon 3 skipping in the DES gene. a: Alignment results of whole exome sequencing reads of the coding regions of the DES gene (exon 2, 3, and 4) are shown as coverage tracks (blue) in Integrative Genomics Viewer (IGV). At the bottom, exon 2, 3, and 4 of the DES gene are shown as RefSeq annotation tracks (deep blue). The line (orange) marks the position of the splicing site mutation, DES: c.735+1G>A. Zoom-in IGV view of the WES coverage tracks shows the mutation is near the 3′ donor splice site at position +1 in the intron between exons 2 and 3 (left). Sanger sequencing chromatogram of c.735+1G>A of the DES gene (right). b,c: IGV-generated Sashimi plot of the splicing events of the DES gene in the proband and the control (red and blue, respectively). d: Gel electrophoresis of reverse transcription polymerase chain reaction product from the patient’s and control’s muscle tissues. e: Sanger sequencing of the products retrieved from the gel.
To verify the pathogenicity of the splicing variant, ectopectoralis specimens from the patient and a 40-year-old male volunteer without myopathy were collected while they were undergoing pacemaker implantation. We found thousands of splicing events from transcriptome sequencing and when we focused on DES, abnormal junction from exon 2 to exon 3 was found in the patient (Figure 2c). So we concluded that NM_001927.3:c.735+1G>A led to abnormal DES mRNA loss of exon 3. To further confirm the results, we examined the expression of DES mRNA in the patient’s and the control’s ectopectoralis specimen by reverse transcription polymerase chain reaction. Gel electrophoresis showed 2 different bands (Figure 2d). Sanger sequencing of the products elucidated complete exon 3 skipping in the lower band while the upper band was shown to be normal DES mRNA (Figure 2e). Transcriptome sequencing showed about 363 upregulated genes and 213 downregulated genes in the patient’s ectopectoralis specimen; more differentially expressed genes were found when we set P < .05 (Supplemental Figure 3a and b).
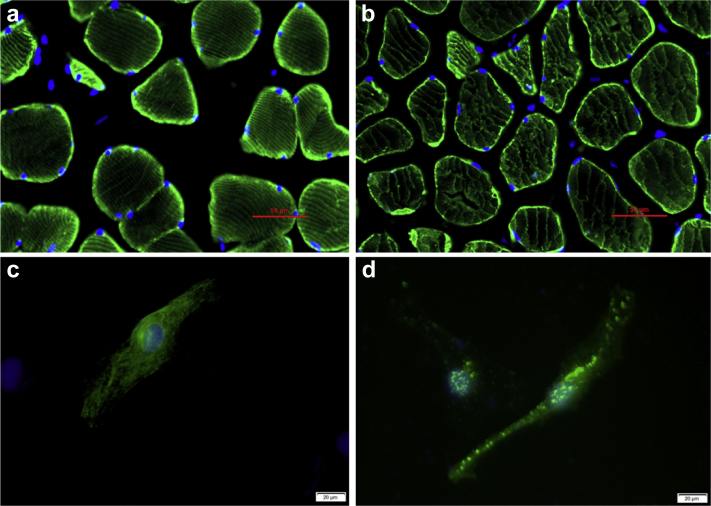
To explore the impact of mutant desmin protein, immunofluorescence of the patient’s and the control’s ectopectoralis specimen was carried out (Supplemental methods and materials). Compared with the control, the patient’s ectopectoralis specimen showed sparse and disorganized desmin network in each myocyte (Figure 3a and b). Wild-type desmin and mutant desmin expression plasmid were constructed using pcDNA3.1- and then transfected into HeLa cells, which expressed very little endogenous desmin. HeLa cells transfected with pcDNA3.1-WTDES formed an extensive and well-organized cytoplasmic network (Figure 3c). In contrast, mutant desmin in cells transfected with pcDNA3.1-mutDES produced sporadic desmin-positive granules and aggregates (Figure 3d).
Figure 3.
