Abstract
We previously reported efficient precision targeted integration of reporter DNA in zebrafish and human cells using CRISPR/Cas9 and short regions of homology. Here, we apply this strategy to isolate zebrafish Cre recombinase drivers whose spatial and temporal restricted expression mimics endogenous genes. A 2A-Cre recombinase transgene with 48 bp homology arms was targeted into proneural genes ascl1b, olig2 and neurod1. We observed high rates of germline transmission ranging from 10 to 100% (2/20 olig2; 1/5 neurod1; 3/3 ascl1b). The transgenic lines Tg(ascl1b-2A-Cre)is75, Tg(olig2-2A-Cre)is76, and Tg(neurod1-2A-Cre)is77 expressed functional Cre recombinase in the expected proneural cell populations. Somatic targeting of 2A-CreERT2 into neurod1 resulted in tamoxifen responsive recombination in the nervous system. The results demonstrate Cre recombinase expression is driven by the native promoter and regulatory elements of the targeted genes. This approach provides a straightforward, efficient, and cost-effective method to generate cell type specific zebrafish Cre and CreERT2 drivers, overcoming challenges associated with promoter-BAC and transposon mediated transgenics.
Subject terms: Genetics, Functional genomics
Introduction
The Cre/lox recombinase system has been widely used in zebrafish for spatiotemporal control of gene expression and lineage tracing1–4. The effectiveness of the system is dependent on drivers that provide precise spatial and temporal Cre expression, and recent technical advances combining multiple recombinases have increased the ability to restrict activity to defined cell populations5,6. However, the approach is still limited by methods used to isolate zebrafish transgenics which are susceptible to position effect and multigenerational silencing. An improved method to isolate recombinase transgenics that recapitulate endogenous gene expression patterns would increase the robustness and reproducibility of Cre/lox lineage tracing and gene function studies.
In organisms such as zebrafish that lack embryonic stem cell methodology, Tol2-mediated transgenesis7,8 has traditionally been used to isolate Cre drivers9. In one approach, a gene promoter is cloned or BAC-engineered into a promoter-Cre fusion inside a Tol2 vector4,10. Alternative strategies using Tol2 transgenesis exploit the nearly unbiased, genome wide random integration of the transposon to isolate novel cell type and tissue-specific drivers. Enhancer trap Tol2 vectors contain a minimal promoter-Cre-2A-reporter cassette and are designed to drive expression under the control of local enhancer elements at the integration site11,12. Tol2 vectors engineered with a splice acceptor-mCherry-2A-Cre-ERT2 gene trap are driven by endogenous promoter and regulatory elements after in-frame integration within a gene13. Although each Tol2-based method allows for efficient recovery of transgenics, difficulties associated with defining and cloning a complete promoter and associated regulatory elements, combined with multicopy transgene integration, position effects and gene silencing, limits the effectiveness of creating drivers with consistent restricted expression patterns3,14,15.
Recent advances in gene editing with engineered site-specific nucleases may solve a number of the limitations associated with Tol2 transgenic approaches used to isolate zebrafish transgenic Cre drivers. The ability of TALENs and CRISPR/Cas9 to direct a double stand break to a specific location in the genome and generate single stranded overhangs for DNA repair has been exploited to introduce exogenous DNA at the target site via non-homologous end-joining (NHEJ) and homology directed repair (HDR) pathways16–20. Early studies in zebrafish demonstrated the addition of complementary homology arms flanking a donor transgene significantly increased the frequency and precision of targeted integration21–23. One report in zebrafish described using CRISPR and 1 kb of homology cloned in front of a targeting cassette to integrate Cre-ERT2 upstream of the otx2 translation initiation codon24. However, the efficiency of precision integration at multiple loci using this approach is not known.
To streamline isolation of zebrafish Cre drivers for specific cell lineages and cell types, we applied our recently reported method for CRISPR/Cas9 short homology directed targeted integration25 to generate Cre transgenic lines. As proof of principle we targeted a 2A-Cre cassette in frame into a coding exon in the proneural transcription factor genes ascl1, olig2 and neurod1 that define neural stem, progenitor and post-mitotic cell lineages26–32. We chose to target ascl1b and olig2 since both are differentially expressed in highly aggressive pediatric brain cancers characterized by embryonal-like, poorly differentiated tumors33,34, consistent with their role in progenitor cell function during neural development. We and others had shown previously ascl1b and olig2 are overexpressed in zebrafish embryonal brain tumor models, reinforcing the conservation of molecular mechanisms driving neuroectodermal tumor types35,36. Together with neurod1, a marker of early neural commitment and differentiation, ascl1b and olig2 are good candidates to generate Cre drivers that would be useful for functional studies in neurogenesis and brain tumor pathogenesis. The 2A-Cre and 2A-CreERT2 targeting vectors used for integration are part of our pPRISM vector series that contains cassettes with linked fluorescent secondary marker for allele tracking (Welker et al., in preparation). Recovery of 2A-Cre integration alleles was highly efficient, with frequencies ranging from 10 to 100%, similar to other cargos as reported previously25. As expected, the expression of functional Cre recombinase was restricted to cell populations defined by ascl1b, olig2 and neurod1. Somatic targeting of Cre-ERT2 into neurod1 shows tamoxifen inducible recombinase activity in the nervous system. Together, our results demonstrate CRISPR/Cas9 directed targeted integration is an efficient and robust method for isolating endogenous Cre drivers that reflect the endogenous promoter activity of the targeted genes.
Results and discussion
CRISPR/Cas9 mediated Knock-in of 2A-Cre into ascl1b, olig2, and neurod1
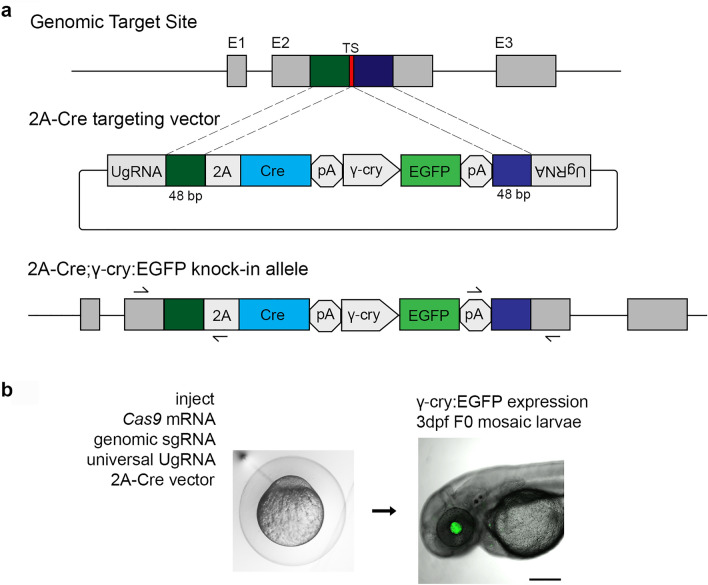
Proneural Cre drivers were generated by integration of a 2A-Cre recombinase cDNA cassette in frame into a coding exon of the zebrafish ascl1b, olig2 and neurod1 genes (Fig. 1). We used our recently published strategy for CRISPR/Cas9 precision targeted integration with short homology arms that is likely to drive integration by homology mediated end joining (HMEJ)25. The 2A-Cre targeting construct contains a secondary fluorescent marker cassette with the γ-crystallin (γ-cry) promoter driving EGFP expression (Fig. 1a). Efficient targeted integration after injection into embryos is driven by in vivo liberation of the homology arms flanking the cassette, which are released by Cas9 induced double strand breaks at the universal gRNA (UgRNA) sites in the vector (Fig. 1a).
