Abstract
A proper number of oligodendrocytes in the nerve system is essential for neuronal functions. In the olfactory bulb (OB), enriched oligodendrocytes are crucial for olfactory information processing. However, how the precise number of oligodendrocytes in the OB is regulated remains elusive. Here we identified that the transcription factor 4 (Tcf4)-mediated cell death is essential for generating an appropriate number of oligodendrocyte progenitor cells (OPCs) and thereby oligodendrocytes in the OB. We showed that Nkx2.1-positive progenitors in the medial ganglionic eminence (MGE) and anterior entopeduncular area (AEP) provide the first source of OPCs in the OB. Conditional depletion of Tcf4 leads to an increase of OPCs in the OB, which is mediated by the suppression of programmed cell death. Furthermore, we showed that Tcf4 mediated OPC survival is cell-autonomous by transplantation assay. Mechanistically, we identified Bax/Bak as a potential key pathway to promote OPC elimination during OB development. Depletion of Bax/Bak in Nkx2.1 lineage results in an increase of OPCs in the OB. Mutations in TCF4 causes Pitt-Hopkins syndrome, a severe neurodevelopmental disorder. Thus, our findings reveal an important intrinsic mechanism underlying the survival control of OPCs in the OB and provide new insights into the pathogenesis of Pitt–Hopkins syndrome.
Subject terms: Developmental neurogenesis, Glial development
Introduction
A precise number of oligodendrocytes is essential for neuronal functions through the myelination of axons in the central nervous system (CNS)1–3. Oligodendrocytes develop from oligodendrocyte precursor cells (OPCs), which originate from the radial glial cells (RGCs) of the ventricular zone in several regions of the embryonic brain4. In contrast to mature oligodendrocytes, OPCs maintain their proliferative capacity and continue to differentiate into oligodendrocytes in the adult brain5. However, OPCs derived from distinct origins are functionally redundant and populate the cortex in sequential waves6,7, suggesting that the number of OPCs produced from distinct origins is tightly controlled. Programmed cell death is one of the key mechanisms in controlling the precise number of cells, including neurons and glial cells by eliminating the excess during brain development8–11.
RGCs expressing the homeodomain transcription factor Nkx2.1 in the ventricular zone of the MGE and AEP generates most of the interneurons in the cortex12. After differentiation, these interneurons migrate tangentially to the dorsal cortex to form connections with local projection neurons13–15. In addition to the production of interneurons, Nkx2.1-positive RGCs also contribute to the generation of the first wave of OPCs which further differentiate into mature oligodendrocytes in the neocortex6,16. While extensive studies have focused on how OPCs are generated from Nkx2.1-positive RGCs in the neocortex, the contribution of these RGCs to the production of OPCs in the OB, the essential structure involved in olfaction, is largely unknown.
Tcf4 (also known as E2-2 and ITF2), a basic helix–loop–helix (bHLH) transcription factor, is highly expressed in both developing and adult brains17–19. TCF4 mutations cause Pitt–Hopkins syndrome, a neurodevelopmental disorder characterized by developmental delay and intellectual disability and is highly associated with schizophrenia20,21. Tcf4 plays essential roles in hippocampal development and cortical lamination in the developing brain22–24. Perturbations of Tcf4 in mouse models disrupt synaptic function which leads to deficits in learning and memory25–27. The expression of Tcf4 has been detected in various cell types, including neurons, astrocytes, and oligodendrocytes18. However, the role of Tcf4 in regulating oligodendrocyte development remains largely unknown. Here, we conditionally inactivated Tcf4 in the Nkx2.1 lineage and showed that Tcf4 plays an essential role in regulating OPC survival during OB development.
Results
Nkx2.1-positive RGCs generate OPCs in early OB developmental stages
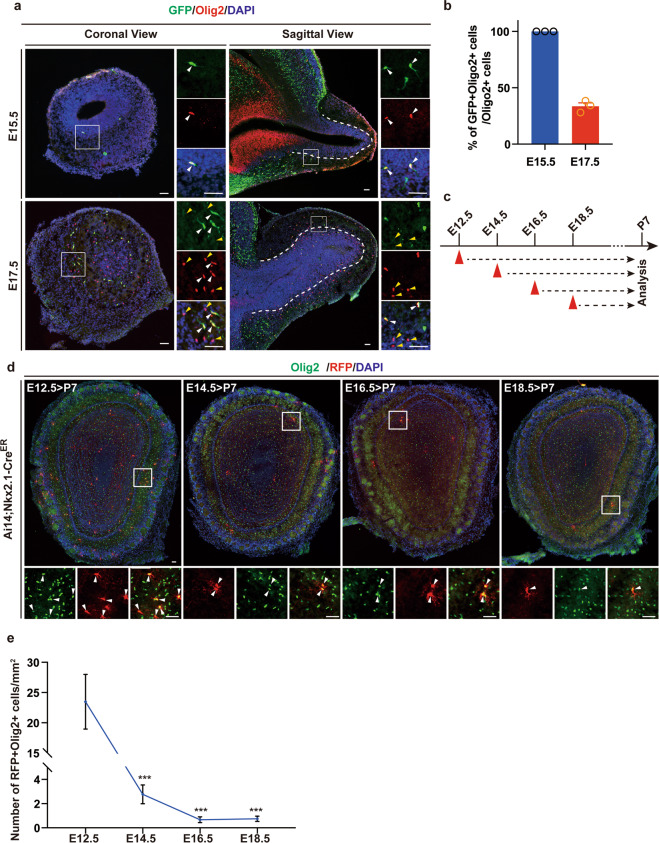
To examine how Nkx2.1-positive RGCs generate OPCs of the OB, we first crossed Nkx2.1Cre mice with Ai3 reporter mice, which allows us to label Nkx2.1-positive progenitors of the MGE and AEP, and thereby their progenies starting from the early stage of cortical development around embryonic day 10.5 (E10.5)14. At E15.5, we observed a small number of GFP-positive cells in the OB (Fig. 1a). Remarkably, when stained the sections for Olig2, a marker for OPCs and mature oligodendrocytes, we found that all Olig2-positive cells were positive for GFP (Fig. 1a, b), suggesting that Nkx2.1-positive RGCs generate the first wave of OPCs in the OB. At E17.5, we observed that around 30% of Olig2-positive cells were expressing GFP (Fig. 1a, b). To further examine the time course of Nkx2.1-positive RGCs generating OPCs in the OB, we crossed Ai14 reporter mice with Nkx2.1CreER mice and induced the Cre expression through tamoxifen induction at different developmental stages (Fig. 1c). When analyzed the OB at postnatal day 7 (P7), we found that most OPCs were derived from RGCs labeled at E12.5 (Fig. 1d, e). Taken together, these data demonstrated that Nkx2.1 lineage contributed to the generation of early OPCs in the OB.