Desmin filament assembly of recombinant wild-type and mutant desmin. a: Immunofluorescence-detected desmin in the control’s skeletal muscle tissue; scale bar corresponds to 50 μm. b: Immunofluorescence-detected desmin in the patient’s skeletal muscle tissue; scale bar corresponds to 50 μm. c: HeLa cells transfected with pcDNA3.1-WTDES formed an extensive organized cytoplasmic network; scale bar corresponds to 20 μm. d: Cells transfected with pcDNA3.1-mutDES plasmid produced scattered desmin-positive aggregates; scale bar corresponds to 20 μm.
Discussion
We identified a de novo mutation, NM_001927.3:c.735+1G>A, which had been discovered but not published and whose pathogenicity remained unknown.7 The patient presented with ABBB, which then progressed to complete AVB, mild cardiac hypertrophy, and left ventricular diastolic dysfunction, while no skeletal myopathy or neurological symptom was observed. According to several case reports published, ABBB may appear in patients with coronary artery disease, adrenergic stimulation, and even lymphoma undergoing chemotherapy; on the other hand, ABBB may present with complicated cardiac arrhythmias such as atrial fibrillation or finally progress into AVB.8,9 In our case, the clinical phenotype was related with desmin mutation. Desmin is enriched in Purkinje cells, and desmin knockout mice showed slower conduction velocities, which may account for the high morbidity of cardiac conduction defect in mutation carriers.10,11 Boulé and colleagues12 demonstrated that in a desminopathy case with mutation (p.Ser13Phe) in the head domain of desmin, the Purkinje system was involved in the pathogenesis of ABBB and AVB. The desminopathy in our case resulted in a truncated protein, Del(Asp214-Glu245). The missing peptide was located in the 1B segment of the alpha helix in desmin protein; we infer that the mutation might also result in abnormal Purkinje system and clinically lead to cardiac conduction defect. The pacemaker programming report recorded intermittent AVB, providing evidence for residual AV node function. So we infer that the mutation leads to abnormal Purkinje system but not the superior AV node.
Since the majority of DES mutations were missense mutations, splice site mutation is relatively rare. In our case, the mutation occurs at the highly conserved GT-AG splicing site. Aberrant splicing leads to deletion of exon 3 in desmin mRNA, which finally translates into mutant desmin lacking 32 amino acids. Several DES mutations in splice site causing exon 3 skipping have been reported.13,14 The mutant desmin protein aggregates in cytoplasm instead of forming a functional cytoskeleton network, which is the typical pathologic characteristic of DES mutation and is thought to be toxic for cellular function.2 Since desmin works as a connection between membranous organelles and nucleus or cell membrane, mitochondria dysfunction and impaired cell communication may also contribute to desminopathy, which has been proved by in vivo and vitro studies.15
In this case, cardiac magnetic resonance imaging showed fibrosis in the interventricular septum and the patient’s serum high-sensitive troponin I remained elevated during hospitalization, suggesting constant cardiac damage and repair. Fibrosis in the interventricular septum might impair cardiac conduction systems, thus causing ABBB. Nonsustained ventricular tachycardia occurred even with the implantation of a pacemaker, although it lasted for only about 7 beats. More frequent follow-up was needed to avoid adverse outcome.
Conclusion
In summary, we report a DES mutation related with mild cardiac hypertrophy and ABBB. Integrated whole exome and transcriptome sequencing can be highly effective and sensitive to uncover gene mutation with its outcomes, thus they should be considered in clinical work for potential patients.
Acknowledgments
We thank the patients for their participation in this study.
Ethics statement: The patient’s family and control have given written informed consent to participating in this study. The Ethical Committee of the Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China, reviewed and approved our study protocol in compliance with the Helsinki declaration.
Footnotes
Funding: This work was supported by grants from the National Natural Science Foundation of China (No. 81570348). Conflict of interest statement: None.
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.hrcr.2020.10.003.