Figure 1.
CRISPR/Cas9 short homology directed targeted integration strategy for efficient recovery of Cre knock-in alleles. (a) Schematic of the genomic target site, Cre donor vector, and final knock in allele. A CRISPR sgRNA (red) was chosen in the coding sequence. 48 bp upstream (dark green) and 48 bp downstream (dark blue) of the cut site are included in the donor vector as homology arms. The donor vector has UgRNA sequences flanking the homology arms for in vivo liberation of the knock in cassette. The knock-in cassette contains an in frame 2A-Cre for expression of Cre recombinase, and a secondary marker driving γ-cry:EGFP expression in the lens. Arrows indicate PCR primers for 5′ and 3′ genome/vector junction analysis. (b) 1 cell stage zebrafish embryos are injected with Cas9 mRNA (300 pg), genomic sgRNA (25 pg), UgRNA (25 pg), and 2A-Cre targeting vector (10 pg). At 3 days post fertilization, embryos expressing EGFP in the lens are selected and raised to adulthood. TS genomic sgRNA target site, UgRNA universal short guide RNA target site, γ-cry gamma crystallin promoter, EGFP green fluorescent protein, pA transcription termination and polyadenylation sequence.
sgRNAs were designed to CRISPR/Cas9 sites in exon 1 of ascl1b and exon 2 of olig2 and neurod1 (Table 1). The sgRNA mutagenesis efficiency was tested by co-injection with Cas9 mRNA into 1-cell zebrafish embryos, followed by PCR amplification of the targeted exon and analysis of heteroduplex formation in the PCR product by gel electrophoresis. After confirmation of efficient mutagenesis, 48 bp of sequence on either side of the genomic target Cas9 was used to design and clone 5′ and 3′ homology arms into the 2A-Cre targeting vector. Zebrafish embryos were injected at the 1-cell stage with the genomic target site sgRNA, the UgRNA, Cas9 mRNA, and the 48 bp homology arm 2A-Cre targeting vector. At 3 days post-fertilization (dpf), larvae were screened for expression of the lens specific γcry:EGFP secondary marker (Fig. 1b), and positive larvae selected to raise to adulthood to test for germline transmission. Targeting of ascl1b, olig2 and neurod1 resulted in 58%, 48% and 37%, respectively, of F0 injected embryos showing EGFP expression in the lens (Table 1), demonstrating the presence of the injected targeting vector. Evidence of somatic targeted integration at the genomic site was confirmed by junction fragment PCR on injected embryo.
Table 1.
Efficient recovery of endogenous neurogenic Cre driver lines by 2A-Cre-γcry:EGFP targeted integration.
| Neurogenic gene | ascl1b | olig2 | neurod1 |
|---|---|---|---|
| Targeted exon | 1 | 2 | 2 |
| 5′ and 3′ homology arms (bp/bp) | 48/48 | 48/48 | 48/48 |
| F0 embryos expressing secondary marker | 58% (61/105) | 48% (52/109) | 37% (21/57) |
| F0 adults transmitting secondary marker | 100% (3/3) | 10% (2/20) | 20% (1/5) |
| F0 adults transmitting precise integration allele | 33% (1/3) | 5% (1/20) | 20% (1/5) |
To test whether somatic integration of the 2A-Cre cassette led to expression of functional Cre recombinase in the expected neural cell populations, we injected the genomic sgRNA, UgRNA, Cas9 mRNA, and the donor vector into embryos from the transgenic ubi:Switch floxed reporter line Tg(ubi:loxP-EGFP-loxP-mCherry) (Mosimann et al., 2011). Injected F0 mosaic Tg(ubi:loxP-EGFP-loxP-mCherry) embryos showed a switch from EGFP to mCherry expression in cells throughout the brain and retina, while EGFP expression remained in cells outside of the central nervous system (Supplementary Fig. S1). These results suggested that Cre expression was controlled by the endogenous regulatory elements of the targeted proneural genes, leading to recombination in the specific neural progenitor populations defined by those genes. Furthermore, the results suggest that the expressed Cre has recombinase activity that leads to recombination at loxP sites and excision of the floxed EGFP cassette. All neural progeny descended from the neural progenitor populations inherit the recombination event and express mCherry. The observed recombination in F0 injected animals indicated on-target integration of the 2A-Cre cassette was relatively efficient. Together, these results suggested targeted integration would be an effective method to generate endogenous Cre drivers that can promote spatially restricted cell-type specific Cre-mediated recombination.
Stable germline 2A-Cre knock-in alleles were established from adult F0s outcrossed to wildtype WIK to identify individuals transmitting the γcry:EGFP secondary marker to their progeny. We first measured the frequency of germline transmission of the γcry:EGFP secondary marker and found rates for ascl1b, olig2 and neurod1 of 100%, 10% and 20%, respectively (Table 1). F1 embryos positive for EGFP expression were tested for on-target precise integration at the genomic target site by PCR amplification and sequencing of the 5′ and 3′ genome/cassette junctions (Fig. 1, Supplementary Fig. S2). We found F0 transmission of precise integration alleles through the germline was relatively high, with frequencies at ascl1b, olig2 and neurod1 of 33% (1/3), 5% (1/20) and 20% (1/5), respectively (Table 1). These results at 3 independent loci demonstrate 2A-Cre in-frame targeted integration alleles can be efficiently recovered after screening a minimum of 20 F0 adults.
Single F1 adults harboring precise integration alleles were outcrossed to recover F2 adults, and single F2 adults were used to establish Tg(ascl1b-2A-Cre)is75, Tg(olig2-2A-Cre)is76, and Tg(neurod1-2A-Cre)is77 F3 families. Because the 2A-Cre cassette was integrated in the proneural gene coding sequence, a loss of function mutation should result; therefore, the transgenic lines are maintained as heterozygotes. The F3 heterozygous ascl1b-2A-Cre, olig2-2A-Cre, and neurod1-2A-Cre larvae and adults did not exhibit anatomical or behavioral phenotypes.