Fig. 1. Nkx2.1 lineage generates OPCs in the OB at early stages.
a Coronal and sagittal sections of the OB from Ai3;Nkx2.1Cre mice stained for GFP and Olig2 at E15.5 and E17.5. White arrowheads indicate Nkx2.1-positive RGCs derived OPC lineage cells (GFP+Olig2+). Yellow arrowheads show other source derived OPC lineage cells (Olig2+GFP−). The dashed lines represent the boundary of the olfactory ventricle zone and the granule cell layer. The small mono-channels are the higher magnification of the solid square boxes. Scale bar: 50 μm. b Quantification of Nkx2.1-positive RGCs derived Olig2-positive cells at E15.5 and E17.5. Data are presented as mean ± SEM. At least 3 mice were analyzed for each time point. c Experimental paradigm shows that tamoxifen (TAM) was injected at E12.5, E14.5, E16.5, and E18.5 and sacrificed at P7. d Confocal images of sections of the OB from Ai14;Nkx2.1-CreER mice stained for Olig2 and RFP. Arrowheads indicate the RFP- and Olig2-double-positive cells. Scale bar: 50 μm. e Quantification of the number of Nkx2.1-derived OPC lineage cells generated at E12.5, E14.5, E16.5, and E18.5. One-way ANOVA, data are presented as mean ± SEM. ***p < 0.001 (n = 3).
Depletion of Tcf4 leads to an increase of OPCs in the OB
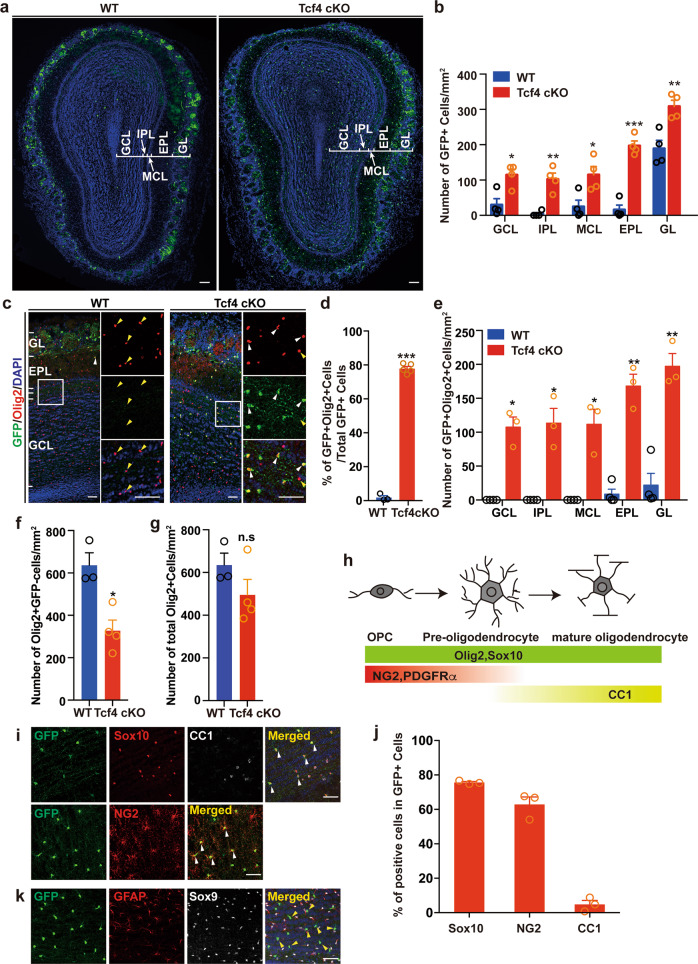
By performing in situ hybridization, we found that the expression of Tcf4 was highly abundant in the ventral cortical regions, such as MEG and AEP (Fig. S1). In addition, it has been reported that Tcf4 is expressed in OPCs by analyzing RNA sequencing data although its role in OPC development has not been addressed28. To ask whether Tcf4 regulates OPC development, we conditionally inactivated Tcf4 in Nkx2.1-lineage specifically by crossing Tcf4f/f mice with Nkx2.1Cre line to generate Tcf4 conditional knockout mice (Tcf4f/f;Nkx2.1Cre, Tcf4 cKO). To visualize the Nkx2.1 progeny, we crossed either Tcf4 cKO or WT (Nkx2.1Cre) mice with Ai3 or Ai14 reporter mice29. When analyzed at P28, we found a significant increase of GFP-positive cells in the OB of Tcf4 cKO compared to that of WT (Fig. 2a, b). As these cells were different from neurons and astrocytes morphologically, we reasoned that these cells might be oligodendrocytes. Indeed, when stained the sections of both WT and Tcf4 cKO for markers expressed in the oligodendrocyte lineage (Fig. 2c), we found that nearly 80% of GFP-positive cells were labeled by Olig2 in the OB of Tcf4 cKO at P28, while very few Olig2- and GFP-double positive cells in the OB of WT were detected (Fig. 2d). The distribution of these Olig2-positive cells was found in all laminar layers in the OB of Tcf4 cKO (Fig. 2e). Interestingly, when analyzed the number of Olig2-positive and GFP-negative cells which were originated from another source, we found a reduction of this population in the OB of Tcf4 cKO (Fig. 2f), suggesting a compensation mechanism that maintains oligodendrocyte homeostasis. Indeed, the total number of Olig2-positive cells was not altered upon the loss of Tcf4 (Fig. 2g). The expression of Olig2 in the oligodendrocyte lineage begins from the OPC stage and persists to the mature stage (Fig. 2h)30. Similarly, Sox10 is also expressed in different developmental stages during oligodendrocyte maturation (Fig. 2h)5. The increase of oligodendrocytes in the OB of Tcf4 cKO was further confirmed by the staining of Sox10 (Fig. 2i). We then asked whether the increased Olig2-positive cells were OPCs or mature oligodendrocytes (Fig. 2h) and stained for NG2, a marker for OPC31. We found that most cells were positive for NG2 and not positive for mature oligodendrocyte marker CC1 (Fig. 2i, j), suggesting that they are OPCs. Even in the aged adult mouse OB, they were positive for NG2, but not CC1 (Fig. 2S). We also confirmed that these cells were not astrocytes as they were negative for GFAP and Sox9, markers for astrocytes (Fig. 2k). Collectively, these data demonstrated that loss of Tcf4 led to the increase of OPCs in the OB.
Fig. 2. Conditional deletion of Tcf4 in Nkx2.1 lineage leads to an increase of OPCs in the OB.
a Representative images showing GFP-positive cells from WT and Tcfk4 cKO OB. Scale bar: 100 μm. b Quantification of the distribution of GFP-positive cells. Student’s t test was used to compare two groups. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 (n = 4). c Representative images of RFP and Olig2 staining in WT and Tcf4 cKO OBs at P28. Small monochannels are the higher magnification of the solid square boxes. White arrowheads represent the GFP+Olig2+ cells and yellow arrowheads mark GFP−Olig2+ cells. Scale bar: 50 μm. d–g Quantifications of Nkx2.1-derived Olig2-positive cells (d) and their distribution in each layer of the OB (e), the number of non-Nkx2.1 derived Olig2-positive cells (f), and the total number of Olig2-positive cells in the OB (g). Student’s t test was used to compare two groups. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, n.s. nonsignificant. At least three samples were analyzed for each condition. h Diagram shows molecular markers expressed during OPC development. i Representative images show the immunostaining for GFP, Sox10, CC1, and NG2. White arrowheads indicate GFP+Sox10+ cells negative for CC1. j Quantification of cells is positive for either Sox10 or NG2 or CC1. k Representative images show the immunostaining for GFP, GFAP, and Sox9. Yellow arrowheads indicate GFP-positive cells which are negative for GFAP and Sox9. Scale bar: 50 μm.