Appendix. Supplementary data
References
- 1.Capetanaki Y., Bloch R.J., Kouloumenta A., Mavroidis M., Psarras S. Muscle intermediate filaments and their links to membranes and membranous organelles. Exp Cell Res. 2007;313:2063–2076. doi: 10.1016/j.yexcr.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 2.Capetanaki Y., Papathanasiou S., Diokmetzidou A., Vatsellas G., Tsikitis M. Desmin related disease: a matter of cell survival failure. Curr Opin Cell Biol. 2015;32:113–120. doi: 10.1016/j.ceb.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemen C.S., Herrmann H., Strelkov S.V., Schroder R. Desminopathies: pathology and mechanisms. Acta Neuropathol. 2013;125:47–75. doi: 10.1007/s00401-012-1057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schroder R., Schoser B. Myofibrillar myopathies: a clinical and myopathological guide. Brain Pathol. 2009;19:483–492. doi: 10.1111/j.1750-3639.2009.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Spaendonck-Zwarts K.Y., van Hessem L., Jongbloed J.D. Desmin-related myopathy. Clin Genet. 2011;80:354–366. doi: 10.1111/j.1399-0004.2010.01512.x. [DOI] [PubMed] [Google Scholar]
- 6.Gearhart A.S., Batra A.S. Isolated left bundle branch block progressing to complete heart block and asystole: A novel presentation of a desmin mutation. HeartRhythm Case Rep. 2018;4:184–186. doi: 10.1016/j.hrcr.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldfarb L.G., Vicart P., Goebel H.H., Dalakas M.C. Desmin myopathy. Brain. 2004;127:723–734. doi: 10.1093/brain/awh033. [DOI] [PubMed] [Google Scholar]
- 8.Mitrega K., Lenarczyk R., Pruszkowska P., Kalarus Z., Sredniawa B. Alternating left and right bundle branch block. Kardiologia Polska. 2014;72:987. doi: 10.5603/KP.2014.0198. [DOI] [PubMed] [Google Scholar]
- 9.Saini A., Padala S.K., Koneru J.N., Ellenbogen K.A. Alternating bundle-branch block: what is the mechanism? Circulation. 2018;137:1192–1194. doi: 10.1161/CIRCULATIONAHA.118.033637. [DOI] [PubMed] [Google Scholar]
- 10.Price M.G. Molecular analysis of intermediate filament cytoskeleton--a putative load-bearing structure. Am J Physiol. 1984;246:H566–H572. doi: 10.1152/ajpheart.1984.246.4.H566. [DOI] [PubMed] [Google Scholar]
- 11.Schrickel J.W., Stockigt F., Krzyzak W. Cardiac conduction disturbances and differential effects on atrial and ventricular electrophysiological properties in desmin deficient mice. J Interv Card Electrophysiol. 2010;28:71–80. doi: 10.1007/s10840-010-9482-8. [DOI] [PubMed] [Google Scholar]
- 12.Boulé S., Richard P., de Groote P., Renaud F., Charron P. Recurrent suspected myocarditis combined with infrahisian conduction disturbances revealing a desminopathy. HeartRhythm Case Rep. 2015;1:305–309. doi: 10.1016/j.hrcr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalakas M.C., Park K.Y., Semino-Mora C., Lee H.S., Sivakumar K., Goldfarb L.G. Desmin myopathy, a skeletal myopathy with cardiomyopathy caused by mutations in the desmin gene. N Engl J Med. 2000;342:770–780. doi: 10.1056/NEJM200003163421104. [DOI] [PubMed] [Google Scholar]
- 14.Clemen C.S., Fischer D., Reimann J. How much mutant protein is needed to cause a protein aggregate myopathy in vivo? Lessons from an exceptional desminopathy. Hum Mutat. 2009;30:E490–E499. doi: 10.1002/humu.20941. [DOI] [PubMed] [Google Scholar]
- 15.Alam S., Abdullah C.S., Aishwarya R. Aberrant mitochondrial fission is maladaptive in desmin mutation-induced cardiac proteotoxicity. J Am Heart Assoc. 2018;7 doi: 10.1161/JAHA.118.009289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.