Endogenous proneural 2A-Cre transgenic lines express Cre in a pattern that recapitulates the targeted gene
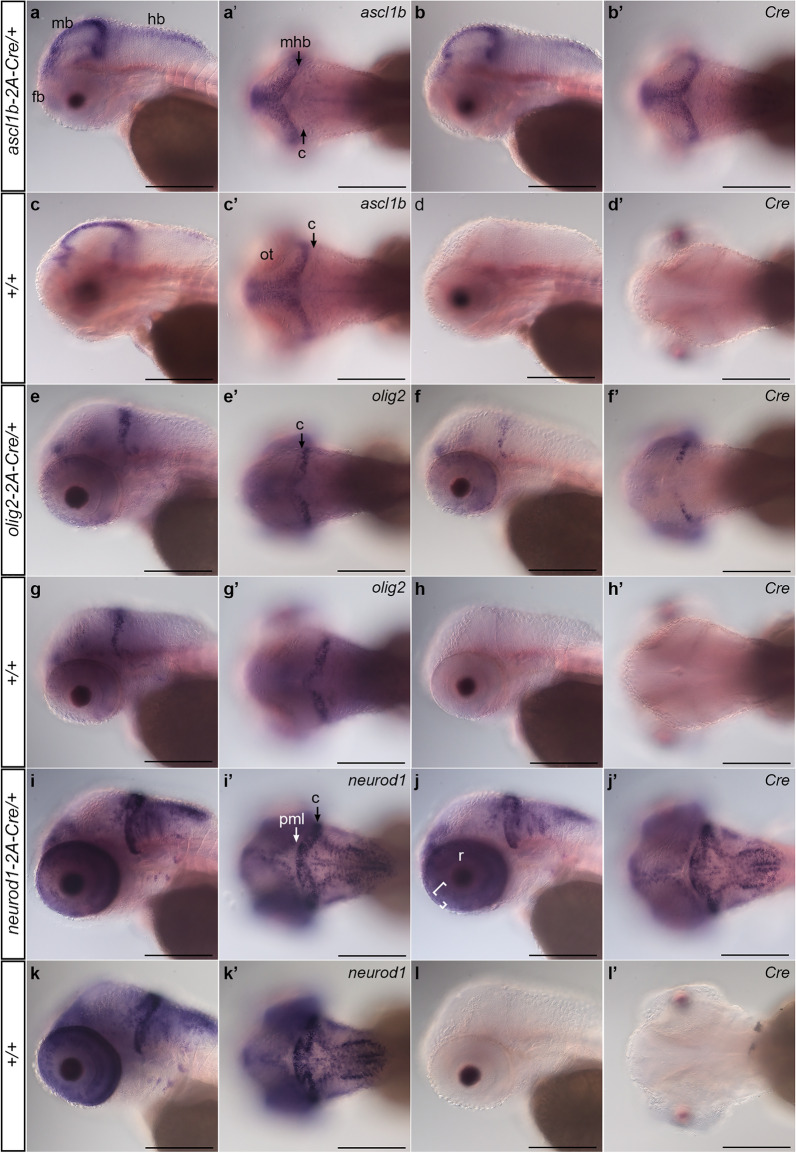
To confirm the ascl1b-2A-Cre, olig2-2A-Cre, and neurod1-2A-Cre integration lines express Cre in a pattern that recapitulates the targeted gene, we used whole mount in situ hybridization to examine Cre transcript localization. Adult F2 or F3 heterozygotes were outcrossed to wild type WIK and 3 dpf sibling larvae were hybridized with a probe complementary to Cre or to the target gene mRNA. Each of the lines showed that the pattern of Cre expression was identical to the endogenous gene mRNA (Fig. 2).
Figure 2.
Expression of 2A-Cre integration alleles recapitulates ascl1b, olig2, and neurod1 expression pattern. Whole mount in situ hybridization for endogenous genes and Cre was performed in 3 dpf larvae obtained from outcrossing ascl1b-2A-Cre/+, olig2-2A-Cre/+, and neurod1-2A-Cre/+ lines to wild type WIK. Sibling larvae from the same clutch were sorted into lens EGFP-positive and -negative groups before fixation. For each genotype and probe 3 individual larvae were photographed. (a–d) ascl1b and Cre expression in ascl1b-2A-Cre/+ larvae show similar patterns in the forebrain, and along the midbrain ventricle and midbrain-hindbrain border. (e–g) olig2 and Cre expression in olig2-2A-Cre/+ larvae were restricted to the forebrain, the posterior half of the cerebellum, and a subset of cells in the retina. (i–k) neurod1 and Cre expression in neurod1-2A-Cre/+ larvae were detected in the forebrain, adjacent to the midbrain and hindbrain ventricles and enriched in the cerebellum ((i′), C black arrow), and in the retina inner (large bracket) and outer (small bracket) nuclear layers (j). neurod1 and Cre expression were not detected in the posterior peripheral midbrain layer ((i′), PML white arrow). Cre expression was not detected in wild type +/+ sibling larvae (d,h,l). c cerebellum, fb forebrain, hb hindbrain, mb midbrain, mhb midbrain-hindbrain border, nt neural tube, ot optic tectum, pml peripheral midbrain layer, r retina. Scale bar 250 μm.
ascl1b-2A-Cre/+ larvae hybridized with ascl1b or Cre probes showed the same pattern of signal in the forebrain, along the midbrain ventricle and midbrain-hindbrain border in the optic tectum, and on the dorsal surface of the hindbrain (Fig. 2a–d), as previously reported37. olig2-2A-Cre/+ larvae showed Cre expression matched olig2 in wild type larvae (Fig. 2e–h)38, and was restricted to the posterior half of the cerebellum, a subset of cells in the neural retina, and the ventral forebrain. Like the endogenous neurod1 in wild type (Fig. 2i,j)39,40, Cre expression in neurod1-2A-Cre/+ was observed in the forebrain, adjacent to the midline in the midbrain and hindbrain, and absent from the peripheral midbrain layer of the optic tectum, consistent with neurod1 expression in committed progenitors and newborn post-mitotic neurons. It was highly expressed in the cerebellum and in the inner and outer neural layers of the retina (Fig. 2i–l). For all three lines, the consistency between the Cre and endogenous gene expression patterns suggested that both were controlled by the same regulatory elements, providing gene specific spatial and temporal Cre expression in neural progenitor populations defined by the proneural transcription factors.