Differentiation of neural progenitor cells remains unaffected upon the loss of Tcf4
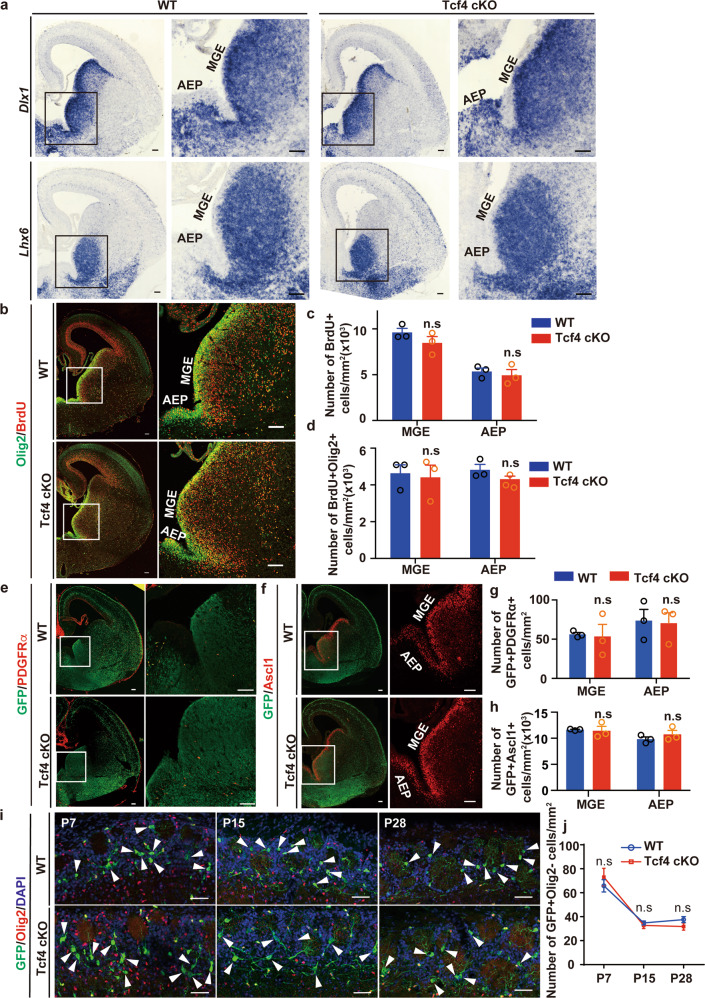
To ask whether the increased OPCs was due to the decreased differentiation of RGCs toward the neuronal lineage upon the loss of Tcf4 in the MGE and AEP, we first examined the expression of Dlx1 and Lhx6, markers for differentiated neural progenitors, by performing the in situ hybridization (Fig. 3a). We found that the expression of Dlx1 and Lhx6 was not altered in the MGE and AEP of Tcf4 cKO compared to these of WT (Fig. 3a). To examine the proliferation capacity of RGCs in the MGE and AEP, we administrated bromodeoxyuridine (BrdU) to mouse embryos at E15.5 and then analyzed them 2 h later. We found that the number of BrdU-positive progenitor cells was not altered in Tcf4 cKO compared to WT (Fig. 3b, c). Particularly, the proliferation of OPCs remained unchanged upon the loss of Tcf4 (Fig. 3b, d). This was further confirmed by staining the sections for PDGFRα and Ascl1, markers for differentiated OPCs32 (Fig. 3e–h). Therefore, the increased OPCs in the OB of Tcf4 cKO did not result from the increased differentiation of RGCs toward the oligodendrocyte lineage. Nkx2.1-positive RGCs of MGE and AEP not only generate oligodendrocytes but also interneurons of the OB located at the glomerular layer of the OB with distinct morphology14,33. We then asked whether the number of interneurons was also altered in the OB of Tcf4 cKO. As no specific markers to label interneurons of the OB derived from Nkx2.1-positive RGCs14,33, we quantified the number of cells positive for GFP but negative for Olig2. At P7, we found that the number of interneurons in the OB of Tcf4 cKO was comparable to that of WT (Fig. 3i, j). However, the number of interneurons was declined at P15 and P28 in the OB of both WT and Tcf4 cKO (Fig. 3j). Taken together, our results showed that the increased OPCs in the OB of Tcf4 cKO was not due to the increase of neural progenitor differentiation toward oligodendrocyte lineage.
Fig. 3. Proliferation and differentiation of Nkx2.1-RGCs were not altered upon the loss of Tcf4.
a In situ hybridization with Dlx1and Lhx6 probes from WT and Tcf4 cKO mice at E15.5. Higher magnification images of the inserts showed on the right panels. Scale bar: 100 μm. b Representative images of immunostaining for GFP, Olig2, and BrdU. The right panel shows the higher magnification of the inserts. Scale bar: 100 μm. c, d Quantification of the proliferating progenitors (c) and proliferating OPCs (d). Student’s t test was used to compare two groups. Data are presented as mean ± SEM. n.s. nonsignificant. e, f Immunostaining for PDGFRα (e) and Ascl1 (f). Scale bar: 100 μm. g, h Quantification of PDGFRα-positive (g) and Ascl1-positive (h) progenitors. Student’s t test was used to compare two groups. Data are presented as mean ± SEM. n.s. nonsignificant. i Representative images show immunostaining for GFP and Olig2 at P7, P15, and P28 from both WT and Tcf4 cKO OBs. Neurons are negative for Olig2 and display distinct morphology (arrowheads). Scale bar: 50 μm. j Quantification of neurons at different developmental stages of OB as indicated. Student’s t test was used to compare two groups. Data are presented as mean ± SEM. n.s. nonsignificant.