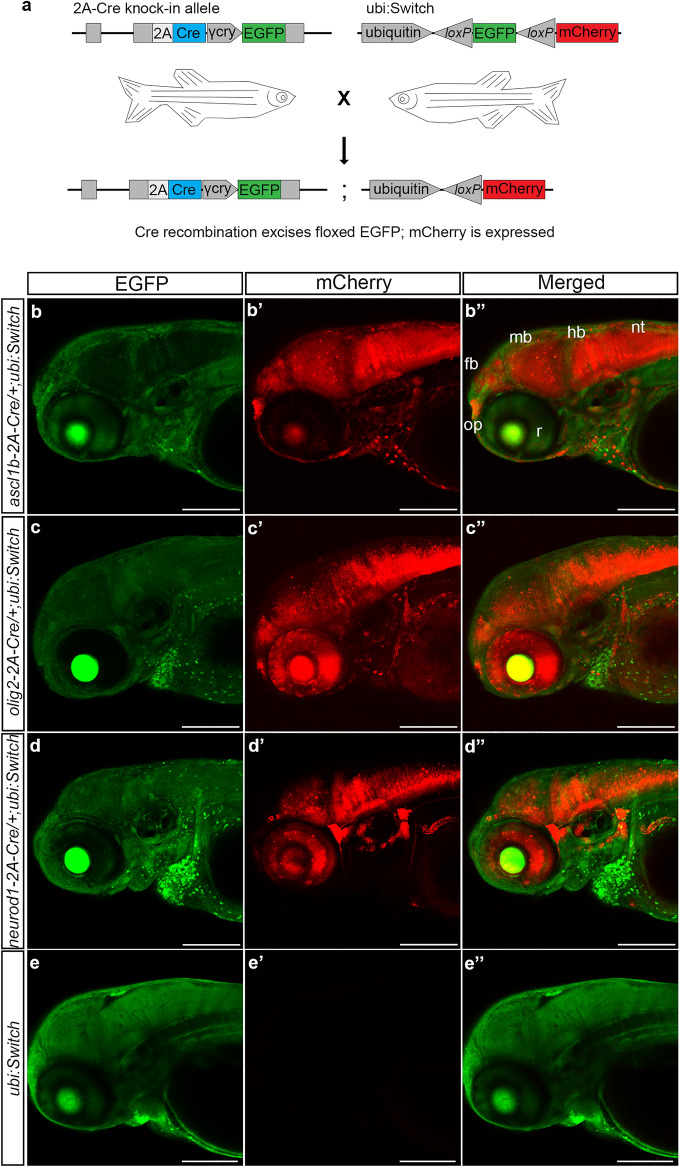
Endogenous neurogenic 2A-Cre transgenic lines lead to efficient Cre-loxP recombination in the expected neural cell populations defined by proneural gene expression
To confirm the stable ascl1b-2A-Cre, olig2-2A-Cre, and neurod1-2A-Cre driver lines express functional Cre recombinase in the expected neural progenitor cell populations, F0 adults were mated to the ubi:Switch line (Fig. 3a). The offspring were imaged at 3 dpf and showed expression had switched from EGFP to mCherry in neurons in the developing forebrain, midbrain, hindbrain and retinas of the double heterozygous 2A-Cre; ubi:switch larvae (Fig. 3b–d). ascl1b-2A-Cre led to near complete switching of EGFP to mCherry along the anterior–posterior axis of CNS, from the olfactory placode extending to the neural tube, with the exception of the neural retina (Fig. 3b). This is consistent with the early expression of ascl1b in the midbrain and hindbrain regions of the neural plate in 4–10 somite stage embryos, which broadens to cells in the anterior telencephalon, diencephalon, midbrain tegmentum and hindbrain by 25 somites/22 h post fertilization, but is completely absent from the developing retina41,42. Little switching occurred in cells outside of the CNS. These results indicated Cre expressed by the ascl1b-2A-Cre allele led to efficient recombination at the ubi:Switch transgene loxP sites and excision of the floxed EGFP cassette.
Figure 3.
Transgenic ascl1b-2A-Cre, olig2-2A-Cre and neurod1-2A-Cre lines express functional Cre that promotes recombination at loxP sites in the expected neural progenitor populations. (a) F0 adults harboring precise integration alleles were mated to the recombination reporter line ubi:Switch to generate double transgenic 2A-Cre driver; ubi:Switch embryos. (b–d) Confocal imaging of 3 dpf double transgenic ascl1b-2A-Cre; ubi:switch (b), olig2b-2A-Cre; ubi:switch (c), and neurod1-2A-Cre; ubi:switch (d) larvae shows a switch from GFP to mCherry expression in neural cells derived from ascl1b, olig2, and neurod1 progenitors. (e) A switch from GFP to mCherry expression isn’t detected in single transgenic ubi:Switch larvae. fb forebrain, γ-cry gamma crystallin promoter, EGFP green fluorescent protein, hb hindbrain, mb midbrain, nt neural tube, op olfactory placode, r retina. Scale bar 200 μm.
Similar to ascl1b-2A-Cre, the olig2-2A-Cre transgenic line led to recombination and switching from EGFP to mCherry expression throughout the CNS, including the retina (Fig. 3c). The pattern was consistent with early expression of olig2 in the neural plate presumptive ventral diencephalon at 8 hpf43; forebrain dorsal thalamus, subpallium, posterior ventral thalamus and posterior tuberculum at 24–30 hpf44; and along the forebrain ventricles, retina, midbrain/hindbrain border and cerebellum at 48 hpf45,46. The pattern of switching induced by the neurod1-2A-Cre line extended throughout neurons in the forebrain, midbrain, hindbrain, retina, and the PNS (Fig. 3d), as expected, given neurod1 is expressed in committed neural progenitors and early post mitotic neurons throughout the nervous system47. In single transgenic ubi:Switch larvae switching does not occur and only EGFP expression is detected (Fig. 3e).
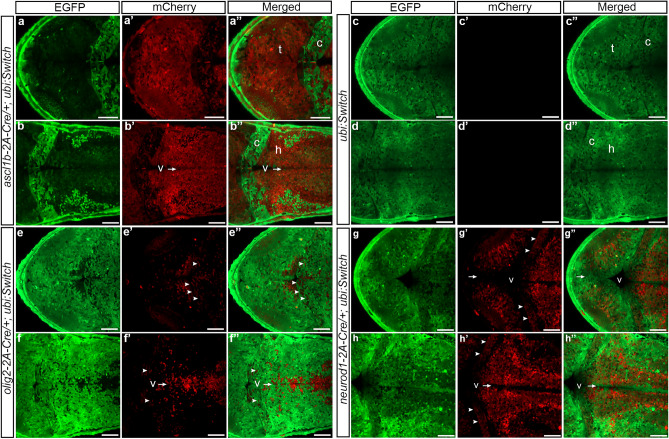
Higher resolution analyses of recombinase switching in the ascl1b-2A-Cre, olig2-2A-Cre, and neurod1-2A-Cre 3 dpf larval midbrain and hindbrain were consistent with cell-specific expression of Cre recombinase (Fig. 4). F3 ascl1b-2A-Cre, F2 olig2-2A-Cre, and F2 neurod1-2A-Cre adults were crossed to the ubi:Switch line and RFP expression was examined in the 3 dpf larval dorsal tectum, cerebellum and hindbrain. In ascl1b-2A-Cre; ubi:Switch nearly all cells throughout the tectum (Fig. 4a–a′′), including cells lining the ventricle, expressed mCherry, consistent with the high levels of ascl1b and Cre transcripts detected in the ventricular zone of the midbrain by in situ hybridization (Fig. 3a–d). A subset of cells in the cerebellum had switched from EGFP to mCherry expression (Fig. 4a′′,b′′), which may be from the small number of cells expressing ascl1b in the cerebellum (Fig. 3a′,b′,c′), or the result of earlier recombination in progenitors during embryonic brain development. mCherry expression was detected throughout the hindbrain, as expected given the earlier widespread expression of ascl1b in stem/progenitors in the neural plate and embryonic brain regions, and the expression of ascl1b and Cre transcripts detected by in situ hybridization in a subset of cells lining the ventricles and located laterally in the dorsal 3 dpf hindbrain (Fig. 3a–d). A switch from EGFP to mCherry expression wasn’t detected in sibling single transgenic ubi:Switch larvae (Fig. 4c–c′′,d–d′′). In olig2-2A-Cre; ubi:Switch the switch from EGFP to mCherry was detected in a subset of cells at the posterior region of the tectum (Fig. 4e–e′′), in the cerebellum (Fig. 4e,f arrowheads), in cells lining the hindbrain ventricle (Fig. 4f–f′′ V arrow), and in cells scattered throughout the hindbrain (Fig. 4e–e′′,f–f′′). In contrast, in neurod1-2A-Cre; ubi:Switch larval brains mCherry wasn’t detected in cells lining the ventricle of the tectum or hindbrain (Fig. 4g,h, arrows). mCherry was present in cells throughout the cerebellum (Fig. 4g′,h′ arrowheads) and the midbrain and hindbrain parenchyma, as expected given neurod1 expression is induced after committed progenitors migrate away from the ventricle and transition to the neuronal state (Fig. 4g–g′′,h–h′′). The mCherry expression was consistent with the strong pattern of in situ hybridization signal for neurod1 and Cre in the 3dpf midbrain and hindbrain (Fig. 3i–l). Together, these results show the 2A-Cre knock-in lines express functional Cre recombinase in the pattern and cell type defined by the targeted proneural gene, leading to progenitor cell descendants inheriting the recombined floxed allele.
Figure 4.
ascl1b-2A-Cre, olig2-2A-Cre and neurod1-2A-Cre Cre recombinase activity in the 3 dpf larval midbrain and hindbrain. F3 or F2 adults were mated to the recombination reporter line ubi:Switch to generate double transgenic embryos. Confocal imaging of ascl1b-2A-Cre; ubi:Switch (a–a′′,b–b′′), control sibling ubi:Switch (c,d), olig2-2A-Cre; ubi:Switch (e–e′′,f–f′′) and neurod1-2A-Cre; ubi:Switch (g–g′′,h–h′′) larval midbrain tectum (t), cerebellum (c) and hindbrain (h). Arrows point to the ventricular zone lining the midbrain and hindbrain ventricles (v). Expression of ascl1b-2A-Cre earlier in brain development leads to nearly all descendant neurons expressing mCherry (a,b). No switching occurs in the absence of Cre (c–c′′,d–d′′). olig2-2A-Cre leads to mCherry expression in a subset of cells in the cerebellum (arrowheads), and neural progenitors lining the hindbrain ventricular zone and their descendants (e,f). In neurod1-2A-Cre mCherry expression is absent from the ventricular zone (v arrows) but present throughout neurons in the tectum, cerebellum (arrowheads) and hindbrain (g,h). Scale bars 50 μm.
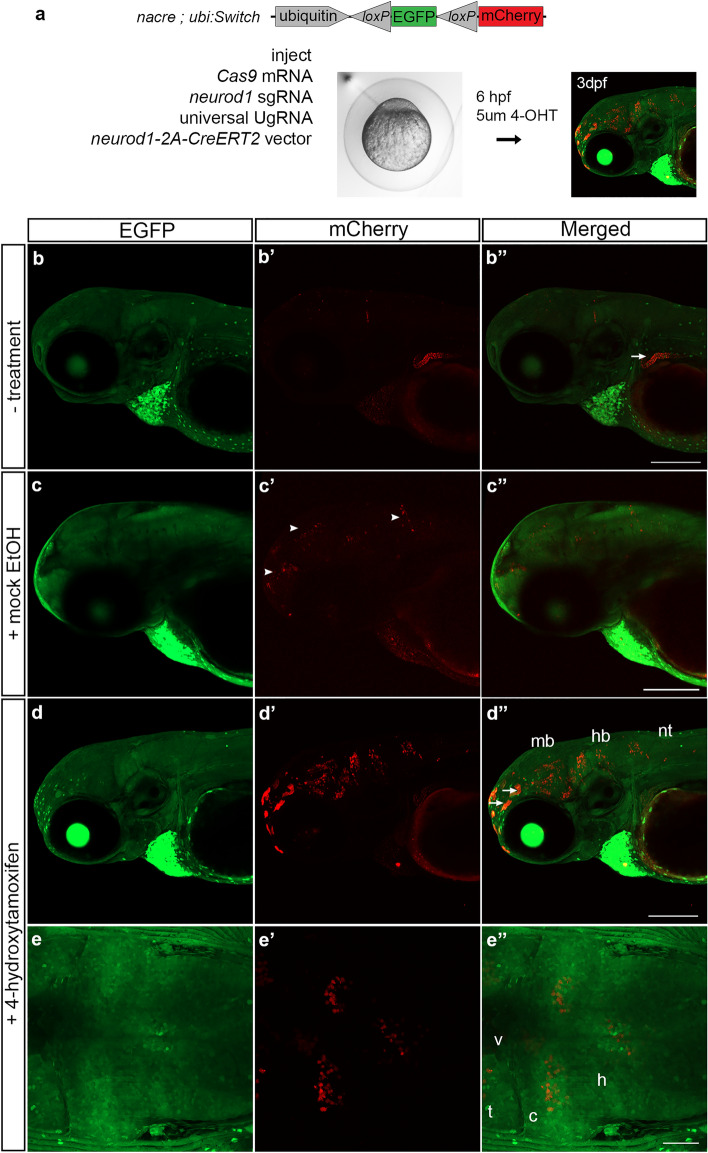
We next replaced the Cre cDNA in the 2A-Cre-PRISM vector with CreERT2, which was used previously by Mosimann et al., 2011 to generate the ubiquitin:CreERT2 transgenic by Tol2 mediated transgenesis4. To test whether somatic integration of CreERT2 would lead to expression of tamoxifen responsive Cre recombinase, the 2A-CreERT2 cassette was targeted to the neurod1 exon 2 sgRNA site in nacre; ubi:Switch embryos that lack melanin pigment (Fig. 5a). At 6hpf injected embryos were treated with 5 µm 4-hydroxytamoxifen (4-OHT) in ethanol or ethanol alone and placed in a light-blocking container. On day 2 and day 3 the embryos were placed in fresh solution. At 3dpf, mCherry expression was detected in 17% (4/23) of untreated injected embryos (Fig. 5b–b′′) and 11% (2/19) of mock treated injected embryos (Fig. 5c–c′′) (Table 2), however the amount of switching was reduced compared to treated embryos. Recombination in the absence of treatment (Fig. 5b–b′′) or tamoxifen (Fig. 5c–c′′) may be caused by a stress response from injection, that alters hsp70 ability to tether CreERT2 at the cell membrane and in the cytoplasm. 45% (10/22) of injected embryos treated with 4-hydroxytamoxifen showed widespread mosaic mCherry expression throughout the brain and spinal cord (Fig. 5d–d′′,e–e′′; Table 2). Some examples of off targeting were detected in the gut (Fig. 5b′′ arrowhead) and skin (Fig. 5d′′ arrows). As expected, uninjected control embryos did not show mCherry expression (0/24) (Table 2). neurod1-2A-CreERT2-genomic DNA junction fragment analysis showed evidence of integration that correlates with switching in targeted embryos (Supplementary Fig. S3). Together, these results show targeted integration of Cre-ERT2 leads to endogenous expression of tamoxifen inducible Cre recombinase.