Tcf4 promotes OPC programmed cell death during OB development
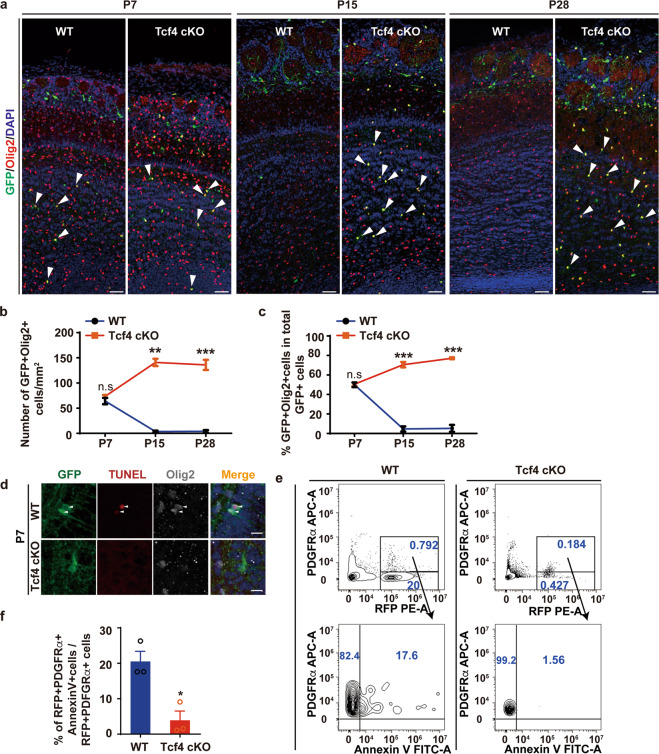
Another possibility for the increase of OPCs in the OB of Tcf4 cKO is the reduction of OPC apoptosis. Due to the transient expression of the classical apoptotic markers and quick clearance of dying cells, it is difficult to directly quantify the dying cells by immunostaining for apoptotic markers6,10. Therefore, we first examined the number of OPCs at different developmental stages (Fig. 4a). At P7, we found that the number of OPCs in the OB was comparable between WT and Tcf4 cKO (Fig. 4a, b). However, in the WT OB, the number of OPCs declined after the first postnatal week and became nearly undetectable at P15 and at P28 (Fig. 4a, b). In contrast, the number of OPCs increased after P7 and persisted till adulthood at P28 in Tcf4 cKO OB (Fig. 4a–c), indicating a reduction of the OPC apoptosis in the lack of Tcf4. Indeed, we detected more DNA breaks, formed in the last phase of apoptosis, in the OB of WT compared to that of Tcf4 cKO by performing TUNEL assay, although the number is low due to the quick clearance most likely (Fig. 4d)10. To further confirm the reduction of the cell death when Tcf4 is depleted, we sorted OPCs from the OB of both WT and Tcf4 cKO by fluorescence activating cell sorting using the surface marker PDGFRα while examining the expression of Annexin V, a surface marker for cells undergoing apoptosis34 (Fig. 4e). Indeed, we found that the number of OPCs expressing Annexin V was significantly decreased in the OB of Tcf4 cKO (Fig. 4f). Taken together, these data demonstrated that OPCs in the WT OB underwent apoptosis after the first postnatal week and were nearly undetectable in adulthood.
Fig. 4. Loss of Tcf4 results in programmed cell death in OPCs.
a Representative images of immunostaining for GFP and Olig2 at various developmental stages in both WT and Tcf4 cKO OBs. Arrowheads show OPCs. Scale bar: 50 μm. b, c Quantification of the density of OPCs (b) and the proportion of Nkx2.1-derived OPCs (c). Student’s t test was used to compare two groups. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. At least three OBs were analyzed for each genotype. d Representative images of immunostaining of GFP, TUNEL, and Olig2 in WT and Tcf4 cKO OBs at P7. Arrowheads indicate the OPCs undergoing cell death. Scale bar: 25 μm. e FACS analysis of OPCs undergoing cell death. f Quantification of the proportion of AnnexinV-positive OPCs in both WT and Tcf4 cKO OBs. Student’s t test was used to compare two groups. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. At least three OBs were analyzed for each genotype.
Tcf4 regulates the survival of OPCs cell-autonomously
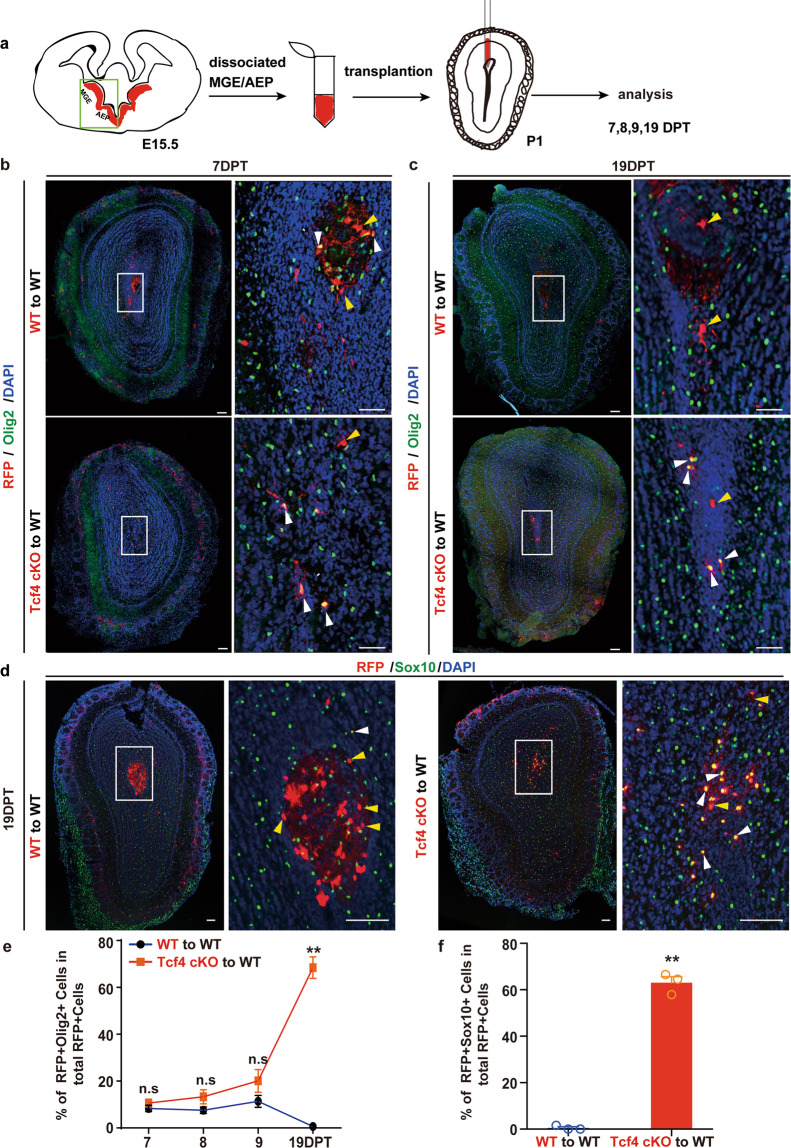
To further examine whether Tcf4 acts cell-autonomously or non-autonomously to regulate OPC survival, we performed transplantation experiments (Fig. 5). In this assay, we dissociated RGCs of MGE and AEP from either control (Ai14;Nkx2.1Cre) or Tcf4 cKO (Tcf4f/f;Ai14;Nkx2.1Cre) embryos at E15.5 and then transplanted these cells into the OB of the WT mice at P1 (Fig. 5a). Transplanted animals were analyzed at various days post-transplantation (DPT). After transplantation, RGCs differentiated into OPCs and neurons which were visualized by RFP reporter. We stained sections for Olig2 to examined the number of OPCs. When analyzed at 7 DPT, the number of OPCs of both WT and Tcf4 cKO OBs was similar (Fig. 5b, e). At 19 DPT, however, we hardly observed Olig2-positive OPCs when WT progenitor cells were transplanted (Fig. 5c, e), suggesting that WT OPCs underwent apoptosis as endogenous WT OPCs did. Interestingly, we observed that OPCs survived when progenitor cells lacking Tcf4 were transplanted into the WT OB (Fig. 5c, e). The identity of OPCs was also confirmed by the staining for Sox10, another marker for OPCs (Fig. 5d, f). A series of analysis at different time points after the transplantation showed that WT OPCs persisted in the host OB till 9 DPT and then underwent apoptosis while OPCs lacking Tcf4 survived till 19 DPT (Fig. 5e), which is similar to what we observed for endogenous OPCs. Thus, these data showed that Tcf4 regulated OPCs survival in a cell-autonomous manner.
Fig. 5. Tcf4 regulates OPC survival cell-autonomously.
a Schematic diagram illustrating the transplantation experiment. b–d Representative images showing that the WT or Tcf4 cKO progenitors of MGE differentiate into OPCs 7DPT (b), and OPCs lacking Tcf4 survived 19DPT (c). The survived OPCs expressing Sox10 (d), an OPC marker. The enlarged images were shown on the right panels. Yellow arrowheads indicate Olig2-negative cells and white arrowheads indicate OPCs. Scale bar: 100 μm. e, f Quantifications of OPCs after transplantation (e) and the number of Sox10-positive OPCs (f). Student’s t test was used to compare two groups. Data are presented as mean ± SEM. **p < 0.01, n.s. nonsignificant. At least three OBs were analyzed for each genotype at each time point.
Increased proliferation of survived OPCs lacking Tcf4
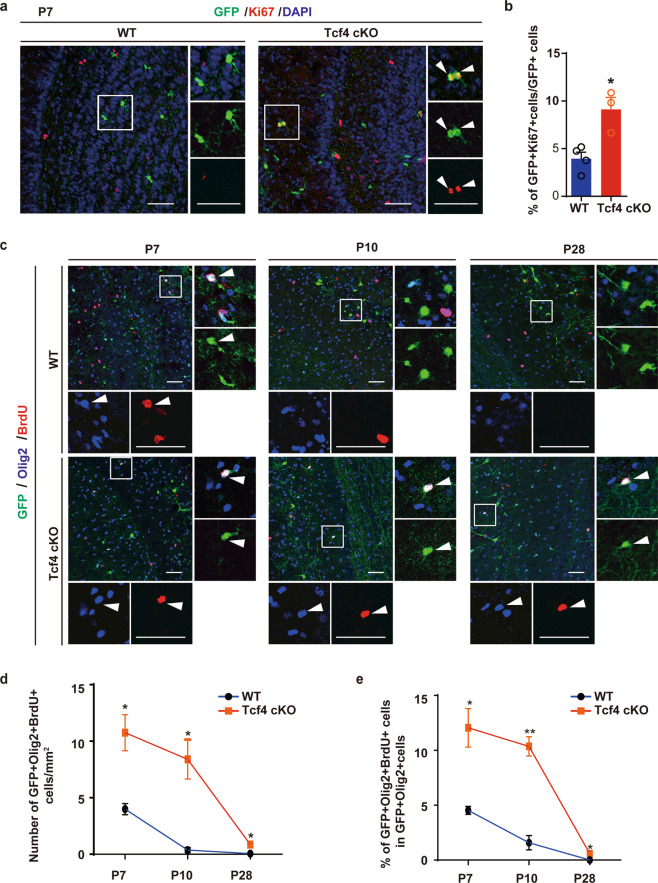
We observed that the number of OPCs in the Tcf4 cKO OB increased gradually from P7 to P28 (Fig. 4c). Likely, this was due to the proliferation of survived OPCs in the Tcf4 cKO OB as OPCs are known to proliferate till adulthood5. To test this hypothesis, we first stained the sections for Ki67, a marker for cell proliferation. We found that most progenies derived from Nkx2.1-positive RGCs were not positive for Ki67 in the WT OB (Fig. 6a). However, we found many pairs of Ki67-positive cells in the Tcf4 cKO OB, suggesting they were proliferating (Fig. 6a, b). To further examine the proliferative status of these OPCs, we administrated BrdU into the postnatal mice and then analyzed 2 h later. We found that some of the Olig2-positive OPCs of both WT and Tcf4 cKO were positive for BrdU at P7 (Fig. 6c), although the number of proliferating OPCs in the WT OB was significantly lower than in the Tcf4 cKO OB (Fig. 6c–e). As OPCs underwent apoptosis in the WT OB after the first postnatal week, the number of OPCs positive for BrdU was dramatically reduced at P10 and P28 (Fig. 6d, e). In contrast, many OPCs that survived in the Tcf4 cKO OB were positive for BrdU even after P7 (Fig. 6c–e), indicating that these survived OPCs were capable of proliferation. However, the number of proliferating OPCs in the Tcf4 cKO OB declined during OB development (Fig. 6e). Therefore, the persisted proliferation of survived OPCs in the Tcf4 cKO OB may also contribute to the increased number of OPCs.
Fig. 6. Proliferation of OPCs in Tcf4 cKO OB.
a Sections of OB from WT and Tcf4 cKO stained for GFP and Ki67 at P7. Scale bar: 50 μm. b Quantification of proliferating OPCs in both WT and Tcf4 cKO OBs. Student’s t test was used to compare two groups. Data are presented as mean ± SEM. *p < 0.05. At least three samples were analyzed for each genotype. c Representative images show sections of OB from WT and Tcf4 cKO stained for Olig2 and BrdU at P7, P10, and P28. Arrowheads indicate proliferating OPCs which are BrdU-positive. Scale bar: 50 μm. d, e Quantification of the density of proliferating OPCs (d) and the number of proliferating OPCs derived from the Nkx2.1 lineage (e). Student’s t test was used to compare two groups. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01. At least three OBs were analyzed for each genotype at each time point.