Figure 5.
Tamoxifen regulated CreERT2 recombinase activity in F0 somatic targeted embryos. (a) nacre; ubi:Switch embryos injected with Cas9mRNA, sgRNAs and the neurod1-2A-CreERT2 targeting vector. At 6 hpf embryos were mock treated or treated with 5um 4-hydroxytamoxifen and imaged at 3 dpf. (b–b′′) Injected embryos without treatment or (c–c′′) mock treated show a low level of expression in the brain. (d–d′′) 4-hydroxytamoxifen treatment resulted in switching from EGFP to mCherry expression in cells in the midbrain, hindbrain and neural tube. Arrows point to off-target expression. (e–e′′) Dorsal imaging of the brain in the larvae shown in d–d′′ revealed mCherry expression was detected in cells throughout the midbrain and hindbrain parenchymal tissue. 4-OHT 4-hydroxytamoxifen, c cerebellum, EGFP enhanced green fluorescent protein, hb hindbrain, mb midbrain, nt neural tube, t optic tectum, v ventricle. Scale bars (b′′,c′′,d′′) 200 μm; (e′′) 50 μm.
Table 2.
Tamoxifen responsive Cre recombinase activity in neurod1-2A-CreERT2 targeted embryos.
| Condition | # viable 3dpf larvae | # mcherry + 3dpf larvae |
|---|---|---|
| Injected | 23/30 | 4/23 (17%) |
| Injected + mock ethanol treatment | 19/30 | 2/19 (11%) |
| Injected + 5 µm 4-OHT treatment | 22/60 | 10/22 (45%) |
| Uninjected | 24/30 | 0/24 (0%) |
In summary, in this study we applied our CRISPR/Cas9 short homology directed targeted integration strategy25 to generate a set of zebrafish proneural Cre drivers that express functional Cre recombinase in the pattern of the endogenous targeted ascl1b, olig2, and neurod1 genes. Somatic targeting of 2A-CreERT2 into neurod1 in ubi:Switch embryos showed switching in the nervous system after treatment with tamoxifen, suggesting our approach can be used to generate endogenous CreERT2 lines that express inducible Cre recombinase activity. We were able to efficiently recover precise in-frame 2A-Cre recombinase germline integration alleles after screening a minimum of only 20 adult F0 fish. The frequencies of allele recovery were similar to our previous report describing efficient knock in of 2A-RFP and 2A-EGFP reporter cassettes at eight zebrafish loci25. Although ascl1b, olig2, and neurod1 do not show haploinsufficiency, for other loci it may be beneficial to maintain the wild type endogenous gene expression in the targeted allele. Previous work showed homozygous otx2:CreERT2 embryos generated by CreERT2 integration upstream of the zebrafish otx2 initiating ATG were morphologically normal, but endogenous otx2 expression was reduced24. In contrast to our method, the frequency of on target integration using 1000 bp of 5′ homology was 3%, and the targeted cassette did not contain a secondary marker for rapid visual genotyping. Nagy et al. (2019) inserted Cre before the translation termination codon in the mouse megakaryocyte/platelet-specific Gp1ba gene, which led to a decrease in GPIbα protein expression and multiple platelet defects48. These studies suggest the potential for targeted integration to negatively impact endogenous gene expression will be specific to gene and location, and may require testing alternative design strategies. Our efficient and simplified method for generating zebrafish transgenic Cre driver lines by CRISPR targeted integration can be readily applied to other loci, expanding opportunities to develop new recombinase genetic tools for investigation of specific cell types, developmental stages, or disease states.
Methods
Ethics declarations and approval for animal experiments
The zebrafish research in this study was performed according to the Guidelines for Ethical Conduct in the Care and Use of Animals49. All zebrafish experiments were carried out in accordance with Iowa State University Animal Care and Use Committee IACUC-18-279 and IACUC-20-058 approved protocols. All methods involving zebrafish were in compliance with ARRIVE guidelines50, and the American Veterinary Medical Association (2020) and NIH guidelines for the humane use of animals in research.
Zebrafish strains and maintenance
Zebrafish (Danio rerio) were maintained on an Aquatic Habitats (Pentair) or Aquaneering aquaculture system at 27 °C on a 14 h light/10 h dark cycle. The wild type strain WIK was used to generate the knock in lines and was obtained from the Zebrafish International Resource Center (https://zebrafish.org/home/guide.php). The casper mutant fish and ubiquitously expressed floxed EGFP-mCherry line ubi:Switch (Tg(ubi:loxP-EGFP-loxP-mCherry)) (Mosimann et al. 2011) were obtained from the lab of Dr. Leonard Zon (Harvard). casper; ubi:switch and nacre; ubi:switch fish were established by crossing ubi:switch into casper, followed by additional backcrosses to casper.
pPRISM-2A-Cre and -CreERT2 vectors and targeted integration
The pPRISM-2A-Cre vector was designed to be compatible with our previously described short homology-based CRISPR/Cas9 knock in strategy25. The cassette in the pPRISM-2A-Cre vector has two functional units: (1) a 2A-Cre cDNA cassette and (2) a secondary fluorescent reporter. The 2A-Cre unit is composed of a self-cleaving 2A peptide from porcine teschvirus-1, the Cre recombinase cDNA, and an ocean pout transcriptional termination and polyadenylation sequence (Zoarces americanus, Gibbs and Schmale, 2000). The viral 2A peptide has been used extensively in zebrafish for production of multicistronic messenger RNAs51, allowing release of Cre protein from the short endogenous gene’s polypeptide. The secondary reporter cassette for visually track transgenic embryos contains the gamma crystallin (γ-cry) promoter, mini-intron, nls-EGFP, bovine growth hormone transcriptional termination and two SV40 polyadenylation sequences (Yang and Cvekl 2005; Clark et al. 2011; Kim et al. 2011). Flanking the entire sequence are universal sgRNA (UgRNA) sites for Cas9 induced double strand breaks to liberate the short homology arms and drive integration of the Cre-reporter targeting cassette25. The pENTR_D_CreERT2 vector was a gift from Dr. Christian Mosimann (University of Colorado Anschutz Medical Campus). Using a combination of restriction enzyme digestion, PCR, and New England Biolab Hi-Fi cloning, the Cre cDNA in the pPRISM-2A-Cre vector was removed and replaced with CreERT2.