Tcf4 acts through Bax/Bak pathway to promote the cell death of OPCs
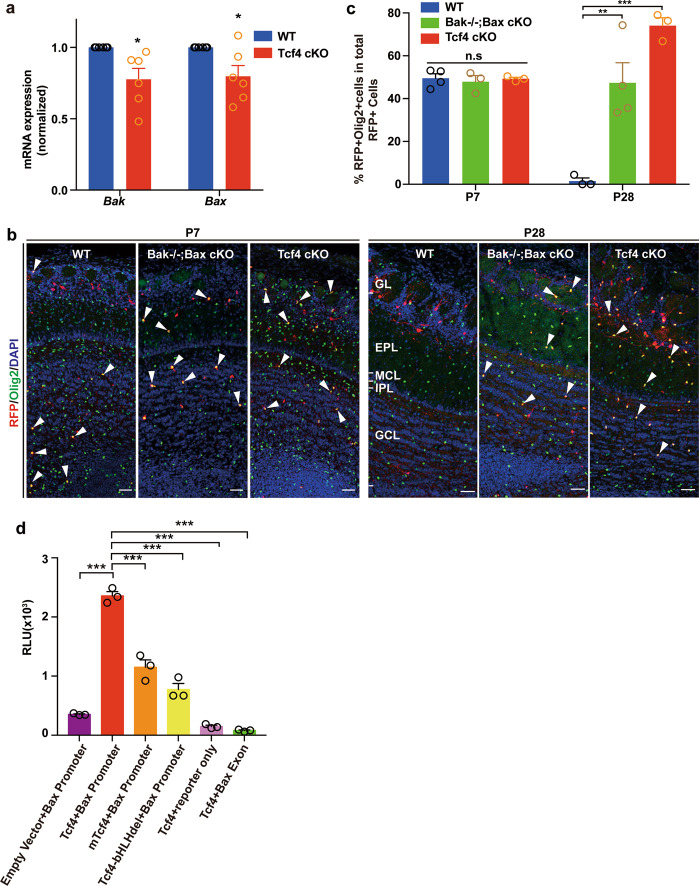
The pro-apoptotic Bcl2 family members Bcl2-associated X protein (Bax) and Bcl2 antagonist/killer 1 (Bak) are key regulators of the apoptosis signaling pathway35,36 and play essential roles in regulating neuronal cell death8. We reasoned that Tcf4 could potentially regulate Bax/Bak pathway to control OPC survival. To test this hypothesis, we first examined the expression of Bax and Bak in Tcf4 cKO OBs by real-time quantitative polymerase chain reaction (qPCR). We found that the mRNA expression level of Bax and Bak was reduced in Tcf4 cKO OBs compared to that of WT OBs (Fig. 7a). To further examine whether Tcf4 acts though Bax/Bak to control the OPC survival, we generated conditional knockout mice in which Bax and Bak were removed in Nkx2.1 lineage specifically by crossing Bak−/−;Baxf/f;Ai14 mice with Nkx2.1Cre line (Bak−/−;Bax cKO). At P7, the number of OPCs was similar in the OBs of WT, Bak−/−;Bax cKO and Tcf4 cKO (Fig. 7b, c). However, we found a significant increase of OPC when Bak and Bax were removed in the OB of Bak−/−;Bax cKO (Fig. 7b, c). Therefore, the suppression of cell death in Nkx2.1 lineage resulted in an increase of OPCs in the OB. We then performed a dual-luciferase transcriptional activation assay to examine whether Tcf4 regulates Bax expression (Fig. 7d). Indeed, we found that Tcf4 activated the transcription of the Bax promoter while no transcription activation was observed when Bax promoter was replaced with a fragment of its exon region (Fig. 7d). The activation of Bax transcription was dependent on the bHLH domain of Tcf4 as the transcription activation was abolished when the bHLH domain of Tcf4 was removed (Fig. 7d, Tcf4-bHLHdel). Interestingly, when Tcf4 carried point mutations at R578H, R580W, and R582P within the bHLH domain, which were found in patients with Pitt–Hopkins syndrome37, the transcription activation of Bax was reduced significantly (Fig. 7d, mTcf4). Taken together, Bax/Bak pathway is a potential key downstream target of Tcf4 to control OPC survival.
Fig. 7. Tcf4 regulates OPC survival through Bax/Bak pathway.
a Relative expression level of Bax and Bak in WT and Tcf4 cKO OBs. The expression level of WT was normalized to 1. Student’s t test was used to compare two groups. Data are presented as mean ± SEM. *p < 005. Six OBs were analyzed for each genotype. b Representative images show immunostaining for Olig2 and RFP at P7 and P28 in OBs of WT, Bak1−/−;Bax cKO and Tcf4 cKO. Arrowheads indicate OPCs. Scale bar: 50 μm. c Quantification of the number of OPCs from (b). Two-way ANOVA was used for statistical analysis. Data are presented as mean ± SEM. **p < 0.01, ***p < 0.001, n.s. nonsignificant. At least three OBs were analyzed for each genotype at each time point. d Tcf4 regulates Bax transcription examined by dual-luciferase assay. Tcf4 with disease-related point-mutations (mTcf4) and Tcf4 lacking the bHLH domain (Tcf4-bHLHdel) were used for examining the function of Tcf4 bHLH domain. Data are shown as mean ± SEM. One-way ANOVA, ***p < 0.001.
Discussion
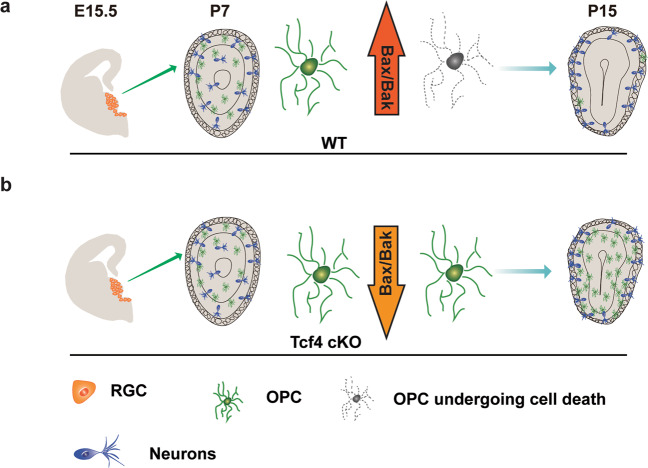
In this study, we revealed a novel role for Tcf4 in regulating OPC survival during OB development (Fig. 8). We showed that Nkx2.1-positive RGCs contribute to the first wave of OPCs in the OB. When Tcf4 was conditionally removed in the Nkx2.1 lineage, the number of OPCs in the OB was significantly increased, which was not due to the enhanced OPC differentiation. Unexpectedly, we found that Tcf4 acts on Bax/Bak pathway to control the OPC survival during OB development. The increased OPCs in the OB could potentially impact neuronal functions, which provides insights into TCF4-related brain disorders.
Fig. 8. A model that illustrates how the OPC number is controlled by Tcf4 mediated cell death.
a MGE- and AEP-derived OPCs undergo cell death through Bax/Bak pathway after the first postnatal week. b However, when Tcf4 is removed, the downregulation of Bax/Bak leads to an increase of OPC survival.
Dysregulation of oligodendrocyte production has been implicated in the disruption of axonal myelination, which leads to severe neurological disorders38,39. Particularly, impairments in olfaction are often associated with autism spectrum disorders40,41, highlighting the importance of neural circuits in the olfactory system. Oligodendrocyte mediated myelination could play essential roles in regulating neuronal functions in the OB. Indeed, disruption of OPC differentiation toward oligodendrocytes has been found in CHD7/CHD8 mutations, which result in CHARGE syndrome and autism spectrum disorders42. Therefore, tight control of the number of OPCs, thereby oligodendrocytes in the OB is important for myelination related disorders. We found that programmed cell death plays a pivotal role in maintaining a precise number of OPCs during OB development, which could be an essential survival control of OPCs and potentially guarantee proper olfactory functions.