CRISPR sgRNAs target sites in the coding sequence were selected and tested for efficient indel formation by co-injection of 25 pg sgRNA plus 300 pg Cas9 mRNA into 1 cell stage wild type WIK zebrafish embryos, followed by PCR amplification of the targeted exon and analysis of heteroduplex formation by gel electrophoresis. 48 bp 5′ and 3′ homology arms were designed and cloned into the pPRISM-2A-Cre vector as described25. sgRNA and homology arm oligonucleotide sequences are listed in Supplementary Table S1. For targeted integration the injection mix contained 25 pg of genomic sgRNA, 25 pg of UgRNA, 10 pg of targeting vector, and 300 pg Cas9 mRNA. Wild type WIK zebrafish embryos at the 1-cell stage were injected with 2 nl of the injection mix and screened at 3 dpf for fluorescent secondary marker expression. All injected embryos showing EGFP expression in the lens were selected and raised to adulthood.
neurod1-2A-CreERT2 somatic targeting and tamoxifen treatment
The neurod1 48 bp 5′ and 3′ homology arms were cloned into the pPRISM-2A-CreERT2 vector. nacre; ubi:Switch embryos were injected with 25 pg of genomic sgRNA, 25 pg of UgRNA, 10 pg of targeting vector, and 300 pg Cas9 mRNA. At 6 hpf the embryos were placed in 5 um 4-hydroxytamoxifen (Sigma H6278) in ethanol or ethanol alone and placed in a light-blocking container at 28 °C. The tamoxifen solution was replaced on day two and day three with fresh solution.
Isolation of stable integration alleles and genome/vector junction analysis
To identify adult F0 founders, adult fish were outcrossed to wild type WIK and at least 75 embryos were screened for lens-specific EGFP expression. EGFP positive F1 embryos were selected for genome/vector junction analysis. Genomic DNA was extracted by digestion of single embryos in 50 mM NaOH at 95 °C for 30 min and neutralized by addition of 1/10th volume 1 M Tris–HCl pH 8.0. Both 5′ and 3′ junctions were amplified by PCR using the primers listed in Supplementary Table S2, TOPO-TA cloned and sequenced. EGFP positive F1 embryos from transmitting founders were raised to adulthood and confirmed by fin clip. F1 adult animals with precise integration events were outcrossed to wild type WIK and single F2 adults were used to establish independent transgenic lines.
In situ hybridization and live embryo imaging
Embryos were obtained from ascl1b-2A-Cre/+, olig2-2A-Cre+ and neurod1-2A-Cre/+ outcrossed to wild type WIK, and placed in embryo media with 0.003% 1-phenyl 2-thiourea (PTU) at 24 h post fertilization to block pigment production. Whole mount in situ hybridization was performed as described previously52. 3 dpf larvae were fixed overnight at 4 °C in 4% paraformaldehyde or 4% paraformaldehyde/4% sucrose in PBS. neurod1, olig2 and ascl1b cDNAs were cloned by RT-PCR using total RNA isolated from 3 dpf embryos and SuperScript III (Invitrogen). Primers for reverse transcription and PCR are listed in Supplementary Table S2. Digoxigenin-labeled probes were generated from linear plasmid DNA using DIG RNA Labeling Mix (Roche) and hybridized probes were detected with anti-digoxigenin antibody (Anti-Digoxigenin-AP, Roche) and NBT/BCIP (Roche). Larvae were imaged on a Zeiss Axioskop 2 and photographed with a Cannon Rebel T3 camera. Larvae for live imaging were placed in embryo media with 0.003% 1-phenyl 2-thiourea (PTU) at 24 h post fertilization to block pigment production. At 2 or 3 dpf the PTU treated embryos were anesthetized in 160ug/ml tricaine methane sulfonate and mounted in 1.2% low-melting agarose/160ug/ml tricaine methane sulfonate. Images were captured on a Zeiss LSM 700 or Zeiss LSM 800 laser scanning confocal microscope.
Supplementary Information
Acknowledgements
The authors thank Dr. Leonard Zon (Harvard University) for the Tg(ubi:loxP-EGFP-loxP-mCherry) ubi:Switch transgenic and casper mutant zebrafish line, and Dr. Christian Mosimann (University of Colorado Anschutz Medical Campus) for the CreERT2 cDNA. This study was supported by the Office of The Director of the National Institutes of Health under Award Number R24OD020166 (MM, JJE, KJC, SCE), and by a graduate scholarship from the CNPq Brazilian National Council for Scientific and Technological Development (MPA).
Author contributions
M.P.A., J.J.E. and M.M. conceived and designed the study. M.P.A. and M.M. wrote the main manuscript text and prepared figures and supplementary figures. M.P.A., S.S. and J.L. performed and documented experiments. J.M.W. constructed DNA targeting vectors. S.C.E. and K.J.C. contributed to vector construction. All authors reviewed and contributed to editing of the manuscript.
Funding
NIH R24OD020166 (MM, JJE, SCE, KJC), CNPq Brazilian National Council for Scientific and Technological Development (MPA).
Data availability
All DNA constructs and transgenic zebrafish lines reported in this study are available on request.
Competing interests
JJE has competing interests with Recombinetics Inc., Immusoft Inc., LifEngine and LifEngine Animal Health. KJC has competing interests with Recombinetics Inc., Lifengine and Lifengine Animal Health. SCE has competing interests with LifEngine and LifEngine Animal Health. MPA, JMW and MM declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-81239-y.
References
- 1.Sanchez-Iranzo H, et al. Tbx5a lineage tracing shows cardiomyocyte plasticity during zebrafish heart regeneration. Nat. Commun. 2018;9:428. doi: 10.1038/s41467-017-02650-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirchgeorg L, Felker A, van Oostrom M, Chiavacci E, Mosimann C. Cre/lox-controlled spatiotemporal perturbation of FGF signaling in zebrafish. Dev. Dyn. 2018;247:1146–1159. doi: 10.1002/dvdy.24668. [DOI] [PubMed] [Google Scholar]
- 3.Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-controlled site-specific recombination in zebrafish. PLoS ONE. 2009;4:e4640. doi: 10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosimann C, et al. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu K, Jin H, Zhou B. Genetic lineage tracing with multiple DNA recombinases: A user's guide for conducting more precise cell fate mapping studies. J. Biol. Chem. 2020;295:6413–6424. doi: 10.1074/jbc.REV120.011631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poulin JF, et al. PRISM: A progenitor-restricted intersectional fate mapping approach redefines forebrain lineages. Dev. Cell. 2020;53:740–753. doi: 10.1016/j.devcel.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balciunas D, et al. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carney TJ, Mosimann C. Switch and trace: Recombinase genetics in zebrafish. Trends Genet. 2018;34:362–378. doi: 10.1016/j.tig.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Forster D, et al. Genetic targeting and anatomical registration of neuronal populations in the zebrafish brain with a new set of BAC transgenic tools. Sci. Rep. 2017;7:5230. doi: 10.1038/s41598-017-04657-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquart GD, et al. A 3D searchable database of transgenic zebrafish Gal4 and Cre lines for functional neuroanatomy studies. Front. Neural Circuits. 2015;9:78. doi: 10.3389/fncir.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tabor KM, et al. Brain-wide cellular resolution imaging of Cre transgenic zebrafish lines for functional circuit-mapping. Elife. 2019 doi: 10.7554/eLife.42687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jungke P, Hammer J, Hans S, Brand M. Isolation of novel CreERT2-driver lines in zebrafish using an unbiased gene trap approach. PLoS ONE. 2015;10:e0129072. doi: 10.1371/journal.pone.0129072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang XW, Gong S. An overview on the generation of BAC transgenic mice for neuroscience research. Curr. Protoc. Neurosci. 2005 doi: 10.1002/0471142301.ns0520s31. [DOI] [PubMed] [Google Scholar]
- 15.Mukherjee K, Liao EC. Generation and characterization of a zebrafish muscle specific inducible Cre line. Transgenic Res. 2018;27:559–569. doi: 10.1007/s11248-018-0098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beumer KJ, et al. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedell VM, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson DF, et al. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. U.S.A. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki K, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin J, Chen J, Solnica-Krezel L. Efficient homologous recombination-mediated genome engineering in zebrafish using TALE nucleases. Development. 2014;141:3807–3818. doi: 10.1242/dev.108019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoshijima K, Jurynec MJ, Grunwald DJ. Precise editing of the zebrafish genome made simple and efficient. Dev. Cell. 2016;36:654–667. doi: 10.1016/j.devcel.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hisano Y, et al. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Sci. Rep. 2015;5:8841. doi: 10.1038/srep08841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kesavan G, Hammer J, Hans S, Brand M. Targeted knock-in of CreER (T2) in zebrafish using CRISPR/Cas9. Cell Tissue Res. 2018;372:41–50. doi: 10.1007/s00441-018-2798-x. [DOI] [PubMed] [Google Scholar]
- 25.Wierson WA, et al. Efficient targeted integration directed by short homology in zebrafish and mammalian cells. Elife. 2020 doi: 10.7554/eLife.53968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 28.Takebayashi H, et al. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr. Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- 29.Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: Conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci. Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Kim EJ, Ables JL, Dickel LK, Eisch AJ, Johnson JE. Ascl1 (Mash1) defines cells with long-term neurogenic potential in subgranular and subventricular zones in adult mouse brain. PLoS ONE. 2011;6:e18472. doi: 10.1371/journal.pone.0018472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkinson G, Dennis D, Schuurmans C. Proneural genes in neocortical development. Neuroscience. 2013;253:256–273. doi: 10.1016/j.neuroscience.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 32.Aprea J, Nonaka-Kinoshita M, Calegari F. Generation and characterization of Neurod1-CreER(T2) mouse lines for the study of embryonic and adult neurogenesis. Genesis. 2014;52:870–878. doi: 10.1002/dvg.22797. [DOI] [PubMed] [Google Scholar]
- 33.Picard D, et al. Markers of survival and metastatic potential in childhood CNS primitive neuro-ectodermal brain tumours: an integrative genomic analysis. Lancet Oncol. 2012;13:838–848. doi: 10.1016/S1470-2045(12)70257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturm D, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164:1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultz LE, et al. Epigenetic regulators Rbbp4 and Hdac1 are overexpressed in a zebrafish model of RB1 embryonal brain tumor, and are required for neural progenitor survival and proliferation. Dis. Model Mech. 2018 doi: 10.1242/dmm.034124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modzelewska K, et al. MEK inhibitors reverse growth of embryonal brain tumors derived from oligoneural precursor cells. Cell Rep. 2016;17:1255–1264. doi: 10.1016/j.celrep.2016.09.081. [DOI] [PubMed] [Google Scholar]
- 37.Wagle M, et al. EphrinB2a in the zebrafish retinotectal system. J. Neurobiol. 2004;59:57–65. doi: 10.1002/neu.10340. [DOI] [PubMed] [Google Scholar]
- 38.McFarland KA, Topczewska JM, Weidinger G, Dorsky RI, Appel B. Hh and Wnt signaling regulate formation of olig2+ neurons in the zebrafish cerebellum. Dev. Biol. 2008;318:162–171. doi: 10.1016/j.ydbio.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hozumi S, et al. DEAD-box protein Ddx46 is required for the development of the digestive organs and brain in zebrafish. PLoS ONE. 2012;7:e33675. doi: 10.1371/journal.pone.0033675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forbes-Osborne MA, Wilson SG, Morris AC. Insulinoma-associated 1a (Insm1a) is required for photoreceptor differentiation in the zebrafish retina. Dev. Biol. 2013;380:157–171. doi: 10.1016/j.ydbio.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allende ML, Weinberg ES. The expression pattern of two zebrafish achaete-scute homolog (ash) genes is altered in the embryonic brain of the cyclops mutant. Dev. Biol. 1994;166:509–530. doi: 10.1006/dbio.1994.1334. [DOI] [PubMed] [Google Scholar]
- 42.Thisse, C. & Thisse, B. High throughput expression analysis of ZF-models consortium clones. ZFIN Direct Data Submission (2005).
- 43.Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev. Biol. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- 44.Borodovsky N, Ponomaryov T, Frenkel S, Levkowitz G. Neural protein Olig2 acts upstream of the transcriptional regulator Sim1 to specify diencephalic dopaminergic neurons. Dev. Dyn. 2009;238:826–834. doi: 10.1002/dvdy.21894. [DOI] [PubMed] [Google Scholar]
- 45.Luo N, et al. Syndecan-4 modulates the proliferation of neural cells and the formation of CaP axons during zebrafish embryonic neurogenesis. Sci. Rep. 2016;6:25300. doi: 10.1038/srep25300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kani S, et al. Proneural gene-linked neurogenesis in zebrafish cerebellum. Dev. Biol. 2010;343:1–17. doi: 10.1016/j.ydbio.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 47.Rauch, G. J. Submission and Curation of Gene Expression Data. ZFIN Direct Data Submission (2003).
- 48.Nagy Z, et al. The Gp1ba-Cre transgenic mouse: A new model to delineate platelet and leukocyte functions. Blood. 2019;133:331–343. doi: 10.1182/blood-2018-09-877787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.American Psychological Association Guidelines for ethical conduct in the care and use of animals. J. Exp. Anal. Behav. 1986;45:127–132. doi: 10.1901/jeab.1986.45-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Percie du Sert N, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. J. Physiol. 2020 doi: 10.1113/JP280389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Provost E, Rhee J, Leach SD. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis. 2007;45:625–629. doi: 10.1002/dvg.20338. [DOI] [PubMed] [Google Scholar]
- 52.McGrail M, et al. Expression of the zebrafish CD133/prominin1 genes in cellular proliferation zones in the embryonic central nervous system and sensory organs. Dev. Dyn. 2010;239:1849–1857. doi: 10.1002/dvdy.22307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All DNA constructs and transgenic zebrafish lines reported in this study are available on request.