TCF4 mutations cause Pitt-Hopkins syndrome, a rare neurodevelopmental disorder characterized by intellectual disability and autistic behavior22,28,43. In addition, genetic variants in TCF4 have been linked to neuropsychiatric diseases, for example, schizophrenia and autism20,28,44. Loss of Tcf4 in neurons leads to alteration of neuronal activity26 and deficits in learning and memory27. We show here that Tcf4 plays an essential role in OPC development. The proper number of OPC is crucial for oligodendrocyte maturation which is fundamental for axonal myelination. For example, dysregulation of oligodendrocyte development impairs the cognitive function in mouse models45. Therefore, Tcf4-mediated OPC survival may underlie the neurological deficits related to TCF4 mutations. Indeed, disruption of oligodendrocyte development has been implicated as a common feature shared by Pitt–Hopkins syndrome and autism spectrum disorder28. Therefore, our studies revealed the importance of Tcf4 in controlling OPC survival during OB development, which provides novel insights into the mechanisms underlying TCF4-associated brain disorders.
Material and methods
Animals
All animal work was approved by the Animal Care and Use Committee of Shanghai Medical College of Fudan University. The following mice were used: Tcf4flox/flox mice22, Nkx2.1-Cre mice14, Bak−/−;Baxflox/flox mice46, Ai14 or Ai3 reporter mice. Animals were bred under the condition of a 12-h light/dark cycle.
Immunohistochemistry
Deeply anesthetized neonatal mice were transcardially perfused with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA). Then brains were then post-fixed overnight at 4 °C. Embryonic brains were harvested and fixed in 4% PFA overnight at 4 °C. After incubation with blocking buffer (0.05% Triton X-100, 5% normal donkey serum in PBS), cryosections were incubated with desired antibodies overnight at 4 °C and then with appropriate fluorescence-conjugated secondary antibodies at room temperature for 1 h before mounting.
Image acquisition and statistical analysis
Confocal images were acquired using Nikon A1R confocal microscope. Images were processed and analyzed using NIS—Elements AR (Nikon) and ImageJ software. For data analysis, Prism7 or Excel was used. All data are presented as mean ± SEM. Significances are marked as *p < 0.05, **p < 0.01, ***p < 0.001. Unless specified, two comparisons were tested using two-tailed unpaired Student’s t tests. Multiple comparisons were analyzed using ANOVA.
Flow cytometry analysis
Neural tissue dissociation kit (P) (Miltenyi Biotec;130–093-231) was used to dissociate the OB from Tcf4 cKO and WT mice at P7. The cell suspension was incubated with antibody anti-PDGFRα (1:500, 558774, BD-Bioscience), followed by the incubation with anti-rat with Alexa647 (1:600, 712-605-153, Jackson ImmunoResearch) in dark for 30 min on ice. Cells were then incubated with Annexin V-FITC (640945, Biolegend) for 20 min at 37 °C. After washing, cells were analyzed by flow cytometry (MoFloAstrios EQ) according to the manufacturer’s instructions.
Transplantation
Embryonic MGE and AEP (E15.5) were harvested from both WT and Tcf4 cKO mice. After dissociation, a single-cell suspension was injected into the OBs of WT host mice at P1 guided by stereotaxic apparatus (position: anteroposterior to bregma (AP) 1.3 mm, lateral to midline (L) 0.6 mm, ventral to the dura (V) 0.5 mm) according to a protocol described previously47.
Tamoxifen and BrdU injection
Tamoxifen (Sigma, T5648) was dissolved in corn oil to prepare the stock solution at a concentration of 20 mg/ml. Tamoxifen was injected intraperitoneally with a dose of 10 µg/kg body weight. BrdU (Sigma, B5002) was dissolved in PBS to prepare the stock solution at the concentration of 10 mg/ml. BrdU was injected intraperitoneally at a concentration of 0.1 μg/kg body weight.
Dual-luciferase assay
The Bax promoter was amplified by PCR and cloned into luciferase reporter pGL4.0 vectors (Promega). Point mutations of hTCF4 (human TCF4) were made through PCR amplification. Gene sequences were confirmed by Sanger sequencing. The Dual-Luciferase Reporter Assay System (Promega) was performed according to the manufacturer’s instructions.
Supplementary information
Acknowledgements
We thank all members of the Xie lab for discussions. We thank Dr. Z. Yang for providing Dlx1 and Lhx6 probes, Dr. M. He for providing Bak−/−;Baxf/f mice. We thank the IOBS Imaging Core facility for expert microscopy.
Funding information
This study was supported by the National Key Research and Development Program of China (grant No. 2018YFA0108000), the National Natural Science Foundation of China (grant Nos. 31872763, 31471038, and 81828008), Shanghai Municipal Science and Technology Major Project (Nos. 2018SHZDZX01 and 19JC1411003) and ZJ Lab to Y. Xie. The National Natural Science Foundation of China (grant No. 31600842) to Y.C.
Author contributions
Yilan Z. and Y.X. conceived and designed the experiments. Yilan Z., Y.C., Y.W., X.D., Yifan Z., and Yubin Z. performed the experiments. Yilan. Z. and Yubin Z. analyzed the data. Y.X. wrote the paper with the inputs from other authors.
Ethics statement
All animal work was approved by the Animal Care and Use Committee of Shanghai Medical College of Fudan University.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Verkhratsky
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-03371-3).
References
- 1.Gibson EM, et al. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science. 2014;344:1252304. doi: 10.1126/science.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao L, et al. Rapid production of new oligodendrocytes is required in the earliest stages of motor-skill learning. Nat. Neurosci. 2016;19:1210–1217. doi: 10.1038/nn.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steadman PE, et al. Disruption of oligodendrogenesis impairs memory consolidation in adult mice. Neuron. 2020;105:150–164 e156. doi: 10.1016/j.neuron.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowitch DH. Glial specification in the vertebrate neural tube. Nat. Rev. Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- 5.Elbaz B, Popko B. Molecular control of oligodendrocyte development. Trends Neurosci. 2019;42:263–277. doi: 10.1016/j.tins.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kessaris N, et al. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat. Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orduz D, et al. Developmental cell death regulates lineage-related interneuron-oligodendroglia functional clusters and oligodendrocyte homeostasis. Nat. Commun. 2019;10:4249. doi: 10.1038/s41467-019-11904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Southwell DG, et al. Intrinsically determined cell death of developing cortical interneurons. Nature. 2012;491:109–113. doi: 10.1038/nature11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong FK, Marin O. Developmental cell death in the cerebral cortex. Annu Rev. Cell Dev. Biol. 2019;35:523–542. doi: 10.1146/annurev-cellbio-100818-125204. [DOI] [PubMed] [Google Scholar]
- 10.Wong FK, et al. Pyramidal cell regulation of interneuron survival sculpts cortical networks. Nature. 2018;557:668–673. doi: 10.1038/s41586-018-0139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun LO, et al. Spatiotemporal control of CNS myelination by oligodendrocyte programmed cell death through the TFEB-PUMA Axis. Cell. 2018;175:1811–1826 e1821. doi: 10.1016/j.cell.2018.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang ZJ, Paul A. The diversity of GABAergic neurons and neural communication elements. Nat. Rev. Neurosci. 2019;20:563–572. doi: 10.1038/s41583-019-0195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim L, Mi D, Llorca A, Marin O. Development and functional diversification of cortical interneurons. Neuron. 2018;100:294–313. doi: 10.1016/j.neuron.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- 15.Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 16.van Tilborg E, et al. Origin and dynamics of oligodendrocytes in the developing brain: Implications for perinatal white matter injury. Glia. 2018;66:221–238. doi: 10.1002/glia.23256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung M, et al. Analysis of the expression pattern of the schizophrenia-risk and intellectual disability gene TCF4 in the developing and adult brain suggests a role in development and plasticity of cortical and hippocampal neurons. Mol. Autism. 2018;9:20. doi: 10.1186/s13229-018-0200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H, Berens NC, Ochandarena NE, Philpot BD. Region and cell type distribution of TCF4 in the postnatal mouse brain. Front. Neuroanat. 2020;14:42. doi: 10.3389/fnana.2020.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Pontual L, et al. Mutational, functional, and expression studies of the TCF4 gene in Pitt-Hopkins syndrome. Hum. Mutat. 2009;30:669–676. doi: 10.1002/humu.20935. [DOI] [PubMed] [Google Scholar]
- 20.Forrest MP, Hill MJ, Quantock AJ, Martin-Rendon E, Blake DJ. The emerging roles of TCF4 in disease and development. Trends Mol. Med. 2014;20:322–331. doi: 10.1016/j.molmed.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Blake DJ, et al. TCF4, schizophrenia, and Pitt-Hopkins syndrome. Schizophr. Bull. 2010;36:443–447. doi: 10.1093/schbul/sbq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, et al. Transcription factor 4 safeguards hippocampal dentate gyrus development by regulating neural progenitor migration. Cereb. Cortex. 2020;30:3102–3115. doi: 10.1093/cercor/bhz297. [DOI] [PubMed] [Google Scholar]
- 23.Page SC, et al. The schizophrenia- and autism-associated gene, transcription factor 4 regulates the columnar distribution of layer 2/3 prefrontal pyramidal neurons in an activity-dependent manner. Mol. Psychiatry. 2018;23:304–315. doi: 10.1038/mp.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, et al. Disruption of TCF4 regulatory networks leads to abnormal cortical development and mental disabilities. Mol. Psychiatry. 2019;24:1235–1246. doi: 10.1038/s41380-019-0353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thaxton C, et al. Common pathophysiology in multiple mouse models of Pitt-Hopkins syndrome. J. Neurosci. 2018;38:918–936. doi: 10.1523/JNEUROSCI.1305-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rannals MD, et al. Psychiatric risk gene transcription factor 4 regulates intrinsic excitability of prefrontal neurons via repression of SCN10a and KCNQ1. Neuron. 2016;90:43–55. doi: 10.1016/j.neuron.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy AJ, et al. Tcf4 regulates synaptic plasticity, DNA methylation, and memory function. Cell Rep. 2016;16:2666–2685. doi: 10.1016/j.celrep.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phan BN, et al. A myelin-related transcriptomic profile is shared by Pitt-Hopkins syndrome models and human autism spectrum disorder. Nat. Neurosci. 2020;23:375–385. doi: 10.1038/s41593-019-0578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/S0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 31.Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE. NG2+ CNS glial. progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68:668–681. doi: 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng K, et al. Molecular and genetic evidence for the PDGFRalpha-independent population of oligodendrocyte progenitor cells in the developing mouse brain. J. Neurosci. 2018;38:9505–9513. doi: 10.1523/JNEUROSCI.1510-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lledo PM, Merkle FT, Alvarez-Buylla A. Origin and function of olfactory bulb interneuron diversity. Trends Neurosci. 2008;31:392–400. doi: 10.1016/j.tins.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Engeland M, Nieland LJ, Ramaekers FC, Schutte B, Reutelingsperger CP. Annexin V-affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry. 1998;31:1–9. doi: 10.1002/(SICI)1097-0320(19980101)31:1<1::AID-CYTO1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 35.Green DR. Apoptotic pathways: the roads to ruin. Cell. 1998;94:695–698. doi: 10.1016/S0092-8674(00)81728-6. [DOI] [PubMed] [Google Scholar]
- 36.Lindsten T, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol. Cell. 2000;6:1389–1399. doi: 10.1016/S1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sepp M, Pruunsild P, Timmusk T. Pitt-Hopkins syndrome-associated mutations in TCF4 lead to variable impairment of the transcription factor function ranging from hypomorphic to dominant-negative effects. Hum. Mol. Genet. 2012;21:2873–2888. doi: 10.1093/hmg/dds112. [DOI] [PubMed] [Google Scholar]
- 38.Yeung MSY, et al. Dynamics of oligodendrocyte generation in multiple sclerosis. Nature. 2019;566:538–542. doi: 10.1038/s41586-018-0842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenzie IA, et al. Motor skill learning requires active central myelination. Science. 2014;346:318–322. doi: 10.1126/science.1254960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennetto L, Kuschner ES, Hyman SL. Olfaction and taste processing in autism. Biol. Psychiatry. 2007;62:1015–1021. doi: 10.1016/j.biopsych.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ashwin C, et al. Enhanced olfactory sensitivity in autism spectrum conditions. Mol. Autism. 2014;5:53. doi: 10.1186/2040-2392-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marie C, et al. Oligodendrocyte precursor survival and differentiation requires chromatin remodeling by Chd7 and Chd8. Proc. Natl Acad. Sci. USA. 2018;115:E8246–E8255. doi: 10.1073/pnas.1802620115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sweatt JD. Pitt-Hopkins syndrome: intellectual disability due to loss of TCF4-regulated gene transcription. Exp. Mol. Med. 2013;45:e21. doi: 10.1038/emm.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gould EA, et al. Mild myelin disruption elicits early alteration in behavior and proliferation in the subventricular zone. Elife. 2018;7:e34783. doi: 10.7554/eLife.34783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeuchi O, et al. Essential role of BAX,BAK in B cell homeostasis and prevention of. autoimmune disease. Proc. Natl Acad. Sci. USA. 2005;102:11272–11277. doi: 10.1073/pnas.0504783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vergano-Vera E, et al. Generation of GABAergic and dopaminergic interneurons from endogenous embryonic olfactory bulb precursor cells. Development. 2006;133:4367–4379. doi: 10.1242/dev.02601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.